Abstract
Apicomplexan protists such as Plasmodium and Toxoplasma contain a mitochondrion and a relic plastid (apicoplast) that are sites of protein translation. Although there is emerging interest in the partitioning and function of translation factors that participate in apicoplast and mitochondrial peptide synthesis, the composition of organellar ribosomes remains to be elucidated. We carried out an analysis of the complement of core ribosomal protein subunits that are encoded by either the parasite organellar or nuclear genomes, accompanied by a survey of ribosome assembly factors for the apicoplast and mitochondrion. A cross-species comparison with other apicomplexan, algal and diatom species revealed compositional differences in apicomplexan organelle ribosomes and identified considerable reduction and divergence with ribosomes of bacteria or characterized organelle ribosomes from other organisms. We assembled structural models of sections of Plasmodium falciparum organellar ribosomes and predicted interactions with translation inhibitory antibiotics. Differences in predicted drug–ribosome interactions with some of the modelled structures suggested specificity of inhibition between the apicoplast and mitochondrion. Our results indicate that Plasmodium and Toxoplasma organellar ribosomes have a unique composition, resulting from the loss of several large and small subunit proteins accompanied by significant sequence and size divergences in parasite orthologues of ribosomal proteins.
Keywords: ribosomes, Apicomplexa, organelles, large subunit (LSU) proteins, small subunit (SSU) proteins, antibiotics
2. Introduction
Plasmodium parasites have three genomes [1]: a 23 Mb nuclear genome distributed on 14 linear chromosomes [2], a 35 kb circular genome found in the relic plastid (the apicoplast) [3] and a 6 kb linear genome in the mitochondrion [4,5]. Each of these genomes is transcribed by its own apparatus [6–8] and each compartment possesses a suite of unique ribosomes for its translation [9–11]. Recent reports have provided insights into the partitioning, function and antibiotic interactions of organellar translation factors in Plasmodium spp. [12–16].
Eukaryotic ribosomes consist of one large (60S) and one small (40S) subunit which come together during translation to form an 80S particle. By contrast, ribosomes of bacterial origin consist of a large (50S) and small (30S) subunit that assemble to form a 70S particle. Consistent with their endosymbiotic origins, the apicoplast and mitochondria contain 70S ribosomes that are distinguishable in size (around 20 nm) from the 80S eukaryotic-type ribosomes (around 25–30 nm) found in the cytosol and rough endoplasmic reticulum (ER) [9,17,18]. In addition to ultrastructural characterization, early sequencing of organellar DNA revealed bacterial-type rRNA molecules on the mitochondrial and apicoplast genomes [19,20], although the unexpected presence of the apicoplast understandably gave rise to confusion between apicoplast and mitochondrial DNA in some of the earliest analyses [21]. Further sequencing of the 35 kb apicoplast genome revealed the presence of a complete set of rRNAs as well as a cluster of ribosomal proteins of clear plastid and bacterial origins [3]. Complete sequencing of the 6 kb mitochondrial genome revealed a collection of fragmented rRNA molecules, but no ribosomal proteins [19,22].
Initial analysis of sequenced Plasmodium nuclear DNA fragments and expressed sequence tags (ESTs), then later assembly of the entire Plasmodium falciparum nuclear genome, revealed many more ribosomal proteins with apicoplast and mitochondrial targeting sequences [2,23] that are post-translationally processed for targeting to organelles. The subsequent sequencing of organellar and nuclear genomes from a large number of other apicomplexans has expanded our picture of ribosomal and other translation components in organelles. Here, we attempt to clarify the complement of the core protein translation components by performing a cross-species survey of ribosomal proteins and ribosome assembly factors required for organellar translation in apicomplexans.
Our survey identifies considerable divergence between the organellar ribosomes of apicomplexan parasites and the ribosomes characterized in bacteria or other endosymbiotic organelles. In addition to very significant sequence and size divergences in identified orthologues of ribosomal proteins, several ribosomal proteins are either missing or sufficiently divergent to be unrecognizable. Within the phylum, we also detect several differences in ribosomal protein composition, both in those encoded by apicoplast genomes and those found in the nucleus.
Using the conserved ribosomal proteins and rRNA species identified, we have assembled structural models of the sections of the apicoplast and mitochondrial ribosomes to predict interactions of those ribosomes with parasite-killing drugs predicted to bind to bacterial ribosomes. We find considerable differences in these predicted drug–ligand interactions, with several of the modelled structures suggesting specificity of inhibition between apicoplast and mitochondrial ribosomes.
3. Results and discussion
3.1. Compositional analysis of apicoplast and mitochondrial ribosomes of Plasmodium falciparum
We conducted a survey of available sequences of apicomplexan apicoplast genomes, comparing ribosomal proteins encoded by different species. A list of ribosomal proteins was first assembled, based particularly on the well-annotated nuclear and organellar genomes of the red alga Cyanidioschyzon merolae [24–26] and the diatom Thalassiosira pseudonana [27,28]. We used several search strategies—genome projects were interrogated by text searches to find all annotated ribosomal proteins, and these were manually examined, gene models and predicted proteins were subject to blastp searches, whereas genome nucleotide data were subjected to tblastn searches. The OrthoMCL database of orthology groups [29] was also searched to find relevant homologues of ribosomal proteins.
3.2. Organellar genomes
These searches revealed several ribosomal proteins on the apicomplexan organellar genomes that had previously been missed as open reading frames (ORFs), or annotated as hypothetical ORFs. The 50S ribosomal protein L11 had previously been annotated on the Toxoplasma gondii apicoplast genome [30] but the syntenic protein on the P. falciparum genome had been hitherto annotated as orf129 [3]. This protein can now be assigned as the missing 50S L11 (table 1; gene IDs detailed in the electronic supplementary material, table S1). Similarly, the apicoplast 30S ribosomal proteins S4, S7 and S19 and the 50S proteins L4 and L36 had previously not been annotated in Babesia—we found ORFs on the B. bovis apicoplast genome that correspond to S19 and L36 at similar positions as on the P. falciparum apicoplast genome (table 1; electronic supplementary material, table S1).
Table 1.
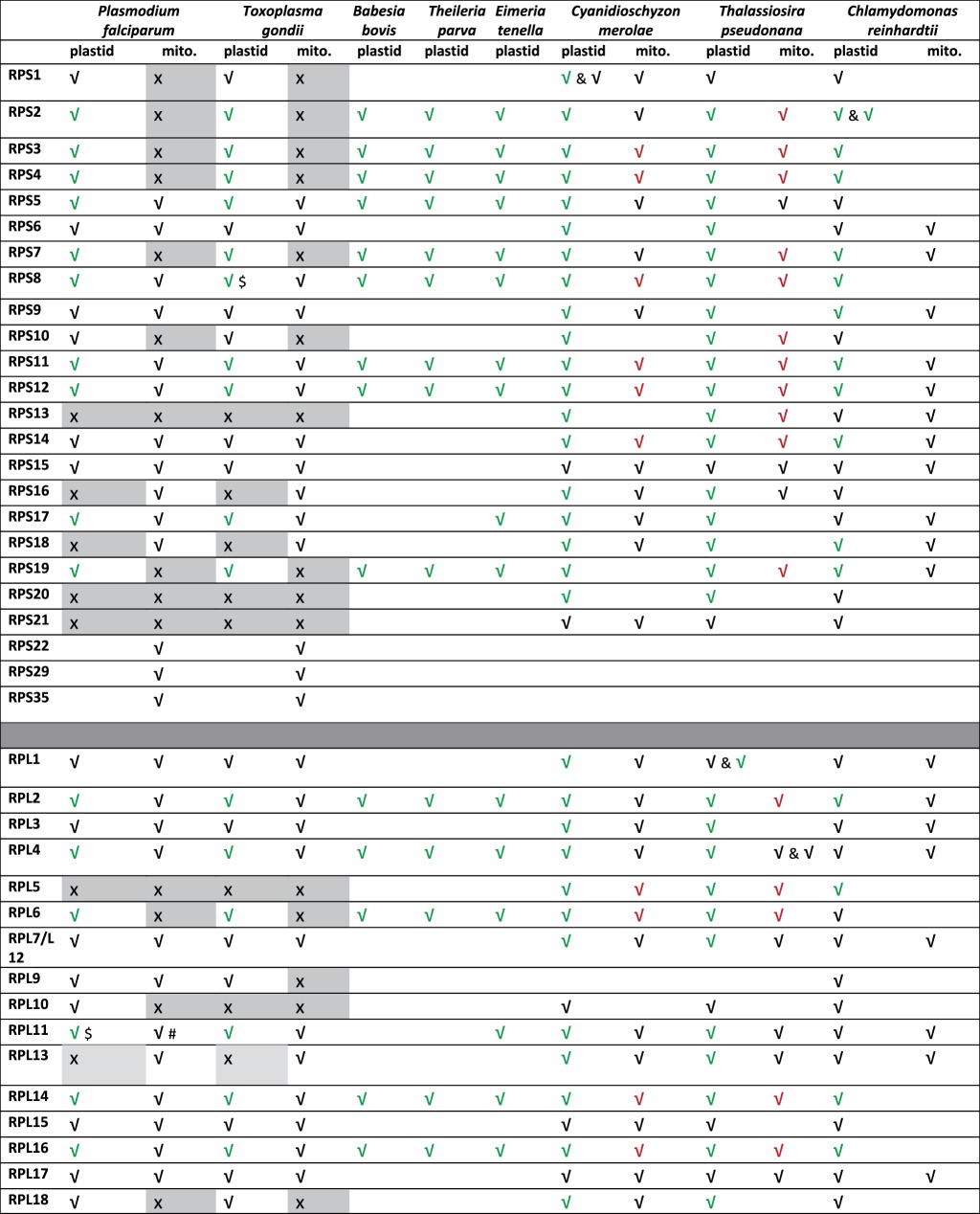
Plastid and mitochondrial ribosome large subunit (LSU) and small subunit (SSU) proteins identified for apicomplexan parasites (P. falciparum, T. gondii, B. bovis, T. parva and E. tenella), red alga (C. merolae), green alga (C. reinhardtii) and diatom (T. pseudonana). #, assigned by sequence similarity or by excluding other organellar counterpart, but targeting leader is non-obvious; $, contains an internal stop codon that may be suppressed. The L7–L12 dimer in eukaryotes is referred to as L8, but L7 and L12 are represented by a single gene in bacteria and organelles. Ticks in black correspond to nuclear-encoded proteins, ticks in red correspond to mitochondrial-encoded proteins and ticks in green correspond to plastid-encoded proteins. Crosses on grey background correspond to proteins for which a comprehensive search was performed on organellar and nuclear genomes and failed to detect any orthologue. Only plastid-encoded ribosomal proteins are listed for the Apicomplexans Babesia, Theileria (Piroplasmida) and Eimeria (Coccidia).
