Abstract
The broad binding repertoire of antibodies has permitted their use in a wide range of applications. However, some uses of antibodies are precluded due to limitations in the efficiency of antibody generation. In vitro evolved binding proteins, selected from combinatorial libraries generated around various alternative structural scaffolds, are promising alternatives to antibodies. We have solved the crystal structure of a complex of an all α-helical in vitro selected binding protein (affibody) bound to protein Z, an IgG Fc-binding domain derived from staphylococcal protein A. The structure of the complex reveals an extended and complementary binding surface with similar properties to protein–antibody interactions. The surface region of protein Z recognized by the affibody is strikingly similar to the one used for IgG1 Fc binding, suggesting that this surface contains potential hot-spots for binding. The implications of the selected affibody binding-mode for its application as a universal binding protein are discussed.
The combinatorial strategy used by the immune system for the generation of molecular variability has proven immensely successful in recognizing a broad range of foreign molecules. Antibodies (Abs) directed to specific molecular targets have also found important applications in research and medicine. However, emerging large-scale applications of Abs in proteomics, such as the generation of Ab-based protein chips, highlight limitations in traditional Ab production strategies. In vitro selection schemes based on combinatorial libraries are now challenging immunological methods for generating specific binding proteins (1). These in vitro methods potentially allow much more rapid selection of binders with good affinities and also bypass problems with immunological tolerance. They also allow selection at appropriately controlled conditions, which can be critical for the generation of binders directed to more labile or complex molecular structures. The usefulness of Abs in large-scale applications is also limited by the problems of producing them in recombinant expression systems, due to the disulphide bond formation required for the folding and stability of the Ig domains. Therefore, a further potential advantage of the in vitro-based selection methods is that alternative nonimmunoglobulin scaffolds can be used where factors such as potential bioavailability, intrinsic stability, and ease of production can also be considered (2, 3).
Protein Z is a 58-residue three-helix bundle domain derived from staphylococcal protein A (SPA), which binds to the Fc portion of IgG from different species (4). By simultaneously randomizing 13 amino acid positions located at the two helices making up the Fc-binding face of protein Z, binding proteins (affibodies) capable of binding to desired targets have been selected by using phage display technology (5, 6). In a previous study, we used SPA, corresponding to the parental structure for affibody library constructions, as the selection target, resulting in the identification of a protein A-binding affibody (7).
Here, we have solved the crystal structure of the affinity protein pair consisting of protein Z and the antiprotein A affibody ZSPA-1 at 2.3-Å resolution.
Previously, other structures of synthetic binders have been determined, but they are either limited to short peptides or based on natural interactions that have been improved in vitro (8–12). The present structure, therefore, to the best of our knowledge, constitutes the first determined structure of an artificially evolved protein–protein complex of two globular proteins.
Materials and Methods
Protein Production, Crystallization, and X-Ray Data Collection.
Affibody library construction, selection of the ZSPA-1 affibody, and production of the ZSPA-1 and Z proteins have been described (5, 7).
Protein Z and the ZSPA-1 affibody were mixed in a 1:1 ratio; the complex was crystallized by the sitting drop vapor diffusion method in 96-well crystallization plates sealed with tape. Protein concentration was 72 mg⋅ml−1 in 50 mM Tris⋅HCl at pH 7.5. Protein solution (0.6 μl) was mixed with 0.5 μl of the reservoir solution consisting of 1.6 M MgSO4 and 100 mM Mes, pH 6.5. Crystals grew after ≈4 months, probably due to the additional increase in concentration resulting from evaporation through the tape and/or plastic. The crystals had a boat-like shape, pronounced birefringence, and a size of ≈0.5 × 0.1 × 0.1 mm.
Diffraction data were collected at 100 K on a 165-mm charge-coupled device area detector (MAR-Research, Hamburg, Germany) at beam line I711 at the MAXII synchrotron in Lund, Sweden; the data were processed and scaled by using denzo and scalepack (13).
