Abstract
Background
Tuberculosis (TB) infections caused by multidrug-resistant Mycobacterium tuberculosis (MDR MTB) remain a significant public health concern worldwide. Georgia has a high prevalence of MDR MTB. The genetic mechanisms underlying the emergence of MDR MTB strains in this region are poorly understood and need to be determined for developing better strategies for TB control. This study investigated the frequency of major drug resistance mutations across rpoB, katG and inhA loci of Georgian MDR MTB strains and explored differences between new and previously treated patients.
A total of 634 MTB strains were examined for which an MDR phenotype had been previously determined by the proportions method. The GenoType®MTBDRplus system was applied to screen the strains for the presence of rpoB (S531L, H526D, H526Y, and D516V), katG (S315T) and inhA promoter region (C15T and T8C) mutations. The target loci were amplified by PCR and then hybridized with the respective site-specific and wild type (control) probes.
Results
Out of the 634 isolates tested considered by phenotypic testing to be resistant to RIF and INH, this resistance was confirmed by the GenoType®MTBDRplus assay in 575 (90.7%) isolates. RIF resistance was seen in 589 (92.9%) and INH resistance was seen in 584 (92.1%); 67.2% and 84.3% of MDR strains harbored respectively rpoB S531L and katG S315T mutations (generally known as having low or no fitness cost in MTB). The inhA C15T mutation was detected in 22.6% of the strains, whereas rpoB H526D, rpoB H526Y, rpoB D516V and inhA T8C were revealed at a markedly lower frequency (≤5.2%). The specific mutations responsible for the RIF resistance of 110 isolates (17.4%) could not be detected as no corresponding mutant probe was indicated in the assay. There was no specific association of the presence of mutations with the gender/age groups. All types of prevailing mutations had higher levels in new cases.
A great majority of the Georgian MDR MTB strains have a strong preference for the drug resistance mutations carrying no or low fitness cost. Thus, it can be suggested that MDR MTB strains with such mutations will continue to arise in Georgia at a high frequency even in the absence of antibiotic pressure.
Keywords: M. tuberculosis, Mutation, Multidrug resistant tuberculosis, GenoType®MTBDRplus, Fitness cost
Introduction
Tuberculosis is one of the leading infectious killers in the world today, second only to HIV. An estimated 2 billion people are infected with the bacteria that cause TB, and each year 8 million people are newly diagnosed with the disease. In spite of adequate therapy, an estimated 2–3 million people die of the disease every year [1]. Adding to the heavy burden of TB-related morbidity and mortality are drug-resistant strains of the disease. Multidrug-resistant TB is defined as strains of Mycobacterium tuberculosis expressing in vitro resistance to at least Rifampicin (RIF) and Isoniazid (INH) – two of the most powerful anti-tuberculous agents available. Resistance to these agents leads to longer, more complicated and more costly therapy. The estimates of the global burden of disease caused by TB in 2009 are as follows: 9.4 million incident cases, 14 million prevalent cases, and 1.3 million deaths among HIV-negative people and 0.38 million deaths among HIV-positive people [1]. Developing countries account for 95% of all TB cases and 98% of all TB deaths worldwide [2]. Among TB patients notified in 2009 (5.8 million), an estimated 250,000 (range, 230,000–270,000) had multidrug-resistant TB (MDR-TB). Of these, slightly more than 30,000 (12%) were diagnosed with MDR-TB and notified [1].
Georgia is a country in the South Caucasus which regained its independence from the Soviet Union in 1991. Tuberculosis is a significant health problem in Georgia with an estimated incidence of 107 per 100,000 population, making it the fifth highest burden country in the European region [3]. MDR-TB has emerged as a serious public health problem in Georgia; in the period 2001–2004 MDR-TB strains were isolated in 28.1% of all TB cases [4]. A study of MDR-TB among hospitalized patients at the National Centre for Tuberculosis and Lung Diseases (Tbilisi, Georgia) showed that in the period 2006–2008 the rates of MDR-TB were very high: 23% among new cases and 55% among previously treated cases [5]. Out of 4732 TB cases in 2009, MDR-TB was found in 10.3% of newly diagnosed patients and in 31.1% of previously treated patients [6,7]. These high rates of MDR-TB have made the timely identification of resistant MTB strains extremely important both in achieving effective disease management and in preventing their spread [8].
