Abstract
In regenerative medicine, the prospect of stem cell therapy holds great promise for the recovery of injured tissues and effective treatment of intractable diseases. Tracking stem cell fate provides critical information to understand and evaluate the success of stem cell therapy. The recent emergence of in vivo noninvasive molecular imaging has enabled assessment of the behavior of grafted stem cells in living subjects. In this review, we provide an overview of current optical imaging strategies based on cell- or tissue-specific reporter gene expression and of in vivo methods to monitor stem cell differentiation into neuronal lineages. These methods use optical reporters either regulated by neuron-specific promoters or containing neuron-specific microRNA binding sites. Both systems revealed dramatic changes in optical reporter imaging signals in cells differentiating into a neuronal lineage. The detection limit of weak promoters or reporter genes can be greatly enhanced by adopting a yeast GAL4 amplification system or an engineering-enhanced luciferase reporter gene. Furthermore, we propose an advanced imaging system to monitor neuronal differentiation during neurogenesis that uses in vivo multiplexed imaging techniques capable of detecting several targets simultaneously.
Keywords: Reporter-based cell tracking, Differentiation imaging, Neuronal differentiation, Neuronal microRNA, Multiplex imaging
Introduction
Over the last decade, stem cell therapy has been intensively studied in regenerative medicine to make possible the replacement of lost tissues with fresh tissue for functional recovery [1–3]. Amid numerous trials involving the therapeutic use of pluripotent stem cells, a number of techniques to elucidate the behavior of transplanted stem cells were developed for potential clinical use. Recently, the rapid progress of molecular imaging techniques has enabled noninvasive evaluation of the characteristics of transplanted stem cells and, in particular, of their viability. Molecular imaging allows real-time visualization of survival and quantitative viability measurements of stem cells implanted into an injured site by using stem cells labeled with proper radiotracers or reporter-based gene expression schemes [4–9].
However, to be successful with stem cell therapy, we should note that the implanted stem cells should be differentiated into an appropriate cell lineage with high efficacy, and in this sense, we need an evaluation system capable of tracking changes in cell fate to indicate successful or unsuccessful replacement of damaged tissues. Molecular imaging strategies are particularly suitable for this purpose because of their capability for noninvasive repetitive tracking of stem cell characteristics to monitor the differentiation of transplanted stem cells.
A variety of imaging modalities, such as nuclear, optical, and magnetic resonance (MR) imaging, can be easily applied to visualize the process of cellular differentiation in vivo. Radionuclide imaging approaches offer superior detection sensitivity (nM-pM) that enables imaging of deeper areas. Nevertheless, monitoring small differences between undifferentiated and differentiated cells with radionuclide imaging may be difficult because of its low resolution and high background noise. In comparison, MR imaging has substantial value for soft tissue contrast with high resolution. However, this technique is challenging to use because of its low sensitivity and false-positive rate, resulting from nonspecific uptake of magnetic nanomaterials into phagocytic cells. Optical imaging represents potential merits such as low background and high sensitivity. These properties allow for a suitable imaging modality to track the changes of cell fate during the differentiation process.
In this review, we focused on optical imaging tools for in vivo evaluation of grafted stem cells’ fate commitment into neuronal lineages in vivo, representing (1) lineage-specific promoter-mediated optical reporter gene techniques, (2) technologies to improve signal intensity of optical reporters for enhanced visualization of cellular differentiation, (3) imaging strategies for neuron-specific microRNA (miRNA)-targeted neuronal differentiation, and (4) suggestions concerning the usefulness of multiplex imaging to evaluate differentiation patterns both in vitro and in vivo.
From Innovative Reporter Imaging to Tissue-specific Reporter Gene Studies
To realize the rapid advances of molecular imaging that enable visualization of changes in biological processes in vivo, the concept of the reporter gene was a real necessity. The reporter-based imaging strategy endowed us with the ability to localize or monitor the migration of implanted cells, to evaluate therapeutic responses of cells and tissues to drugs, and to visualize the endogenous expression of transgenes in living subjects [10–12].
Since then, amid huge efforts to develop various reporters to monitor biological phenomena in vivo, a variety of imaging reporter genes have been developed. In particular, to develop an efficient in vivo reporter, the red-shifted variant of the Renilla luciferase reporter was constructed using a mutagenesis screening technique [13]. This new luciferase variant exhibited high-sensitivity quantum yield and greater enzymatic activity than the native luciferase. Taking advantage of this improved reporter system, the location, survival, and proliferation of implanted stem cells could be easily evaluated. While ubiquitin promoter-driven reporter genes made in vivo cell tracking easier, in vivo monitoring of cell differentiation needs reporters with different characteristics than those for cell tracking. This desired characteristic was ‘cell- or tissue-specificity’ associated with cellular differentiation.