The crystals belong to the hexagonal crystal system, scaled well in P622, and could be assigned to space group P6122 or P6522 based on the systematic absences. One complex per asymmetric unit gives a calculated solvent content of 47%. The Wilson B was unusually high at 55 Å2. Data statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Data statistics | Z–ZSPA-1 complex |
|---|---|
| Spacegroup | P6122 |
| Cell parameters, Å: a = b | 55.55 |
| c | 155.75 |
| Resolution, Å (outer shell) | 20–2.3 (2.38–2.30) |
| No. of observations | 57198 |
| Unique reflections | 6847 |
| Rsym* (outer shell) | 0.050 (0.290) |
| Completeness, % (outer shell) | 99.7 (99.8) |
| Refinement | |
| Rcryst, %† | 22.4 |
| Rfree (5% of data), %‡ | 25.5 |
| Non-H atoms | 1072 |
| Solvent molecules | 182 |
| rms deviation bonds, Å | 0.006 |
| rms deviation angles, ° | 1.17 |
| Ramachandran plot, % of residues§ | |
| Most favored | 93.9 |
| Allowed | 6.1 |
| Generously allowed and disallowed | 0.0 |
Rsym = ∑j∑h|〈Ihj − Ih〉|/∑j∑hIjh, where Ihj is the jth observation of reflection h.
Rcryst = ∑∥Fobs − Fcalc∥/|∑obs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
Rfree is equivalent to Rcryst for a 5% subset of reflections not used in the refinement.
Calculated by using PROCHECK (28).
Phase Determination and Structure Refinement.
Molecular replacement searches were initially unsuccessful, probably due to the problem of separating the Patterson self and cross vectors for such a small protein when running cross rotation and translation searches separately. A correct solution was, however, obtained with the program epmr (14), which uses a real-space evolutionary search method, with a polyserine model of domain D from SPA (PDB ID code , chain G) (15). The solution was found in space group P61 with four molecules per a.u. and an initial R value of 51%. The space group could subsequently be reduced to P6122 with one complex per a.u.
Extensive rounds of model building and refinement were performed. Interpretation of maps and model building were done by using the program quanta (Molecular Simulations). Model refinement was done with cns (16). The free R value was calculated from 5% of the data. See Table 1 for statistics.
Figures were created by using either the swiss pdb viewer (17) and pov-ray, molscript (18) or bobscript, Robert Esnouf's extended version of molscript, and RASTER3D (19).
Results and Discussion
Overall Structure.
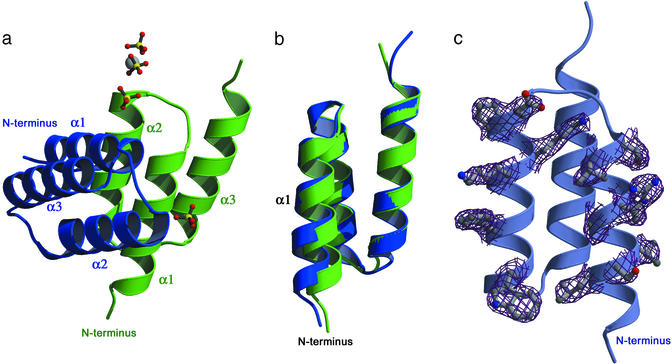
The structure was solved by molecular replacement and refined to good stereochemistry and R values (Table 1). The electron density is well defined, and the model includes residues four to the C-terminal residue 58 in the ZSPA-1 affibody and residues 4–57 in protein Z (Fig. 1a). Despite the relatively high average B factor (61 Å2 for the protein), the side chain conformations are clearly visible except for a few surface side chains. Both protein Z and the affibody have the three-helix bundle topology, as previously reported for protein Z in solution (NMR, PDB ID code , 10 models) (20). A hydrophobic core is formed by small hydrophobic residues and one aromatic residue (Phe-30), resulting in a close packing between the helices.
Fig 1.
Structure of the in vitro evolved complex. (a) Structure of the complex, the ZSPA-1 affibody in blue and protein Z in green. The ordered sulfate ions with partial occupancy and putative magnesium ion from the mother liquor are also shown; however, they do not seem to influence the interaction surface. (b) Superposition of the two molecules. Notice the shift of helix 1 in the affibody (blue) compared with protein Z (green). (c) Electron density (2 Fobs − Fcalc map contoured at 1 σ) for all 13 mutated residues in the affibody; for clarity, only the electron density around the side chains is displayed.
