Abstract
The structural repertoire and kinetic threshold distinguishing legitimate signaling substrates are fundamental questions in proteolytic networks and pathways. We used N-terminal proteomics to address these issues by identifying cleavage-sites within the Escherichia coli proteome driven by the apoptotic signaling protease caspase-3 and the bacterial protease GluC. Defying the dogma that proteases cleave primarily in natively unstructured loops, we found that both caspase-3 and GluC cleave in α-helices nearly as frequently as extended loops. Strikingly, biochemical and kinetic characterization revealed that E. coli caspase-3 substrates were greatly inferior to natural substrates, suggesting protease/substrate co-evolution. Engineering an E. coli substrate to match natural catalytic rates defined a kinetic threshold depicting a signaling event. This unique combination of proteomics, biochemistry, kinetics and substrate engineering reveals new insights into the structure-function relationship of protease targets and their validation from large-scale approaches.
Proteases are prominent components of biological systems in health and disease, often functioning as signaling molecules in networks and pathways – reviewed in 1,2. In these settings, proteases signal through limited proteolysis of discrete substrate pools, transmitting information by altering substrate localization, regulation, or activity. Identifying endogenous protease substrates and their cleavage-sites is paramount to delineating their downstream molecular signaling pathways 3,4. However, the critical substrate repertoire of most proteases is incompletely known or in many cases completely lacking. Unlike the stringent specificity of restriction endonucleases, proteases are enzymes with varying degrees of specificity and selectivity not solely influenced by a substrate’s cleavage-site amino acid sequence.
There are four main determinants influencing which substrates a protease cleaves and where cleavage-sites are. Spaciotemporal co-localization: proteases and substrates must be simultaneously present in the same sub-cellular compartment or physical space to be biologically relevant. Exosite interactions: some proteases and substrates utilize surfaces distinct from the active site and cleavage-site to drive affinity and selectivity. Sub-site specificity: many proteases display some degree of amino acid specificity in positions adjacent to the scissile bond, which provide varying levels of substrate cleavage-site selectivity. Motifs have been proposed for many proteases, based on information from synthetic peptide libraries or phage display technology 5–12. One of the best examples of this is the caspase family that maintains a strict requirement for aspartate in the P1 position, with distinctions in the P4, P3, P2, and P1’ positions 7,8. Structural presentation: proteases are thought to cleave substrates in flexible solvent exposed loops 13,14, and have been used historically to map protein domain limits. Crystal structures of protease inhibitors with prominent cleavage-site loops have also reinforced this view 15,16; however, disproportionately few substrate structures have been solved that include residues encompassing the cleavage-site. The likely explanation for this is that cleavage-sites are in flexible and unstructured regions of proteins, also known as disordered regions, and are inherently difficult to crystallize. Thus the key is to identify protease cleavage-sites from a library that samples both structural conformation and amino acid distribution to discriminate between sub-site amino acid preference and cleavage-site structure.
Based on these current concepts, we hypothesized that limited proteolysis of any proteome should be dominated by cleavages in unstructured regions. We endeavored to assess the contributions of both amino acid composition and structural presentation to substrate proteolysis using N-terminal proteomics (N-terminomics) on the non co-evolved, structurally intact, and well-characterized E. coli proteome, which allowed us to identify cleavage-sites on a global scale. We established two experimental paradigms. In the unbiased paradigm – the E. coli proteome challenged with the human protease caspase-3 and the Staphylococcal protease glutamyl endopeptidase (GluC) - there could be no pre-selection of substrates and proteases during evolution. In the biased paradigm - specific human caspase substrates challenged with human caspase-3 – we expect co-evolution of substrate sequences and structural requirements to fit the biological role of the caspase. An additional advantage of the E. coli proteome is that many of the potential substrates have previously reported structures, which we used to elucidate the structural conformation of the cleavage-sites found. Finally, we were able to propose threshold criteria of protease/substrate co-evolution. These studies offer fundamental insights into the structure-function relationship of proteases and substrates, and reveal a critical catalytic threshold distinctive of natural substrates, which non-evolved substrates fail to meet.
RESULTS
EXPERIMENTAL APPROACH & WORKFLOW
To elucidate the thee-dimensional conformation of protease cleavage-sites, we assembled an in vitro system comprising two purified proteases: human caspase-3 and Staphylococcal GluC; and an evolutionarily unbiased library of protein substrates composed of soluble E. coli lysate. Many measures were taken to maintain the structural integrity of proteins within the E. coli lysate, yet we saw no indication at any point in the process that proteins were denatured. We screened for cleavage-sites by combining our proteases-of-interest with the E. coli lysate, and assayed for cleavage-site peptides using N-terminomics, a method we recently reported to identify protease cleavage-sites in complex biological samples 17. Briefly, proteases cleave substrates to expose free N-terminal α-amines, which can be specifically labeled with a cleavable biotin tag. After digestion with trypsin, biotinylated peptides are enriched on neutravidin-linked agarose, eluted, and analyzed by LC-MS/MS. The proteome-wide coverage of N-terminomics, coupled with its lack of any structural bias, suits it perfectly to identify protease cleavage-sites, which can then be mapped back onto the structures of substrates residing in the Protein Data Bank (PDB), thereby elucidating the structure encompassing the cleavage-site (Fig. 1). Experiments were conducted using a range of protease concentrations spanning two orders of magnitude.
Figure 1.
Experimental approach. Samples of a library of structured proteins (E. coli soluble lysate) is treated with a range of exogenous protease concentrations (see text for details), and screened for cleavage-sites using N-terminomics. The location of cleavage sites is determined, and compared to available structures deposited in the Protein Data Bank, revealing the relationship between proteolysis and secondary structure.
