Abstract
Salmonella enterica is the leading etiologic agent of bacterial food-borne outbreaks worldwide. This ubiquitous species contains more than 2,600 serovars that may differ in their host specificity, clinical manifestations, and epidemiology. To characterize salmonellosis epidemiology in Israel and to study the association of nontyphoidal Salmonella (NTS) serovars with invasive infections, 48,345 Salmonella cases reported and serotyped at the National Salmonella Reference Center between 1995 and 2012 were analyzed. A quasi-Poisson regression was used to identify irregular clusters of illness, and pulsed-field gel electrophoresis in conjunction with whole-genome sequencing was applied to molecularly characterize strains of interest. Three hundred twenty-nine human salmonellosis clusters were identified, representing an annual average of 23 (95% confidence interval [CI], 20 to 26) potential outbreaks. We show that the previously unsequenced S. enterica serovar 9,12:l,v:− belongs to the B clade of Salmonella enterica subspecies enterica, and we show its frequent association with extraintestinal infections, compared to other NTS serovars. Furthermore, we identified the dissemination of two prevalent Salmonella enterica serovar Typhimurium DT104 clones in Israel, which are genetically distinct from other global DT104 isolates. Accumulatively, these findings indicate a severe underreporting of Salmonella outbreaks in Israel and provide insights into the epidemiology and genomics of prevalent serovars, responsible for recurring illness.
INTRODUCTION
Human infections by Salmonella enterica are a global public health concern, leading to approximately 93.8 million cases of gastroenteritis and 155,000 deaths each year (1). In the United States and worldwide, Salmonella is the leading bacterial pathogen responsible for food-borne outbreaks (2). S. enterica is a highly versatile pathogen that can infect a broad range of hosts and causes different clinical outcomes (3). Despite the high genetic similarity between S. enterica serovars, these may vary significantly in their host specificity, clinical manifestation, and epidemiology. For example, most serotypes cause gastroenteritis in healthy individuals, while host-specific serovars such as Salmonella enterica serovar Typhi or S. enterica serovar Paratyphi induce life-threatening enteric fever. The majority of nontyphoid Salmonella (NTS) infections in humans present as gastroenteritis; however, about 5% may be invasive and manifest as bacteremia or other extraintestinal focal infections (4). A few S. enterica serovars, such as Choleraesuis or Dublin, are more likely to cause bacteremia than are others (5), and recent studies found significant differences in disease outcomes between other serovars (6, 7). Nevertheless, the link between salmonellosis epidemiology, genetic content, and the severity of disease caused by different NTS serovars is poorly understood.
Recently developed whole-genome sequencing (WGS) technologies and the ability to compare genomes of different serovars have provided important insights into host specificity and clinical manifestation. Genome degradation (inactivated or missing genes), characterizing the genomes of S. Typhi and S. Paratyphi, and the presence of unique virulence genes and pathogenicity islands are believed to play a role in their human-restricted tropism (reviewed in reference 8).
Early detection of outbreaks is crucial for effective public health intervention and remains a fundamental challenge for health authorities. A few European countries have successfully implemented laboratory-based biosurveillance systems since the early 1990s (9). In Israel, there is currently no statistical surveillance of laboratory-confirmed salmonellosis at the national level. To better understand the association of NTS serovars with invasive disease and to characterize irregular salmonellosis clusters in Israel, we have analyzed 48,345 Salmonella cases reported to the National Salmonella Reference Center (NSRC) between 1995 and 2012. By applying a national laboratory-based surveillance, pulsed-field gel electrophoresis (PFGE), genomics, and phylogenetic approaches, we demonstrated a severe underreporting of putative outbreaks in Israel and showed that S. enterica 9,12:l,v:− has the highest invasive index among all NTS serovars in Israel. Moreover, we analyzed the phylogenetic relationship of multidrug-resistant (MDR) Salmonella Typhimurium DT104 isolates and demonstrated the circulation of two prevalent, endemic, and distinct clones of this strain in Israel.
MATERIALS AND METHODS
Identification of irregular time-based salmonellosis clusters.
Salmonellosis is a reportable disease in Israel, by law. All microbiology laboratories countrywide are required to passively submit clinical and food Salmonella isolates from all sources to the NSRC at the Government Central Laboratories, Israel Ministry of Health in Jerusalem, where serological identification is performed according to the Kauffmann-White-Le Minor scheme (10). All of the serotyped isolates, their source, sending laboratory, date of isolation, and patient identifiers (IDs) are documented. Antibiogram profiling is performed for some of the isolates according to a routine scheme.
In this study, 69,156 Salmonella isolates, which were reported to the NSRC from 1995 to 2012, were analyzed. Salmonella isolates that were found to be contaminated, that were from the Typhi or Paratyphi serovar, were sent from the Palestinian territories, were not linked to a valid ID number, or were repeated isolates from the same patient were all excluded from the analysis. Finally, the analysis included 48,345 single cases of NTS that were linked to distinct IDs. Detection of irregular clusters (statistical alerts) was performed using the R Language and Environment for Statistical Computing, version 2.15.1 (http://www.R-project.org). All computations were executed with the package “surveillance” that was developed for monitoring count data of infectious diseases (11). Each week in the studied period was compared against historical serotype counts extending 4 years back, using a 9-week time window centered on the middle week. Thus, the actual detection of statistically significant deviations from a random pattern started at the fifth week of 1999 and ended at the last week of 2012. Following the model's requirements, in a 53-week year, the counts of the last week were combined with the 52nd week. As proposed by Farrington et al. (12), to approximate data symmetry, the threshold values were calculated using 2/3 power transformation and α = 0.05 was chosen to ensure sufficient sensitivity. This robust parametric generalized linear model (GLM) is based on overdispersed Poisson regression with a log-link. It has many merits, including the ability to adjust seasonal effects and time trends. Moreover, data skewness is handled and past outbreaks are assigned lower weights to avoid artificial inflation of the following upper bounds.
To avoid overrepresentation of outbreak alarms generated by rare serovars, no alarm was generated if the examined week had fewer than three cases of the investigated serovar. The analysis was performed independently for the 37 most prevalent Salmonella serovars in Israel, accounting for 95% of all reported NTS cases between 1995 and 2012. The remaining 5% were grouped and analyzed together as “all others.” Alarms that were separated by 8 weeks or more, during which there was no significant alarm, were counted as unrelated clusters.
Antibiotic resistance.
Antibiotic resistance of outbreak isolates was tested using the Kirby-Bauer disk diffusion method and included the following antibiotics: amoxicillin (20 μg/ml), ampicillin (10 μg/ml), chloramphenicol (30 μg/ml), ciprofloxacin (5 μg/ml), nalidixic acid (30 μg/ml), nitrofurantoin (300 μg/ml), spectinomycin (100 μg/ml), streptomycin (10 μg/ml), sulfamethoxazole (23.75 μg/ml), tetracycline (30 μg/ml), and trimethoprim (1.25 μg/ml).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed according to the PulseNet International Standardized Protocol (13). The clusters were analyzed with BioNumerics software (Applied Maths, Kortrijk, Belgium). The UPGMA (unweighted-pair group method using average linkages) clustering method and Dice similarity coefficients with 1% optimization and 0.85% tolerance parameters were applied. Isolates were defined as genetically related if they presented ≥96% PFGE similarity.