The main chain structures of the ZSPA-1 affibody, and protein Z are very similar and can be superimposed with an rms deviation of 0.64 Å for the α carbons of residues 5–56. Interestingly, helices 2 and 3 can be superimposed with an rms deviation of only 0.33 Å for the α carbons (residues 20–56) despite the mutation of glutamate 25 to proline in helix 2. This superposition shows that the N-terminal end of helix 1 shifts away ≈1.5 Å from helix 3 in the affibody compared with protein Z (Fig. 1b).
The Selected Binding Mode.
In the interaction surface, helices 1 and 2 of the affibody mainly pack against helix 1 in the Z domain but also interact with a large part of helix 2. This creates a somewhat distorted four-helix bundle at the dimer interface with a tilt angle of ≈60° between the helices of the different monomers. The dimer interface and the core of the proteins are the most rigid regions of the structure, as judged from the B values, and all interacting residue side chains are well defined (Fig. 1c). The terminal residues, the loops connecting helices 2 and 3 and helix 3 of the Z domain, are the regions with the highest B values. Four sulfate ions with partial occupancy and one potential magnesium ion from the mother liquor are also visible in the electron density but do not seem to influence the interaction surface (Fig. 1a). Two of the sulfate ions coordinate the magnesium bound to Glu-25 in the beginning of helix 2 of protein Z, and another sulfate is seen at the N-terminal end of helix 2 in the affibody. The last sulfate is situated close to Arg-14 in the affibody and may also interact somewhat with Arg-27 of protein Z. Because of the clear side chain densities and partial occupancy of the ions, it is unlikely that their presence alters side chain conformations significantly.
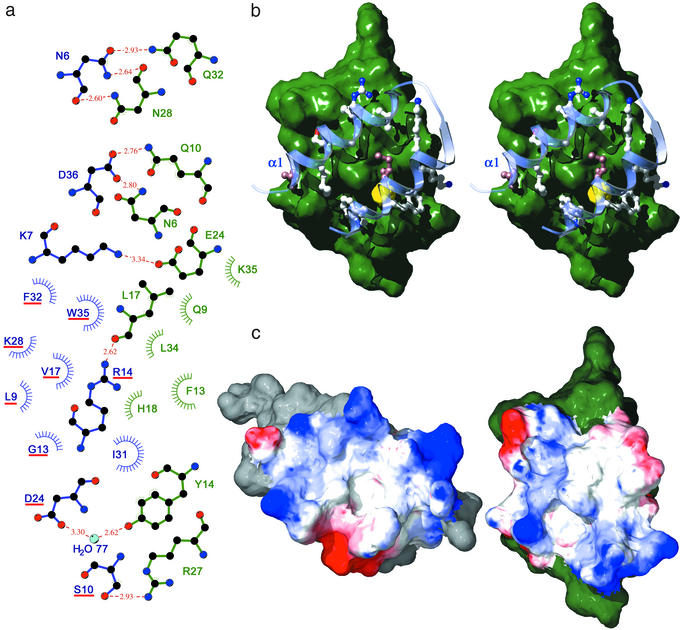
The binding surface is constituted by a central hydrophobic patch, dominated by Phe-13 and Leu-17 of protein Z and Gly-13, Ile-31, and Trp-35 of the affibody. This patch is lined by polar and charged residues that contribute seven short (<3 Å) H bonds and two weaker H bonds, of which one is mediated by a water molecule on the border of the interaction surface. Two of the H bonds are to the backbone of protein Z, and one is to the backbone of the affibody (Fig. 2a).
Fig 2.
The interaction between protein Z and the ZSPA-1 affibody. (a) Ligplot (21) representation of the interaction, ZSPA-1 affibody on the left in blue and protein Z on the right in green. H bonding residues are drawn out, and residues that contribute hydrophobic interactions are indicated. Mutated residues in the affibody are underlined in red. There is one peripheral water molecule (H2O77) contributing H bonds to both proteins. (b) Stereo view of the interaction; the ZSPA-1 affibody helices 1 and 2 over the surface of protein Z. The beginning of helix 1 is indicated. Closely interacting residues are drawn out; nonmutated residues are in brown. A small cavity is shown in yellow; the cavity is very hydrophobic and no electron density is seen inside. (c) The complex is opened up to show the electrostatics of the interaction surface. Red is negative, and blue is positive. ZSPA-1 affibody is on the left, and protein Z is on the right. The complex is formed by moving the figures together, like closing a book.