N-TERMINOMICS RESULTS & CLEAVAGE-SITE IDENTIFICATION
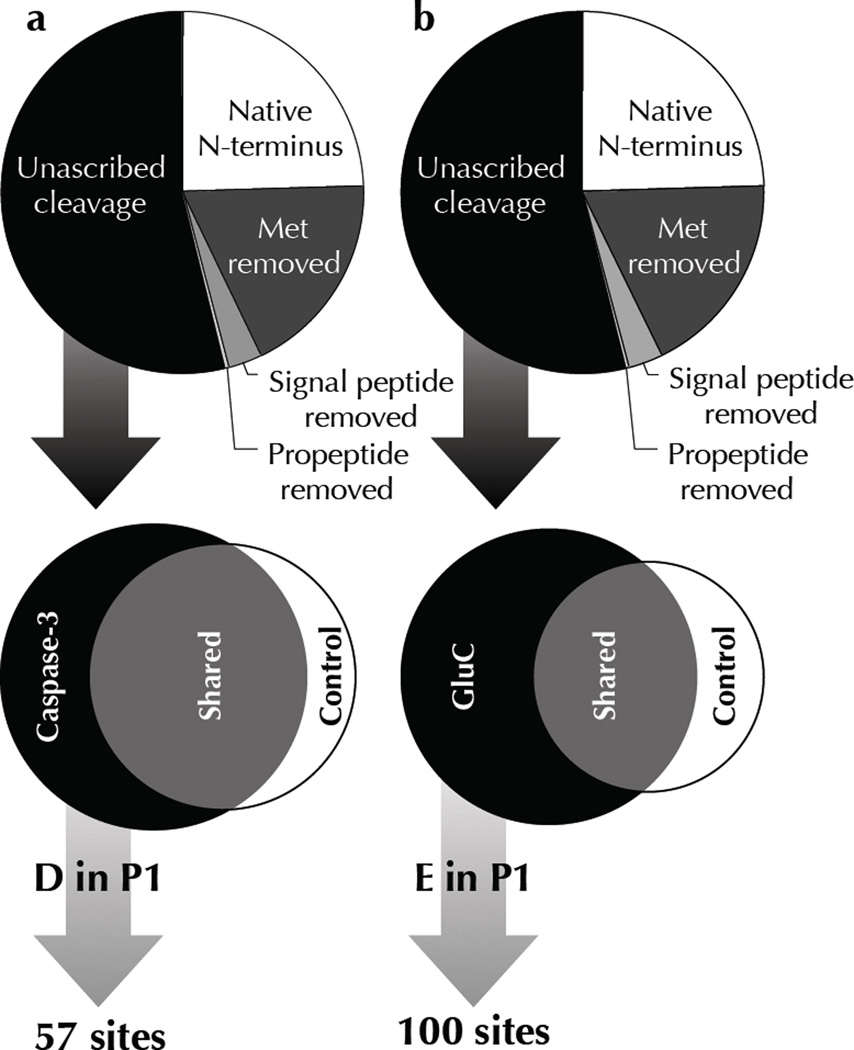
High quality peptides from each experiment were determined by searching a concatenated forward and reverse E. coli semi-tryptic database, resulting in peptide false discovery rate of less than 2% 18. The resulting peptide lists corresponded to protease cleavage-sites in E. coli proteins as well as unblocked protein N-termini (Supplementary Tables 1,2). We used a three-pronged approach to discern cleavage-sites of the purified proteases from the substantial background of endogenous proteolytic events (Fig. 2, Table 1). Cleavage-sites produced from the addition of exogenous proteases to E. coli lysate would differ from annotated cleavage-sites of endogenous proteases, falling into the category of unascribed cleavage-sites. This step was primarily used to exclude cleavage-sites originating from co-translational protein processing, such as initiator methionine excision and signal peptide removal. Protease-of-interest cleavage-sites were also excluded if they were concurrently found in control samples representing lysate alone, or in the case of caspase-3, treated with a catalytically inactive mutant of the protease. The last stipulation for identifying protease-of-interest cleavage-sites was that the strict P1 amino acid specificity for aspartate (Asp) for caspase-3, or glutamate (Glu) for GluC, must be preserved. Indeed, the stringent primary (P1) specificity of these proteases greatly simplifies the task of discerning true protease-of-interest cleavage-sites from other unascribed cleavage-sites not found in control samples due to incomplete sampling. Lists of caspase-3 and GluC cleavage-sites in E. coli proteins are shown in Supplementary Tables 3,4.
Figure 2.
N-terminomics reveals protease specific cleavage-sites. N-terminomic analysis of E. coli lysate treated with (a) human caspase-3 and (b) GluC reveals specific substrates and corresponding cleavage-sites. A tripartite criterion was used to identify genuine cleavage-sites of exogenous proteases from background proteolytic events inherent in E. coli lysate: Cleavage-sites must (1) not already be annotated as an endogenous site of proteolysis, (2) be only found in protease treated samples, and never in control samples, and (3) maintain the P1 aspartate/glutamate characteristic of each protease. See Table 1 for definitions.
Table 1.
Total and non-redundant peptide spectra identified by N-terminal proteomics. N-terminal peptides corresponding to protease cleavage-sites as well as unblocked protein N-termini are sub-divided based on their proteolytic processing. Protein N-termini retaining the initiator Met are designated “Native N-terminus”, whereas those processed by Methionine Aminopeptidase are termed “Met removed”. Periplasmic secreted proteins that have been processed by signal peptidases are grouped as “Signal peptide removed”, and likewise proteins with annotated propeptide cleavages are shown as “Propeptide removed”. All other N-termini correspond to internal cleavage-sites that have not been annotated, and thus are termed “Unascribed cleavage”. Protease-of-interest cleavage-sites were found by sieving this last group for cleavage-sites found only in the protease treated samples and cleaved after an Asp for human caspase-3, or a Glu for GluC.
| human caspase-3 | Staphylococcal GluC | |||
|---|---|---|---|---|
| Category | NR N-term | Spectra | NR N-term | Spectra |
| Native N-terminus | 253 | 12,533 | 196 | 8,066 |
| Met removed | 209 | 9,991 | 148 | 7,448 |
| Signal peptide removed | 37 | 1,178 | 26 | 659 |
| Propeptide removed | 1 | 24 | 1 | 5 |
| Unascribed cleavage | 598 | 8,133 | 434 | 4,735 |
| Total | 1,098 | 31,859 | 805 | 20,913 |
| Protease only | 162 | 947 | 263 | 2,195 |
| D or E in P1 | 57 | 490 | 100 | 767 |
To verify the purity and activity of the proteases added to E. coli lysate, we assessed the frequency of amino acids in the P1 position for all unascribed cleavage-sites comparing control and protease treated samples (Fig. 3). These data confirm the P1 specificity of human caspase-3 is enriched for Asp, and GluC is enriched for Glu with no other obvious enrichments. The frequency of P1 amino acids was normalized to the total number of cleavage-sites in each sample to allow for the unequal numbers of cleavage-sites originating from control and protease treated samples. An anticipated consequence of the normalization is a decrease in the frequency of protease treated P1 amino acids other than the enhanced Asp or Glu residues. This accounts for the noticeable decrease in arginine and lysine from control to protease treated samples, with the implication that most of the constitutive proteolytic cleavages in E. coli are at arginine and lysine.
Figure 3.
Distribution of amino acids in the P1 position of unascribed cleavage-sites. Protease only cleavage-sites (black bars) reveal the hallmark P1 specificity of human caspase-3 (a) and GluC (b). Cleavage-sites found in control samples (grey bars) suggest endogenous proteolysis at Ala, Lys, and Arg residues in the P1 position, and confirm the lack of endogenous Asp or Glu specific protease activity in E. coli.