Differentiation is defined as a cellular process by which dividing or growing cells become fully specialized. The differentiation process includes changes in cell morphology, cell size, and cellular metabolic activity that are triggered by the highly organized expression of specific genes. Lineage-specific transcription factors are involved in inducing these specialized cells. Additionally, 5′-regulatory regions containing specific promoters located upstream of the coding gene play important roles in determining cell lineage commitment. From this concept, the tissue-specific promoter-based transgene imaging strategy was proposed and extensively investigated to make the reporter gene be expressed only in the cell or tissue of interest. This in vivo imaging using tissue-specific reporters can elucidate spatially distributed reporter activities in small animals and reveal tissue-specific differentiation of implanted stem cells [14, 15]. Yong et al. used a β-cell–specific insulin promoter-driven tri-fusion reporter gene to track the endogenous β-cell population in a transgenic mouse model for diabetes [14]. This investigation documented a progressive decrease of bioluminescence signals in the pancreatic tissue of a streptozotocin-induced type 1 diabetes model. In addition, insulin-specific promoter-driven luciferase activity was used to monitor cell-specific signals for 40-60 days in a type 2 diabetes model. These studies might be easily extended to monitor responses of the endogenous β-cell population to therapeutic drugs and to evaluate the survival of transplanted islet β-cells in a model of type 1 diabetes. Successful study regarding the tissue-restricted reporter was also reported in in vivo imaging of transgenic mouse heart using cardiomyocyte-specific transgene [15]. The transgenic model mouse carrying the sodium/iodide symporter (NIS) reporter gene in their cardiomyocytes controlled by α-MHC (alpha myosin heavy chain) promoter disclosed that these tissue-specific transgenes were expressed in mature cardiomyocytes in a cell-specific fashion. 99mTc-Pertechnetate scan and 124I PET imaging demonstrated prominent uptakes in the myocardium of these adult mice [15]. This transgenic mouse model provides myocardial stem or progenitor cells helpful to the visualization of cardiac differentiation processes if dissected and implanted into other mouse models of human diseases.
This cell- or tissue-specific promoter-based strategy can confer substantial advantages as an imaging tool to track cell differentiation in living subjects. We will discuss in vivo imaging of stem cell differentiation into neuronal lineages further in the next section.
Optical Reporter-based Differentiation Imaging into Neurons
The co-immunostaining of proliferative (bromodeoxyuridine staining) or stem cell markers (nestin staining) with neuron-specific markers (e.g., β-tubulin III) upon brain tissue sections has been the approach commonly used to verify the extent of endogenous neurogenesis in the central nervous system [16]. However, because it is too invasive, this method prevents repetitive experiments on the same animal. A molecular imaging approach capable of evaluating various biological processes noninvasively was therefore introduced to overcome this limitation in tracking neurogenesis.
Couillard-Despres et al. reported noninvasive determination of early neuronal differentiation of endogenous neural stem cells in the adult brain [17]. The authors developed a transgenic mouse model that enabled in vivo evaluation of neuronal cell generation. They monitored neurogenesis using an optical transgene reporter regulated by the early neuron-specific doublecortin promoter (DCX) that was transiently expressed in the stage of neuronal precursors. However, DCX expression was confined to the early stage of neuronal precursors, thus proving inadequate for monitoring late-stage neurogenesis in the adult brain. In our previous study, the neuron-specific enolase (NSE) gene was chosen as a potential alternative target because its expression is maintained in the adult mouse brain even at a late stage. NSE expression undergoes a gradual increase during brain development, which makes it ideal as a marker of neuronal differentiation in imaging studies [18–20]. This pattern of expression is also recapitulated in the GENSAT (Gene Expression Nervous System Atlas) database (available at http://www.gensat.org). The GENSAT database was established to map gene expression patterns in the developing and adult central nervous system of the mouse based on transgenic mouse models and gene mapping techniques such as in situ hybridization. This database allows for an easy approach to evaluating expression patterns of interesting genes during the developmental process in the mouse brain.
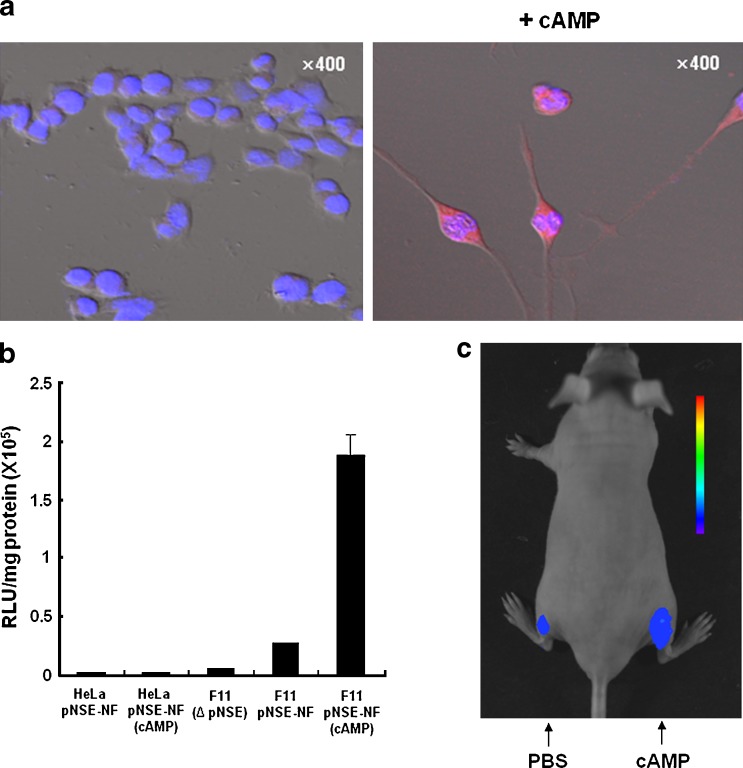
In the neuron-specific NSE promoter-driven reporter system for evaluation of cell fate change, activation of the NSE promoter by endogenous neuron-specific transcriptional factors simultaneously enhanced expression of the transgene reporter located downstream of the NSE promoter. Figure 1 depicts an approximately 4-fold increase in the luciferase signal in neuronal precursor F11 cells transfected with plasmid vector carrying sodium iodide symporter (NIS) and luciferase gene under the control of the NSE promoter (pNSE). This increased signal was detected after cyclic adenosine monophosphate (cAMP) induction of neuronal differentiation in F11 cells [21, 22]. This pNSE-driven reporter system was also applied to an animal model. The results showed a higher bioluminescence signal after cAMP treatment of an F11-bearing mouse model. We suggest that an imaging strategy using lineage-specific promoter-driven reporters is broadly applicable to monitoring the process of differentiation into diverse cell types such as osteoblasts or cardiomyocytes.