 |
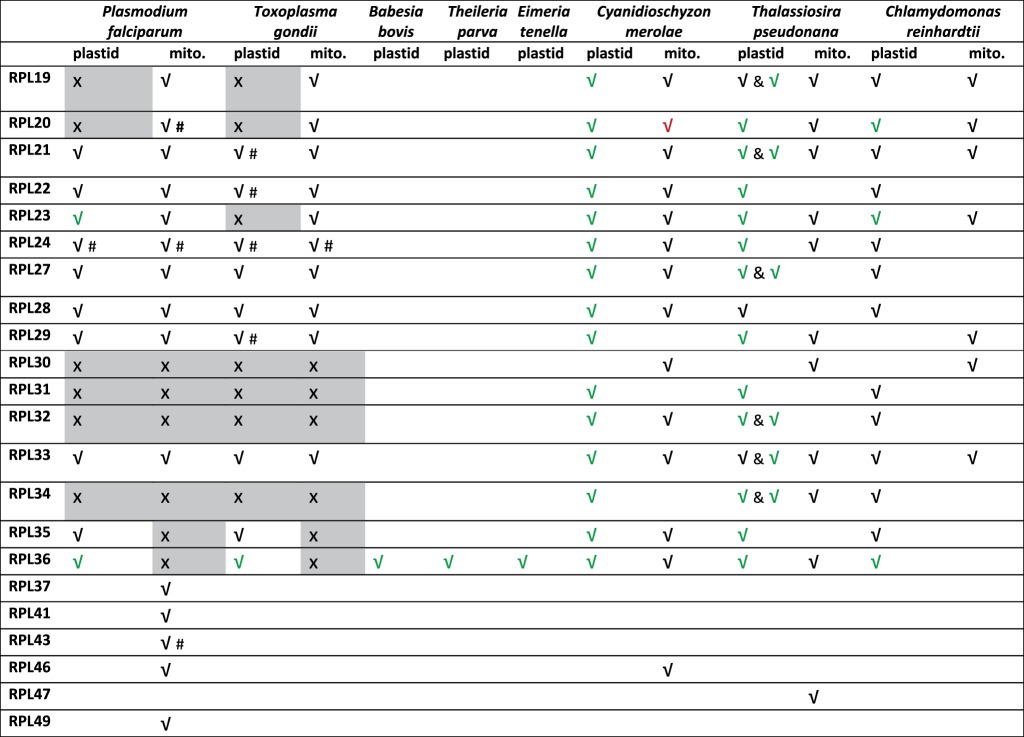 |
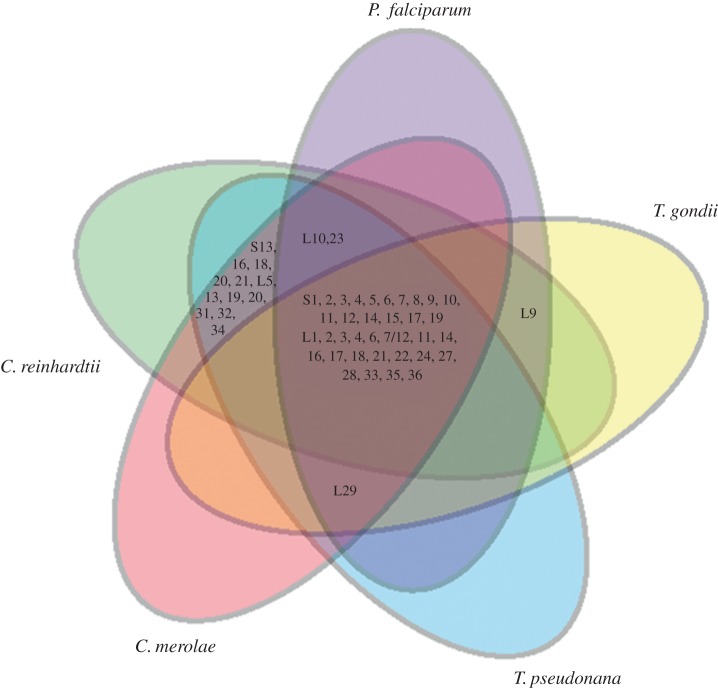
Several differences are seen between the apicoplast ribosomal complements of apicomplexan parasites. Of those proteins encoded by the apicoplast genome itself, the 50S protein L23 is present in Plasmodium, but absent from the Toxoplasma apicoplast genome, and undetectable in its nuclear genome. Rpl23 has also been previously noted as missing from the Eimeria apicoplast genome [31] and is not apparent in other apicomplexan genomes (table 1). This protein, thought to be involved in chaperone docking, is non-essential for growth in Bacillus subtilis [32], and eukaryotes, eubacteria and archaea have divergent ribosomal structures around the L23 site [33] so its absence in some apicomplexan parasites is plausible. Some chloroplast genomes lack L23; and in spinach, the role of L23 has been postulated to be replaced by chloroplast targeting of a eukaryotic 60S type L23a/L25 [34]. However, no N-terminal targeting sequences are apparent on the corresponding Toxoplasma genes.
Another difference between apicoplast genomes within Apicomplexa is the presence or absence of the ribosomal protein S17 (table 1). Plasmodium, Toxoplasma and Eimeria apicoplast genomes carry this gene, but it appears to have been lost from the apicoplast genomes of the piroplasm parasites Theileria and Babesia. We found no evidence for transfer of this apicoplast gene to the nucleus in these parasites (though mitochondrial S17 representatives are present), but S17 is small and relatively poorly conserved at a primary sequence level, so may simply be undetectable in the order Piroplasmida.
3.3. Missing large subunit ribosomal proteins
A number of large subunit (LSU) organellar ribosomal proteins appear to have been lost altogether from apicomplexan genomes. A striking apparent absence in Apicomplexa is the organellar Rpl5. L5 is a 5S rRNA-binding protein that is essential for assembly of the 50S central protuberance in bacteria [35], most of which is clearly retained in apicomplexan ribosomes; however, L5 is missing from mammalian mitochondrial ribosomes [36], so is a plausible absence from apicomplexan organellar ribosomes as well. Another 50S ribosomal protein that binds the 5S rRNA, L25, is missing from other plastid and mitochondrial ribosomes [36,37] and is also absent from apicomplexan genomes.
The 50S ribosomal protein Rpl10 is an apicoplast-targeted protein readily detected in Plasmodium, Babesia and Theileria, but not in Toxoplasma. L10 is relatively large and well conserved with clear apicoplast-targeted orthologues in Plasmodium spp. but no equivalent is obvious in Toxoplasma. Mitochondrial L10 homologues are not obvious for any apicomplexan species. In some other organelles, L10 appears to have been replaced by a nuclear L10 [38], but no L10 with an N-terminal targeting sequence is apparent in Toxoplasma.
The L19 and L20 proteins have orthologues in Plasmodium and Toxoplasma with probable mitochondrial targeting sequences, but we found no orthologues of these with apicoplast targeting leaders (table 1 and figure 1). These are small proteins (approx. 120 amino acids each) and may hence be missed in a sequence similarity search. Several other 50S ribosomal proteins—L30, L31, L32 and L34—have mixed distributions in other organellar ribosomes [37,39,40], and we found no apicoplast or mitochondrial representatives of any of these proteins in apicomplexans.
Figure 1.
A five-set Venn diagram showing the distribution of nuclear- or plastid-encoded ribosomal proteins that would constitute the plastid ribosomes of apicomplexans P. falciparum and T. gondii, red alga C. merolae, green alga C. reinhardtii and diatom T. pseudonana.