SUB-SITE AMINO ACID PREFERENCES OF HUMAN CASPASE-3 AND GLUC
We investigated the specificity of both human caspase-3 and GluC using the WebLogo tool that identifies sequence conservation and amino acid frequency at positions flanking the cleavage-site (http://weblogo.berkeley.edu/logo.cgi) 19. Search parameters were extended from position P10 to P10’ in order to identify potential sub-site preferences not previously assessed by positional scanning peptide libraries. WebLogos were generated from the 57 caspase-3 cleavage-sites and 94 of the 100 GluC cleavage-sites identified by N-terminomics, whose sequences span P10 to P10’ (Fig. 4a,b). Six of the GluC cleavage-sites were located within 10 residues of substrate N- or C-termini, and thus could not be included in the WebLogo analysis. Surprisingly, our WebLogo analysis of human caspase-3 revealed no preferences in position P3, despite the widely accepted notion that Glu is preferred. The WebLogo results also suggest a minor preference for small and uncharged amino acids in the P2’ position, which is similar to the P1’ preference. To control for potential amino acid bias surrounding the primary Asp and Glu residues that characterize the protease cleavage specificity, we analyzed 1087 Asp-containing and 1220 Glu-containing sequences from E. coli proteins by WebLogo, showing no inherent amino acid bias (Supplementary Fig. 1a, 2a). The lack of specificity for caspase-3 in position P3 could also be accounted for by a proteome-wide depletion of Glu residues in position P3 in relation to Asp sequences. Therefore, we performed a Two-Sample-Logo analysis of these control Asp and Glu sequences from E. coli with 1000 sequences containing an equal distribution of each amino acid at every position (http://www.twosamplelogo.org/cgi-bin/tsl/tsl.cgi) (Supplementary Fig. 1b, 2b) 20. Indeed, Glu in P3 was not depleted in the E. coli proteome in relation to Asp in P1. However, on the whole these results support earlier specificity data for human caspase-3 and GluC based on substrates with no structure to them, although the contribution of the P3 sub-site in caspase-3 is inconsequential in the context of our structured substrate dataset. This means that the cleavage-site recognition in the immediate vicinity of the scissile bond (P4-P2’ in the case of caspase-3) is largely dependent on the properties of the protease, not the substrate.
Figure 4.
Specificity of caspase-3 and GluC. WebLogo representations of protease cleavage-sites depict the amino acid conservation and frequency at each position. The classic human caspase-3 consensus sequence DEVD↓G based on peptide positional scanning libraries 7,8 is recapitulated with two notable exceptions: there is no conservation at P3 and a weak additional preference for small uncharged amino acids at P2’ (a). GluC is not known to possess any extended specificity 32; however, we see a weak preference for hydrophobic residues in the P1’ position (b).
STRUCTURES PREFERRED BY HUMAN CASPASE-3 AND GLUC
Many of the E. coli proteins we identified as protease substrates had solved structures due to the efforts of the Joint Centers for Structural Genomics and individual researchers depositing protein crystal and NMR structures into the PDB. Mapping the cleavage-sites that we identified back to reported structures revealed the three-dimensional conformations that encompassed sites of proteolysis (Fig. 5, Fig. 6, and Supplementary Fig. 3). Cleavage-sites were manually assessed and visualized using PyMOL software 21, and secondary structure assignments were retrieved from the Dictionary of Protein Secondary Structure (DSSP) 22. Secondary structures were also predicted using the PSIPRED algorithm (http://128.16.10.201/psipred/psiform.html) 23, allowing us to also assess cleavage-site conformations of structurally unresolved substrates. As expected, WebLogo analysis of cleavage-site secondary structures revealed frequent cleavage in extended loop structures or regions of no electron density; however, there were numerous cleavage-sites residing in α-helices for both proteases tested (Fig. 5) contradicting a central dogma of proteolysis. Although the majority of cleavage-sites are from crystal-derived structures, we also found cleavage-sites in solution structures solved by NMR lessening the possibility of helix formation induced by crystal growth. Thus the notion that cleavage-sites can reside in α-helices is strongly supported by the prevalent examples in both structural methods. Scattered reports of this phenomenon exist in the literature; however, the magnitude and relative prevalence compared to loop conformations has never been directly addressed in a structurally unbiased system like ours 24. As expected, extended β-strands were almost never tolerated in cleavage-sites from human caspase-3 or GluC. An analysis of control Asp and Glu site secondary structures (Supplementary Fig. 4) showed a tendency toward loop and helical structures; however, this does not conflict with our finding that both caspase-3 and GluC cleaved E. coli substrates in α-helices as well as loops.
Figure 5.
Secondary structure preferences of human caspase-3 and GluC. WebLogo representations of secondary structures from protease cleavage-sites with the scissile bond indicated by the grey arrow. Secondary structure assignments were determined from (a,c) protein structures residing in the PDB as defined by DSSP, or (b,d) predicted by the PSIPRED algorithm. Both human caspase-3 and GluC cleaved substrates in loops as well as helices, but almost never in strands. The intolerance of human caspase-3 for β-strands appears to be restricted around the scissile bond. However, the strand intolerance for GluC is shifted toward the substrates N-terminus. Secondary structure assignments are as follows: L = loop, A = α-helix, B = β-strand.
Figure 6.
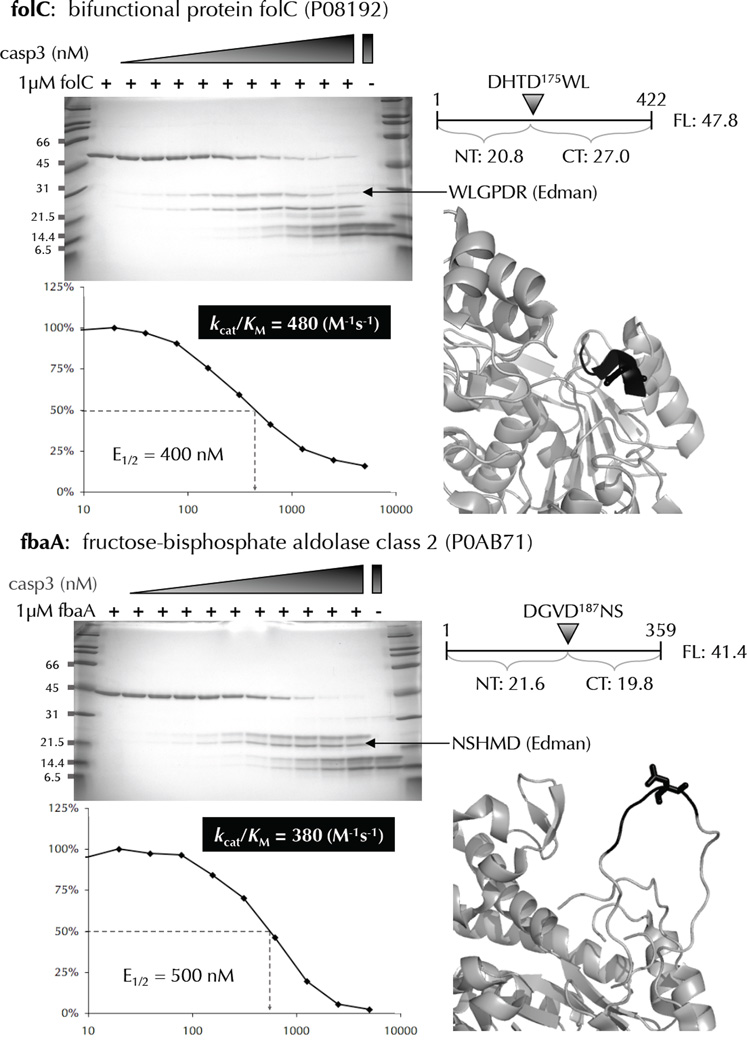
Biochemical and kinetic analysis of cleavage-sites identified by N-terminomics. Many E. coli proteins that were identified as substrates of human caspase-3, and containing only 1 cleavage-site were recombinantly expressed, purified, and subjected to in vitro cleavage by human caspase-3. The cleavage-site P4-P1’ amino acids are colored black with the P1 residue in stick format. Substrate cleavage-sites were identified by N-terminal sequencing of the proteolytic fragments using Edman degradation. The E1/2 values were measured based on the disappearance of the full-length substrate using densitometry. These values were used to calculate kcat/KM for each substrate.