Despite the limited size of the affibody library used for selection, ≈5 × 107, the evolved interaction surface is remarkably complementary in shape (Fig. 2b). Several large side chains from the affibody penetrate cavities on the Z domain (Ser-10, Phe-32, and Trp-35). Phe-13 on Protein Z forms a hydrophobic knob that is buried efficiently between the helices in the affibody. Nine of the 13 randomized residues and four of the nonmutated residues take part in the interaction; the contacts are thus very much dominated by the mutated residues. The good surface complementarity is verified by the absence of water molecules in the interaction surface and the presence of only one small cavity with a size of ≈16 Å3 (Fig. 2b, shown in yellow).
When the complex is opened up to show the electrostatics of the interaction surface (Fig. 2c), one can see that there is also a fairly good charge complementarity evolved, even though only one interprotein salt bridge is formed.
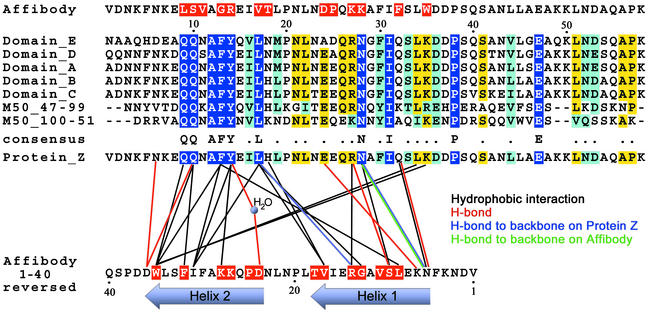
The ZSPA-1 affibody has been shown to bind all five of the individual domains of SPA with similar affinities (KD 2–6 μm) (7). This is consistent with the observation that only 3 of 13 interacting residues of protein Z are nonconserved between the different protein A domains, and that the three nonconserved residues are found in the periphery of the binding surface (Fig. 3 Bottom).
Fig 3.
Sequences. (Top) Sequence of the ZSPA-1 affibody, mutated residues in red. (Middle) Sequence alignment of the different Ig-binding domains of protein A and homologous domains from the Staphylococcus aureus Mu50 genome (gene 15925408); completely conserved residues are shown in blue. (Bottom) Interactions between protein Z and the ZSPA-1 affibody are drawn as lines; the sequence of the affibody is reversed for clarity.
Conformational Changes on Binding.
In addition to the present structure, the structure of protein Z free in solution is available (NMR, PDB ID code , 10 models). The B domain of protein A (an Ala-29-Gly difference compared with protein Z) has also been structurally characterized in complex with Fc of human IgG1 (2.8-Å resolution, PDB ID code ) (22). To assess the determinants of the interaction and the conformational changes that occur in the Z domain surface on binding, we have compared side chain conformations among the three models. Because of the limited resolution of the protein B-IgG1 Fc crystal structure and the disagreement between the different models in the NMR structure, it is hard to address the relevance of small conformational changes. However, on analysis of the conformational distribution in the 10 NMR structures, a few significant conformational changes of side chains on binding are observed. Compared with the solution structures, Gln-10, Phe-13, and Tyr-14 have different side chain conformations in the affibody complex. In the IgG1 Fc complex Phe-13 and Tyr-14 adopt almost the same conformations as in the affibody complex, whereas Gln-10 assume a different conformation, more similar to some of the NMR structures. Phe-13 makes hydrophobic interactions, whereas Gln-10 and Tyr-14 contribute with hydrogen bonds as well as hydrophobic contacts in both complexes. A sequence alignment (Fig. 3) of the different Ig-binding domains of SPA (SWISS-PROT) with homologous proteins found in other staphylococcal species shows that these residues are part of a completely conserved region in the beginning of helix 1. It is tempting to speculate that this part of helix 1 constitutes a hot spot for the IgG1 interaction (see below). Interaction hot spots have been observed in other protein–protein complexes, where only a small fraction of the interacting residues contribute critically to the binding constant (for recent discussions see refs. 23 and 24).