Fig. 1.
Visualization of neuronal differentiation in neuronal precursor cells exposed to cAMP. (a) Differentiation of F11 cells into neuronal cells was induced by cAMP. A 4-day cAMP induction promoted neurite outgrowth and expression of MAP2 (red color in cytoplasm), a neuronal marker. DAPI staining was expressed as blue color in the nucleus. (b) A reporter vector system containing luciferase and the sodium-iodide symporter (NIS) gene under the control of the NSE promoter were used to confirm the neuronal differentiation pattern. After induction of neuronal differentiation by cAMP, a significant increase in the luciferase signal was noted in the F11 cells. When the NIS- and luciferase-expressing vector regulated by NSE promoter (pNSE-NF) was transfected with HeLa cells that are not affected by exposure of cAMP, no significantly increased luciferase signal was observed in HeLa cells after treatment of cAMP. Δ pNSE represents reporter plasmid vector without NSE promoter. (c) Subcutaneous injection of F11 cells containing a differentiation reporter system was conducted. Reporter systems in the mice that received cAMP to the right thigh displayed a brighter bioluminescence signal in comparison to the luciferase signal of the group treated with phosphate-buffered saline. Reprinted with permission from [21]
Improvement of In Vivo Sensitivity of Optical Imaging for Tracking Neuronal Differentiation
Optical imaging techniques, especially bioluminescence-based approaches, generate low background noise and high sensitivity that is suitable for in vivo follow-up of differentiation processes. However, their sensitivity is insufficient to assess and interpret details of relevant biological events at the molecular level. To overcome this, a wide variety of signal amplification approaches has been proposed to enhance in vivo detection limits. Rabinovich and colleagues [23] developed an improved optical reporter vector system containing an engineered luciferase enzyme obtained through codon optimization techniques, such as insertion of Woodchuck hepatitis virus in the viral backbone to enhance exportation of mRNA to the cytoplasm and addition of a (G3S)2 linker located at the 3′ portion of the internal ribosomal entry site sequence to avoid abnormal RNA folding. In this result, an increase of more than 100-fold in the luciferase activity was found in T cells infected with an enhanced firefly luciferase retrovirus (v-effLuc), compared with general firefly luciferase retroviral vector (v-ffLuc)-infected T cells. Especially an optical in vivo imaging device placed in the subcutaneous area allowed detection of cell numbers as low as three. This improved optical reporter vector will facilitate highly sensitive detection of a broad variety of biological processes, ranging from efficient tracking of a few stem cells to possible monitoring of the differentiation of grafted stem cells.
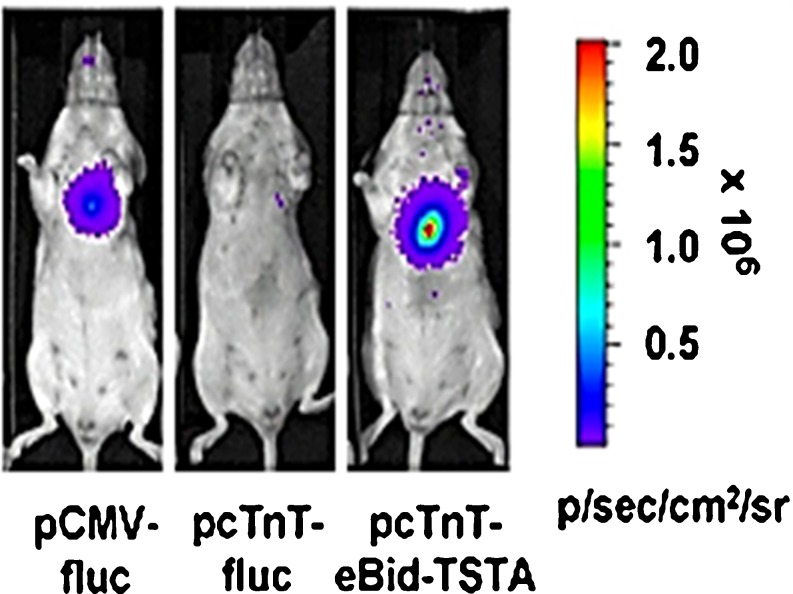
With respect to differentiation imaging, low differentiation efficiency and weak promoter activity thereof, in the case of using cell-specific promoter-driven reporters, was the bottleneck in acquiring sufficient imaging signals from inefficient differentiation processes. While grafted stem cells are differentiated into specific-lineage-committed cells with very low differentiation efficacy, high in vivo sensitivity is required to monitor these differentiated cells. Moreover, most cell lineage-specific promoters have weak promoter activity, which has further limited the application of the usual reporter transgene system for imaging stem cell differentiation in vivo. As an alternative method to overcome this, another amplification scheme of the reporter signal was introduced as a GAL4 amplification vector based on a yeast two-hybrid system called two-step transcriptional amplification (TSTA) [24, 25]. An amplifiable optical reporter system using the GAL4-VP16 fusion reporter gene was developed a decade ago by the Gambhir group. This optical reporter system, based on TSTA, was used recently in imaging studies of an orthotopic hepatocarcinoma rat model carrying a cancer-specific reporter gene controlled by a surviving promoter. Expression of this transgene reporter was higher and more specific than that of the cytomegalovirus promoter (pCMV)-driven luciferase reporter gene [26]. Optical reporter systems driven by the cardiac troponin T promoter (pcTnT) or the granzyme B promoter (pGB) were also based on a similar TSTA strategy. These systems exhibited heart-specific expression or highly amplified reporter activity during T cell activation [27, 28]. Figure 2 illustrates significantly stronger bioluminescence signals in pcTnT-eBid-TSTA–mediated cardiac reporter gene expression 28 days after intramyocardial injection compared to the pCMV-fluc- and pcTnT-fluc-mediated gene reporters.
