Abstract
The exceptional species diversity of flowering plants, exceeding that of their sister group more than 250-fold, is especially evident in floral innovations, interactions with pollinators and sexual systems. Multiple theories, emphasizing flower–pollinator interactions, genetic effects of mating systems or high evolvability, predict that floral evolution profoundly affects angiosperm diversification. However, consequences for speciation and extinction dynamics remain poorly understood. Here, we investigate trajectories of species diversification focusing on heterostyly, a remarkable floral syndrome where outcrossing is enforced via cross-compatible floral morphs differing in placement of their respective sexual organs. Heterostyly evolved at least 20 times independently in angiosperms. Using Darwin's model for heterostyly, the primrose family, we show that heterostyly accelerates species diversification via decreasing extinction rates rather than increasing speciation rates, probably owing to avoidance of the negative genetic effects of selfing. However, impact of heterostyly appears to differ over short and long evolutionary time-scales: the accelerating effect of heterostyly on lineage diversification is manifest only over long evolutionary time-scales, whereas recent losses of heterostyly may prompt ephemeral bursts of speciation. Our results suggest that temporal or clade-specific conditions may ultimately determine the net effects of specific traits on patterns of species diversification.
Keywords: angiosperm evolution, heterostyly, phylogenetic methods, plant breeding system, speciation
1. Introduction
Explaining why life is so diverse and how some lineages became more species-rich than others are central objectives in evolutionary biology [1,2]. Large differences in species numbers between lineages occur across disparate organismal groups [3–6], yet our understanding of the causes of these asymmetries remains fragmentary [7,8]. Flowering plants are one of the most successful evolutionary lineages, with an estimated 352 000 species [9], representing more than 250 times more species than all other seed plants, and dominate the world's terrestrial biomes [10]. Ever since Darwin described the rapid rise and early diversification of the angiosperms in the Cretaceous as ‘an abominable mystery’ ([11], pp. 20–21), many theories have been proposed to explain how and why flowering plants became so diverse and ecologically successful [10,12,13].
Recent studies suggest that the higher species diversity of flowering plants is explained by numerous, independent increases in rates of diversification, rather than by a single increase concomitant with their origin and associated with the evolution of a unique trait or set of traits [10,14,15]. One overarching hypothesis proposed to explain the evolutionary success of angiosperms posits that they have inherently higher phenotypic evolvability than other land plants, allowing them to ‘reinvent themselves’ time and again ([10], p. 377). The idea of evolutionary reinvention is epitomized by the spectacular diversity of flowers and inflorescences, their associated pollination syndromes and sexual systems and the prevalence of parallel and convergent evolution of similar floral features [16,17].
The origin and evolvability of flowers have been proposed as key to angiosperm diversification [10,18,19]. Flowers often ensure sexual reproduction via outcrossing in dispersed populations of relatively few individuals [20] and allow for efficient seed production and dispersal [21]. The evolutionary plasticity of floral traits has enabled angiosperms to evolve intricate relationships with biotic pollinating agents, prompting the evolution of pollinator specialization [22] and reproductive isolation [23]. However, very few studies have explicitly linked floral trait evolution with macro-evolutionary patterns of diversification. Clades with bilaterally symmetrical flowers and/or floral nectar spurs consistently contain more species than their sister clades, suggesting that the evolution of these traits may be repeatedly driving rapid species diversification [24,25]. However, beyond these salient examples, little is known about the impacts of floral trait innovations on the evolutionary dynamics of species diversification.
Clearly, recurrent origins of a specific floral feature alone do not necessarily confer evolutionary success over longer evolutionary time-scales [26]. For example, the transition from outcrossing to selfing is very common in plant evolution [27], but, owing to negative genetic effects [28,29], selfing lineages may represent evolutionary dead-ends [30]. Indeed, lineages with systems enforcing outcrossing may have higher long-term net-diversification rates than lineages without such systems [31,32], even though associations between a lack of outcrossing and increased diversification have also been demonstrated [33]. Thus, the macro-evolutionary outcomes of gains and losses of mechanisms that enforce outcrossing remain poorly understood.