Conformational flexibility in helix 3 of protein Z has previously been implied from problems to trace this helix in the protein B–IgG complex (22). A higher mobility of helix 3 in protein Z is also supported by the high B value of this region in the present structure.
The shift seen in helix 1 of the affibody compared with the Z domain in the present structure is most likely mainly related to the introduction of mutations where, for example, the introduction of a glycine residue (Gly-13) in the middle of the helix allows it to bend slightly toward protein Z. The three-helix bundle of the affibody thus appears to provide some conformational plasticity, leading to slight rearrangements of the two helices of the binding surface. However, most interhelical interactions are conserved when comparing the affibody and protein Z, indicating that the conformational plasticity is still limited by the conservation of the basic interhelical interactions.
Comparison to IgG Binding.
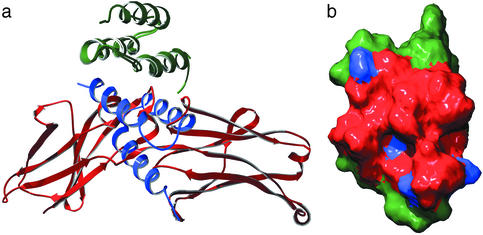
The availability of two cocomplex structures in which single SPA-derived domains (B or Z domain) interact with two very different protein partners, IgG1 Fc, or the ZSPA-1 affibody, now allows for a detailed comparison of their protein–protein interaction characteristics (Fig. 4a). When the Z:ZSPA-1 affibody complex is compared with the protein B:IgG1 Fc complex, there is a shift of the backbone conformation moving the N-terminal residues 5 and 6 in protein (Z/B) closer to IgG1. Despite the two completely different folds of the binding surfaces of IgG1 Fc (loops and β-strands) and the affibody (α-helical), the surface of protein Z involved in the interaction with the ZSPA-1 affibody is strikingly similar to the surface recruited for the interaction with IgG1 Fc (Fig. 4b). The previously discussed cavity is also present at the same position on this surface (19 Å3), and there is another cavity of similar size (16 Å3) in the vicinity of Ile-31. The Z–IgG complex buries a surface area of about 1,300 Å2; in the Z–affibody complex, it is actually somewhat larger at 1,665 Å2.
Fig 4.
Comparison to IgG interaction. (a) Superposition of the two structurally determined protein Z/B complexes; protein Z/B in green, ZSPA-1 affibody in blue, and IgG1 Fc in red. (b) The interacting surfaces on the superpositioned proteins Z/B defined as residues that lose solvent accessibility on binding. Red, IgG1 Fc complex; the ZSPA-1 affibody complex buries all of the same residues and in addition the blue areas.
The reasons for the very good agreement between these interacting surfaces are not obvious. It is, however, not unlikely that the selection procedure shows some bias for an overlapping surface for at least two reasons. First, the elution of binders was done through competition with an excess of human polyclonal IgG, and therefore affibodies bound to very different surface areas may not have been isolated. Second, targeting of the affibody to regions of protein Z that are also conserved in the affibody could lead to affibody/affibody self recognition during the selection procedure, thus reducing the amount of available binders. However, both of these effects would be eliminated by only a slight overlap in the binding surface. The very similar binding surfaces observed in the structures suggest a structural reason for the convergence of binding modes in the two protein complexes. In recent in vitro selection experiments from phage display peptide libraries, preferential selection for binding hot spots has been observed (8). In analogy, the protein Z surface may contain such a hot spot where the conserved residues of helix 1, discussed above, are the prime candidates.
Comparison to the NMR Structure.