In recent years, the development of new molecular methods based on PCR has allowed the rapid detection and identification of genetic mutations related to resistance, specifically resistance to RIF and INH [10,11,14]. These methods are based on the targeting of mutations in the rpoB, katG, and inhA genes, the mutations that account for the highest frequency of documented M. tuberculosis genetic diversity. Within the last few decades, several chromosomal mutations in MTB responsible for resistance to most of the major drugs, including Rifampin and Isoniazid, have been discovered [9].
Point mutations in rpoB, a gene encoding the β-subunit of DNA-dependent RNA polymerase, have been shown to account for a strong majority of RIF resistance worldwide. As RIF mono-resistance is relatively rare, detection of RIF resistance is a good indicator of MDR-TB [9]. More specifically, 95% of these RIF resistance-causing mutations are located within an 81 base pair hotspot region of rpoB, spanning codons 507–533, a region known as the RIF resistance determining region (RRDR) [10]. More than 35 resistant alleles have been identified in this region [11,12]. Mutations in codons 516, 526 and 531 of rpoB are most commonly associated with high-level RIF resistance [13,14], but the frequency with which these mutations are observed varies by geographic location. INH resistance in MTB is more complex than RIF resistance in that a number of genes are implicated. However, up to 95% of INH resistance may be due to mutations in katG [15]. The most frequently observed alteration in katG is a serine-to-threonine substitution at codon 315 (S315T), located within the active site of the catalase moiety of katG. The S315T alteration in this proposed binding site of INH prevents katG-mediated activation of INH [16]. Additionally, mutations in the promoter region of inhA account for 8% to 20% of INH resistance in MTB. A C-to-T substitution at nucleotide 215 results in the over-expression of inhA, an NADH-dependent enoylacyl reductase involved in mycolic acid synthesis, and INH resistance arises as a result of drug titration [15].
The aim of the present study is to determine the frequency of major drug resistancemutations across rpoB, katG and inhA loci of Georgian MDR MTB isolates using a molecular test.
Materials and methods
Clinical strains
A total of 634 strains of MTB from pulmonary MDR-TB diagnosed cases registered during the period 2010–2011 at the National Centre for Tuberculosis and Lung Diseases (NCTLD) of Georgia were examined. The strains were recovered from 259 new and 375 retreatment pulmonary MDR-TB cases. Cultures of these strains were previously examined and confirmed for M. tuberculosis complex (MTBC) using the standard microbiological method [17]. The strains were additionally confirmed for M. tuberculosis/M. canetti by the GenoType®MTBC assay (Lifescience GmbH, Nehren, Germany). For these strains, MDR phenotypes were predetermined using the method of proportions with LÖ wenstein-Jensen solid medium [18,19] GenoType®MTBDRplus assay; 634 MDR-MTB strains of M. tuberculosis were screened for the presence of the most common drug resistance mutations of rpoB, katG and inhA using the GenoType®MTBDRplus assay, which was performed according to the manufacturer’s instructions (Hain Lifescience GmbH, Nehren, Germany). Briefly, the following PCR conditions were applied for the amplification of target rpoB, katG and inhA loci: 15 min of initial denaturation at 95 °C; 10 cycles involving subsequent denaturation for 30 s at 95 °C, and annealing for 2 min. at 58 °C; additional 20 cycles with denaturation for 25 s at 95 °C, annealing for 40 s at 53 °C, and elongation for 40 s at 70 °C; and a final extension step for 8 min at 70 °C. Hybridization and detection were performed in an automated TwinCubator (Hain Lifescience GmbH, Nehren, Germany) using the following procedures: the PCR amplification products were denatured at room temperature for 5min; the single-stranded biotin-labeled