BIOCHEMICAL VERIFICATION OF N-TERMINOMIC CLEAVAGE-SITES
To directly validate the cleavage-sites revealed by N-terminomics and kinetically characterize them, we selected several representative substrates cleaved at low, intermediate, and high human caspase-3 concentration for biochemical characterization. Substrates were selected that contained only 1 cleavage-site and had previously been shown to express well from a dataset of all cloned E. coli genes 25. We were able to rapidly evaluate these 14 proteins due to the availability of the Genobase ASKA collection of E. coli open reading frames cloned into N-terminal 6 histidine-tagged expression vectors. A substantial advantage of using this tagged protein collection is that the proteins are expressed in a homologous system and are purified under similar conditions used to produce the original E. coli lysate, enhancing the likelihood that the expressed proteins are conformationally similar to those in the original lysate. Potential substrates of human caspase-3 were expressed, purified, and evaluated in an in vitro cleavage assay. Rates of cleavage (kcat/KM) were measured by treating 1 µM substrate with a dilution series of protease for 1 hour at 37°C, followed by SDS-PAGE detection of cleavage products. The enzyme concentrations corresponding to 50% cleavage of the full-length substrates were determined by densitometry. KM values for these substrates were determined by competition with a fluorogenic substrate (Supplementary Table 5) and were all above 10 µM, so substrates in our assay are at least 10 fold below the KM. This allowed us to determine their respective kcat/KM values from the following half-life equation:
E1/2 is the concentration of protease that produces 50% substrate cleavage in time = t. The half-life equation used to determine kcat/KM values is based on a single cleavage-site; therefore substrates with two or more sites are overestimates of each cleavage-site kcat/KM value. We also verified that the cleavage-products observed in our cleavage assay matched the expected sizes based on the cleavage-sites identified in the N-terminomics screen. In addition, we sequenced many of the C-terminal cleavage products by Edman degradation to confirm the exact cleavage-sites. Examples of the validation and kinetic analysis are shown in Figure 6, with a full dataset in Supplementary Figure 1. Several of the substrates assayed revealed multiple cleavage-sites, despite our selection of substrates that were identified from a single N-terminomics hit. These additional cleavage-sites were identified through Edman degradation and for the most part would produce tryptic peptides too short to be unambiguously identified by MS. We tested 14 recombinant E. coli proteins in our cleavage assay, and only 3 (21%) could not be confirmed as human caspase-3 substrates either because the cleavage-products co-migrated with the human caspase-3 large and small subunits on the gel, or because no cleavage was observed at the concentrations tested. Lack of biochemically confirm of these substrate cleavage-sites in the purified system does not necessarily mean that these substrates were not cleaved in E. coli lysate. Possible reasons for this include altered substrate conformation in complexes with other proteins in the E. coli lysate, or partial unfolding due to the action of cellular chaperones. Our kinetic validation of protease cleavage-sites identified through N-terminomics is the first of its kind and serves as a general proof-of-principle that high quality MS/MS data from single peptides accurately identifies protein N-termini and sites of proteolytic cleavage.
SUBSTRATE ENGINEERING
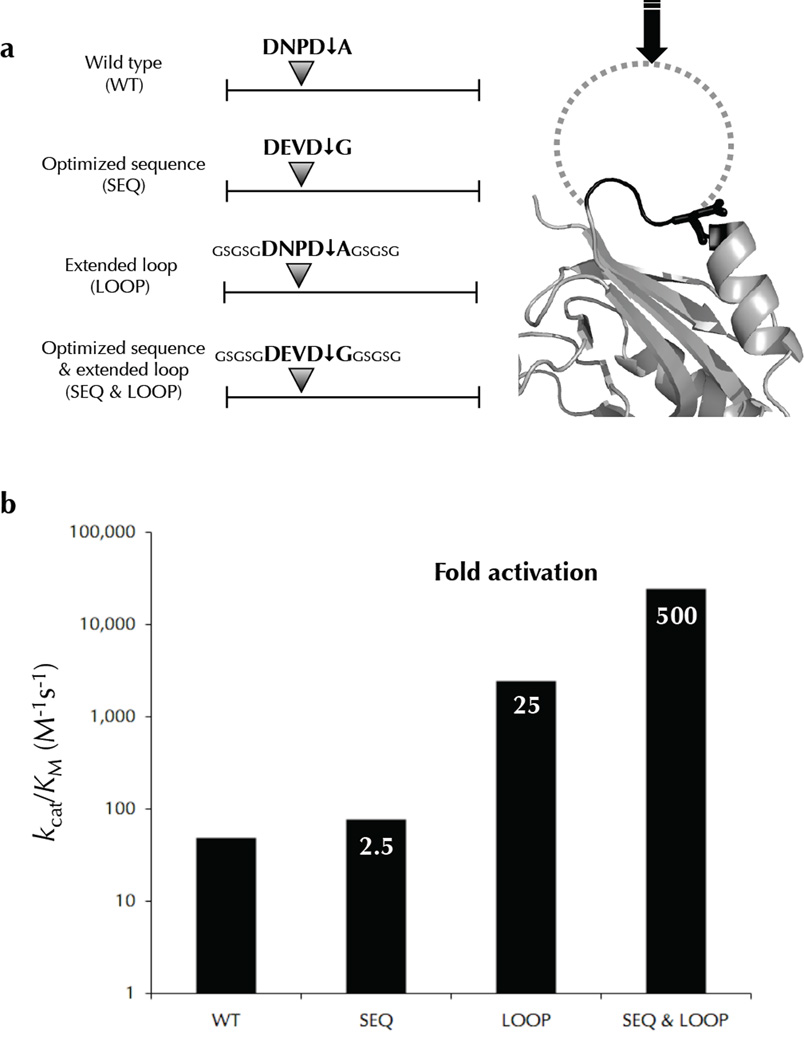
We next designed a panel of mutants to dissect how substrate features affect the rate of proteolysis. The E. coli protein carA was chosen as a template for engineering. The protein expressed well and the cleavage products were clearly distinct from the full-length precursor allowing for accurate quantitation in our cleavage assay. The wild type carA is cleaved at the sequence DNPD↓A in a short loop linking a beta-strand to an α-helix. Mutants were designed to optimize the cleavage-site amino acid sequence to DEVDG 7,8, to extend the cleavage-site loop making it more flexible (GSGSGDNPD↓AGSGSG), and both optimize the amino acid sequence and extend the loop (GSGSGDEVD↓GGSGSG). These carA mutants were evaluated in our cleavage assay to determine their respective kcat/KM values (Fig. 7). The optimized sequence was cleaved 2.5 times better than wild type, and the extended loop mutant was cleaved over 25 times better than wild type. Importantly, combining the optimized sequence with the extended loop dramatically increased the kcat/KM to over 500 fold above wild type making it comparable to some natural caspase substrates, and defines a threshold that bona fide substrates should satisfy or exceed (see below). This suggests that an extended loop is more influential than an optimal sequence in sensitizing a substrate to proteolytic cleavage, and flexible extended sites are cleaved dramatically better with an optimized amino acid sequence. These results strongly support the idea that substrates co-evolve with proteases to present both an extended loop and an optimal cleavage-site sequence.