3.4. Missing small subunit ribosomal proteins
Compared to the 50S subunit, the 30S subunit retains proportionally more members in the apicoplast genome rather than transfers to the nuclear genome (table 1; gene IDs detailed in the electronic supplementary material, table S1). Several proteins are also missing or undetected among the 30S proteins of the mitochondria and apicoplast. The mitochondrion in particular appears to be missing a large number of subunits, and we were unable to find mitochondrial targeted orthologues of S1, S2, S3, S4, S7, S10, S13, S19, S20 or S21. Most of these are retained on the mitochondrial genomes of diatoms (table 1) and are widely conserved among other organellar ribosomes, so their complete absence in apicomplexan mitochondria is unexpected and not easily explained. One possibility is that the mitochondrial ribosomes employ prokaryotic subunits encoded by the apicoplast (though no mechanism is obvious for this) or may use proteins dually targeted to the mitochondrion and apicoplast. Several apparently mitochondrial targeted proteins are annotated as 30S ribosomal proteins in apicomplexans, including S22, S29 and S35. These are not widespread members of mitochondrial ribosomes, so their presence here may be linked to the possible absence of other canonical members.
Several 30S proteins are also apparently lacking in the apicoplast ribosomes. Despite the presence of clear orthologues in red algal and diatom organelles (table 1), no apicoplast (or mitochondrial) S13 ribosomal proteins are apparent in any apicomplexan species. This protein interacts with the 50S subunit and the P-site tRNA during translocation [41] and is essential for translation in other bacterial ribosomes [42], so its apparent absence is puzzling [42].
Apicomplexan parasites also appear to have lost their apicoplast version of S16, which is encoded on the plastid genomes of diatoms and of red and green alga, but have retained mitochondrial targeted S16 proteins (table 1 and figure 1). In bacteria, S16 is essential and plays a central role in 30S ribosomal assembly [43]. S16 is dual targeted between mitochondria and chloroplasts in many plants [44] but we see no evidence for the presence of a possible apicoplast leader upstream of the mitochondrial S16 in Plasmodium or Toxoplasma. Another 30S protein, S18 is absent from all the apicomplexan apicoplasts we surveyed, though mitochondrial S18 s are present (table 1). This protein has no obvious orthologue in archaeal or eukaryotic ribosomes [45], although it is essential in tobacco plastids [46].
S20 and S21 are also missing from the apicomplexans we surveyed. S20 is not essential in Salmonella [47], knockout of S21 impairs but does not ablate translation in other plastids [48], and neither is essential in B. subtilis [32] so these are plausible absences from the organellar ribosomes of Apicomplexa.
3.5. Ribosome assembly proteins for the apicoplast and mitochondrion
Ribosome biogenesis involves multiple steps of ribosomal RNA (rRNA) processing and association of rRNA with ribosomal proteins [49–51]. As with any complex RNA molecule, the rRNA in parasite organelles is prone to the formation of numerous local non-native secondary structures. A set of cofactors known as ribosomal biogenesis/assembly factors prevents formation of these stable, misfolded regions in the rRNA and promotes ribosome assembly [50]. These factors serve as check points during the assembly process where they mediate proper rRNA folding and protein–RNA interactions by creating specific nucleotide modifications in rRNA or by acting as RNA/protein chaperones. This ultimately results in the assembly of mature ribosomal subunits. We performed an extensive search for putative ribosomal biogenesis factors targeted to the apicoplast and mitochondrion in the PlasmoDB genome database using current annotations as well as new assignments based on targeting prediction algorithms.
Ribosome assembly factors belong to the following broad categories—GTPases, chaperones/maturation factors and DEAD-box proteins [52]. GTP hydrolysis by proteins of the GTPase superclass is involved at different stages of ribosome biogenesis mediating subunit assembly. Era, Der, Obg and YihA are known to interact with either the mature subunits or the 70S ribosome while YlqF also exhibits interaction with a ribosomal subunit intermediate [53]. Sequence analysis indicates the presence of multiple P-loop GTPases in P. falciparum that contain highly conserved motifs (table 2). The Der protein is conserved among eubacteria but not in archaea or eukaryotes [52]; two Der homologues, with predicted targeting to the apicoplast and mitochondrion, respectively, could be identified. A search for homologues of organellar Era and YihA proteins yielded putative candidates with mitochondrial localization while the single YlqF homologue had apicoplast targeting elements. Two candidates were found for Obg, one of which was predicted to be mitochondrial while the other appears to be targeted to the apicoplast.
Table 2.
Organellar ribosome assembly proteins of P. falciparum and their predicted targeting.
| s. no. | ribosome assembly proteins | putative interactions and functions | PlasmoDB annotation | PlasmoDB gene ID | apicoplast targeting |
mitochondrial targeting |
probable organellar destination | |||
|---|---|---|---|---|---|---|---|---|---|---|
| PlasmoAP | PATS | signal peptide (TargetP) | PlasMit | MitoProt II | ||||||
| GTPases | ||||||||||
| 1 | Era/Bex | involvement in 16S rRNA processing and 30S subunit biogenesis | GTPase, putative | PF3D7_1435800 | −/++ | no (0.048) | no (0.024) | mito. (91%) | (0.550) | mitochondrion |
| 2 | Der/EngA/YfgK/YphC | association with 50S subunit and involvement in its maturation | GTP-binding protein, putative | PF3D7_1217300 | 0/++ | yes (0.955) | yes (0.929) | non-mito. (99%) | (0.570) | apicoplast |
| GTP-binding protein EngA, putative | PF3D7_0313500 | −/++ | no (0.062) | no (0.051) | mito. (91%) | (0.409) | mitochondrion | |||
| 3 | Obg/CgtAE/YhbZ/ObgE | association with 30S and 50S subunit; also co-sediments with 16S and 25S rRNA | GTP-binding protein, putative | PF3D7_1411600 | +/++ | yes (0.970) | yes (0.806) | non-mito. (99%) | (0.860) | apicoplast |
| GTP-binding protein, putative | PF3D7_0824300 | −/++ | no (0.392) | no (0.037) | mito. (91%) | (0.924) | mitochondrion | |||
| 4 | YihA/EngB/YsxC | interaction with Der protein and activation of its GTP activity. Involvement in 50S subunit assembly | GTP-binding protein, putative | PF3D7_0513400 | −/++ | no (0.058) | no (0.030) | mito. (91%) | (0.842) | mitochondrion |
| GTP-binding protein, putative | PF3D7_1442200 | −/++ | no (0.061) | no (0.029) | mito. (91%) | (0.674) | mitochondrion | |||
| 5 | YlqF/RbgA | involvement in 50S subunit assembly; co-sediments with 45S intermediate | GTPase, putative | PF3D7_0410700 | ++/++ | yes (0.976) | yes (0.928) | non-mito. (99%) | (0.526) | apicoplast |
| maturation factors and chaperones | ||||||||||
| 6 | RimM | interaction with RP-S19 in the free 30S subunit and involvement in 16S rRNA processing | mitochondrial preribosomal assembly protein rimM precursor, putative | PF3D7_1032000 | ++/++ | yes (0.992) | yes (0.971) | non-mito. (99%) | (0.560) | apicoplast |
| 7 | RlmE/RrmJ/FtsJ | specific methylation at uridine of 23S rRNA in the fully assembled 50S subunit | rRNA methyltransferase, putative | PF3D7_1309600 | −/+ | no (0.451) | no (0.087) | non-mito. (99%) | (0.213) | |
| large subunit rRNA methyltransferase, putative | PF3D7_1354300 | −/++ | no (0.022) | no (0.035) | mito. (91%) | (0.145) | mitochondrion | |||
| rRNA methyltransferase, putative | PF3D7_0908600 | −/++ | no (0.023) | no (0.031) | non-mito. (99%) | (0.149) | ||||
| 8 | RsmB/Sun/RrmB/Fmu | specific methylation at cytosine of 16S rRNA | methyltransferase, putative | PF3D7_1020400 | −/++ | no (0.030) | no (0.120) | mito. (91%) | (0.525) | mitochondrion/apicoplast |
| 9 | KsgA/RsmA/Dim1 | specific di-methylation at two adjacent adenosines near 3′ end of 16S rRNA in the 30S particle | small subunit rRNA dimethylase, putative | PF3D7_1415800 | −/++ | no (0.185) | no (0.086) | mito. (91%) | (0.981) | mitochondrion [54] |
| apicoplast dimethyladenosine synthase, putative | PF3D7_1249900 | ++/++ | yes (0.999) | yes (0.941) | non-mito. (99%) | (0.996) | apicoplast | |||
| 10 | DnaJ/HSP40 | chaperone | heat shock protein 40 (DnaJ) | PF3D7_0409400 | −/++ | yes (0.900) | no (0.032) | non-mito. (99%) | (0.474) | apicoplast targeting demonstrated [55] |
| DnaJ protein, putative | PF3D7_0629200 | 0/0 | yes (0.502) | yes (0.982) | non-mito. (99%) | (0.450) | apicoplast | |||
| 11 | DnaK/HSP70 | heat shock protein 70 (Hsp70-3) | PF3D7_1134000 | −/++ | no (0.046) | no (0.027) | mito. (91%) | (0.443) | mitochondrion | |
| 12 | GroEL/Cpn60 | heat shock protein 60 (HSP60) | PF3D7_1015600 | −/++ | no (0.019) | no (0.043) | mito. (91%) | (0.951) | mitochondrial targeting demonstrated [56] | |
| 60 kDa chaperonin (CPN60) | PF3D7_1232100 | ++/++ | yes (0.979) | yes (0.776) | non-mito. (99%) | (0.824) | apicoplast targeting demonstrated [56] | |||
| 13 | GroES/Cpn10 | 10 kDa chaperonin (CPN10) | PF3D7_1215300 | −/− | no (0.317) | no (0.110) | non-mito. (99%) | (0.493) | mitochondrial targeting demonstrated [56] | |
| 14 | Cpn20 | 20 kDa chaperonin (CPN20) | PF3D7_1333000 | ++/++ | no (0.944) | yes (0.931) | non-mito. (99%) | (0.665) | apicoplast targeting demonstrated [56] | |
Chaperones assist in proper folding/unfolding and assembly/disassembly of ribosomal proteins and rRNA. We identified seven chaperones, five of which are already annotated in previous reports as being targeted to the apicoplast (DnaJ, Cpn60 and Cpn20) or mitochondrion (Cpn60 and Cpn10) [55,56]. Two other putative chaperones—DnaJ and DnaK—that might be apicoplast- and mitochondrion-targeted, respectively, were also identified (table 2). In addition to chaperones, RNA maturation factors play a vital role in the rRNA modifications during ribosome biogenesis. RrmJ, RsmB and KsgA are methyltransferases that methylate specific nucleotides in rRNA during their maturation [57–59]. KsgA methylates two adjacent adenosine residues at the 3′ terminal helix of small subunit (SSU) rRNA that are two of three nucleotide modifications that are known to be conserved in nearly all known ribosomes throughout evolution [60] with few exceptions [61–64]. Two homologues of RlmE/RrmJ were identified in the P. falciparum genome, one of which had a predicted mitochondrial targeting signal while the location of the other cannot be clearly predicted. Two KsgA/RsmA were predicted, one for the mitochondrion and the other with possible dual targeting to both organelles. A single RsmB with possible dual targeting to the apicoplast and mitochondrion was also identified. Further, a homologue of the ribosomal maturation factor RimM that is involved in SSU biogenesis [65] is predicted for the apicoplast (table 2).