amplicons were hybridized to specific probes attached to the MTBDRplus strip by incubation for 30 min at 45 °C; the strip was stringently washed, and then was treated by a streptavidin–alkaline phosphatase (AP) conjugate. After subsequent 30 min incubation at room temperature, the MTBDRplus strip was subjected to an AP staining reaction to detect colorimetric bands. The MTBDRplus strip contains a total of 27 reaction zones. These include 21 probes for screening of target rpoB, katG, and inhA drug resistance mutations and their corresponding wild type loci, 3 probes specific to rpoB, katG, and inhA genes, 1 probe specific to MTBC (TUB), and 1 conjugate control (CC) and 1 amplification control (AP) probes. The probes, rpoB MUT1, rpoB MUT2A, rpoB MUT2B, and rpoB MUT3 specifically target respectively the most common rpoB mutations D516V, H526Y, H526D and S531L that confer RIF resistance in M. tuberculosis. The probes katG MUT1, katG MUT2 specifically recognize the most common katG mutation S315T that confers INH resistance. The probes inhA MUT1, inhA MUT2, inhA MUT3A and inhA MUT3B allow the screening respectively for inhA promoter region mutations C15T, A16G, T8C, and T8A contributing to INH resistance. The CC and the AP probes serve for verification of the test procedures.
Statistical analysis was performed using χ2, Fisher exact, and McNemar tests with the aid of EpiInfo (version 3.4; CDC, Atlanta, GA); 95% confidence intervals and P values were also calculated. P values ≤0.05 were considered statistically significant.
Results
Out of the 634 isolates tested, considered by phenotypic testing to be resistant to RIF and INH, this resistance was confirmed by the Genotype®MTBDRplus assay in 575 (90.7%) isolates. RIF resistance was seen in 589 (92.9%) isolates and INH resistance was indicated in 584 (92.1%) isolates. INH resistance due to katG S315T was found in 535 (84.3%) isolates; due to inhA C (−15) Twas found in 143 (22.6%) isolates; and due to inhA T (−8) C was found in 8 (1.3%) isolates. Both katG S315T and inhA mutations were detected in 126 (19.9%) cases.
Single mutations
The results of this study indicate the most common drug resistance mutations are rpoB, katG and inhA among MDR-MTB isolates from Georgia (Table 1). The most common mutation responsible for RIF resistance in MDR strains was rpoB S531L (426/67.2%). The most common mutations responsible for INH resistance were katG S315T (535/84.3%) and inhA C (−15) T (143/22.6%). Table 1 also shows the distribution of single mutations among new and retreatment cases. The mutation rpoB S531L accounted for resistance measured in 208 (80.3%) new cases versus 218 (58.1%). The mutation katG S315T accounted for resistance measured in 252 (97.3%) new cases versus 283 (75.5%). The rest of the mutations (rpoB D516V, rpoB H526Y, rpoB H526D, inhAT (−8) C showed decreasing levels respectively 33 (5.2%), 13 (2.1%), 7 (1.1%), and 8 (1.3%). There was no statistically significant differences between new and retreatment cases as well as between genders for these mutations.
Table 1.
The most common drug resistance mutations of rpoB, katG and inhA of M. tuberculosis MDR isolates fromnew versus previously treated TB cases.
| Mutation | No. (detection %) of MDR isolates from | P value* | ||
|---|---|---|---|---|
| All cases (n = 634) | New cases (n = 259) | Previously treated cases (n = 375) | ||
| rpoB S531L | 426 (67.2) | 208 (80.30) | 218 (58.1) | ≤0.000001 |
| rpoB H526Y | 13 (2.1) | 4 (1.5) | 9 (2.4) | 0.45 |
| rpoB H526D | 7 (1.1) | 4 (1.5) | 3 (0.8) | 0.37 |
| rpoB D516V | 33 (5.2) | 16 (6.2) | 17 (4.5) | 0.35 |
| katG S315T | 535 (84.3) | 252 (97.3) | 283 (75.5) | ≤0.000001 |
| inhA C (−15) T | 143 (22.6) | 69 (26.6) | 74 (19.7) | 0.02 |
| inhA T (−8) C | 8 (1.3) | 2 (0.8) | 6 (1.6) | 0.48 |
Total – 465, 169 isolates not including in the table: No mutation on rpoB – 110, Single mutation only on rpoB – 14, Single mutation only on katG – 7, Single mutation only on inhA – 2, Without any mutations – 36.