Figure 7.
Engineered carA mutants are cleaved more efficiently than wild type. The E. coli caspase-3 substrate was engineered to dissect the contribution of sequence and structure on cleavage efficiency. The various constructs with are shown in (a) with an optimized sequence, an extended loop, and a combination of both. Relative rates of cleavage were measured for these mutants revealing that an extended loop conformation improves the kcat/KM more dramatically than an optimized sequence (b). However, an optimized sequence in parallel with an extended loop synergizes to elicit efficient cleavage.
KINETIC COMPARISON OF E. COLI SUBSTRATES WITH NATURAL HUMAN CASPASE-3 SUBSTRATES
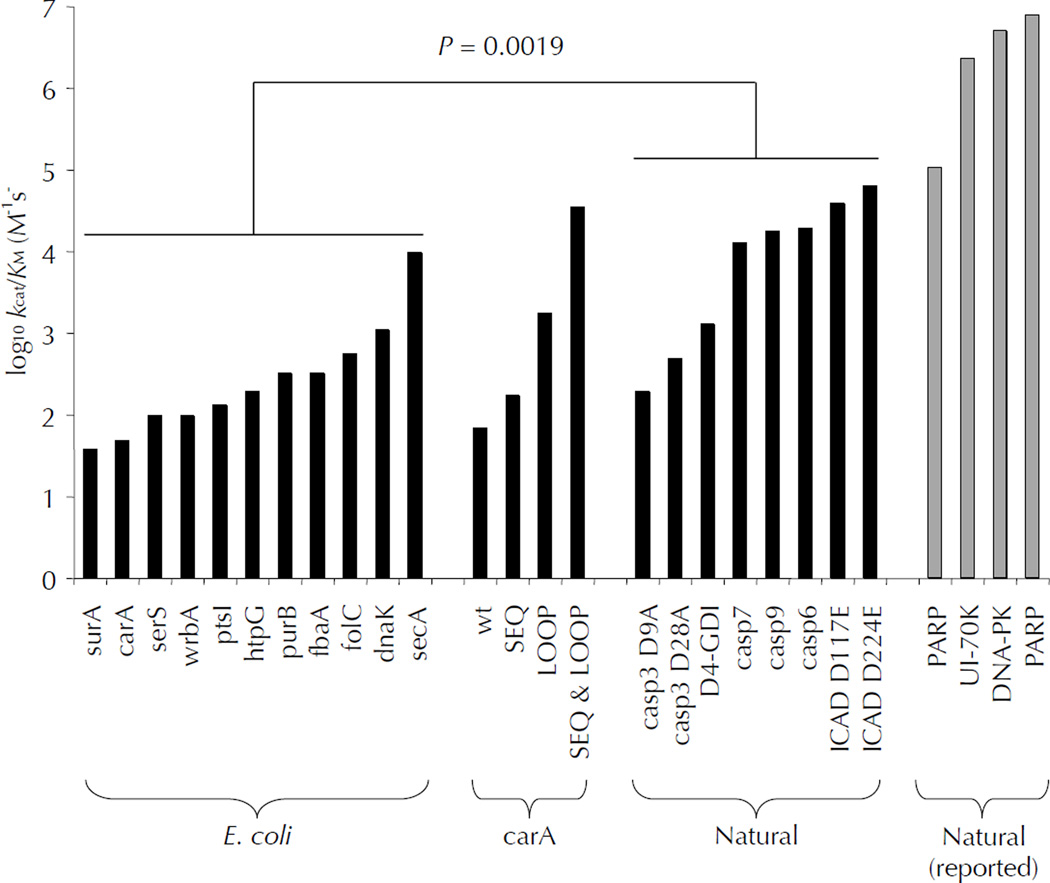
A goal of this study was to compare the specificity of caspase-3 on an unbiased natively folded substrate proteome (E. coli) with a naturally evolved one (human) containing caspase signaling targets. The extensive characterization of apoptotic caspase substrates in the literature is a valuable resource from which we chose several well-characterized substrates for our study 4,26. Kinetic values were determined experimentally in our cleavage assay, or collected from reported literature values 27,28 (Fig. 8). Natural caspase-3 substrates measured in our assay had significantly higher kcat/KM values than non-natural E. coli substrates (p = 0.0019). Only secA and the optimized carA mutant were able to approach values comparable to natural substrates. We cannot rationalize how some natural substrates are cleaved more efficiently than the optimized carA mutant due to the amino acid sequence or local structure of their cleavage-sites. Despite minor variations in experimental conditions between our study and others in the literature, their reported kinetics show extremely efficient cleavage of natural substrates, which reiterates our findings and suggests that these natural caspase-3 substrates may have evolved exosites to increase their rates of cleavage. Importantly, it is not certain that limited proteolysis of substrates that results in gain-of-function needs to be rapid, but there is a good chance that loss-of-function cleavages should be rapid enough to remove biological activity, as proposed earlier 4. However, all of the activating cleavages of natural substrates in our study do maintain elevated cleavage kinetics.
Figure 8.
Natural human caspase-3 substrates are kinetically superior to E. coli substrates. Human caspase-3 cleaves most E. coli substrates with kcat/KM between 50 and 2,000 M−1s−1, while several biologically relevant human caspase-3 substrates were cleaved with values greater than 10,000 M−1s−1. The propensity of natural substrates to be kinetically superior to E. coli substrates was shown to be statistically significant (p=0.0019). Engineering the E. coli substrate carA to contain an optimized cleavage-site sequence in an extended loop improved the kcat/KM value to over 30,000 M−1s−1. Some natural substrates have kcat/KM values greater than 30,000 M−1s−1, implying additional mechanisms for enhancing catalysis, discussed in the text.