DEAD-box proteins, which are conserved across bacteria and viruses to humans [66], belong to a large family of RNA helicases that possess RNA-dependent ATPase activity. They act as RNA chaperones, mediate RNA–protein interaction and unwind local RNA structures [67–69]. A number of putative proteins that may belong to the DEAD-box family and are predicted to contain sequence elements for organellar import (PF3D7_1445900, PF3D7_0218400, PF3D7_1332700, PF3D7_1418900, PF3D7_0504200, PF3D7_1021500 and PF3D7_1251500) were identified. These proteins have a conserved DEAD-box motif and RNA helicase domain but could not be unambiguously classified as a specific member (SrmB, CsdA, DbpA, RhlE or RhlB) of the Escherichia coli DEAD-box helicase family [70–74].
3.6. Structure modelling of Plasmodium falciparum organellar ribosome subunits and drug interaction sites
Several antibiotics, including clindamycin, chloramphenicol and the macrolides erythromycin and azithromycin, bind in the vicinity of the ribosome LSU peptidyl transferase centre or the peptide exit tunnel and inhibit parasite growth. This group also includes thiostrepton that contacts ribosomal protein L11 and the GTPase region of 23S rRNA [75]. Translation inhibitory antibiotics have two putative target organelles, the apicoplast and mitochondrion, of the parasite. Some antibiotics (e. g. clindamycin, azithromycin, chloramphenicol and tetracycline) have been demonstrated to have a delayed-death effect, a phenotype associated with apicoplast-specific action [76,77]. A single point mutation in the LSU rRNA gene of the T. gondii apicoplast confers clindamycin resistance in vitro [78] and resistance to azithromycin in P. falciparum has been attributed to two point mutations: one in the P. falciparum apicoplast LSU rRNA and a second in the apicoplast-encoded Rpl4 [79]. Thiostrepton causes immediate parasite killing and is proposed to have additional targets in P. falciparum [80,81]. In order to understand the differential interaction of these drugs with apicoplast and mitochondrial ribosomes, we carried out in silico modelling of LSU rRNA and relevant ribosomal proteins (L4, L11 and L22) involved in interactions with antibiotics in bacteria. This was followed by docking of antibiotics in order to estimate their relative specificity for mitochondrial and apicoplast ribosomes.
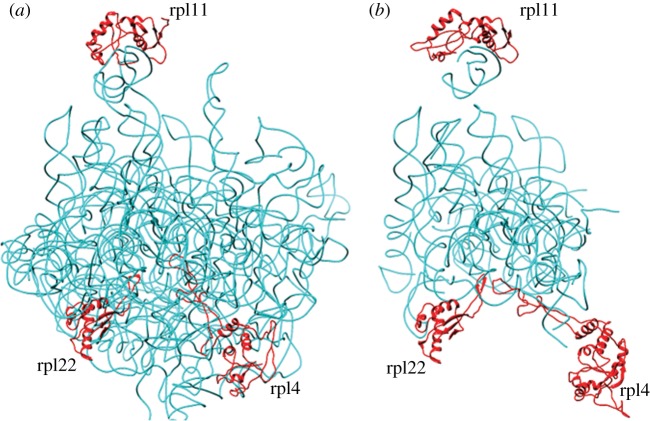
Prediction of the three-dimensional structures of parasite organelle ribosomes is demanding due to the difficulty in obtaining high-resolution experimental models. This is further complicated by the presence of highly fragmented rRNA encoded by the P. falciparum mitochondrial genome [11]. A stand-alone version of the RNA prediction tool ModeRNA was used for the comparative modelling of rRNA, whereas modelling of ribosomal proteins L4, L11 and L22 was performed by Modeller v. 9.10. For modelling of the mitochondrial ribosome, different fragments of mitochondrial LSU rRNA were aligned manually on the basis of conserved secondary structure topology, modelled separately and then superimposed together on the E. coli template to obtain a complex RNA model structure. All the modelled subunits (rRNA and protein) were superimposed on the E. coli ribosome template to generate the apicoplast and mitochondrial ribosome complexes (figure 2). The fragmented rRNA comprising the core of the mitochondrial ribosome is highly reduced, though retains conservation of the peptidyl transferase centre and the peptide exit tunnel where most antibiotics bind.
Figure 2.
Structure models of P. falciparum apicoplast (a) and mitochondrial (b) LSU rRNA and proteins L11, L4 and L22. The rRNA and protein subunits were modelled separately and superimposed on the E. coli ribosome template to generate the ribosome complexes. LSU rRNA is shown in cyan and proteins in red.
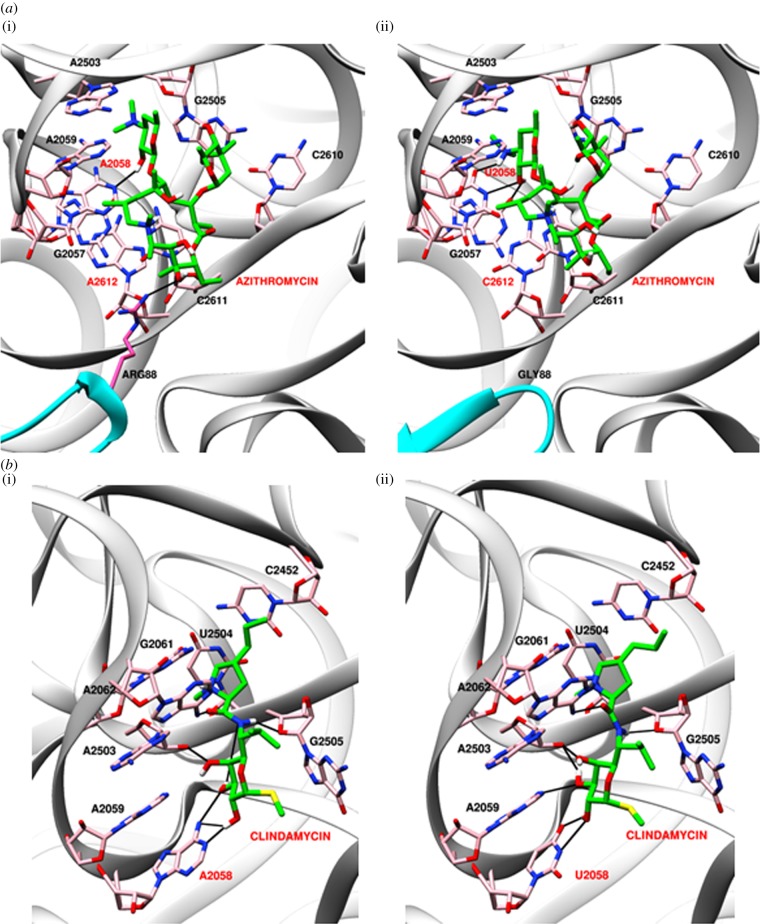
Molecular docking of antibiotics was performed on P. falciparum apicoplast and mitochondrial ribosome models using Autodock4. Autodock uses grid-based energy evaluation for docking, where ligands are treated as flexible entities by exploring torsional degrees of freedom of ligand molecules. The first step of the Autodock algorithm involves conformational sampling of ligands followed by prediction and ranking of free energy of binding of these conformations. One hundred Autodock runs were performed for each inhibitor. To validate the reproducibility and sensitivity of the docking program, Autodock4 was used to dock the inhibitor co-complexed with the E. coli template. The inhibitor dock scores obtained for apicoplast and mitochondrial ribosomes are given in table 3. In the apicoplast, L22 located at the binding site for azithromycin [82] contains Arg88 that is predicted to form an H-bond with the inhibitor (figure 3a). Arg88 is replaced by Gly88 in mitochondrial L22 that does not form an H-bond with azithromycin. In addition, the rRNA sequence at the binding site also differs at two positions: A2612 and A2058 (E. coli number) in the apicoplast are replaced by C2612 and U2058, respectively, in the mitochondrion, a change that would alter the hydrophobic environment at the site. This might explain the differential specificity of azithromycin for organellar ribosomes. The higher affinity of the antibiotic for the apicoplast ribosome is also reflected in the lower dock scores obtained for azithromycin and the related macrolide erythromycin (table 3). Together with the LSU rRNA, L22 and L4 are predicted to form the peptide exit tunnel on the ribosome. The G76V mutation of apicoplast L4 has been reported to contribute to azithromycin resistance in P. falciparum lines [79] and modelling on the ribosome–azithromycin structure predicted a conformational shift in the side chain of Leu75 of L4 that could interfere with the azithromycin binding pocket. However, this model was constructed on the Deinococcus radiodurans (an extremophile bacterium) model that proposed the binding of two azithromycin residues at the site, one that interacted with the LSU rRNA and the other with L4, L22 and LSU rRNA [83]. Structures of the Haloarcula marismortui (an archaeon) and Thermus thermophilus large ribosomal subunits complexed with azithromycin have since led to the conclusion that a single molecule of the antibiotic binds to the ribosome [82]. This is supported by biochemical experiments that indicate that only one azithromycin molecule is bound to the E. coli ribosome [84]. No direct role for L4 in the interaction of azithromycin with P. falciparum apicoplast and mitochondrial ribosomes was detected in our model.