Combinations of mutations
The most common combinations of mutations responsible for MDR in Georgia were: rpoB S531L + katG S315T (311/49.1%) and rpoB S531L + katG S315T + inhA C (−15) T (89/14.0%). The remainder of existing combinations did not exceed the level of 3.5%. Their distribution among new and retreatment cases is shown in Table 2. The rpoB S531L + katG S315T combination accounted for resistance measured in 155 new cases (59.8%) versus 156 previously treated cases (41.6%), which is statistically significant (p-value ≤0.05). For the rest of the combinations, no statistically significant differences were found.
Table 2.
The frequency of combinations of mutations responsible for MDR tuberculosis in Georgia.
| Mutation profiles | No. (frequency %) of MDR isolates from | P value* | ||
|---|---|---|---|---|
| All cases (n = 634) | New cases (n = 259) | Prev. treated cases (n = 375) | ||
| rpoB S531L + katG S315T | 311(49.1) | 155(59.8) | 156(41.6) | 0.000006 |
| rpoB S531L + inhA C (−15) T | 12(1.9) | 6(2.3) | 6(1.6) | 0.51 |
| rpoB S531L + katG S315T + inhA C (−15)T | 89(14.0) | 42(16.2) | 47(12.5) | 0.189 |
| rpoB H526Y + katG S315T | 7(1.1) | 2(0.8) | 5(1.3) | 0.5 |
| rpoB H526T + katG S315T + inhA T (−8) C | 6(0.9) | 2(0.8) | 4(1.1) | 0.7 |
| rpoB H526D + katG S315T + inhA C (−15) T | 7(1.1) | 4(1.5) | 3(0.8) | 0.3 |
| rpoB D516V + inhA C (−15) T | 11(1.7) | 5(1.9) | 6(1.6) | 0.75 |
| rpoB D516V + katG S315T | 22(3.5) | 11(4.2) | 11(2.9) | 0.37 |
Total – 465, 169 isolates not including in the table: No mutation on rpoB – 110, Single mutation only on rpoB – 14, Single mutation only on katG – 7, Single mutation only on inhA – 2, Without any mutations – 36.
Comparison of genotype and phenotype
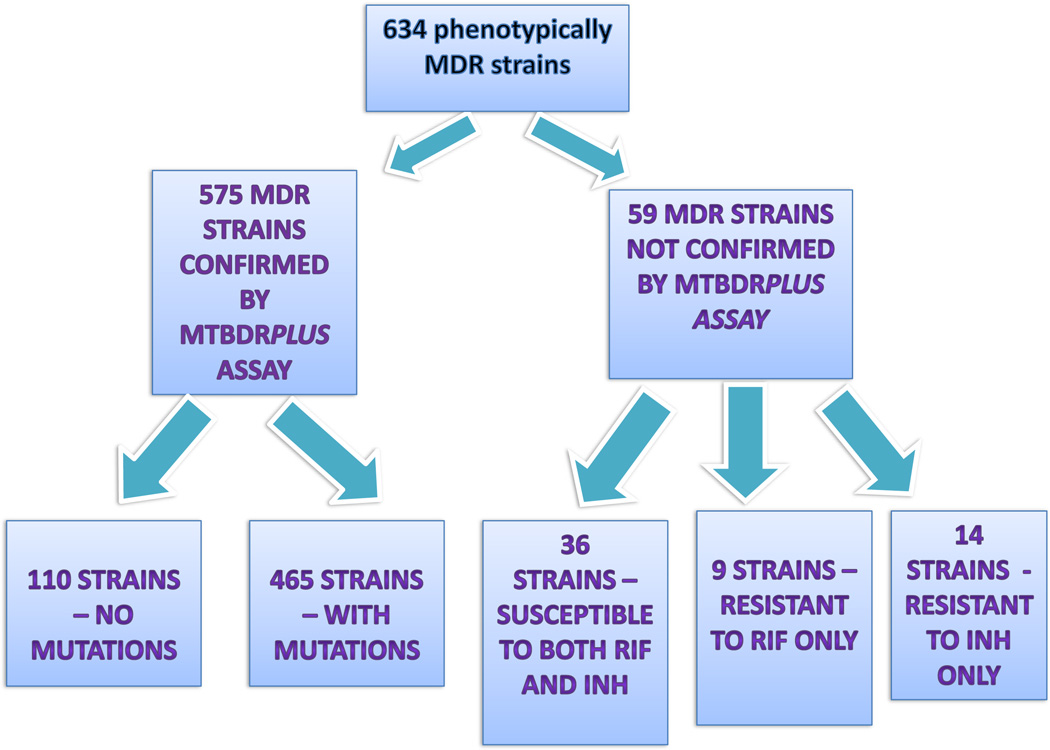
The results of genotype and phenotype comparisons are shown in Fig. 1. Comparative analysis performed on DST results of all 634 isolates demonstrated that they were divided into two groups depending on wild type signal presence or absence.
Fig. 1.
M. tuberculosis strains phenotype and genotype comparison.
The group of 59 (9.3%) MTB isolates did not show MDR profiles by Genotype®MTBDRplus assay while they were indicated as MDR by gold standard DST.