Fig. 2.
In vivo luciferase imaging of cardiac-specific reporter gene expression in normal mice. Intramyocardial delivery of pCMV-fluc, pcTnT-fluc, and pcTnT-eBid-TSTA vector was performed in three representative mice. eBid stands for the enhanced bidirectional TSTA vector. Bioluminescence images 28 days after myocardial DNA delivery revealed much greater luciferease intensity in the transcriptional amplification vector group (pcTnT-eBid-TSTA) than that in pTnT-fluc-injected group. The TSTA system generated greater luciferase signal than the constitutive promoter-driven luciferase system (pCMV-fluc). Reprinted with permission from [27]
Also, in our previous study, to augment the weak transcriptional activity of the NSE promoter, the TSTA system using GAL4-VP16 fusion protein was introduced to improve sensitivity in monitoring neuronal differentiation in vivo [21]. Transient transfection of F11 cells with pNSE-TSTA-luciferase exhibited an increase of more than 100-fold in luciferase signals compared to the conventional pNSE-luciferase system. Signal amplification was reconfirmed through in vivo studies with F11-bearing mice. Specifically, in vivo neuronal induction exhibited dramatically higher bioluminescence activity in cAMP-treated than in untreated F11 implants. The signal amplification strategy using the TSTA system is thus well suited to various applications involving cellular differentiation imaging using promoter-based transgene reporter methods. If the retroviral vector containing an enhanced firefly luciferase can be combined with a neuron-specific promoter-driven TSTA system, highly improved optical images will be obtained for in vivo detection of the neuronal fates of transplanted stem cells.
Optical Imaging for Neuron-specific miRNA Action on Neuronal Differentiation
miRNA is an endogenously expressed RNA molecule belonging to a large class of small noncoding RNAs. miRNAs regulate most biological processes, including self-renewal and proliferation, cell differentiation and development, aging, and apoptosis at the posttranscriptional level [29–32]. Tissue- or disease-specific miRNAs were recently identified by microarray analysis. Since then, miRNAs have been considered a critical therapeutic target for neurodegenerative disorders and other gene-related diseases [33–37]. Application of a neuronal differentiation imaging based on miRNA expression and processing can be accomplished by targeting neuronal miRNAs expressing differentially between undifferentiated and differentiated cells. A reporter system can be constructed to target a neuron-related miRNA using miRNA-binding sequences located in the 3′-untranslated regions of optical reporter gene mRNAs (e.g., green fluorescent protein or luciferase) [38–40]. This system simulates the real functional actions of miRNA, including translational repression and mRNA degradation. In the presence of miRNA in cells, reporter activity is suppressed by the interaction of miRNA with the target mRNAs’ miRNA-binding sequences. The amount of miRNA can even be quantified by examining the degree of suppression of the transduced miRNA-specific optical reporter genes. Neuron-related miRNAs, such as miR-124, miR-132, miR-9, and miR-128, have been identified through real-time reverse transcription-polymerase chain reaction (RT-PCR) or microarray data. Furthermore, the expression levels of miR-124 and miR-9 were reported to increase continuously during neurogenesis [41–43]. These miRNAs were therefore considered ideal targets to be measured during neuronal differentiation in both in vivo and in vitro investigations.
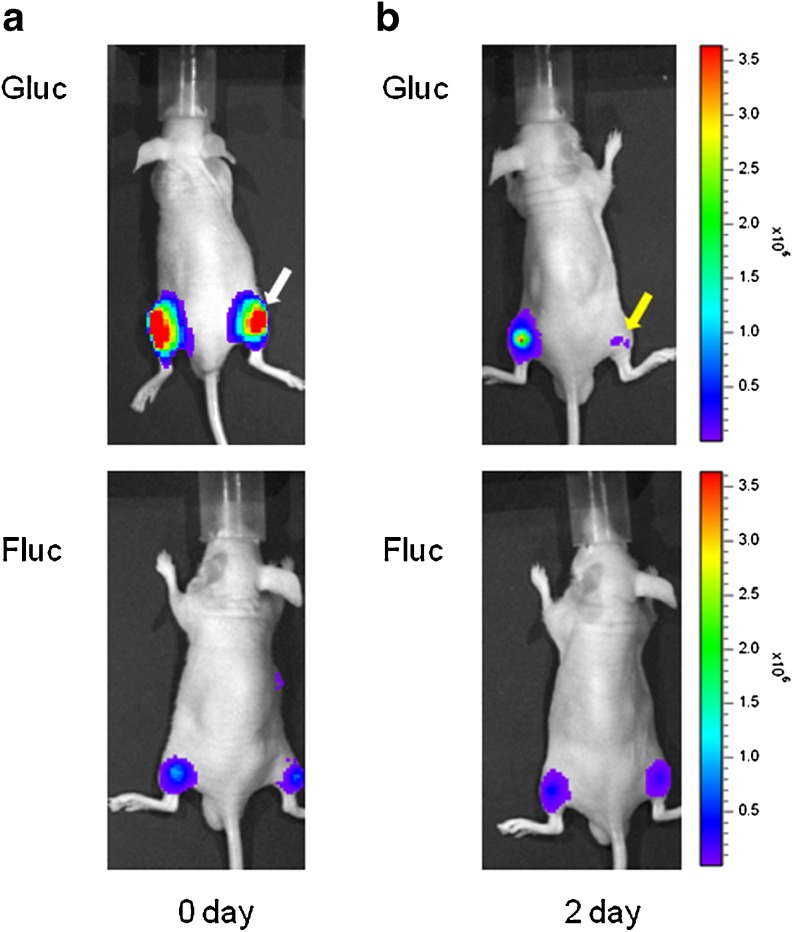
Ko et al. [44] suggested the monitoring of neuronal differentiation using a reporter imaging system to target neuron-related miRNAs. For this, they cloned a Gaussia luciferase reporter vector containing a perfectly matching miR-124a sequence. The system detects the level of miR-124a action in different cell lineages. In their experiment, reporter-transfected P19 cells, which were known to be differentiated into neuronal cells by treatment with retinoic acid, were implanted into both thighs of mice. Dual optical reporter genes were used to simultaneously track the miR-124a action during neurogenesis and survival of the implanted cells in vivo. P19 cells were co-transfected with the Gaussia luciferase vector containing a miR-124a–binding sequence and the firefly luciferase vector containing no binding sequences. The results revealed 2 days after treatment with retinoic acid, the Gaussia luciferase activity was significantly reduced in the neuronal differentiation-induced group, in which the firefly luciferase signal was the same as that in its counterpart, indicating that the signal reduction was not from the death of implanted cells, as is shown in Fig. 3.