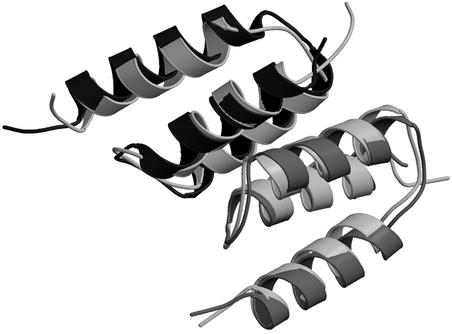
The present structure has also been solved independently by NMR methods, as presented in the accompanying paper by Wahlberg et al. (25). Overall, the structures are very similar, with an rms deviation of 1.18 Å for the α carbons for the entire complex (amino acids 5–56 of both chains) when comparing to the representative NMR structure, as defined in ref. 25 (Fig. 5). The interactions between the affibody and protein Z are generally the same but with a few interesting differences. Asn-6 of the affibody adopts different conformations in the two structures, leading to differences in the hydrogen bonding pattern and interactions with protein Z. In the x-ray structure, this residue makes one H bond to Gln-32 and also exchanges two symmetric H bonds with Asn-28 of protein Z, as seen in Fig. 2a. In the NMR structure, on the other hand, this residue has another conformation and H bonds to Asp-36 on protein Z. Asn-6 of protein Z also shows conformational differences, making a H bond to Asp-36 on the affibody in the x-ray structure, but does not seem to interact with the affibody in the NMR structure. Tyr-14 of protein Z shows a slight shift when the structures are compared; although small, this changes the hydrogen bonding from a water-mediated H bond to Asp-24 of the affibody in the x-ray structure to a direct H bond to the backbone carbonyl oxygen on the same residue in the NMR structure.
Fig 5.
Superposition of the x-ray and NMR structures. In the x-ray, affibody is shown in black and protein Z is shown in dark gray. In the NMR structure, both chains are shown in light gray.
All these differences occur on the edge of the interface between the two molecules. Probably, they represent alternate conformations where the most energetically favorable state changes depending on buffer conditions.
Applicability of in Vitro Evolved Binding Proteins.
The two-helix scaffold that forms the binding surface of the affibody is very different from that of Abs where six complementary determining region (CDR) loops constitute the binding scaffold. The loop structures of the CDRs should in theory be able to access a larger conformational space than the relatively rigid α-helical structures of the affibody, although some interhelical repositioning could take place in the affibody, as observed in the present structure. Also, in the affibodies, only 13 amino acid positions are randomized, whereas the number of variable residues in the complementary determining regions normally constitutes two to four times that number. To what extent is then the more compact affibody concept able to mimic the binding mode of Ab–antigen interactions?
On the basis of several published studies of Ab–antigen interactions (26–28), the average Ab–antigen complex buries 1,600 −1,700 Å2 and has eight to nine H bonds. The Protein Z–ZSPA-1 affibody complex buries 1,665 Å2 and has nine H bonds in the interaction surface. However, there is one distinct difference in the binding mode. No buried water molecules are found in the ZSPA-1–protein Z binding surface, whereas buried water molecules appear to mediate the interactions in all high-resolution Ab–antigen structures in the PDB database.
In conclusion, the affibody binds protein Z as a globular protein with a binding surface that in many respects is similar to what is seen in structurally characterized Ab–antigen complexes. However, the lack of buried water molecules in the affibody complex suggests that the interaction is more complementary than in the typical Ab–antigen complexes. Together with the expected relative rigidity of the α-helix scaffold of the affibody, as compared with complementary determining region loops in Abs, this indicate that the affibody structural framework may provide at least a similar potential for binding specificity.
Whether the binding mode seen in the present structure is common to in vitro evolved binders remains to be seen, but it is clear that they can mimic some of the interaction properties of Abs and will likely be able to generally provide both affinity and specificity. Due to their ease of generation and production, in vitro evolved binding proteins should constitute a valuable tool and provide possibilities for novel applications in biotechnology.
Acknowledgments
We thank Yngve Cerenius for assistance at the I711 beam-line at MAX-Lab, Pål Stenmark for help with data collection, and Matthew Bennet for advice. P.-Å.N. is cofounder and consultant for Affibody AB (Bromma, Sweden). This work was supported by The Swedish Research Council and the Göran Gustafssons Foundation for research in natural science and medicine.
Abbreviations
SPA, staphylococcal protein A
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code ).