According to this assay, out of 59 MTB isolates, 36 (61%) had wild type sequences indicating susceptibility to both RIF and INH; 14 isolates (23.7%) showed wild type profiles for INH only; and 9 isolates (15.3%) to RIF only.
Another group of 575 strains without wild type signals included 110 isolates (17.4%) with RIF resistance. The specific mutations responsible for this resistance, however, could not be detected as no corresponding mutant probe was indicated in the assay.
Discussion
The emergence of drug-resistant isolates of M. tuberculosis poses a serious threat to global TB control, and remains a major problem for healthcare worldwide. Understanding the relationship between antibiotic resistance and the transmissibility and virulence of M. tuberculosis is essential for predicting the future burden of drug-resistant disease [20]. Assays for the rapid detection of resistance, such as the Genotype®MTBDRplus system enable earlier detection of resistance and thereby tailoring of treatment regimens.
In this study the spectrum of mutations associated with the resistance to RIF and INH (dominance of single mutations in the rpoB Ser531Leu, katG Ser315Thr, inhA C [−15] T) was similar or close to previously reported on larger populations from several different geographic locations [8,10,21,22,35]. These patterns are seen more in newly diagnosed patients, and these mutations seem to be increasing over time. This may be due to the fact that these mutations do not seem to affect mycobacterial fitness. This suggests that ongoing transmission of these strains is what is occurring in the community.
The rpoB S531L mutation accounted for RIF resistance in 67.2% of MDR isolates. In contrast, the proportion of mutations at codons 526 (3.2%) and 516 (5.2%) of rpoB is lower. The low fitness cost associated with rpoB S531L may account for the high frequency with which it is observed [23]. The katG S315T mutation was found in 84.3% of MDR isolates. The S531T katG mutation is proposed to lead to clinically significant INH resistance without exacting a significant fitness cost. This hypothesis is consistent with both animal models of virulence and molecular epidemiological cluster studies [15,24,25].