DISCUSSION
We have successfully probed the specificity and structural preferences of human caspase-3 and Staphylococcal GluC in the context of a folded protein substrate library, overcoming the limitation of other methods that unlink amino acid composition from protein structure. New focused proteomic technologies have enabled us to address this question on a proteome-wide level. Although the number of caspase-3 and GluC cleavages-sites in E. coli proteins that we report is not extraordinary, these cleavage-sites are the most relevant for our structure-function study of protease substrates, as multiple cleavages in a protein are likely to destroy its native organization, thereby precluding any structural interpretation. Accordingly, our experimental conditions produced mainly single cleavage-sites in E. coli proteins, thus making our structural interpretation of these cleavage-sites valid.
These techniques are not without caveats relating to sensitivity of detection. However, the signature of proteolysis obtained from N-terminomics gives sufficient data for clear-cut analysis of specificity parameters. N-terminomics cannot reveal what quantity of precursor substrate is cleaved to generate the proteolytic fragments identified, even in conjunction with techniques employing heavy and light isotope methodologies. Consequently, we coupled traditional biochemical techniques to quantitate proteolysis and confirm proteomic results from representative substrates.
As expected, a clear relationship was observed between the number of cleavage-site peptides and the caspase-3 concentration used to generate them (Supplementary Fig. 5). However, the kcat/KM values did not appear to correlate well with the concentration of caspase-3 needed to identify cleavage-sites by N-terminal proteomics (Supplementary Fig. 6). This result is likely explained by two dominant factors: (1) the well-documented phenomenon that peptides do not all ionize equally well, creating bias for cleavage-site peptides that do ionize well, and (2) proteins are not maintained at equal levels in E. coli, and thus the effective substrate concentration varies dramatically for different proteins. We addressed the second concern by correlating the reported abundance levels of the substrates we identified for each concentration of human caspase-3 tested (Supplementary Fig. 7) 29. Understandably, abundant substrates tended to be identified at lower caspase-3 concentration, but the caspase-3 N-terminomics revealed cleavages in proteins varying in concentration by nearly 5 orders of magnitude. This suggests that protein abundance is of modest importance in the acquisition of N-terminomics data, and attests to the sensitivity of our N-terminomics methodology. While preceding proteomic studies have produced lists of numerous substrate cleavage-sites 17,24,30–36, fundamental aspects of kinetics, recognition of structured elements, and other factors influencing proteomic detection have remained unanswered until now. Our results suggest that substrate abundance, cleavage-site kcat/KM, and the capacity of the resultant peptide to ionize all contribute to the identification of cleavage-sites by N-terminal proteomics. This study is the first to biochemically confirm and kinetically evaluate the significance of proteome-wide proteolysis, allowing us to address the sequence/structure relationship that underlies limited proteolysis by signaling proteases such as caspase-3.
The approximately 4500 proteins contained in E. coli lysate display a diversity of structural conformations amino acid sequence combinations. Importantly, E. coli does not possesses any known Asp or Glu specific proteases, reducing the evolutionary pressure to alter the distribution of these amino acids throughout its proteome. The absence of endogenous caspase- or GluC-like proteases also substantially diminished the possibility of confounding exosite interactions. The folding and stability of E. coli proteins should also be compatible with the proteases tested because it normally grows at human body temperature, yielding a proteome selected for stability at 37°C – the temperature we use in our assays. Likewise, the proteases tested are from humans or the commensal skin bacteria Staphylococcus aureus, which produce the protease GluC.
The starting hypothesis stated that we would find cleavage-sites predominantly located distinct from regions of ordered secondary structure (helices and sheets). Our data of cleavages on folded proteins with reported structures falsify this hypothesis. Cleavage is almost as frequently observed in α-helices as in regions without secondary structure. Are the helices unfolding, or can proteases cleave the helices directly? The parsimonious explanation is that helices are cut without unfolding. GluC presents a broad and exposed active site that helical structures may be able to access; however, previous structures of caspase-3 show a deep and narrow active site cleft with an extended tetrapeptide inhibitor bound, which does not appear to accommodate bulkier structured substrates. To reconcile the observed α-helical caspase-3 cleavage-sites, we propose flexible reordering of either the substrate or the active site, and that only the exposed P1 Asp is required to accommodate the S1 sub-site, while other sub-sites need not be occupied. Interestingly, analysis of cleavage-sites located in solved protein structures from the CutDB database (http://cutdb.burnham.org/) also showed a high frequency of cleavage-sites in α-helices and loops, with substantially less cleavage in β-strands (personal communication) 37.
An alternative explanation for helical substrate cleavage is a local helical unfolding concomitant with protease binding, and we find many examples of short helices positioned within larger loops that may be in a dynamic equilibrium fluctuating between helix and loop in solution. Yet, we find no other indications, such as enhanced local temperature factors in the structures, which could account for flexibility of the cleaved helices. Unfolding of substrates is more difficult to envision for cleavage-sites in long stretches of α-helix that may not be as structurally dynamic, and yet these are cleaved also. We found two examples of cleavage-sites in long α-helixes resolved by NMR (Supplementary Fig. 8), which excludes the possibility that α-helixes are formed during protein crystallization. Likely a combination of dynamic α-helices sampling loop conformations, and rigid helices with a protruding P1 amino acid that accommodates the protease specificity, together account for the observed helical cleavage-sites in E. coli proteins.
E. coli cleavage-sites were analyzed for common biophysical features using the DSSP database of standardized secondary structure assignments (ftp://ftp.ebi.ac.uk/pub/databases/dssp/). The solvent accessibility and hydrogen bonding energy of each amino acid for every cleavage-site was collected, and the average values and standard deviations were plotted from P10 to P10’ (Supplementary Fig. 9,10). Interestingly, no obvious biophysical features were associated with cleavage-sites.
We propose that the folded E. coli proteome serves as a baseline of unbiased caspase-3 cleavage specificity and kinetics. We demonstrate that one way to improve substrate hydrolysis from 103 M−1s−1 to 105 M−1s−1 is to extend the cleavage-site loop away from the surface of the protein and incorporate an optimal sequence. Both of these features separately enhance the cleavage efficiency by caspase-3, together synergizing to account for a 500-fold enhancement (kcat/KM). All of the catalytic rates for E. coli proteins were less than this, probably because of suboptimal position and composition of their cleavage-sites. The catalytic enhancement seen in natural substrates is likely the result of co-evolution of human caspase-3 and its substrates, producing cleavage-sites on extended unstructured loops with optimized amino acids in P4, P2, P1, and P1’. Although we have not directly ruled out the possibility that α-helices can be cleaved as efficiently as extended loops, this seems unlikely in light of the carA engineering experiments, which show that a flexible loop is the critical feature of kinetically superior sites. Therefore, even though caspase-3 can cleave α-helices inefficiently, we do not anticipate natural signaling substrates of caspase-3 to be cleaved in α-helices. Secondary exosite interactions at surfaces distinct from the active site may be an additional mechanism for specific and efficient cleavage. This has been convincingly demonstrated for the protease thrombin and its substrate fibrinogen 38, as well as matrix metalloproteases and their cognate substrates 39. This mechanism may account for the extremely efficient cleavage kinetics reported for several natural caspase-3 substrates.