Whole-genome sequencing (WGS) and bioinformatics.
The endemic S. Typhimurium DT104 strain (outbreak isolate 138736) was sequenced to a draft level using the PacBio RS II DNA sequencing system (Pacific Biosciences). The reads (3,003-bp average length) generated from two single-molecule real-time (SMRT) cells provided more than 40.4-fold coverage and were subjected to an assisted assembly using the S. Typhimurium DT104 strain (NCTC 13348) as a reference.
Whole-genome shotgun assemblies for S. Typhimurium DT104 isolates 95799, 98346, 104772, 108402, and 116045 and one blood isolate of the invasive S. 9,12:l,v:− serotype (94293) were generated at Expression Analysis (Durham, NC) using 50 cycles of Illumina's paired-end chemistry to a depth of ∼175-fold coverage and assembled using Velvet (14). Whole-genome alignment and single nucleotide polymorphism (SNP) detection for these six genomes were performed using the Mauve software (15). Reads for all other publically available S. Typhimurium genomes were obtained from the Short Read Archive (http://www.ncbi.nlm.nih.gov/sra). The reads were aligned to the S. Typhimurium DT104 NCTC 13348 genome using Bowtie-2 (16). Variant detection was performed compared to the reference strain using SAMtools (17) and custom scripts. In brief, only bases with a phred score of >30 (0.1% error) were used to call a consensus base, and consensus bases with a score of >60 (0.0001% error) were used to call variants. The resultant SNP matrix was further pruned to remove sites that were not reliably called in 99% of the strains. Maximum likelihood trees were constructed using FastTree (18) and the SNP matrix obtained from the previous step. SnpEff (19) was used to predict the consequences of the variants using the S. Typhimurium DT104 NCTC 13348 genome as reference.
BEAST v1.8 (20) was used for Bayesian phylogenetic inference of the DT104 isolates. The exponential growth coalescent tree priors were used with a general time-reversible nucleotide substitution model. The isolation dates of the samples were used to calibrate the time scale of the tree, and an uncorrelated lognormal relaxed molecular clock was used to account for rate variation among lineages. Two independent Markov chain Monte Carlo (MCMC) analyses were run for 100 million states, with sampling every 10,000 states. The results for the two runs were combined after discarding the first 10,000 states sampled as burn-in. Tree Annotator was used to calculate the maximum clade credibility tree from all the sampled trees. Annotation and comparison between S. 9,12:l,v:− and S. Typhimurium LT2 were performed using RAST (http://rast.nmpdr.org/). All publically available Salmonella genome assemblies were downloaded from PATRIC (http://patricbrc.org). Mauve was used to generate pairwise alignments of each strain with S. Typhimurium LT2 as a reference. The pairwise alignments were parsed using custom scripts to create a pseudomultiple alignment, where putative orthologous bases to each S. Typhimurium LT2 position were determined. This alignment was used to determine an SNP matrix using custom scripts. Maximum likelihood trees were generated using FastTree (18) and the SNP matrix.
Confirmation of DT104 phage type.
S. Typhimurium DT104 strain confirmation was determined by three criteria: (i) the presence of the Salmonella genomic island I and prophage III by PCR using the primers SGI1p12F, 5′ CAACTTCCGTAAGTTCAGCTACAGC 3′; SGI1p12R, 5′ TAGCTCTATCCAGCAATGCGGATTG 3′; DT104p3F, 5′ TGAAGGCTCTCAGCATATCACCCGTA 3′; and DT104p3R, 5′ ATCCACTGCCGAACGTTATCGTGGT 3′ (21); (ii) a PFGE pulsotype similar to that of the S. Typhimurium DT104 strain NCTC 13348; and (iii) antibiotic resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline.
Nucleotide sequence accession numbers.
All seven genome shotgun projects have been deposited at GenBank under the accession numbers PRJNA242139, PRJNA215426, PRJNA215428, PRJNA215434, PRJNA215436, PRJNA215440, and PRJNA215425.
RESULTS
Characterization of irregular salmonellosis clusters.
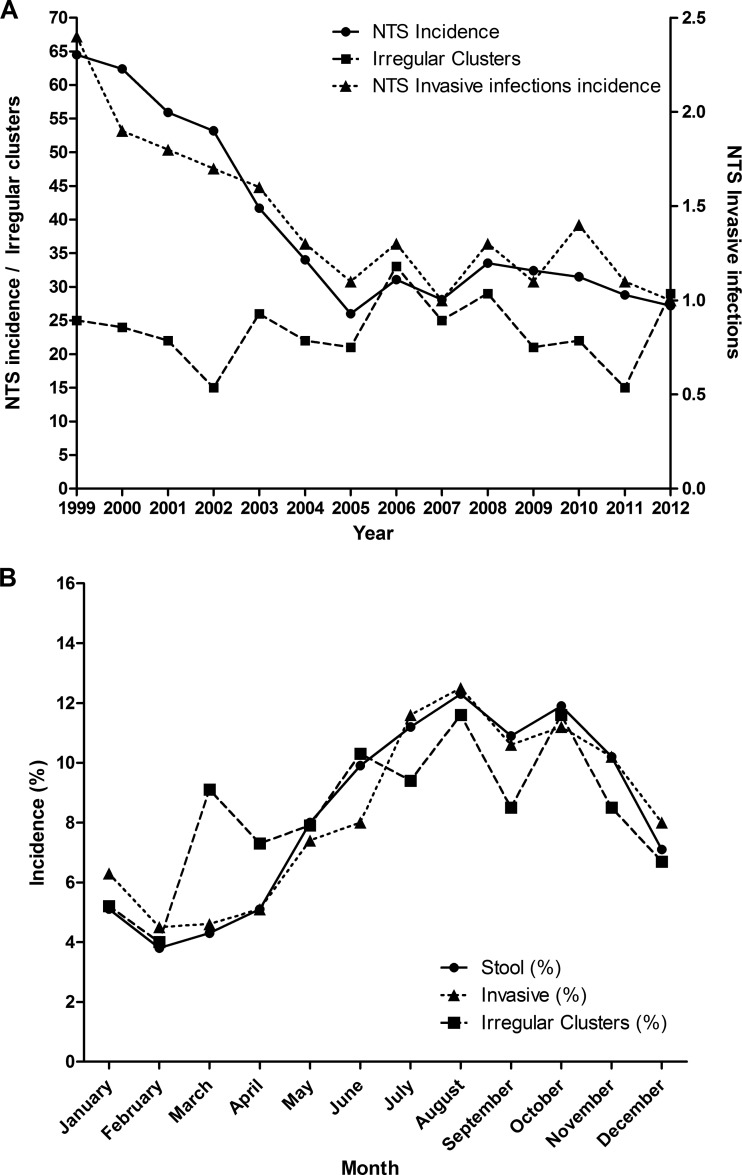
To identify time-based irregular clusters (alarms), which often indicate outbreaks, we have implemented the Farrington algorithm (12) to retrospectively analyze 48,345 cases reported and serotyped at the NSRC between 1995 and 2012. This analysis revealed 329 statistically significant clusters (comprising 3 to 633 associated cases) from 1999 to 2012, with an average of 23 (95% confidence interval [CI], 20 to 26) salmonellosis clusters per year (Fig. 1A). We also found that 7,427 (15.4%) of the reported cases were associated with predicted clusters. The vast majority of these clusters were previously unreported and hence were never investigated by health authorities. Surprisingly, a monotonic correlation between the annual incidence and the number of clusters was not observed (Spearman's rho, −0.02; P value, 0.946). For instance, despite a sharp drop in Salmonella incidence between 1999 and 2005, and a moderate decline between 2008 and 2012, the yearly number of salmonellosis clusters increased in 2003 and in 2012 (Fig. 1A).