In contrast with the data of Gegia et al. [26], in the present study the distribution of single mutations among new and previously treated cases shows that all types of prevailing mutations had higher levels in new cases. In the earlier report [26], drug resistance-related mutations in MTB strains isolated from 196 patients of the Georgian National Centre for Tuberculosis and Lung Diseases were examined and drug resistance-related mutation rates for pretreated and new cases were significantly different. Antimycobacterial drug resistance-related mutations, which included three individual mutations – rpoB S531L, katG S315G and S315T – were detected in significantly higher numbers in pretreated cases than in new ones. The data indicating a higher level of transmission of MDR-TB strains in new cases than in those previously treated may be associated with evidences which some resistance mutations, particularly rpoB S531L and katG S315T appear to confer no or low fitness cost. Thus antibiotic-resistant bacteria will not disappear as a result of restricted use of antibiotics but might instead, as shown by recent clinical studies, persist in the population for a long time even after antibiotic use has been reduced or eliminated [20,23,27,28]. Studies have shown that there seems to be a strong selection for low-cost drug resistance mutations in vivo [29,30].
The cost of the resistance-conferring mutations, in terms of bacterial fitness and the ability of the bacteria to genetically compensate for such costs, are key parameters in determining if resistance-conferring mutations will be maintained within a bacterial population in the absence of antimicrobial therapy. Experimental work conducted in various model systems has established that chromosomal mutations conferring antibiotic resistance are almost invariably associated with a significant cost, and in the absence of drugs are adapted for by accessory compensatory mutations rather than by reversion to the drug-sensitive, high-fitness genotype [31].
The combination of mutations with more prevalent nucleotide changes were observed in codons rpoB S531L and katG S315T. The mentioned combination of mutations had a higher frequency in new cases. In this study, 88.6% (n = 412) of all isolates found to have a combination of mutations involving nucleotide changes in codons 531 (TCG → TTG) demonstrated an association with higher levels of resistance to RIF (MIC, ≥100 µg/ml). The combination of mutations involving katG S315T mutation was 95.1% (n = 442), and this mutation is described as having the association with higher levels of resistance to INH (MIC, ≥100 µg/ml). This data is consistent with earlier reports [21,32–34].
There were resistant strains in this study for which no mutations were detected. The set of DNA probes used in the Genotype®MTBDRplus assay covers most of the RIF-resistance mutations prevailing in Georgia. A caveat in the interpretation of the Genotype®MTBDRplus assay with respect to Rifampicin detection is that resistance may be indicated by the absence of a wild-type hybridization signal alone, without confirmation by a mutant probe signal. However, some of the isolates did seem to demonstrate phenotypic RIF resistance probably based on other types of mutations. There are very few reports in publications about such strains. So, Hauck et al. report two strains from French patients showing weak resistance to Rifampicin (MIC = 1 mg/L) with a wild-type profile using Genotype®MTBDRplus assay [35]. As recent investigations of Rosales-Klintz et al. showed, there are clear geographical differences in the presence and proportion of resistance-related mutations [36]. The present investigation confirms the correctness of this conclusion and the necessity of further research concerning MTB isolate genotypes and their association with the drug resistance in this region.
Conclusions
The results of this study illustrate that the geographical distribution of mutations resulting in drug resistance in M. tuberculosis in Georgia is similar to what is reported elsewhere. This may have important implications for the roll-out of rapid genotypic tests to identify drug-resistant M. tuberculosis. More rapid testing will allow for improved diagnostics and treatment for patients with drug-resistant forms of the disease. If rapid genotypic assays for the detection of drug resistance are to be widely used, there is a need to continually monitor local patterns of drug-resistance mutations to ensure that if clonal groups of M. tuberculosis do emerge, they are properly diagnosed as drug-resistant.
When examining the possible differences in new and retreatment cases, it was found that all types of prevailing mutations (rpoB S531L and katG S315T) had higher levels in new cases. A great majority of the Georgian MDR-MTB strains have a strong preference for the drug resistance mutations carrying no or low fitness cost. This is true for isolates from both new and previously treated cases, but the prevalence of such mutations among new cases allow us to suggest that MDR-MTB strains with such mutations will continue to arise in Georgia at a high frequency even in the absence of antibiotic pressure. Thus, the ongoing transmission of these strains will occur even in the setting of DOTS and further points out the importance of prompt and effective MDR-TB treatment.