Our results have immediate implications in several areas of protease research. First, we present data defining a kinetic threshold that many physiologically relevant substrates of signaling proteases, such as caspase-3, do fulfill. We propose that substrates be challenged with this kinetic litmus test to strengthen claims of biological relevance. Likewise, other proteases could be investigated in a similar manner to define protease-specific kinetic thresholds for natural substrate validation. Second, our specificity and structural analysis of caspase-3 cleavage-sites can be incorporated into substrate prediction algorithms by weighting positions P4, P2, and P1’ with Asp fixed in P1, and limiting predictions to sites in known or predicted loop structures. This dual specificity-structure filter could dramatically diminish the exorbitant number of false positive predictions, making biochemical evaluation possible where it would otherwise be unrealistic. Indeed, the methods and results of this study serve as a broadly applicable template to characterize the amino acid specificity and structural permissiveness of diverse proteases.
METHODS
E. coli lysate preparation
We cultured E. coli K12 strain MG1655 in 3 liters of 2× yeast tryptone medium at 37°C in a shaking incubator. Bacterial cultures were collected at OD600 = 1.0, and pelleted by centrifugation. Cell pellets were resuspended in 1× assay buffer (20 mM PIPES pH 7.2, 100 mM NaCl, 10% (w/v) sucrose, 0.1% (w/v) CHAPS, and 1mM EDTA), and ruptured by sonication on ice for 3 minutes at 50% intensity and 50% duty cycle. Insoluble material was removed by centrifugation, and clarified with a 0.45 µm filter yielding a final concentration of 20 mg ml−1 total protein. The resultant lysate was aliquoted and frozen at −20°C.
Expression and purification of human caspase-3
Human caspase-3 was expressed, purified, and quantitated by active site titration as previously described 40.
Cleavage of E. coli lysate proteins by purified proteases
Aliquots of frozen E. coli lysate were thawed at room temperature and spun down at 13,000 RCF for 5 minutes at 4°C prior to protease treatment. Purified caspase-3 (described above), and GluC (Roche) were prepared at 5× or 10× final concentration in 1× assay buffer (20 mM PIPES pH 7.2, 100 mM NaCl, 10% sucrose, 0.1% CHAPS, 1mM EDTA, and 5 mM fresh DTT) and pre-activated at 37°C for 15 minutes. Active protease was then added to E. coli lysate pre-incubated at 37°C, mixed and the cleavage reaction was maintained at 37°C for 1 hour. Dry guanidine HCl was added to 6 M final concentration, DTT was added to 10 mM, and the samples were boiled for 10 minutes to terminate the cleavage reaction by denaturation.
N-terminomic sample preparation
Samples were prepared as described previously 17 with the following modifications. After labeling with sulfo NHS-SS-biotin (Pierce), unreacted label was quenched by the addition of 50 mM ammonium bicarbonate (AmmBic). Samples were then buffer exchanged into 8M M urea, 50 mM HEPES pH 7.8, 100 mM NaCl. 10 mM AmmBic buffer was used to dilute samples down to 2 M urea. Biotinylated peptides were bound to high capacity neutravidin agarose resin (Thermo). Biotinylated peptides were eluted with the addition of 5 mM tris(2-carboxyethyl)phosphine at 37°C for 30 minutes. Eluted peptides were loaded onto C18 Sep-Pak Vac 6cc cartridges (Waters), washed with 0.1% TFA, and eluted with 50% MeCN, 0.1% TFA. Peptides were dried in a vacuum centrifuge, and solubilized in 50 µL of 0.1% TFA.
Sample analysis by nano LC–MS
One experiment of caspase-3 was analyzed by an Orbitrap LTQ, while the other two caspase-3 and one GluC experiments were analyzed by an LTQ. The Orbitrap LTQ system consisted of a Michrom MS4 MDLC using the 150 × 0.2 mm Magic C18 3 um beads/200 A pores at 2 ul/min, gradient of 2–5% B in 2 min and 5–35% B in the next 118 min; top5 data-dependent MS, MS/MS method with precursor scans in the Orbitrap in the profile mode, resolution of 60,000 MS/MS scans in the LTQ in the centroid mode. The automated NanoLC-LTQ system was previously described17. Each sample run on the Orbitrap LTQ was analyzed once, while the other samples were analyzed 3 or more times on the LTQ.
Database searching
MS/MS spectra were searched as described before. SEQUEST search results were filtered for peptides identified from two or more spectra with a minimum probability score of 0.8, and a cross correlation value (Xcorr) of at least 2.0 41. The peptide false discovery rate was less than 2%.
E. coli protein expression and purification
E. coli proteins were retrieved from the Genobase collection of open reading frames cloned into an N-terminal 6 histidine tagged inducible expression vector and maintained in frozen stock cultures25. Protein expression and purification was performed similar to that of caspase-3. Protein concentration was determined by A280 and purity was verified by SDS-PAGE, and stored at −80°C.
Cloning and engineering of carA
The caspase-3 E. coli substrate, carA was sub-cloned into pET-15b using “carA for” (NdeI) and “carA rev” (BamHI).
carA for: CACACACATATGATTAAGTCAGCGCTATTG
carA rev: CACACAGGATCCTTACTTAGCGGTTTTACG
The optimized cleavage-site mutant was constructed from “carA for” and “opt rev”, and “opt for” and “carA rev”.
opt rev: TCCATCTACCTCATCGCCCGCGATAATGCAGCC
opt for: GATGAGGTAGATGGAGCGGCGCTGGCGTTAGAA
The extended loop mutant was constructed from “carA for” and “ext rev”, and “ext for” and “cara rev”.
ext ref: CGCATCCGGGTTATCACCACTTCCACTACCGCCCGCGATAATGCAGCC
ext for: GATAACCCGGATGCGGGTAGCGGTAGTGGAGCGGCGCTGGCGTTAGAA
The optimized cleavage-site & extended loop mutant was constructed from “carA for” and “o&e rev”, and “o&e for” and “carA rev”.
o&e rev: TCCATCTACCTCATCACCACTTCCACTACCGCCCGCGATAATGCAGCC
o&e for: GATGAGGTAGATGGAGGTAGCGGTAGTGGAGCGGCGCTGGCGTTAGAA
Statistical analysis of substrate kinetics
Statistical analysis of the cleavage kinetics of caspase-3 substrates was performed using the Student’s t-test using an unpaired and two-tailed analysis with equal variance.