FIG 1.
(A) Annual incidence of reported NTS, invasive NTS infections, and predicted salmonellosis clusters in Israel, 1999 to 2012. The annual incidence of NTS Salmonella cases (circles and solid line) and the number of invasive NTS infections (triangles and dotted line) are shown per 100,000 population and were inferred from the laboratory-confirmed Salmonella cases received at the National Salmonella Reference Center. The yearly number of clusters (squares and broken line) was predicted by the Farrington statistical surveillance model as described in Materials and Methods. (B) Seasonal distribution of salmonellosis clusters, 1999 to 2012, and invasive and stool infections with reported NTS Salmonella in Israel, 1995 to 2012.
A clear seasonal pattern of salmonellosis clusters overlapped with the general seasonal Salmonella incidence (Spearman's rho, 0.77; P value, 0.003), demonstrating fewer clusters in the winter, during December to February, and an increased number of clusters in the summer, during June to October (Fig. 1B). Interestingly, additional peaks in salmonellosis clusters were observed in the spring (March), although the general Salmonella incidence at this time is low, suggesting that salmonellosis clusters are not merely the direct outcome of the general Salmonella incidence and that other factors are expected to play a part as well.
Molecular analysis of algorithmically detected clusters.
To better characterize the genetic relatedness of the clusters identified by the Farrington surveillance algorithm, we analyzed by PFGE the clonality of 95 different isolates from 23 salmonellosis clusters (Table 1). For each cluster examined, sporadic nonrelated isolates (one to three) were included for comparison (in total, 136 isolates were analyzed). Molecular confirmation of the salmonellosis cluster caused by Salmonella enterica serovar Enteritidis in August 2004 was reported previously (22). Overall, we demonstrated that except for three predicted clusters (S. enterica serovar Virchow in January 2007, S. Enteritidis in May 2001, and S. enterica serovar Agona in June 2009), 20/23 (86%) identified clusters consisted of related isolates and therefore are likely to be part of monoclonal outbreaks. Three clusters, comprised of genetically unrelated isolates, may represent either a false-positive alarm generated by the algorithm or outbreaks with more than one infecting source (polyclonal outbreaks). This analysis suggests that a major portion of the clusters detected by the implemented Farrington algorithm may represent overlooked outbreaks, which would have been detected and investigated by public health authorities, at the time of occurrence, if this method of surveillance had been in place at the time.
TABLE 1.
PFGE analysis of 23 potential salmonellosis outbreaks, 1999 to 2012a
| Potential outbreak | Serotype | No. of linked cases | Detection date | Antibiotic resistance | Cluster clonality |
|---|---|---|---|---|---|
| 1 | Enteritidis | 6 | Aug 1999 | SMZ | Clonal |
| 2 | Enteritidis | 6 | May 2001 | Nonclonal | |
| 3 | Typhimurium DT104 | 22 | Aug 2001 | AMP, CHL, TET, AMX, SPT, NAL, STR, SMZ | Clonal |
| 4 | Concord | 7 | May 2002 | SMZ | Clonal |
| 5 | Bredeney | 6 | Jun 2003 | SMZ | Clonal |
| 6 | Enteritidis | 10 | Aug 2004 | SMZ | Clonal |
| 7 | Typhimurium | 8 | Jun 2005 | NAL, SMZ | Clonal |
| 8 | Hadar | 3 | Jun 2005 | TET, STR, SMZ | Clonal |
| 9 | Typhimurium DT104 | 4 | Oct 2005 | AMP, CHL,TET, AMX, SPT, NAL, STR, SMZ | Clonal |
| 10 | 16:l,v:− | 6 | Dec 2005 | SMZ | Clonal |
| 11 | Java | 10 | Feb 2006 | AMP, TMP, TET, AMX, NAL, NIT, SMZ | Clonal |
| 12 | Mbandaka | 7 | May 2006 | SMZ | Clonal |
| 13 | Enteritidis | 8 | May–Jun 2006 | NIT, SMZ | Clonal |
| 14 | Hadar | 4 | Nov 2006 | AMP, TET, AMX, STR, SMZ | Clonal |
| 15 | Virchow | 4 | Jan 2007 | Nonclonal | |
| 16 | Enteritidis | 3 | Aug 2007 | AMP, AMX, SMZ | Clonal |
| 17 | Infantis | 10 | Jun 2009 | TET, SPT, NAL, STR, NIT, SMZ | Clonal |
| 18 | Agona | 3 | Jun 2009 | Nonclonal | |
| 19 | Enteritidis | 12 | Sep 2009 | SMZ | Clonal |
| 20 | Schwarzengrund | 4 | Aug 2009 | SMZ | Clonal |
| 21 | Havana | 99 | Mar–May 2011 | SMZ | Clonal |
| 22 | 9,12:l,v:− | 16 | Jun 2011 | TET, SMZ | Clonal |
| 23 | Typhimurium DT104 | 5 | Jun 2011 | AMP, CHL, TET, AMX, SPT, STR, SMZ | Clonal |
Abbreviations: Jan, January; Feb, February; Mar, March; Jun, June; Aug, August; Sep, September; Oct, October; Nov, November; Dec, December; AMX, amoxicillin; AMP, ampicillin; CHL, chloramphenicol; NAL, nalidixic acid; NIT, nitrofurantoin; SPT, spectinomycin; STR, streptomycin; SMZ, sulfamethoxazole; TET, tetracycline; TMP, trimethoprim.
Extraintestinal NTS infection analysis.
Next, we sought to analyze the site of infection in the 48,345 Salmonella cases. Altogether, 1,731 extraintestinal NTS infections were identified, comprising 3.6% of all cases. The infection sites from which extraintestinal salmonellae were isolated included blood (n = 1,171; 67.6%), urine (n = 413; 24%), wound (n = 38; 2.2%), abscess (n = 19, 1%), and other sites (n = 90, 5.2%). In agreement with the general salmonellosis incidence, the incidence of invasive NTS (iNTS) has decreased from about 3 cases per 100,000 population in 1999 to 1 case per 100,000 in 2005 to 2012 (Fig. 1A). The seasonality of invasive salmonellosis was also found to be tightly correlated with the general Salmonella incidence (Spearman's rho, 0.98; P value, 0.0001), presenting higher rates during the spring and summer (April to October). Curiously, an unexplained seasonal decline in the reported cases is seen in September for all strata (Fig. 1B).