Table 3.
Docking scores of antimicrobials in the active site of large ribosomal subunit of E. coli, and P. falciparum apicoplast and mitochondrion.
| antibiotic |
P. falciparum apicoplast LSU |
P. falciparum mitochondrial LSU |
E. coli LSU |
||||
|---|---|---|---|---|---|---|---|
| dock score (kcal mol−1) | rmsd (Å) | dock score (kcal mol−1) | rmsd (Å) | dock score (kcal mol−1) | rmsd (Å) | ||
| 1 | chloramphenicol | −3.44 | 1.31 | −3.19 | 1.33 | −3.55 | 1.25 |
| 2 | erythromycin | −13.85 | 1.01 | −11.04 | 1.61 | −12.2 | 0.97 |
| 3 | azithromycina | −21.47 | 0.7 | −18.3 | 1.78 | −18.64 | 0.64 |
| 4 | clindamycin | −15.97 | 1.13 | −14.44 | 1.57 | −14.94 | 1.07 |
| 5 | thiostreptonb | −2.69 | 3.68 | −1.98 | 1.69 | −2.31 | 0.68 |
aModelled on the Thermus thermophilus ribosome–azithromycin crystal structure.
bModelled on the Deinococcus radiodurans ribosome–thiostrepton crystal structure.
Figure 3.
Modelling of antibiotic interactions with P. falciparum organelle ribosomes. (a) Azithromycin docked onto apicoplast (i) and mitochondrial (ii) ribosomes. As in the Thermus thermophilus ribosome–azithromycin structure, a single azithromycin molecule was docked at the binding site. (b) Interaction of clindamycin with apicoplast (i) and mitochondrial (ii) LSU rRNA. Bases that differ between the apicoplast and mitochondrial rRNA are shown in red and H-bonds as black lines. rRNA is in grey, L22 in cyan and antibiotics are in green.
The only difference in the interaction site for clindamycin between the organelle ribosomes was an A2058U (E. coli number) transversion in the LSU rRNA of the parasite mitochondrion (figure 3b). This residue forms H-bonds with the antibiotic in E. coli [85]. The LSU rRNA residue G2061, whose mutation in the apicoplast is associated with clindamycin resistance in T. gondii [78] and which is critical to the transpeptidation reaction, was conserved in the LSU rRNA of both organelles in Plasmodium. The binding site for chloramphenicol overlaps with that of clindamycin and no obvious differences could be detected in chloramphenicol interactions predicted for apicoplast and mitochondrial ribosomes; the dock scores for chloramphenicol were also comparable for P. falciparum organelle ribosomes. However, the in silico approach used by us would have inherent weaknesses, and conclusions on actual interactions and affinity of these antibiotics for apicoplast/mitochondrial ribosomes awaits experimental validation.
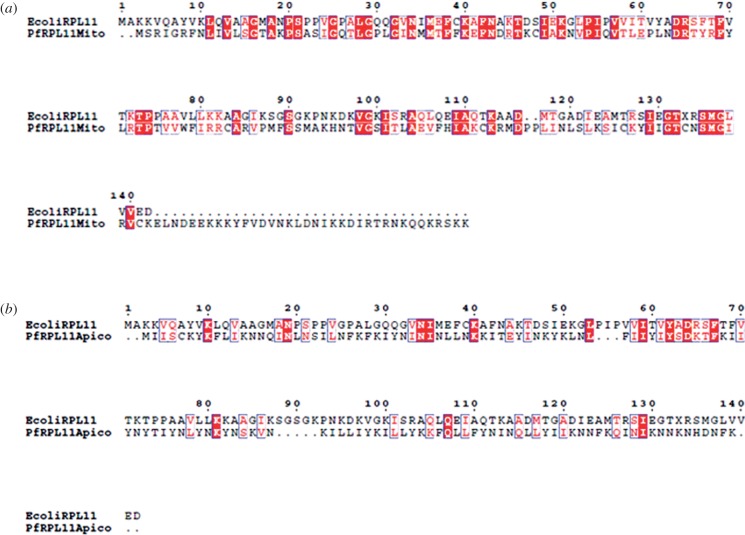
Thiostrepton targets the GTPase associated centre of the 50S ribosome subunit and binds within a cleft between helices 43 and 44 of the LSU rRNA and L11. It overlaps with the position of domain V of elongation factor G (EF-G), thus perturbing the binding of the elongation factor to ribosomes [86]. Plasmodium falciparum organelle LSU rRNAs differ at two residues in the helices: the crucial A1067 site and A1095 (E. coli number) are replaced by G1067 and C1095 in the mitochondrion (figure 4). The former has been shown to alter binding of thiostrepton to the ribosome although introduction of an A1067G mutation in the apicoplast rRNA did not completely abolish in vitro interaction with the antibiotic [87]. It is also important to note the low identity and consequent conformational differences in L11 of the apicoplast and mitochondrion that might influence interaction with thiostrepton. The structural models in figure 4 as well as the ClustalW alignment of E. coli and P. falciparum organelle L11 proteins indicate greater similarity between the bacterial and parasite mitochondrial ribosome–thiostrepton interaction site compared with the apicoplast [86] (figures 4 and 5). The identity between the mitochondrial and apicoplast L11 with the E. coli protein is 24.71% and 10.07%, respectively. In addition to targeting the apicoplast, thiostrepton has also been shown to act on the cytosolic proteasome [80] and has detectable effects on mitochondrial translation [81]. Thiostrepton is also able to partially lock P. falciparum mitochondrial EF-G onto surrogate E. coli ribosomes, an effect not observed with apicoplast EF-G [14].
Figure 4.
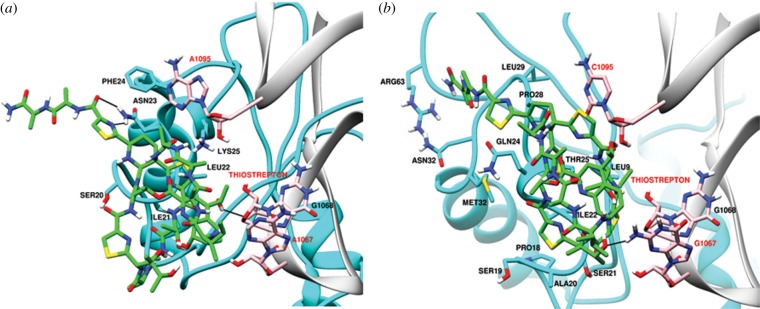
Predicted interaction of thiostrepton with LSU rRNA and L11 of ribosomes of the P. falciparum apicoplast (a) and mitochondrion (b). rRNA is in grey, L11 in cyan and thiostrepton in green.
Figure 5.
ClustalW alignment of E. coli L11 with L11 predicted for the P. falciparum mitochondrion (a) and apicoplast (b).
Except for the macrolide antibiotics whose preferential interaction with P. falciparum apicoplast ribosomes can be explained on the basis of structural differences with ribosomes of the mitochondrion, few obvious structural explanations can be found for differential drug binding to apicoplast and mitochondrial ribosomes by other antibiotics tested by us. Apicoplast-specific inhibitory effects that have been observed with clindamycin and chloramphenicol may thus be due to differential sensitivity attributable to other biological factors such as differences in drug accumulation in the two organelles or reduced rate of translation in the parasite mitochondrion. For thiostrepton, the docking results and structural models reported here support earlier biochemical data that the antibiotic targets both apicoplast and mitochondrial translation thus mediating early parasite death.