The set of DNA probes used in the Genotype®MTBDRplus assay covers most of the RIF-resistance mutations prevailing in Georgia. However, some of the isolates did seem to demonstrate RIF resistance based on other types of mutations. The study shows the necessity of further investigations concerning MTB isolate genotypes and their association with the drug resistance in this region.
Molecular genotyping methods are important in detecting the dominance of transmission or re-infection in a population. Further studies for determination of genotype of Georgian M. tuberculosis isolates are necessary. In addition, the analysis of phylogenetically preserved sequence motifs among members of the M. tuberculosis complex in combination with geographical and epidemiological data will contribute important information for tracing the phylogenetic spread of these pathogens.
Acknowledgments
The authors express their gratitude to Professor Sven Hoffner, Swedish Institute for Communicable Disease Control (Stockholm, Sweden) and Dr. Jennifer Furin from Brigham and Women’s Medical Dept. (Boston, USA) for valuable comments and help in the preparation of this paper.
Footnotes
Conflict of interests
There are no conflicts of interests.
REFERENCES
- 1.World Health Organization. Global TB Control Report. Geneva: WHO; 2010. [Google Scholar]
- 2.US Agency for International Development. Report to congress: health-related research and development activities at USAID – an update on the five-year strategy, 2006–2010. US Agency for International Development. 2009;182(6):1788–1790. [Google Scholar]
- 3.World Health Organization. Global Tuberculosis Control: WHO Report. Geneva: WHO; 2011. [Google Scholar]
- 4.Mdivani N, Zangaladze E, Volkova N, Kourbatova E, Jibuti T, Shubladze N, et al. High prevalence of multidrug-resistant tuberculosis in Georgia. Int. J. Infect. Dis. 2008;12(6):635–644. doi: 10.1016/j.ijid.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vashakidze L, Salakaia A, Shubladze N, Cynamon M, Barbakadze K, Kikvidze M, et al. Prevalence and risk factors for drug resistance among hospitalized TB patients in Georgia. Int. J. Tuberc. Lung Dis. 2009;13(9):1148–1153. [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. MDR-TB & XDR-TB Report. Geneva: WHO; 2010. [Google Scholar]
- 7.World Health Organization. High MDR-TB Burden-Georgia. Geneva: WHO; 2010. [Google Scholar]
- 8.Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for Rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis isolates and clinical specimens. J. Clin. Microbiol. 2007;45:2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillespie SH. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob. Agents Chemother. 2002;46:267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 11.Gingeras TR, Ghandour G, Wang E, Berno A, Small PM, Drobniewski F, et al. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 12.Musser JM. Antimicrobial agent resistance inmycobacteria: moleculargenetics insights. Clin. Microbiol. Rev. 1995;8:451–496. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJ. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312(5782):1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 14.Rigouts L, Nolasco O, de Rijk P, et al. Newly developed primers for comprehensive amplification of the rpoB gene and detection of Rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 2007;45:252–254. doi: 10.1128/JCM.01489-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazbón MH, Brimacombe M, Bobadilla del Valle M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2006;50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu S, Girotto S, Lee C, et al. Reduced affinity for isoniazid in the S315T mutant of Mycobacterium tuberculosis KatG is a key factor in antibiotic resistance. J. Biol. Chem. 2003;278:14769–14775. doi: 10.1074/jbc.M300326200. [DOI] [PubMed] [Google Scholar]
- 17.Beverly GM, Frederick SN, Richard JW., Jr . Mycobacterium: bacteriology. In: Murray PE, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. seventh ed. Washington, DC: ASM Press; 1999. pp. 399–437. [Google Scholar]
- 18.WHO. Guidelines for Surveillance of Drug Resistance in Tuberculosis. Geneva: WHO; 2003. [Google Scholar]