The NTS serovar that presented the highest invasive index (the percentage of extraintestinal infections from all cases) was 9,12:l,v:−, with 7.7% (Fisher's 95% CI, 6.1 to 9.6) of invasive infections (Table 2). This invasive index is more than 2-fold higher than the invasive index of S. Typhimurium (3.4%, P value < 0.001) or the mean invasive index of all NTS serovars (3.6%, P value < 0.001). These results reinforce the notion that NTS serovars may differ in their ability to induce invasive disease and may suggest that some serovar-specific genetic factors are likely to play a role in the manifestation of invasive salmonellosis in addition to many known host factors that were recently reviewed (23).
TABLE 2.
Prevalence of reported S. enterica serovars in Israel (1995 to 2012), their extraintestinal infection frequency, and their representation in predicted salmonellosis clusters (1999 to 2012)
| Serotype | No. (%) of cases | No. (%) of clusters | No. (%) of outbreak-associated cases | No. of invasive infections | Invasive indexa |
|---|---|---|---|---|---|
| Enteritidis | 11,198 (23.2) | 19 (5.8) | 1,279 (11.4) | 514 | 4.6b |
| Virchow | 6,061 (12.5) | 17 (5.2) | 1,066 (17.6) | 289 | 4.8b |
| Typhimurium | 5,733 (11.9) | 18 (5.5) | 574 (10) | 195 | 3.4 |
| Infantis | 5,370 (11.1) | 17 (5.2) | 1,460 (27.2) | 104 | 2b |
| Hadar | 4,084 (8.4) | 14 (4.3) | 325 (8) | 81 | 2b |
| Bredeney | 1,307 (2.7) | 13 (4) | 203 (15.5) | 66 | 5b |
| Montevideo | 1,052 (2.2) | 16 (4.9) | 179 (17) | 33 | 3.1b |
| Agona | 1,005 (2.1) | 16 (4.9) | 159 (15.8) | 14 | 1.4b |
| 9,12:l,v:− | 919 (1.9) | 15 (4.6) | 129 (14) | 71 | 7.7b |
| Blockley | 798 (1.7) | 8 (2.4) | 94 (11.8) | 14 | 1.8b |
| Heidelberg | 787 (1.6) | 5 (1.5) | 364 (46.3) | 31 | 3.9 |
| Newport | 785 (1.6) | 13 (4) | 76 (9.7) | 31 | 3.9 |
| Muenchen | 706 (1.5) | 16 (4.9) | 200 (28.3) | 23 | 3.3 |
| Kentucky | 678 (1.4) | 10 (3) | 122 (18) | 32 | 4.7 |
| Java | 669 (1.4) | 16 (4.9) | 102 (15.2) | 14 | 2.1b |
| Mbandaka | 491 (1) | 13 (4) | 143 (29.1) | 10 | 2 |
| Tennessee | 451 (0.9) | 9 (2.7) | 58 (12.9) | 6 | 1.3b |
| Anatum | 447 (0.9) | 10 (3) | 50 (11.2) | 8 | 1.8b |
| Afula | 383 (0.8) | 14 (4.3) | 67 (17.5) | 3 | 0.8b |
| Senftenberg | 292 (0.6) | 3 (0.9) | 21 (7.2) | 10 | 3.4 |
| 16:l,v:− | 263 (0.5) | 6 (1.8) | 94 (35.7) | 4 | 1.5 |
| Kottbus | 251 (0.5) | 4 (1.2) | 19 (7.6) | 4 | 1.6 |
| Emek | 250 (0.5) | 5 (1.5) | 28 (11.2) | 5 | 2 |
| Havana | 242 (0.5) | 2 (0.6) | 142 (58.7) | 6 | 2.5 |
| Concord | 217 (0.4) | 3 (0.9) | 41 (18.9) | 2 | 0.9b |
| Eastbourne | 164 (0.3) | 1 (0.3) | 3 (1.8) | 9 | 5.5 |
| Saintpaul | 163 (0.3) | 1 (0.3) | 3 (1.8) | 5 | 3.1 |
| Haifa | 143 (0.3) | 0 (0) | 0 (0) | 4 | 2.8 |
| Edinburg | 139 (0.3) | 6 (1.8) | 24 (17.3) | 4 | 2.9 |
| Braenderup | 137 (0.3) | 6 (1.8) | 20 (14.6) | 3 | 2.2 |
| Abony | 130 (0.3) | 5 (1.5) | 19 (14.6) | 1 | 0.8 |
| Stanley | 129 (0.3) | 2 (0.6) | 16 (12.4) | 3 | 2.3 |
| Give | 126 (0.3) | 0 (0) | 0 (0) | 8 | 6.3 |
| Corvallis | 109 (0.2) | 2 (0.6) | 7 (6.4) | 4 | 3.7 |
| Orion | 93 (0.2) | 0 (0) | 0 (0) | 1 | 1.1 |
| Oranienburg | 91 (0.2) | 2 (0.6) | 7 (7.7) | 4 | 4.4 |
| 4,12:−:− | 83 (0.2) | 1 (0.3) | 3 (3.6) | 1 | 1.2 |
| All others | 2,399 (5.0) | 21 (6.4) | 330 (13.8) | 114 | 4.8b |
| Total | 48,345 | 329 (100) | 7,427 (15.4) | 1,731 | 3.6 |
Invasive index = (invasive infections/total cases) × 100.
χ2, P < 0.05 in comparison to the invasive index of all NTS serovars.
Genomic characterization of S. 9,12:l,v:−.
We observed that the previously unsequenced serovar 9,12:l,v:− has the highest invasive index among all NTS serovars in Israel and is associated with multiple salmonellosis clusters (Table 1), which prompted us to further characterize this serovar on the genetic level. This monophasic serovar, which has recently emerged in Bulgaria, Denmark, and the United States (24), was thought to have evolved from a strain of Salmonella enterica serovar Goettingen (9,12:l,v:e,n,z15) (25), and the two serovars share the multilocus sequence type (MLST) 20 (http://mlst.warwick.ac.uk/mlst/dbs/Senterica).