In conclusion, apicoplast and mitochondrial ribosomes of apicomplexan parasites have a unique and reduced composition, a fact that would alter the nature of their interactions with protein translation factors. This survey is a starting point for further functional evaluation of the Plasmodium organellar ribosome machinery, its assembly and interactions with translation factors and translation inhibitory compounds.
4. Material and methods
4.1. Databases and sequence searches
To identify ribosomal proteins and ribosome assembly factors, we searched apicomplexan genomes using the GenBank non-redundant nucleotide and CDS translations [88] using tblastn and blastp, respectively. We additionally performed direct alignments between protein sequences and organellar genomes using blast2seq. Signal peptide portions of apicoplast targeting sequences were sought using SignalP v. 3.0 [89] and by manual inspection of Kyte Doolitle hydropathy plots [90]. Gene models were examined using EuPathDB [91] and evidence for transcription start sites and alternative splicing based on RNAseq data examined using the GBrowse tool [92] implemented at EuPathDB [91]. Putative transit peptide portions of apicoplast targeting sequence were manually inspected or were detected using the PlasmoAP [93] and PATS [94]. Putative mitochondrial transit peptides were manually inspected or were detected using PlasMit [95] or MitoProtII [96].
Where we identified clear ribosomal proteins that lacked clear annotations, or clear organellar trafficking that was not included in earlier annotations, we communicated updated annotations to curation staff. For high confidence assignments the gene names have been changed, for lower confidence assignments relevant comments have been added to gene pages at the EuPathDB [91] and GeneDB genome databases [97].
For ribosome assembly/biogenesis proteins, all predictions were made on the basis of annotations in PlasmoDB as well as assignments made by prediction algorithms—TargetP, PlasmoAP, PATS, PlasMit and MitoProt II.
4.2. Molecular modelling
Prediction of three-dimensional structure of ribosomal structure is highly demanding owing to the difficulty in obtaining high-resolution experimental models. Present work describes the in silico modelling of apicoplast and mitochondrial large subunits of 23S ribosome followed by docking studies with known inhibitors to understand the comparative basis of specificity of these inhibitors. To achieve the modelling of rRNA, an RNA prediction tool ModeRNA [98] was used, whereas modelling of ribosomal proteins L4, L11 and L22 was performed by Modeller v. 9.10 [99]. After model building of large subunit of 23S rRNA, known inhibitors azithromycin, erythromycin, clindamycin, chloramphenicol and thiostrepton were docked into the peptidyl transferase site of modelled apicoplast and mitochondrial ribosome, respectively.
ModeRNA is a comparative modelling tool of RNA which requires a template whose three-dimensional structure is known and which shares sequence similarity with the query sequence, the one to be modelled and pairwise alignment between template and the query sequences [98]. 23S ribosome of E. coli (PDB id: 3OFC) was chosen as the template to model P. falciparum 23S rRNA in apicoplast as well as in mitochondria. As the secondary structures have been published for E. coli and P. falciparum ribosomes, the alignments were performed manually on the basis of conserved secondary structure topology to facilitate the modelling of rRNA of apicoplast and mitochondria in P. falciparum. The RNA models were built with the stand-alone version of the ModeRNA via a Python scripting interface based on the provided alignments. The default ModeRNA modelling procedure was followed. As the P. falciparum mitochondrial RNA is present in fragmented form, different fragments were aligned and modelled separately and then superimposed on the template together to obtain a complex model structure. Simple geometry checks were performed using analyze_geometry function on template and target structures using ModeRNA stand-alone version to ensure the structural integrity of structure.
All the protein models were built with Modeller v. 9.10 based on homologous template structures in E. coli. For each case, 10 different models were produced and the one with the best DOPE score selected. ClustalW was used for alignment between protein templates and the targets to generate comparative models.
All the modelled subunits of each ribosome including RNA and protein were superimposed on template structure and merged together to form the complete ribosome. Although RNA sequences exhibit divergence, the overall structures of modelled ribosomes were found to be well conserved as indicated by the secondary structure topology.
4.3. Molecular docking
Structures of inhibitors were extracted from the Protein Data Bank (PDB) files of large subunit of 70S ribosome co-complexed with the respective inhibitors (PDB IDs: chloramphenicol (3OFC), clindamycin (3OFZ), erythromycin (3OFR), azithromycin (3OHZ) and thiostrepton (3CF5)). Molecular docking was performed on ribosome models used as a receptor to dock our inhibitors of interest using Autodock v. 4 [100]. Kollman charges were assigned with 40 × 40 × 40 grid points of 0.375 Å spacing. One hundred Autodock runs were performed for each inhibitor.
To validate the reproducibility and sensitivity of the docking program, Autodock v. 4 was used to dock the inhibitor co-complexed with template. The limit of Autodock to read maximum atoms of macromolecules was kept constant to default and therefore 35 Å around the ligands was considered only after superimposing the modelled structure on E coli ribosome structure.
Supplementary Material
Funding statement
A.G., P.S. and K.G. received scholarships from the Council for Scientific and Industrial Research, Government of India and A.H. received scholarship from the University Grants Commission, India. This work was supported by the CSIR's Network project Splendid (BSC0104i) to S.H., and by an Australian National Health and Medical Research Council (NHMRC) project grant to S.A.R. S.A.R. is supported by an NHMRC fellowship. This is CDRI communication no. 8670.
References
- 1.Wilson RJM, Gardner MJ, Feagin JE, Williamson DH. 1991. Have malaria parasites three genomes? Parasitol. Today 7, 134–136. (doi:10.1016/0169-4758(91)90276-T) [DOI] [PubMed] [Google Scholar]
- 2.Gardner MJ, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511. (doi:10.1038/nature01097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RJ, et al. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261, 155–172. (doi:10.1006/jmbi.1996.0449) [DOI] [PubMed] [Google Scholar]
- 4.Feagin JE, Gardner MJ, Williamson DH, Wilson RJ. 1991. The putative mitochondrial genome of Plasmodium falciparum. J. Protozool. 38, 243–245. (doi:10.1111/j.1550-7408.1991.tb04436.x) [DOI] [PubMed] [Google Scholar]
- 5.Feagin JE, Werner E, Gardner MJ, Williamson DH, Wilson RJ. 1992. Homologies between the contiguous and fragmented rRNAs of the two Plasmodium falciparum extrachromosomal DNAs are limited to core sequences. Nucleic Acids Res. 20, 879–887. (doi:10.1093/nar/20.4.879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li WB, Bzik DJ, Gu HM, Tanaka M, Fox BA, Inselburg J. 1989. An enlarged largest subunit of Plasmodium falciparum RNA polymerase II defines conserved and variable RNA polymerase domains. Nucleic Acids Res. 17, 9621–9636. (doi:10.1093/nar/17.23.9621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner MJ, Goldman N, Barnett P, Moore PW, Rangachari K, Strath M, Whyte A, Williamson DH, Wilson RJ. 1994. Phylogenetic analysis of the rpoB gene from the plastid-like DNA of Plasmodium falciparum. Mol. Biochem. Parasitol. 66, 221–231. (doi:10.1016/0166-6851(94)90149-X) [DOI] [PubMed] [Google Scholar]
- 8.Li JN, Maga JA, Cermakian N, Cedergren R, Feagin JE. 2001. Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol. Biochem. Parasitol. 113, 261–269. (doi:10.1016/S0166-6851(01)00223-7) [DOI] [PubMed] [Google Scholar]
- 9.McFadden GI, Reith M, Munholland J, Lang-Unnasch N. 1996. Plastid in human parasites. Nature 381, 482 (doi:10.1038/381482a0) [DOI] [PubMed] [Google Scholar]
- 10.Roy A, Cox RA, Williamson DH, Wilson RJ. 1999. Protein synthesis in the plastid of Plasmodium falciparum. Protist 150, 183–188. (doi:10.1016/S1434-4610(99)70020-9) [DOI] [PubMed] [Google Scholar]
- 11.Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS ONE 7, e38320 (doi:10.1371/journal.pone.0038320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas S, Lim EE, Gupta A, Saqib U, Mir SS, Siddiqi MI, Ralph SA, Habib S. 2011. Interaction of apicoplast-encoded elongation factor (EF) EF-Tu with nuclear-encoded EF-Ts mediates translation in the Plasmodium falciparum plastid. Int. J. Parasitol. 41, 417–427. (doi:10.1016/j.ijpara.2010.11.003) [DOI] [PubMed] [Google Scholar]