- 19.Canetti G. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull. World Health Organ. 1963;29:565–578. [PMC free article] [PubMed] [Google Scholar]
- 20.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring Rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2004;48(4):1289–1294. doi: 10.1128/AAC.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian L, Abe C, Lin TP, Yu MC, Cho SN, Wang S, Douglas JT. RpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from east Asian countries. J. Clin. Microb. 2002;40:1091–1094. doi: 10.1128/JCM.40.3.1091-1094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Afanas’ev MV, Ikryannikova LN, Il’ina EN, Sidorenko SV, Kuz’min AV, Larionova EE, et al. Molecular characteristics of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis isolates from the Russian Federation. J. Antimicrob. Chemother. 2007;59(6):1057–1064. doi: 10.1093/jac/dkm086. [DOI] [PubMed] [Google Scholar]
- 23.Billington O, McHugh TD, Gillespie SH. Physiological cost of Rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 1999;43:1866–1869. doi: 10.1128/aac.43.8.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Soolingen D, de Haas PE, van Doorn HR, Kuijper E, Rinder H, Borgdorff MW. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J. Infect. Dis. 2000;182:1788–1790. doi: 10.1086/317598. [DOI] [PubMed] [Google Scholar]
- 25.Pym AS, Saint-Joanis B, Cole ST. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 2002;70(9):4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gegia M, Mdivani N, Mendes RE, Li H, Akhalaia M, Han J, et al. Prevalence of and molecular basis for tuberculosis drug resistance in the Republic of Georgia: validation of a QIAplex system for detection of drug resistance-related mutations. Antimicrob. Agents Chemother. 2008;52(2):725–729. doi: 10.1128/AAC.01124-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enne VI, Livermore DM, Stephens P, Hall LM. Persistence of sulphonamide resistance in E. coli in the UK despite national prescribing restriction. Lancet. 2001;357:1325–1328. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 28.Sjolund M, Wreiber K, Andersson D, Blaser M, Engstrand L. Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Ann. Intern. Med. 2003;139:483–487. doi: 10.7326/0003-4819-139-6-200309160-00011. [DOI] [PubMed] [Google Scholar]
- 29.Bottger EC, Pletschette M, Andersson D. Drug resistance and fitness in Mycobacterium tuberculosis infection. J. Infect. Dis. 2005;191:823–824. doi: 10.1086/427517. [DOI] [PubMed] [Google Scholar]
- 30.Sander P, Springer B, Prammananan T, Sturmfels A, Kappler M, et al. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob. Agents Chemother. 2002;46:1204–1211. doi: 10.1128/AAC.46.5.1204-1211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 32.Bahrmand AR, Titov LP, Tasbiti AH, Yari S, Graviss EA. High-level Rifampin resistance correlates with multiple mutations in the rpoB gene of pulmonary tuberculosis isolates from the Afghanistan border of Iran. J. Clin. Microbiol. 2009;47(9):2744–2750. doi: 10.1128/JCM.r00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Telenti A, Honore N, Bernasconi C, March J, Ortega A, Heym B, et al. Genotypic assessment of isoniazid and Rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J. Clin. Microbiol. 1997;35:3719–3723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohno H, Koga H, Kohno S, Tashiro T, Hara K. Relationship between Rifampin MICs for and rpoB mutations of Mycobacterium tuberculosis strains isolated in Japan. Antimicrob. Agents Chemother. 1996;40:1053–1056. doi: 10.1128/aac.40.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauck Y, Fabre M, Vergnaud G, Soler C, Pourcel C. Comparison of two commercial assays for the characterization of rpoB mutations in Mycobacterium tuberculosis and description of new mutations conferring weak resistance to Rifampicin. J. Antimicrob. Chemother. 2009;64(2):259–262. doi: 10.1093/jac/dkp204. [DOI] [PubMed] [Google Scholar]
- 36.Rosales-Klintz S, Jureen P, Zalutskaya A, Skrahina A, Xu B, Hu Y, et al. Drug resistance-related mutations in multidrugresistant Mycobacterium tuberculosis isolates from diverse geographical regions. Int. J. Mycobacteriol. 2012;1(3):124–130. doi: 10.1016/j.ijmyco.2012.08.001. [DOI] [PubMed] [Google Scholar]