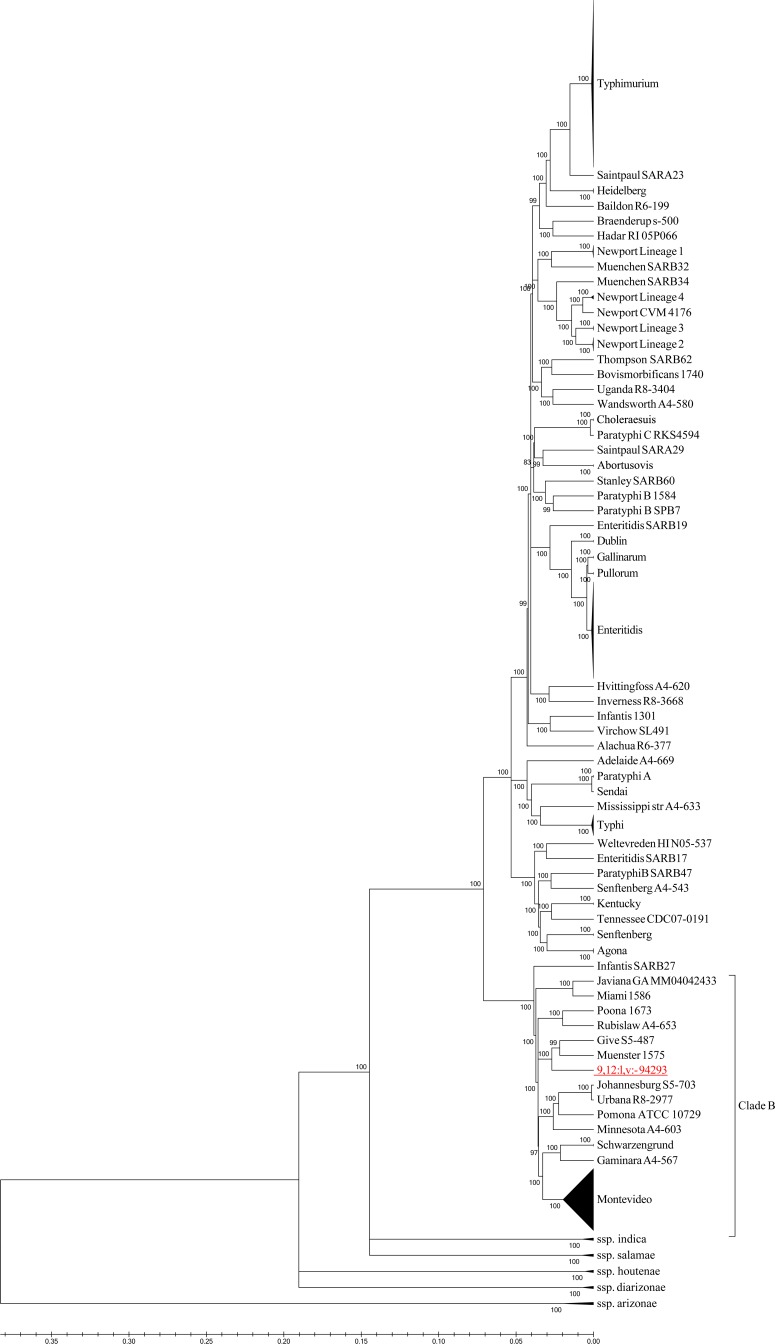
Genome sequence analysis of an invasive blood isolate (no. 94293) of S. 9,12:l,v:− showed that the hin recombinase responsible for flagellar switching was absent, which might explain the monophasic phenotype of this serovar. Figure 2 demonstrates the taxonomic relationship of S. 9,12:l,v:− to 385 other sequenced S. enterica strains, as determined by a concatenated alignment of 3,384 core Salmonella genes. This analysis showed that the newly sequenced S. 9,12:l,v:− serovar is a member of the S. enterica subsp. enterica clade B (26).
FIG 2.
S. 9,12:l,v:− is a member of S. enterica subsp. enterica clade B. The maximum likelihood cladogram of 385 Salmonella strains and S. 9,12:l,v:− was constructed by FastTree using 3,384 orthologous Salmonella genes. Internal nodes show local support values based on the Shimodaira-Hasegawa test. Distances on the tree represent substitutions per variable site.
Genome sequence analysis also showed an interesting composition of virulence genes present in various clade B genomes (26, 27). These include three usher-chaperone fimbrial systems, one of which is the Tcf (Typhi colonization factor), a fimbrial cluster encoded between STM0182-STM0183 homologs and an additional fimbrial operon downstream of the ISSod13 transposase. We also found an uncharacterized type 1 fimbria operon (inserted in STM1197-STM1198 homologs) and a PilV-like adhesin. Besides the fimbriae, the cytolethal distending toxin (CdtB), hemolysin E (HlyE), and the putative typhoid effectors EspN-like (STY1413) and OspB-like (STY1360) were also identified in the S. 9,12:l,v:− genome. An analysis of metabolic genes suggested that S. 9,12:l,v:− carries the Salmonella-uncommon beta-glucuronide operon and the ydj island (9.4 kb). On the other hand, the 9,12:l,v:− genome lacks several metabolic clusters that are present in many other NTS, including the allantoin catabolism island (STM0514-STM0532), the inositol cluster (STM4417-STM4436), l-rhamnonate (STM2288-STM2293), l-tartrate (STM0761-STM0762), the mannose phosphotransferase system (STM4534-STM4540.S), and the aga carbohydrate operon (STM3251-STM3256).
Recurring salmonellosis caused by endemic MDR S. Typhimurium DT104 clones.
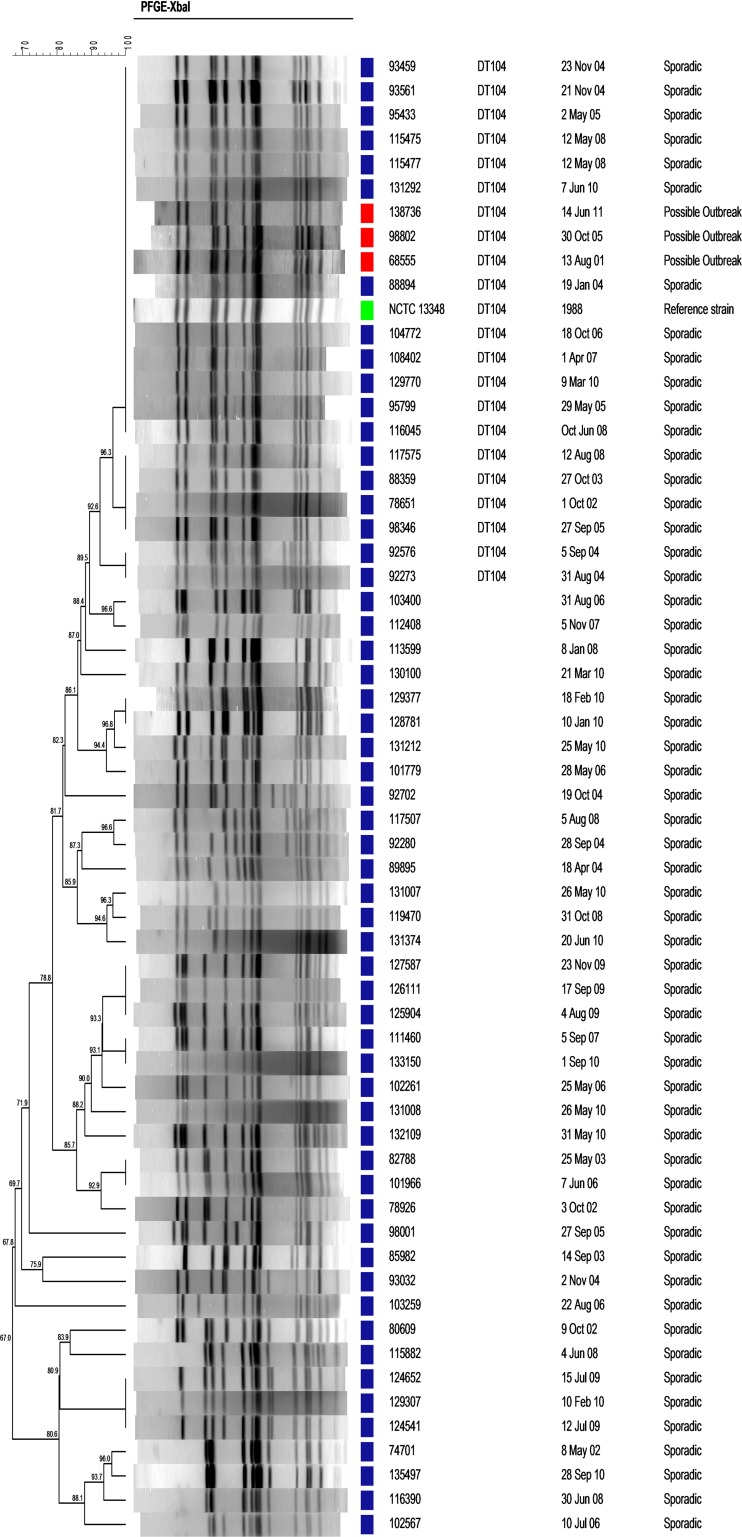
Our analysis showed that three S. Typhimurium clusters (August 2001, October 2005, and June 2011) shared a similar PFGE profile as determined by XbaI and SpeI restriction analysis (data not shown). This particular pattern was also found to be identical to the pulsotype of the sequenced S. Typhimurium DT104 strain NCTC 13348 isolated in the United Kingdom in 1988 (Fig. 3) and to that of an S. Typhimurium DT104 strain from Taiwan (28). PCR analysis demonstrated the presence of the Salmonella genomic island I and prophage III, while antibiotic susceptibility testing showed resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline, confirming a DT104 MDR phenotype.