Fig. 3.
In vivo detection of changes in neuronal miRNA expression level during neurogenesis in P19 cells. The Gaussia luciferase reporter containing the miR-124a binding sequence was transiently transfected into P19 cells that generate neuronal differentiation responsive to retinoic acid (RA). (a) The transfected P19 cells were implanted into each thigh of mouse with or without treatment of RA. RA treatment site is marked as white arrow. (b) Two days after induction of neuronal differentiation by treatment with retinoic acid, a significantly reduced Gaussia luciferase signal was observed (yellow arrow). In vivo normalization was conducted using the cotransfection method with the firefly luciferase gene under the control of cytomegalovirus (CMV) promoter (CMV/Fluc) to track the survival of the implanted P19 cells using CMV/Fluc vector. Reprinted with permission from [44]
The imaging system targeting neuron-specific miRNAs is now a well-established method to monitor the differentiation of implanted stem cells into neuronal lineages. Nevertheless, the complex molecular network of neuronal differentiation cannot be explained by observing a single miRNA target molecule. We think that a simultaneous imaging system capable of observing a variety of neuron-specific target molecules in action is needed to lead us to a deeper understanding of its versatility as an in vivo lineage-monitoring system for implanted cells.
Requisites of Multiplex Imaging
Fate commitment of stem cells into neuronal lineages is determined by regulation of a host of neuron-specific transcription factors and neuronal noncoding RNA expression [45–47]. Imaging methods using neuronal reporters or a neuronal miRNA-targeted reporter system allow us to monitor neuronal differentiation using single specific target molecules. However, the study of imaging fate changes of stem cell differentiation into neuronal lineage using single targeting may limit the understanding of the action of complex neuronal networks during neurogenesis and cannot provide precise information about the process of neuronal differentiation. As the surrounding micro-environment changes around the cells or the cells themselves are changing, the different expression and action pattern of transcription factors and noncoding (miRNA) genes may occur during neurogenesis. Consequently, for a better understanding of tracking the in vivo neuronal fate of cells, simultaneous monitoring of a variety of genes related to neuronal differentiation is essential. To detect diverse markers of neuronal lineage commitment simultaneously, nanomaterials emitting different optical signals might be combined with multiplex imaging techniques. Remarkable progress in the development of nanoparticles for in vitro multiplex analysis has made in vivo multiplex imaging more plausible [48–50].
Nanomaterials such as surface-enhanced Raman scattering (SERS) nanodots using diverse Raman signals or quantum dots having a variety of emission wavelengths with different particle sizes provide practical materials for multiplex analysis in vitro and possibly in vivo. A recent study involving in vivo Raman spectroscopy [48] showed that ten different Raman signals could be separated from the signals acquired in vivo using different types of SERS nanoprobes (120-nm particles) injected into subcutaneous sites. Simultaneous visualization of multiple nanoprobes with single imaging acquisition is therefore possible. Nonetheless, before applying SERS dots to human medicine, research should focus on the successful detection of multiple nanoparticles from grafts to reveal developmental fates or from the target tissues of subjects with diseases.
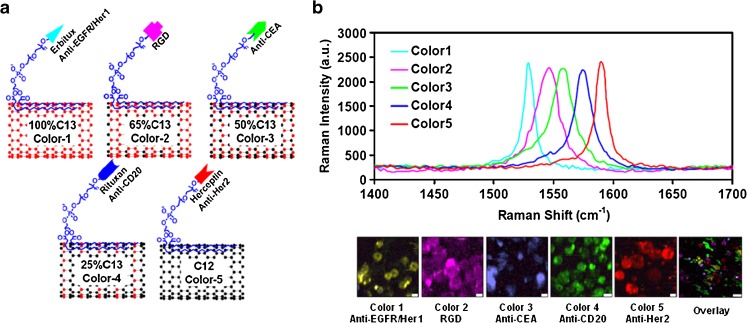
Remarkable research using specific targeting agent-embedded SERS dots has demonstrated the usefulness of fluorescence-tagging SERS nanoprobes for multiplexed imaging. The technique was used to isolate cancer stem cells in vitro by targeting surface and intracellular markers specifically expressed in cancer stem cells or to elucidate biological phenomena, such as apoptosis, by targeting apoptosis-specific markers [49, 50]. Fluorescent SERS (F-SERS) or magnetic SERS (M-SERS) spectroscopic dots containing fluorescent dye (or magnetic core) and Raman chemicals in a silica core can be easily applied for multifunctional studies, providing vital information on fluorescence or magnetic properties that are capable of simply detecting distribution of SERS dots in cells [49–52]. Like this, Raman-based imaging using SERS dot nanoparticles can be a powerful way to represent multiple detection for multiplex imaging on the cellular level. Figure 4 shows one example for multiple Raman color detection of different cancer cells using different Raman-based SWNTs targeting five different cancer cell-specific markers. Five types of cancer cells were specifically and simultaneously labeled with each SWNT after treatment of five different SWNTs having individual cancer-targeting ligands [52]. These results represent the possibility for simultaneous detection of multiple biomarkers related to cell differentiation.