- 13.Gupta A, Mir SS, Jackson KE, Lim EE, Shah P, Sinha A, Siddiqi MI, Ralph SA, Habib S. 2013. Recycling factors for ribosome disassembly in the apicoplast and mitochondrion of Plasmodium falciparum. Mol. Microbiol. 88, 891–905. (doi:10.1111/mmi.12230) [DOI] [PubMed] [Google Scholar]
- 14.Gupta A, Mir SS, Saqib U, Biswas S, Vaishya S, Srivastava K, Siddiqi MI, Habib S. 2013. The effect of fusidic acid on Plasmodium falciparum elongation factor G (EF-G). Mol. Biochem. Parasitol. 192, 39–48. (doi:10.1016/j.molbiopara.2013.10.003) [DOI] [PubMed] [Google Scholar]
- 15.Jackson KE, et al. 2011. Protein translation in Plasmodium parasites. Trends Parasitol. 27, 467–476. (doi:10.1016/j.pt.2011.05.005) [DOI] [PubMed] [Google Scholar]
- 16.Johnson RA, McFadden GI, Goodman CD. 2011. Characterization of two malaria parasite organelle translation elongation factor G proteins: the likely targets of the anti-malarial fusidic acid. PLoS ONE 6, e20633 (doi:10.1371/journal.pone.0020633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepler PK, Huff CG, Sprinz H. 1966. The fine structure of the exoerythrocytic stages of Plasmodium fallax. J. Cell Biol. 30, 333–358. (doi:10.1083/jcb.30.2.333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemgruber L, Kudryashev M, Dekiwadia C, Riglar DT, Baum J, Stahlberg H, Ralph SA, Frischknecht F. 2013. Cryo-electron tomography reveals four-membrane architecture of the Plasmodium apicoplast. Malar. J 12, 25 (doi:10.1186/1475-2875-12-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaidya AB, Akella R, Suplick K. 1989. Sequences similar to genes for two mitochondrial proteins and portions of ribosomal RNA in tandemly arrayed 6-kilobase-pair DNA of a malarial parasite. Mol. Biochem. Parasitol. 35, 97–108. (doi:10.1016/0166-6851(89)90112-6) [DOI] [PubMed] [Google Scholar]
- 20.Gardner MJ, Feagin JE, Moore DJ, Rangachari K, Williamson DH, Wilson RJ. 1993. Sequence and organization of large subunit rRNA genes from the extrachromosomal 35 kb circular DNA of the malaria parasite Plasmodium falciparum. Nucleic Acids Res. 21, 1067–1071. (doi:10.1093/nar/21.5.1067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardner MJ, Bates PA, Ling IT, Moore DJ, McCready S, Gunasekera MBR, Wilson RJM, Williamson DH. 1988. Mitochondrial DNA of the human malarial parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 31, 11–18. (doi:10.1016/0166-6851(88)90140-5) [DOI] [PubMed] [Google Scholar]
- 22.Feagin JE, Mericle B, Werner E, Morris M. 1997. Identification of additional rRNA fragments encoded by the Plasmodium falciparum 6 kb element. Nucleic Acids Res. 25, 438–446. (doi:10.1093/nar/25.2.438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waller RF, et al. 1998. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl Acad. Sci. USA 95, 12 352–12 357. (doi:10.1073/pnas.95.21.12352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohta N, Sato N, Kuroiwa T. 1998. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 26, 5190–5198. (doi:10.1093/nar/26.22.5190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohta N, Matsuzaki M, Misumi O, Miyagishima SY, Nozaki H, Tanaka K, Shin IT, Kohara Y, Kuroiwa T. 2003. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 10, 67–77. (doi:10.1093/dnares/10.2.67) [DOI] [PubMed] [Google Scholar]
- 26.Matsuzaki M, et al. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. (doi:10.1038/nature02398) [DOI] [PubMed] [Google Scholar]
- 27.Armbrust EV, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. (doi:10.1126/science.1101156) [DOI] [PubMed] [Google Scholar]
- 28.Oudot-Le Secq MP, Grimwood J, Shapiro H, Armbrust EV, Bowler C, Green BR. 2007. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277, 427–439. (10.1007/s00438-006-0199-4) [DOI] [PubMed] [Google Scholar]
- 29.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. 2006. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 34, D363–D368. (doi:10.1093/nar/gkj123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denny P, Preisser P, Williamson D, Wilson I. 1998. Evidence for a single origin of the 35 kb plastid DNA in apicomplexans. Protist 149, 51–59. (doi:10.1016/S1434-4610(98)70009-4) [DOI] [PubMed] [Google Scholar]
- 31.Cai X, Fuller AL, McDougald LR, Zhu G. 2003. Apicoplast genome of the coccidian Eimeria tenella. Gene 321, 39–46. (doi:10.1016/j.gene.2003.08.008) [DOI] [PubMed] [Google Scholar]
- 32.Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, Ishizuka M, Sekine Y, Kawamura F. 2012. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J. Bacteriol. 194, 6282–6291. (10.1128/JB.01544-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688. (doi:10.1016/S0092-8674(01)00546-3) [DOI] [PubMed] [Google Scholar]
- 34.Bubunenko MG, Schmidt J, Subramanian AR. 1994. Protein substitution in chloroplast ribosome evolution: a eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J. Mol. Biol. 240, 28–41. (10.1006/jmbi.1994.1415) [DOI] [PubMed] [Google Scholar]
- 35.Korepanov AP, Korobeinikova AV, Shestakov SA, Garber MB, Gongadze GM. 2012. Protein L5 is crucial for in vivo assembly of the bacterial 50S ribosomal subunit central protuberance. Nucleic Acids Res. 40, 9153–9159. (doi:10.1093/nar/gks676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115, 97–108. (doi:10.1016/S0092-8674(03)00762-1) [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi K, Subramanian AR. 2000. The plastid ribosomal proteins. Identification of all the proteins in the 50S subunit of an organelle ribosome (chloroplast). J. Biol. Chem. 275, 28 466–28 482. (doi:10.1074/jbc.M005012200) [DOI] [PubMed] [Google Scholar]
- 38.Kubo N, Arimura S. 2010. Discovery of the rpl10 gene in diverse plant mitochondrial genomes and its probable replacement by the nuclear gene for chloroplast RPL10 in two lineages of angiosperms. DNA Res. 17, 1–9. (doi:10.1093/dnares/dsp024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. 2001. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 276, 43 958–43 969. (doi:10.1074/jbc.M106510200) [DOI] [PubMed] [Google Scholar]
- 40.UniProt C. 2013. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 41, D43–D47. (doi:10.1093/nar/gks1068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng S, Chen Y, Gao YG. 2013. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PLoS ONE 8, e58829 (doi:10.1371/journal.pone.0058829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cukras AR, Green R. 2005. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 349, 47–59. (doi:10.1016/j.jmb.2005.03.075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramaswamy P, Woodson SA. 2009. S16 throws a conformational switch during assembly of 30S 5′ domain. Nat. Struct. Mol. Biol. 16, 438–445. (doi:10.1038/nsmb.1585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda M, Nishikawa T, Fujimoto M, Takanashi H, Arimura S, Tsutsumi N, Kadowaki K. 2008. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 25, 1566–1575. (doi:10.1093/molbev/msn102) [DOI] [PubMed] [Google Scholar]
- 45.Mushegian A. 2005. Protein content of minimal and ancestral ribosome. RNA 11, 1400–1466. (doi:10.1261/rna.2180205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rogalski M, Ruf S, Bock R. 2006. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34, 4537–4545. (doi:10.1093/nar/gkl634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobin C, Mandava CS, Ehrenberg M, Andersson DI, Sanyal S. 2010. Ribosomes lacking protein S20 are defective in mRNA binding and subunit association. J. Mol. Biol. 397, 767–776. (doi:10.1016/j.jmb.2010.02.004) [DOI] [PubMed] [Google Scholar]
- 48.Morita-Yamamuro C, Tsutsui T, Tanaka A, Yamaguchi J. 2004. Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant Cell Physiol. 45, 781–788. (doi:10.1093/pcp/pch093) [DOI] [PubMed] [Google Scholar]
- 49.Holmes KL, Culver GM. 2005. Analysis of conformational changes in 16 S rRNA during the course of 30 S subunit assembly. J. Mol. Biol. 354, 340–357. (doi:10.1016/j.jmb.2005.09.056) [DOI] [PubMed] [Google Scholar]
- 50.Kaczanowska M, Ryden-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71, 477–494. (doi:10.1128/MMBR.00013-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson JR. 2005. Assembly of the 30S ribosomal subunit. Q. Rev. Biophys. 38, 397–403. (doi:10.1017/S0033583506004264) [DOI] [PubMed] [Google Scholar]
- 52.Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80, 501–526. (doi:10.1146/annurev-biochem-062608-160432) [DOI] [PubMed] [Google Scholar]
- 53.Britton RA. 2009. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63, 155–176. (doi:10.1146/annurev.micro.091208.073225) [DOI] [PubMed] [Google Scholar]
- 54.Vedadi M, et al. 2007. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 151, 100–110. (doi:10.1016/j.molbiopara.2006.10.011) [DOI] [PubMed] [Google Scholar]
- 55.Kumar A, Tanveer A, Biswas S, Ram EV, Gupta A, Kumar B, Habib S. 2010. Nuclear-encoded DnaJ homologue of Plasmodium falciparum interacts with replication ori of the apicoplast genome. Mol. Microbiol. 75, 942–956. (doi:10.1111/j.1365-2958.2009.07033.x) [DOI] [PubMed] [Google Scholar]