Determination of KM for caspase-3 substrates
The KM for caspase-3 cleavage of various protein substrates was calculated by using the modified Michaelis-Menten equation when two competitive substrates were present simultaneously in the enzyme reaction 42. The initial velocity (vDEVD) for hydrolysis of Ac-DEVD-AFC (DEVD) in the presence of a competitive non-fluorescent substrate of caspase-3 (Prot) was fit using the equation:
| (Equation 1) |
In Equation 1, [DEVD] and [Prot] are the concentrations for Ac-DEVD-AFC and the protein substrate, respectively, while KM represents the Michaelis-Menten constant for the indicated substrate.
Recombinant proteins were exchanged to standard caspase assay buffer (10 mM Pipes pH 7.2, 100 mM NaCl, 5% sucrose, 0.1% CHAPS, 10 mM DTT) and concentrated. Ac-DEVD-AFC was serially diluted in caspase buffer (2–300 µM), mixed with the protein of interest at constant concentration. Pre-incubated caspase-3 was then added to the substrate mix at 1 nM final concentration, and kinetics of fluorescence generated by AFC was recorded immediately using the SpectraMax Gemini EM plate reader (Molecular Devices). At least three different final concentrations of recombinant protein were used for each KM,Prot determination. The resulting initial velocity was plotted against the Ac-DEVD-AFC concentration and the data was fit using Equation 1. The fit with Equation 1 generated KM,DEVD for DEVD-AFC cleavage when no protein substrate was present, or the KM,Prot for protein substrate cleavage when protein substrate was present.
Supplementary Material
Acknowledgments
This work was supported by NIH Roadmap Initiative National Biotechnology Resource Center grant RR20843 for the Center on Proteolytic Pathways, CA69381 from the NCI, and by Training Grant 5T32CA77109-9 from the NCI.
Footnotes
AUTHOR CONTRIBUTIONS
J.T. designed and performed most experiments and interpreted results, W.Z. performed LC-MS/MS analysis and database searching, C.P designed and performed kM determination experiments, T.R. prepared some N-terminomic samples, S.S. performed Edman degradation, A.E. helped revise the manuscript, S.R. aided in structural interpretation, G.S. interpreted results and together with J.T. and S.R. wrote the manuscript.
REFERENCES
- 1.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 2.Salvesen GS, Abrams JM. Caspase activation - stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 3.Gevaert K, et al. Applications of diagonal chromatography for proteome-wide characterization of protein modifications and activity-based analyses. Febs J. 2007;274:6277–6289. doi: 10.1111/j.1742-4658.2007.06149.x. [DOI] [PubMed] [Google Scholar]
- 4.Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 5.Ding L, et al. Origins of the specificity of tissue-type plasminogen activator. Proc Natl Acad Sci USA. 1995;92:7627–7631. doi: 10.1073/pnas.92.17.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith M, Shi L, Navre M. Rapid identification of highly active and selective substrates for stromelysin and matrilysin using bacteriophage peptide display libraries. J Biol Chem. 1995;270:6440–6449. doi: 10.1074/jbc.270.12.6440. [DOI] [PubMed] [Google Scholar]
- 7.Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 8.Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 9.Deng SJ, et al. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J Biol Chem. 2000;275:31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 10.Harris JL, et al. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci U S A. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazif T, Bogyo M. Global analysis of proteasomal substrate specificity using positional-scanning libraries of covalent inhibitors. Proc Natl Acad Sci U S A. 2001;98:2967–2972. doi: 10.1073/pnas.061028898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- 13.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- 14.Coombs GS, Bergstrom RC, Madison EL, Corey DR. Directing sequence-specific proteolysis to new targets. The influence of loop size and target sequence on selective proteolysis by tissue-type plasminogen activator and urokinase-type plasminogen activator. J Biol Chem. 1998;273:4323–4328. doi: 10.1074/jbc.273.8.4323. [DOI] [PubMed] [Google Scholar]
- 15.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 16.Kelly CA, Laskowski M, Jr, Qasim MA. The role of scaffolding in standard mechanism serine proteinase inhibitors. Protein Pept Lett. 2005;12:465–471. doi: 10.2174/0929866054395383. [DOI] [PubMed] [Google Scholar]
- 17.Timmer JC, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 19.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 21.DeLano WL. The PyMOL Molecular Graphics System. 2002 on the World Wide Web http://www.pymol.org.
- 22.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 23.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 24.Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 26.Fischer U, Janicke RU, Schulze-Osthoff K. Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 2003;10:76–100. doi: 10.1038/sj.cdd.4401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stennicke HR, et al. Pro-caspase-3 is a major physiologic target of caspase-8. J Biol Chem. 1998;273:27084–27090. doi: 10.1074/jbc.273.42.27084. [DOI] [PubMed] [Google Scholar]
- 28.Casciola-Rosen L, et al. Apopain/CPP32 cleaves proteins that are essential for cellular repair: a fundamental principle of apoptotic death. J Exp Med. 1996;183:1957–1964. doi: 10.1084/jem.183.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihama Y, et al. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics. 2008;9:102. doi: 10.1186/1471-2164-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Impens F, et al. Mechanistic insight into taxol-induced cell death. Oncogene. 2008;27:4580–4591. doi: 10.1038/onc.2008.96. [DOI] [PubMed] [Google Scholar]
- 31.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 32.Schilling O, Overall CM. Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat Biotechnol. 2008;26:685–694. doi: 10.1038/nbt1408. [DOI] [PubMed] [Google Scholar]
- 33.Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics. 2007;6:611–623. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Enoksson M, et al. Identification of proteolytic cleavage sites by quantitative proteomics. J Proteome Res. 2007;6:2850–2858. doi: 10.1021/pr0701052. [DOI] [PubMed] [Google Scholar]
- 35.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134:679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat Methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- 37.Igarashi Y, et al. CutDB: a proteolytic event database. Nucleic Acids Res. 2007;35:D546–D549. doi: 10.1093/nar/gkl813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stubbs MT, Bode W. The clot thickens: clues provided by thrombin structure. Trends Biochem Sci. 1995;20:23–28. doi: 10.1016/s0968-0004(00)88945-8. [DOI] [PubMed] [Google Scholar]
- 39.Overall CM. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 40.Denault JB, Salvesen GS. Apoptotic caspase activation and activity. Methods Mol Biol. 2008;414:191–220. doi: 10.1007/978-1-59745-339-4_15. [DOI] [PubMed] [Google Scholar]
- 41.Yates JR, Eng JK, 3rd, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 42.Morrison JF. The slow-binding and slow, tight-binding inhibition of enzyme-catalysed reactions. Trends Biochem. Sci. 1982;3:102–105. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.