Fig. 4.
Five-color multiplex imaging of single-walled carbon nanotubes (SWNTs) to simultaneously target different cancer-related proteins. (a) Schematic illustration of five different SWNTs possessing individual Raman signal peak. FeRu catalyst-based SWNT growth with different C13/C12 growth gas ratios was performed and conjugated with five different cancer-targeting ligands (anti-EGFR/Her1, RGD, anti-CEA, anti-CD20, anti-Her2). (b) Difference in the five colors is defined as individual Raman shift peaks at 1,529 cm-1, 1,546 cm-1, 1,559 cm-1, 1,575 cm-1, and 1,590 cm-1, respectively. Raman imaging was performed after five different SWNTs mixed in phosphate-buffered saline (PBS) were incubated with individual cancer cells for 1 h at 4°C. Each SWNT was specifically targeted to five different cancer cells (MDA-MB-468, human breast cancer cell: positive for EGFR/Her1; U87MG, human brain cancer cell: positive for RGD; LS174T, human colon cancer cell: positive for CEA; Raji, human B-cell lymphoma cell: positive for CD20; BT474, human breast cancer cell: positive for Her2) with distinct Raman color imaging. Reprinted with permission from [52]
Combining these approaches in multiplexed analysis could offer fundamental tools for in vivo imaging of neuronal fate changes by simultaneously targeting various molecules involved in neuronal differentiation.
Discussion and Conclusion
Stem cells containing pluripotent capability are of major importance to the biomedical field. A lack of methods to evaluate cell fate changes in clinical settings and gather detailed information on molecular mechanisms regulating self-renewal and differentiation potency has hampered rapid advances in stem cell therapy geared toward treatment of presently intractable diseases. Development of a molecular imaging system capable of translating in vitro technologies into in vivo preclinical research can resolve this limitation. Furthermore, high-throughput analysis using microarray techniques will improve our knowledge on key biological processes underlying intractability and in vitro rescue feasibility of degenerative diseases.
In this review, we present optical imaging-based techniques for in vivo monitoring of the cellular processes involved in the differentiation of grafted stem cells into neuronal lineages. An imaging method for tracking neuronal differentiation was easily established using optical reporter genes controlled by neuronal lineage-specific promoters. Undifferentiated stem cells generally exhibit weak neuron-specific promoter transcriptional activity. In contrast, differentiated neuronal cells exhibit strong neuronal promoter transcriptional activity, which facilitates visualization by an enhanced optical reporter signal. Unlike virus-derived promoters, for example, the pCMV, the original activity of tissue-specific promoters is generally weak. Introduction of signal amplification systems, such as an enhanced luciferase gene or GAL4 amplification exploiting a yeast two-hybrid system, is helpful to develop an efficient cell differentiation imaging technology. The recently discovered noncoding miRNAs, restricting neuronal fate, are another valuable strategy that can be applied as an ideal imaging system to visualize stem cell differentiation into neurons. These imaging systems are useful as simple tracking methods of neuronal lineages using a single well-known biomarker. However, a single neuron-specific biomarker is insufficient for deeper understanding of differentiation processes in relation to the complex gene networks involved in neuron fate commitment. Thus, the development of a multiplexed imaging method to track many neuron-specific markers simultaneously is essential to assess the neuronal fate of grafted stem cells in vivo.
The tracking imaging system introduced in this review can provide critical information to understand the mechanisms of successful or unsuccessful stem cell therapy by in vivo noninvasive monitoring of the differentiation process. Additionally, it contributes data useful for determining the in vivo differentiation pattern of grafted cells into the tissues of interest in stem cell therapy. Therefore, in vivo imaging techniques for assessing progression to a specific cell fate shall pave the way for the establishment of optimal therapeutic planning of stem cell treatment in clinical fields.
Acknowledgments
This work was supported by the Nanobiotechnology Project (Regenomics, no. 20100002086), the Brain Research Center of the 21st Century Frontier Research Program (2009K001257), the WCU project of the MEST, the NRF (R31-2008-000-10103-0), and the National Research Foundation of Korea grant (2011-0019044) funded by the Korean government (MEST). This research was performed as a collaborative research project of project no. C11007 (Study for Building and Service Implementation of Future Cyber-Infrastructure Resources environment) and supported by the Korea Institute of Science and Technology Information (KISTI).