- 56.Sato S, Wilson RJ. 2005. Organelle-specific cochaperonins in apicomplexan parasites. Mol. Biochem. Parasitol. 141, 133–143. (doi:10.1016/j.molbiopara.2005.01.010) [DOI] [PubMed] [Google Scholar]
- 57.Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275, 16 414–16 419. (doi:10.1074/jbc.M001854200) [DOI] [PubMed] [Google Scholar]
- 58.Formenoy LJ, Cunningham PR, Nurse K, Pleij CW, Ofengand J. 1994. Methylation of the conserved A1518-A1519 in Escherichia coli 16S ribosomal RNA by the ksgA methyltransferase is influenced by methylations around the similarly conserved U1512.G1523 base pair in the 3′ terminal hairpin. Biochimie 76, 1123–1128. (doi:10.1016/0300-9084(94)90040-X) [DOI] [PubMed] [Google Scholar]
- 59.Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. 1999. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry 38, 4053–4057. (doi:10.1021/bi982364y) [DOI] [PubMed] [Google Scholar]
- 60.O'Farrell HC, Pulicherla N, Desai PM, Rife JP. 2006. Recognition of a complex substrate by the KsgA/Dim1 family of enzymes has been conserved throughout evolution. RNA 12, 725–733. (doi:10.1261/rna.2310406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klootwijk J, van den Bos RC, Planta RJ. 1972. Secondary methylation of yeast ribosomal RNA. FEBS Lett. 27, 102–106. (doi:10.1016/0014-5793(72)80419-8) [DOI] [PubMed] [Google Scholar]
- 62.Noon KR, Bruenger E, McCloskey JA. 1998. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 180, 2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steege DA, Graves MC, Spremulli LL. 1982. Euglena gracilis chloroplast small subunit rRNA. Sequence and base pairing potential of the 3′ terminus, cleavage by colicin E3. J. Biol. Chem. 257, 10 430–10 439. [PubMed] [Google Scholar]
- 64.Van Buul CP, Hamersma M, Visser W, Van Knippenberg PH. 1984. Partial methylation of two adjacent adenosines in ribosomes from Euglena gracilis chloroplasts suggests evolutionary loss of an intermediate stage in the methyl-transfer reaction. Nucleic Acids Res. 12, 9205–9208. (doi:10.1093/nar/12.23.9205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lovgren JM, Bylund GO, Srivastava MK, Lundberg LA, Persson OP, Wingsle G, Wikstrom PM. 2004. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA 10, 1798–1812. (doi:10.1261/rna.7720204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. 1989. Birth of the D-E-A-D box. Nature 337, 121–122. (doi:10.1038/337121a0) [DOI] [PubMed] [Google Scholar]
- 67.Jankowsky E, Gross CH, Shuman S, Pyle AM. 2001. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science 291, 121–125. (doi:10.1126/science.291.5501.121) [DOI] [PubMed] [Google Scholar]
- 68.Linder P. 2006. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 34, 4168–4180. (doi:10.1093/nar/gkl468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohr S, Stryker JM, Lambowitz AM. 2002. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell 109, 769–779. (doi:10.1016/S0092-8674(02)00771-7) [DOI] [PubMed] [Google Scholar]
- 70.Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32, 2751–2759. (doi:10.1093/nar/gkh603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48, 1253–1265. (doi:10.1046/j.1365-2958.2003.03513.x) [DOI] [PubMed] [Google Scholar]
- 72.Jagessar KL, Jain C. 2010. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA 16, 1386–1392. (doi:10.1261/rna.2015610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain C. 2008. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA 14, 381–389. (doi:10.1261/rna.800308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharpe Elles LM, Sykes MT, Williamson JR, Uhlenbeck OC. 2009. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 37, 6503–6514. (doi:10.1093/nar/gkp711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McConkey GA, Rogers MJ, McCutchan TF. 1997. Inhibition of Plasmodium falciparum protein synthesis. Targeting the plastid-like organelle with thiostrepton. J. Biol. Chem. 272, 2046–2049. (doi:10.1074/jbc.272.4.2046) [DOI] [PubMed] [Google Scholar]
- 76.Dahl EL, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. 2006. Tetracyclines specifically target the apicoplast of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 50, 3124–3131. (doi:10.1128/AAC.00394-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Budimulja AS, Tapchaisri P, Wilairat P, Marzuki S. 1997. The sensitivity of Plasmodium protein synthesis to prokaryotic ribosomal inhibitors. Mol. Biochem. Parasitol. 84, 137–141. (doi:10.1016/S0166-6851(96)02781-8) [DOI] [PubMed] [Google Scholar]
- 78.Camps M, Arrizabalaga G, Boothroyd J. 2002. An rRNA mutation identifies the apicoplast as the target for clindamycin in Toxoplasma gondii. Mol. Microbiol. 43, 1309–1318. (doi:10.1046/j.1365-2958.2002.02825.x) [DOI] [PubMed] [Google Scholar]
- 79.Sidhu AB, Sun Q, Nkrumah LJ, Dunne MW, Sacchettini JC, Fidock DA. 2007. In vitro efficacy, resistance selection, and structural modeling studies implicate the malarial parasite apicoplast as the target of azithromycin. J. Biol. Chem. 282, 2494–2504. (doi:10.1074/jbc.M608615200) [DOI] [PubMed] [Google Scholar]
- 80.Aminake MN, Schoof S, Sologub L, Leubner M, Kirschner M, Arndt HD, Pradel G. 2011. Thiostrepton and derivatives exhibit antimalarial and gametocytocidal activity by dually targeting parasite proteasome and apicoplast. Antimicrob. Agents Chemother. 55, 1338–1348. (doi:10.1128/AAC.01096-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tarr SJ, Nisbet RE, Howe CJ. 2011. Transcript-level responses of Plasmodium falciparum to thiostrepton. Mol. Biochem. Parasitol. 179, 37–41. (doi:10.1016/j.molbiopara.2011.05.004) [DOI] [PubMed] [Google Scholar]
- 82.Bulkley D, Innis CA, Blaha G, Steitz TA. 2010. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc. Natl Acad. Sci. USA 107, 17 158–17 163. (doi:10.1073/pnas.1008685107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schlunzen F, Harms JM, Franceschi F, Hansen HA, Bartels H, Zarivach R, Yonath A. 2003. Structural basis for the antibiotic activity of ketolides and azalides. Structure 11, 329–338. (doi:10.1016/S0969-2126(03)00022-4) [DOI] [PubMed] [Google Scholar]
- 84.Xiong L, Korkhin Y, Mankin AS. 2005. Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob. Agents Chemother. 49, 281–288. (doi:10.1128/AAC.49.1.281-288.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dunkle JA, Xiong L, Mankin AS, Cate JH. 2010. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA 107, 17 152–17 157. (doi:10.1073/pnas.1007988107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. 2008. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell 30, 26–38. (doi:10.1016/j.molcel.2008.01.009) [DOI] [PubMed] [Google Scholar]
- 87.Clough B, Strath M, Preiser P, Denny P, Wilson IR. 1997. Thiostrepton binds to malarial plastid rRNA. FEBS Lett. 406, 123–125. (doi:10.1016/S0014-5793(97)00241-X) [DOI] [PubMed] [Google Scholar]
- 88.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res. 41, D36–D42. (doi:10.1093/nar/gks1195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. (doi:10.1016/j.jmb.2004.05.028) [DOI] [PubMed] [Google Scholar]
- 90.Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. (doi:10.1016/0022-2836(82)90515-0) [DOI] [PubMed] [Google Scholar]
- 91.Aurrecoechea C, et al. 2013. EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res. 41, D684–D691. (doi:10.1093/nar/gks1113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Podicheti R, Gollapudi R, Dong Q. 2009. WebGBrowse: a web server for GBrowse. Bioinformatics 25, 1550–1551. (doi:10.1093/bioinformatics/btp239) [DOI] [PubMed] [Google Scholar]
- 93.Foth BJ, Ralph SA, Tonkin CJ, Struck NS, Fraunholz M, Roos DS, Cowman AF, McFadden GI. 2003. Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299, 705–708. (doi:10.1126/science.1078599) [DOI] [PubMed] [Google Scholar]
- 94.Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. 2001. Deciphering apicoplast targeting signals - feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene 280, 19–26. (doi:10.1016/S0378-1119(01)00776-4) [DOI] [PubMed] [Google Scholar]
- 95.Bender A, van Dooren GG, Ralph SA, McFadden GI, Schneider G. 2003. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol. Biochem. Parasitol. 132, 59–66. (doi:10.1016/j.molbiopara.2003.07.001) [DOI] [PubMed] [Google Scholar]
- 96.Claros MG, Vincens P. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. (doi:10.1111/j.1432-1033.1996.00779.x) [DOI] [PubMed] [Google Scholar]
- 97.Logan-Klumpler FJ, et al. 2012. GeneDB: an annotation database for pathogens. Nucleic Acids Res. 40, D98–D108. (doi:10.1093/nar/gkr1032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rother M, Rother K, Puton T, Bujnicki JM. 2011. ModeRNA: a tool for comparative modeling of RNA 3D structure. Nucleic Acids Res. 39, 4007–4022. (doi:10.1093/nar/gkq1320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2007. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 50, 2.9.1–2.9.31. (doi:10.1002/0471140864.ps0209s50) [DOI] [PubMed] [Google Scholar]
- 100.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791. (doi:10.1002/jcc.21256) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.