References
- 1.Liu B., Huang, L., Sihlbom, C., Burlingame, A. & Marks, J. D. (2002) J. Mol. Biol. 315, 1063-1073. [DOI] [PubMed] [Google Scholar]
- 2.Skerra A. (2000) J. Mol. Recognit. 13, 167-187. [DOI] [PubMed] [Google Scholar]
- 3.Nygren P. A. & Uhlen, M. (1997) Curr. Opin. Struct. Biol. 7, 463-469. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson B., Moks, T., Jansson, B., Abrahmsen, L., Elmblad, A., Holmgren, E., Henrichson, C., Jones, T. A. & Uhlen, M. (1987) Protein Eng. 1, 107-113. [DOI] [PubMed] [Google Scholar]
- 5.Nord K., Nilsson, J., Nilsson, B., Uhlen, M. & Nygren, P. A. (1995) Protein Eng. 8, 601-608. [DOI] [PubMed] [Google Scholar]
- 6.Nord K., Gunneriusson, E., Ringdahl, J., Stahl, S., Uhlen, M. & Nygren, P. A. (1997) Nat. Biotechnol. 15, 772-777. [DOI] [PubMed] [Google Scholar]
- 7.Eklund M., Axelsson, L., Uhlén, M. & Nygren, P. A. (2002) Proteins 48, 454-462. [DOI] [PubMed] [Google Scholar]
- 8.DeLano W. L., Ultsch, M. H., de Vos, A. M. & Wells, J. A. (2000) Science 287, 1279-1283. [DOI] [PubMed] [Google Scholar]
- 9.Livnah O., Stura, E. A., Johnson, D. L., Middleton, S. A., Mulcahy, L. S., Wrighton, N. C., Dower, W. J., Jolliffe, L. K. & Wilson, I. A. (1996) Science 273, 464-471. [DOI] [PubMed] [Google Scholar]
- 10.Schiffer C., Ultsch, M., Walsh, S., Somers, W., de Vos, A. M. & Kossiakoff, A. (2002) J. Mol. Biol. 316, 277-289. [DOI] [PubMed] [Google Scholar]
- 11.Ay J., Keitel, T., Kuttner, G., Wessner, H., Scholz, C., Hahn, M. & Hohne, W. (2000) J. Mol. Biol. 301, 239-246. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Wiesmann, C., Fuh, G., Li, B., Christinger, H. W., McKay, P., de Vos, A. M. & Lowman, H. B. (1999) J. Mol. Biol. 293, 865-881. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z., (1993) Proceedings of the CCP4 Study Weekend (Science and Engineering Research Council, Daresbury Laboratory, Warrington, U.K.), pp. 56–62.
- 14.Kissinger C. R., Gehlhaar, D. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484-491. [DOI] [PubMed] [Google Scholar]
- 15.Graille M., Stura, E. A., Corper, A. L., Sutton, B. J., Taussig, M. J., Charbonnier, J. B. & Silverman, G. J. (2000) Proc. Natl. Acad. Sci. USA 97, 5399-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D. 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 17.Guex N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714-2723. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 19.Merritt E. A. & Bacon, D. J. (1997) Macromol. Crystallogr. B 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 20.Tashiro M., Tejero, R., Zimmerman, D. E., Celda, B., Nilsson, B. & Montelione, G. T. (1997) J. Mol. Biol. 272, 573-590. [DOI] [PubMed] [Google Scholar]
- 21.Wallace A. C., Laskowski, R. A. & Thornton, J. M. (1995) Protein Eng. 8, 127-134. [DOI] [PubMed] [Google Scholar]
- 22.Deisenhofer J. (1981) Biochemistry 20, 2361-2370. [PubMed] [Google Scholar]
- 23.DeLano W. L. (2002) Curr. Opin. Struct. Biol. 12, 14-20. [DOI] [PubMed] [Google Scholar]
- 24.Ma B. Y., Wolfson, H. J. & Nussinov, R. (2001) Curr. Opin. Struct. Biol. 11, 364-369. [DOI] [PubMed] [Google Scholar]
- 25.Wahlberg E., Lendel, C., Helgstrand, M., Allard, P., Dincbas-Renqvist, V., Hedqvist, A., Berglund, H., Nygren, P.-Å. & Härd, T. (2003) Proc. Natl. Acad. Sci. USA 100, 3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabarti P. & Janin, J. (2002) Proteins 47, 334-343. [DOI] [PubMed] [Google Scholar]
- 27.Lo Conte L., Chothia, C. & Janin, J. (1999) J. Mol. Biol. 285, 2177-2198. [DOI] [PubMed] [Google Scholar]
- 28.Jones S. & Thornton, J. M. (1996) Proc. Natl. Acad. Sci. USA 93, 13-20.8552589 [Google Scholar]
- 29.Laskowski R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]