References
- 1.Hipp J, Atala A. Sources of stem cells for regenerative medicine. Stem Cell Rev. 2008;4:3–11. doi: 10.1007/s12015-008-9010-8. [DOI] [PubMed] [Google Scholar]
- 2.Sharp J, Keirstead HS. Stem cell-based cell replacement strategies for the central nervous system. Neurosci Lett. 2009;456:107–11. doi: 10.1016/j.neulet.2008.04.106. [DOI] [PubMed] [Google Scholar]
- 3.Teo AK, Vallier L. Emerging use of stem cells in regenerative medicine. Biochem J. 2010;28:11–23. doi: 10.1042/BJ20100102. [DOI] [PubMed] [Google Scholar]
- 4.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–14. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang Y, Shah K, Messerli SM, Snyder E, Breakefield X, Weissleder R. In vivo tracking of neural progenitor cell migration to glioblastomas. Hum Gene Ther. 2003;14:1247–54. doi: 10.1089/104303403767740786. [DOI] [PubMed] [Google Scholar]
- 6.Zhou R, Acton PD, Ferrari VA. Imaging stem cells implanted in infarcted myocardium. J Am Coll Cardiol. 2006;48:2094–106. doi: 10.1016/j.jacc.2006.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baril P, Martin-Duque P, Vassaux G. Visualization of gene expression in the live subject using the Na/I symporter as a reporter gene: applications in biotherapy. Br J Pharmacol. 2010;159:61–71. doi: 10.1111/j.1476-5381.2009.00412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucignani G. Molecular imaging is indispensable for the development of stem cell-based myocardial regenerative therapy. Eur J Nucl Med Mol Imaging. 2007;34:422–5. doi: 10.1007/s00259-007-0369-6. [DOI] [PubMed] [Google Scholar]
- 9.Lee Z, Dennis JE, Gerson SL. Imaging stem cell implant for cellular-based therapies. Exp Biol Med (Maywood) 2008;233:930–40. doi: 10.3181/0709-MR-234. [DOI] [PubMed] [Google Scholar]
- 10.Willmann JK, Paulmurugan R, Rodriguez-Porcel M, Stein W, Brinton TJ, Connolly AJ, et al. Imaging gene expression in human mesenchymal stem cells: from small to large animals. Radiology. 2009;252:117–27. doi: 10.1148/radiol.2513081616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serganova I, Blasberg R. Reporter gene imaging: potential impact on therapy. Nucl Med Biol. 2005;32:763–80. doi: 10.1016/j.nucmedbio.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Ponomarev V, Doubrovin M, Shavrin A, Serganova I, Beresten T, Ageyeva L, et al. A human-derived reporter gene for noninvasive imaging in humans: mitochondrial thymidine kinase type 2. J Nucl Med. 2007;48:819–26. doi: 10.2967/jnumed.106.036962. [DOI] [PubMed] [Google Scholar]
- 13.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7:5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong J, Rasooly J, Dang H, Lu Y, Middleton B, Zhang Z, et al. Multimodality imaging of β-cells in mouse models of type 1 and 2 diabetes. Diabetes. 2011;60:1383–92. doi: 10.2337/db10-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang JH, Lee DS, Paeng JC, Lee JS, Kim YH, Lee YJ, et al. Development of a sodium/iodide symporter (NIS)-transgenic mouse for imaging of cardiomyocyte-specific reporter gene expression. J Nucl Med. 2005;46:479–83. [PubMed] [Google Scholar]
- 16.Zhang R, Xue YY, Lu SD, Wang Y, Zhang LM, Huang YL, et al. Bcl-2 enhances neurogenesis and inhibits apoptosis of newborn neurons in adult rat brain following a transient middle cerebral artery occlusion. Neurobiol Dis. 2006;24:345–56. doi: 10.1016/j.nbd.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Couillard-Despres S, Finkl R, Winner B, Ploetz S, Wiedermann D, Aigner R, et al. In vivo optical imaging of neurogenesis: watching new neurons in the intact brain. Mol Imaging. 2008;7:28–34. [PubMed] [Google Scholar]
- 18.Mandel G, McKinnon D. Molecular basis of neural-specific gene expression. Annu Rev Neurosci. 1993;16:323–45. doi: 10.1146/annurev.ne.16.030193.001543. [DOI] [PubMed] [Google Scholar]
- 19.Brecht WJ, Harris FM, Chang S, Tesseur I, Yu GQ, Xu Q, et al. Neuron-specific apolipoprotein e4 proteolysis is associated with increased tau phosphorylation in brains of transgenic mice. J Neurosci. 2004;24:2527–34. doi: 10.1523/JNEUROSCI.4315-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morelli AE, Larregina AT, Smith-Arica J, Dewey RA, Southgate TD, Ambar B, et al. Neuronal and glial cell type-specific promoters within adenovirus recombinants restrict the expression of the apoptosis-inducing molecule Fas ligand to predetermined brain cell types, and abolish peripheral liver toxicity. J Gen Virol. 1999;80:571–83. doi: 10.1099/0022-1317-80-3-571. [DOI] [PubMed] [Google Scholar]
- 21.Hwang do W, Kang JH, Jeong JM, Chung JK, Lee MC, Kim S, et al. Noninvasive in vivo monitoring of neuronal differentiation using reporter driven by a neuronal promoter. Eur J Nucl Med Mol Imaging. 2008;35:135–45. doi: 10.1007/s00259-007-0561-8. [DOI] [PubMed] [Google Scholar]
- 22.Ghil SH, Kim BJ, Lee YD, Suh-Kim H. Neurite outgrowth induced by cyclic AMP can be modulated by the alpha subunit of Go. J Neurochem. 2000;74:151–8. doi: 10.1046/j.1471-4159.2000.0740151.x. [DOI] [PubMed] [Google Scholar]