FIG 3.
Prevalence of endemic MDR S. Typhimurium DT104 pulsotype. The PFGE profile of three S. Typhimurium DT104 isolates responsible for three clusters that occurred in 2001, 2005, and 2011 (red) was compared to the pulsotypes of S. Typhimurium DT104 NCTC 13348 (green) and 57 sporadic isolates (blue) of S. Typhimurium isolated in the past decade. DNA from all isolates was digested with XbaI, and the degree of genetic similarity is shown by the dendrogram at the left.
To get a broader perspective on the distribution of this DT104 strain among S. Typhimurium isolates in Israel, 57 randomly chosen sporadic isolates of S. Typhimurium were analyzed by PFGE. Sixteen isolates (28%) presented a pulsotype similar to that of the epidemic DT104 strain (Fig. 3), indicating a high prevalence of S. Typhimurium DT104 in Israel in the last decade.
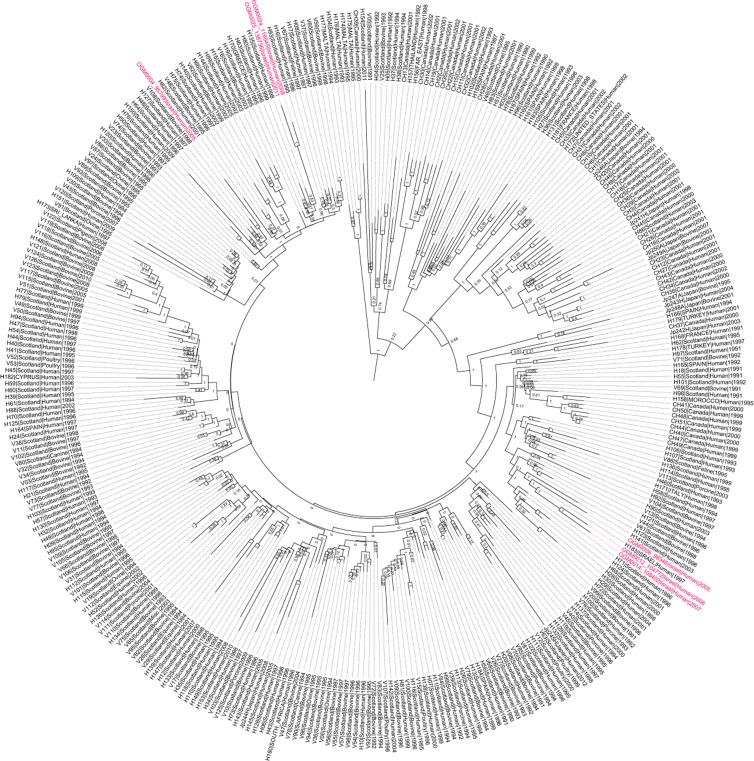
To further characterize the nature of these S. Typhimurium DT104 isolates on the genetic level, we applied whole-genome sequencing to six independent isolates from 2005 (isolates 95799 and 98346), 2006 (104772), 2007 (108402), and 2008 (116045) and one isolate from a 2011 outbreak (138736). Phylogenetic analysis based on 2,230 SNPs retrieved from 335 different S. Typhimurium DT104 genomes (Fig. 4) demonstrated that despite the different isolation times, three newly sequenced Israeli isolates (104772, 108402, and 98346) clustered closely together in one clade. Interestingly, a previously sequenced S. Typhimurium strain (H183) that was isolated from a United Kingdom citizen returning from Israel in 1997 was grouped in this clade as well. Two other Israeli isolates (138736 and 116045) were also clustered in a single but separate clade, indicating the circulation of at least two dominant biotypes of DT104 in Israel. Relaxed molecular clock analysis has shown that these two clades diverged in 1986 (95% highest probability density [HPD], 1985 to 1988). The model also predicted that the hypothetical ancestor of all DT104 strains arose in 1970 (95% HPD, 1962 to 1976), which is consistent with the dates predicted by Mather and colleagues (29).
FIG 4.
Phylogenetic relationship of identified Israeli DT104 isolates with global S. Typhimurium DT104. Maximum clade credibility tree of 335 S. Typhimurium DT104 isolates and six Israeli DT104 isolates (95799, 98346, 104772, 108402, 116045, and 138736, in red) was constructed based on 2,353 SNPs using BEAST v1.80. The tree was calibrated using isolation dates of the strains. The distances represent time, and internal nodes show clade support (bootstrap values) as posterior probabilities.