- 23.Rabinovich BA, Ye Y, Etto T, Chen JQ, Levitsky HI, Overwijk WW, et al. Visualizing fewer than 10 mouse T cells with an enhanced firefly luciferase in immunocompetent mouse models of cancer. Proc Natl Acad Sci USA. 2008;105:14342–6. doi: 10.1073/pnas.0804105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Adams JY, Billick E, Ilagan R, Iyer M, Le K, et al. Molecular engineering of a two-step transcription amplification (TSTA) system for transgene delivery in prostate cancer. Mol Ther. 2002;5:223–32. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Johnson M, Zhang L, Zhang B, Le K, Gambhir SS, et al. Optimization of adenoviral vectors to direct highly amplified prostate-specific expression for imaging and gene therapy. Mol Ther. 2003;8:726–37. doi: 10.1016/j.ymthe.2003.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn BC, Ronald JA, Kim YI, Katzenberg R, Singh A, Paulmurugan R, et al. Potent, tumor-specific gene expression in an orthotopic hepatoma rat model using a Survivin-targeted, amplifiable adenoviral vector. Gene Ther. 2011;18:606–12. doi: 10.1038/gt.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen IY, Gheysens O, Ray S, Wang Q, Padmanabhan P, Paulmurugan R, et al. Indirect imaging of cardiac-specific transgene expression using a bidirectional two-step transcriptional amplification strategy. Gene Ther. 2010;17:827–38. doi: 10.1038/gt.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel MR, Chang YF, Chen IY, Bachmann MH, Yan X, Contag CH, et al. Longitudinal, noninvasive imaging of T-cell effector function and proliferation in living subjects. Cancer Res. 2010;70:10141–9. doi: 10.1158/0008-5472.CAN-10-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–53. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–22. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 31.Saba R, Schratt GM. MicroRNAs in neuronal development, function and dysfunction. Brain Res. 2010;1338:3–13. doi: 10.1016/j.brainres.2010.03.107. [DOI] [PubMed] [Google Scholar]
- 32.Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–87. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 33.Shafi G, Aliya N, Munshi A. MicroRNA signatures in neurological disorders. Can J Neurol Sci. 2010;37:177–85. doi: 10.1017/s0317167100009902. [DOI] [PubMed] [Google Scholar]
- 34.Gao Y, Schug J, McKenna LB, Le Lay J, Kaestner KH, Greenbaum LE. Tissue-specific regulation of mouse MicroRNA genes in endoderm-derived tissues. Nucleic Acids Res. 2011;39:454–63. doi: 10.1093/nar/gkq782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov Today. 2009;14:1123–9. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 36.De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–11. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 37.Erson AE, Petty EM. miRNAs and cancer: New research developments and potential clinical applications. Cancer Biol Ther. 2009;8:2317–22. doi: 10.4161/cbt.8.24.10765. [DOI] [PubMed] [Google Scholar]
- 38.Kim HJ, Chung JK, Hwang do W, Lee DS, Kim S. In vivo imaging of miR-221 biogenesis in papillary thyroid carcinoma. Mol Imaging Biol. 2009;11:71–8. doi: 10.1007/s11307-008-0188-6. [DOI] [PubMed] [Google Scholar]
- 39.Tani S, Kusakabe R, Naruse K, Sakamoto H, Inoue K. Genomic organization and embryonic expression of miR-430 in medaka (Oryzias latipes): insights into the post-transcriptional gene regulation in early development. Gene. 2010;449:41–9. doi: 10.1016/j.gene.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Ko MH, Kim S. Hwang do W, Ko HY, Kim YH, Lee DS. Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 2008;75:605–16. doi: 10.1111/j.1742-4658.2008.06408.x. [DOI] [PubMed] [Google Scholar]
- 41.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:57–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–77. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 43.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13.1-11. [DOI] [PMC free article] [PubMed]
- 44.Ko HY. Hwang do W, Lee DS, Kim S. A reporter gene imaging system for monitoring microRNA biogenesis. Nat Protoc. 2009;4:1663–9. doi: 10.1038/nprot.2009.119. [DOI] [PubMed] [Google Scholar]
- 45.Fukuda S, Taga T. Cell fate determination regulated by a transcriptional signal network in the developing mouse brain. Anat Sci Int. 2005;80:12–8. doi: 10.1111/j.1447-073x.2005.00097.x. [DOI] [PubMed] [Google Scholar]
- 46.Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–8. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;1:29–38. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- 48.Zavaleta CL, Smith BR, Walton I, Doering W, Davis G, Shojaei B, et al. Multiplexed imaging of surface enhanced Raman scattering nanotags in living mice using noninvasive Raman spectroscopy. Proc Natl Acad Sci USA. 2009;06:3511–6. doi: 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu KN, Lee SM, Han JY, Park H, Woo MA, Noh MS, et al. Multiplex targeting, tracking, and imaging of apoptosis by fluorescent surface enhanced Raman spectroscopic dots. Bioconjug Chem. 2007;8:155–62. doi: 10.1021/bc070011i. [DOI] [PubMed] [Google Scholar]
- 50.Woo MA, Lee SM, Kim G, Baek J, Noh MS, Kim JE, et al. Multiplex immunoassay using fluorescent-surface enhanced Raman spectroscopic dots for the detection of bronchioalveolar stem cells in murine lung. Anal Chem. 2009;81:1008–15. doi: 10.1021/ac802037x. [DOI] [PubMed] [Google Scholar]
- 51.Noh MS, Jun BH, Kim S, Kang H, Woo MA, Minai-Tehrani A, et al. Magnetic surface-enhanced Raman spectroscopic (M-SERS) dots for the identification of bronchioalveolar stem cells in normal and lung cancer mice. Biomaterials. 2009;30:3915–25. doi: 10.1016/j.biomaterials.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 52.Liu Z, Tabakman S, Sherlock S, Li X, Chen Z, Jiang K, et al. Multiplexed five-color molecular imaging of cancer cells and tumor tissues with carbon nanotube Raman tags in the near-infrared. Nano Res. 2010;3:222–33. doi: 10.1007/s12274-010-1025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]