DISCUSSION
Salmonella spp. remain a leading bacterial cause of food-borne outbreaks in developed countries (30). Most people infected with Salmonella do not seek medical care or submit a stool sample. Moreover, even after the sample has been submitted to the laboratory, several weeks may pass between the submission date and the actual linkage of the case to a specific outbreak, following serotyping and molecular fingerprinting. Hence, it is of great importance for health authorities to be able to effectively recognize and analyze statistically distinct clusters of laboratory-confirmed cases. The potential benefit of an algorithm-based surveillance can be appreciated in the light of the relatively low number of reported Salmonella outbreaks in Israel, with only a single outbreak recognized in 2010 (31), while our analysis estimated that each year in Israel there are on average 23 potential outbreaks. If one takes into account the EU multiplier for the ratio of underdiagnosis of 58 for every laboratory-confirmed case (32), it is likely that the actual number of outbreaks in the community is even higher. Statistical surveillance methods such as the Farrington algorithm provide an economical, efficient, and valid tool for early detection of potential outbreaks that require further epidemiological investigation and confirmation. Furthermore, improved metadata collection, including patient's residential postal code, age, and sex, can contribute to better outbreak detection and investigation, without compromising patient privacy. These recommendations have been presented to officials in the Israeli Ministry of Health and are currently under consideration.
Overall, the profile of salmonellosis clusters corresponded with the general seasonal Salmonella incidence, with fewer clusters in the winter and higher numbers of clusters in the summer months (Fig. 1B). Nevertheless, an additional peak of salmonellosis clusters was observed in the spring, although the general Salmonella incidence at this time of year is low. While we do not fully understand this phenomenon, an intriguing possibility is that the source of these outbreaks might be migrating birds that extensively pass through Israel during the spring migration that reaches its peak from the beginning of March to mid-May. Several recent reports indicating Salmonella transmission between wild birds and meat-production animals (33, 34) may support this hypothesis.
Our analysis indicated that S. 9,12:l,v:− is frequently associated with extraintestinal infections (about 8% of cases). Since the prevalences of S. 9,12:l,v:− are similar among human and poultry samples (about 2% from all Salmonella isolates), we believe that the source of S. 9,12:l,v:− infections is probably poultry products that are often the reservoir for NTS serovars in industrialized countries (35). A similar observation that attributed a high blood invasiveness ratio to S. 9,12:l,v:− in Israel was reported by Weinberger and Keller in a 5-year study of 1997 to 2002 (36). To the best of our knowledge, the relatively high proportion of extraintestinal infections associated with serovar 9,12:l,v:− found in Israel has not been reported in other countries. Comparisons among countries of the invasive indices of NTS serovars show that there are often substantial differences (7). These data support the existence of geographically related pathovars that may vary in their virulence potential and invasiveness. For example, 13.4% of Salmonella enterica serovar Heidelberg infections are invasive in the United States (6), but only 3.9% of those in Israel are invasive (P value < 0.001). Such differences may substantially reflect different pathogen lineages rather than variation in human populations, such as immunological or nutritional status, genetic background, etc. Future study of genome comparisons between multiple isolates of the same serovar from countries with different invasive indices may help in understanding the nature of these differences.
Recently, we demonstrated, using comparative genomic hybridization (CGH), the presence of several typhoid-associated virulence genes (tcfA, cdtB, hlyE, taiA, STY1413, and STY1360) in S. 9,12:l,v:− and showed that this serotype can induce prolonged systemic infection in the mouse model (37). In the present study, using whole-genome sequencing, we demonstrated the presence of a particular composition of additional adhesion factors and an uncommon metabolic profile. It is possible that this unique repertoire of virulence genes and possibly other genetic features, such as degradation of metabolic pathways, may contribute to the relatively high invasive index of this nontyphoid serovar. In agreement with this notion, a phylogenetic analysis clustered S. 9,12:l,v:− in the same clade with other NTS serovars that were reported as associated with high invasiveness (compared to other NTS [Fig. 2]) including S. Schwarzengrund, S. Urbana, S. Muenster, S. Pomona, and S. Poona. All are characterized by a high proportion (>10%) of invasive disease in the United States (6, 38). It can be speculated therefore that common virulence factors and genomic degradations shared by at least some members of clade B may play a role in their extraintestinal predisposition.
One of the main Salmonella pandemics in the last 2 decades is the emergence of an MDR S. Typhimurium phage type DT104 (39). In Israel, S. Typhimurium DT104 emerged during the 1990s and became the most prevalent phage type among S. Typhimurium strains. In 2008, S. Typhimurium DT104 accounted for 35.1% of all S. Typhimurium isolates (40). In recent years, there has been a decline in the prevalence of DT104 in Israel, similar to the trend reported in other countries. Nevertheless, despite the documented decrease in the incidence of S. Typhimurium DT104, we identified the dissemination of at least two distinct endemic S. Typhimurium DT104 clones responsible for recurring clusters of infection in Israel (Fig. 4).
To summarize, in this study we have analyzed the epidemiology of salmonellosis clusters and extraintestinal infections caused by NTS serovars in Israel. We found that each year there are dozens of unrecognized, and therefore not investigated, clusters of illness. Routine implementation at the national level of statistical surveillance could greatly improve early detection of outbreaks and facilitate effective intervention by health authorities. We showed that the clade B serovar 9,12:l,v:− is responsible for a relatively high proportion of extraintestinal infections compared to other prevalent NTS serovars. S. 9,12:l,v:− and endemic biotypes of S. Typhimurium DT104 are previously underappreciated agents of morbidity in Israel.
ACKNOWLEDGMENTS
We thank Gordon Dougan and Derek Pickard from the Wellcome Trust Sanger Institute for sending us the S. Typhimurium DT104 NCTC 13348 strain. We thank Steffen Porwollik for helpful discussions.
The work in the Gal-Mor laboratory is supported by GIF research grant 1096.39.11/2010 and by grant 249241 from the European Community's Seventh Framework program (PF7/2007-2013). M.M. and P.T.D. acknowledge support from NIH grants AI039557 AI052237, AI073971, AI075093, AI077645 and AI083646; USDA grants 2009-03579 and 2011-67017-30127; the Binational Agricultural Research and Development Fund; and a grant from the Center for Produce Safety.
Footnotes
Published ahead of print 9 April 2014
REFERENCES
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50:882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2013. Surveillance for foodborne disease outbreaks—United States, 2009–2010. MMWR Morb. Mortal. Wkly. Rep. 62:41–47 [PMC free article] [PubMed] [Google Scholar]
- 3.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, Casadesus J, Platt DJ, Olsen JE. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229–255. 10.1017/S0950268899004379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hohmann EL. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263–269. 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- 5.Wilkins EG, Roberts C. 1988. Extraintestinal salmonellosis. Epidemiol. Infect. 100:361–368. 10.1017/S095026880006711X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D'Angelo M, Hurd S, Medus C, Cronquist A, Angulo FJ. 2008. Salmonellosis outcomes differ substantially by serotype. J. Infect. Dis. 198:109–114. 10.1086/588823 [DOI] [PubMed] [Google Scholar]
- 7.Langridge GC, Nair S, Wain J. 2009. Nontyphoidal Salmonella serovars cause different degrees of invasive disease globally. J. Infect. Dis. 199:602–603. 10.1086/596208 [DOI] [PubMed] [Google Scholar]
- 8.Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305:1–13. 10.1111/j.1574-6968.2010.01904.x [DOI] [PubMed] [Google Scholar]
- 9.Hulth A, Andrews N, Ethelberg S, Dreesman J, Faensen D, van Pelt W, Schnitzler J. 2010. Practical usage of computer-supported outbreak detection in five European countries. Euro Surveill. 15(36):19658 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19658 [PubMed] [Google Scholar]
- 10.Le Minor L, Veron M, Popoff M. 1982. A proposal for Salmonella nomenclature. Ann. Microbiol. 133:245–254 [PubMed] [Google Scholar]
- 11.Höhle M. 2007. Surveillance: an R package for the monitoring of infectious diseases. Comput. Stat. 22:571–582. 10.1007/s00180-007-0074-8 [DOI] [Google Scholar]
- 12.Farrington CP, Andrews NJ, Beale AD, Catchpole MA. 1996. A statistical algorithm for the early detection of outbreaks of infectious disease. J. R. Stat. Soc. Ser. A Stat. Soc. 159:547–563. 10.2307/2983331 [DOI] [Google Scholar]
- 13.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7 Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59–67. 10.1089/fpd.2006.3.59 [DOI] [PubMed] [Google Scholar]
- 14.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–U354. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu K, Linder CR, Warnow T. 2011. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS One 6:e27731. 10.1371/journal.pone.0027731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29:1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooke FJ, Wain J, Fookes M, Ivens A, Thomson N, Brown DJ, Threlfall EJ, Gunn G, Foster G, Dougan G. 2007. Prophage sequences defining hot spots of genome variation in Salmonella enterica serovar Typhimurium can be used to discriminate between field isolates. J. Clin. Microbiol. 45:2590–2598. 10.1128/JCM.00729-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein-Zamir C, Tallen-Gozani E, Abramson N, Shoob H, Yishai R, Agmon V, Reisfeld A, Valinsky L, Marva E. 2009. Salmonella enterica outbreak in a banqueting hall in Jerusalem: the unseen hand of the epidemiological triangle? Isr. Med. Assoc. J. 11:94–97 [PubMed] [Google Scholar]
- 23.Gordon MA. 2011. Invasive non-typhoidal Salmonella disease—epidemiology, pathogenesis and diagnosis. Curr. Opin. Infect. Dis. 24:484. 10.1097/QCO.0b013e32834a9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrov P, Hendriksen RS, Kantardjiev T, Asseva G, Sorensen G, Fields P, Mikoleit M, Whichard J, McQuiston JR, Torpdahl M, Aarestrup FM, Angulo FJ. 2009. Occurrence and characterization of Salmonella enterica subspecies enterica serovar 9,12:l,v:− strains from Bulgaria, Denmark, and the United States. Eur. J. Clin. Microbiol. Infect. Dis. 28:473–479. 10.1007/s10096-008-0653-9 [DOI] [PubMed] [Google Scholar]
- 25.Burnens AP, Stanley J, Sechter I, Nicolet J. 1996. Evolutionary origin of a monophasic Salmonella serovar, 9,12:l,v:−, revealed by IS200 profiles and restriction fragment polymorphisms of the fljB gene. J. Clin. Microbiol. 34:1641–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, Degoricija L, Hoelzer K, Rodriguez-Rivera LD, Brown S, Bolchacova E, Furtado MR, Wiedmann M. 2011. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genomics 12:425. 10.1186/1471-2164-12-425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Clifton SW, Weinstock GM, McClelland M. 2013. Evolutionary Genomics of Salmonella enterica subspecies. mBio 4(2):e00579–12. 10.1128/mBio.00579-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu CH, Su LH, Chu CH, Wang MH, Yeh CM, Weill FX, Chu C. 2006. Detection of multidrug-resistant Salmonella enterica serovar Typhimurium phage types DT102, DT104, and U302 by multiplex PCR. J. Clin. Microbiol. 44:2354–2358. 10.1128/JCM.00171-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mather AE, Reid SW, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, de Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. 10.1126/science.1240578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harker KS, Lane C, Gormley FJ, Adak GK. 2014. National outbreaks of Salmonella infection in the UK, 2000–2011. Epidemiol. Infect. 142:601–607. 10.1017/S0950268813001210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Israeli Ministry of Health PHS. 2011. 2010 annual summary of reported food-poisonings, periodic epidemiological report. Department of Epidemiology, Israeli Ministry of Health PHS, Jerusalem, Israel [Google Scholar]
- 32.Havelaar AH, Ivarsson S, Lofdahl M, Nauta MJ. 2013. Estimating the true incidence of campylobacteriosis and salmonellosis in the European Union, 2009. Epidemiol. Infect. 141:293–302. 10.1017/S0950268812000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skov MN, Madsen JJ, Rahbek C, Lodal J, Jespersen JB, Jorgensen JC, Dietz HH, Chriel M, Baggesen DL. 2008. Transmission of Salmonella between wildlife and meat-production animals in Denmark. J. Appl. Microbiol. 105:1558–1568. 10.1111/j.1365-2672.2008.03914.x [DOI] [PubMed] [Google Scholar]
- 34.Velarde R, Porrero MC, Serrano E, Marco I, Garcia M, Tellez S, Dominguez L, Aymi R, Lavin S. 2012. Septicemic salmonellosis caused by Salmonella Hessarek in wintering and migrating song thrushes (Turdus philomelos) in Spain. J. Wildl. Dis. 48:113–121. 10.7589/0090-3558-48.1.113 [DOI] [PubMed] [Google Scholar]
- 35.Hoadley AW, Kemp WM, Firmin AC, Smith GT, Schelhorn P. 1974. Salmonellae in the environment around a chicken processing plant. Appl. Microbiol. 27:848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberger M, Keller N. 2005. Recent trends in the epidemiology of non-typhoid Salmonella and antimicrobial resistance: the Israeli experience and worldwide review. Curr. Opin. Infect. Dis. 18:513–521. 10.1097/01.qco.0000186851.33844.b2 [DOI] [PubMed] [Google Scholar]
- 37.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, Agmon V, McClelland M, Rahav G, Gal-Mor O. 2013. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PLoS One 8(3):e58449. 10.1371/journal.pone.0058449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arshad MM, Wilkins MJ, Downes FP, Rahbar MH, Erskine RJ, Boulton ML, Younus M, Saeed AM. 2008. Epidemiologic attributes of invasive non-typhoidal Salmonella infections in Michigan, 1995–2001. Int. J. Infect. Dis. 12:176–182. 10.1016/j.ijid.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 39.Helms M, Ethelberg S, Molbak K, DT104 Study Group 2005. International Salmonella Typhimurium DT104 infections, 1992–2001. Emerg. Infect. Dis. 11:859–867. 10.3201/eid1106.041017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassal R, Reisfeld A, Andorn N, Yishai R, Nissan I, Agmon V, Peled N, Block C, Keller N, Kenes Y, Taran D, Schemberg B, Ken-Dror S, Rouach T, Citron B, Berman E, Green MS, Shohat T, Cohen D. 2012. Recent trends in the epidemiology of non-typhoidal Salmonella in Israel, 1999–2009. Epidemiol. Infect. 140:1446–1453. 10.1017/S095026881100197X [DOI] [PubMed] [Google Scholar]