Here, we examine the impacts of the floral syndrome ‘heterostyly’, one of the more remarkable outcrossing mechanisms in angiosperms, on the dynamics of diversification. With occurrences in 199 genera distributed over 28 families in 15 orders [34,35], and at least 20 independent evolutionary origins, heterostyly is exemplary of how angiosperm flowers have repeatedly reinvented themselves (sensu [10]). The function of heterostyly as a mechanism to promote outcrossing was first elucidated by Darwin [36] in a series of studies of the primrose family (Primulaceae s.str.; subfamily Primuloideae sensu APG3). Heterostylous populations consist of two (distyly) or three (tristyly) genetic morphs that differ in the reciprocal placement of sexual organs (figure 1). In primroses, as in most other heterostylous groups, the syndrome also encompasses a physiological sporophytic-incompatibility system that prevents self- and intra-morph fertilization (Ferrero et al. [37] document an exception) and ancillary characters, including differences in pollen size and shape of stigmatic papillae [34,38]. The inheritance of heterostyly in primroses conforms to the Mendelian pattern: a single, di-allelic locus (the so-called ‘heterostyly supergene’) controls both morphological and physiological aspects, explaining their tight association in heterostylous species (reviewed in [38,39]). Throughout this paper, ‘heterostyly’ refers jointly to both the morphological and physiological aspects of the heterostylous syndrome.
Figure 1.
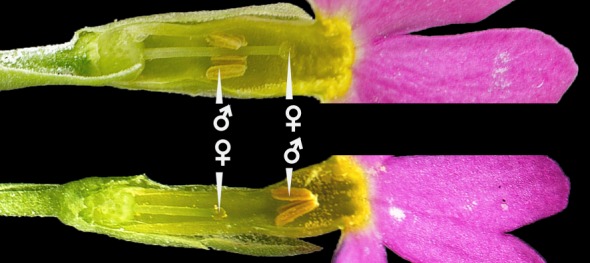
Heterostyly in Primula farinosa: the position of the stigma (♀) in each morph corresponds to the position of the anthers (♂) in the other morph. In Primulaceae, the morphological aspect of heterostyly is almost always associated with di-allelic, sporophytic self-incompatibility, rendering only crosses between morphs fully fertile.
Heterostyly promotes cross-pollination between morphs via delivery and uptake of pollen on distinct parts of the pollinator's body, while preventing or reducing inbreeding via the incompatibility system [40]. The resulting outcrossing mating system [34,38] potentially confers both evolutionary advantages and disadvantages for diversification. For example, precise pollen transfer could facilitate speciation via small changes in floral morphology that might contribute to reproductive isolation [41,42]; additionally, high outcrossing rates could buffer heterostylous lineages from extinction [30,31]. On the other hand, heterostylous plants depend on pollinators and compatible mates for successful reproduction. This may hamper population establishment and allopatric speciation following long-distance dispersal [43], potentially increasing extinction risks when pollinators are unreliable or populations are small. These considerations suggest that temporal or clade-specific factors may ultimately determine the net effects of floral traits on diversification, but such effects have scarcely been investigated to date.
Using a phylogenetic approach that allows the impacts on speciation rates to be disentangled from those on extinction rates, we test the hypothesis that the evolution of heterostyly increased the rate of species accumulation over time, while investigating whether the impacts of heterostyly on speciation, extinction and net-diversification rates may differ over short and long evolutionary time-scales. To this end, we use a range of statistical methods on a newly inferred phylogeny for the primrose family, a monophyletic group [44] of ca 700 species with predominantly generalized hymenopteran pollination syndromes, that is notable for its variation in breeding systems, including both heterostylous (distylous, self-incompatible) and non-heterostylous (self-compatible) taxa.
2. Results
Our divergence-time analyses inferred a highly resolved and strongly supported dated phylogeny (i.e. with high posterior probabilities for three main clades), consistent with previous results [45–49] (figure 2; electronic supplementary material, figure S1). These three clades differ markedly in species numbers: /Primula (slashes indicate clade names [45]), with 190 species (151 heterostylous); /Soldanella, nine species (one heterostylous) and /Androsace, 66 species (one heterostylous). The percentage of sampled, extant species was similar for each clade (32%, 34% and 42%, respectively; electronic supplementary material, table S4).
Figure 2.
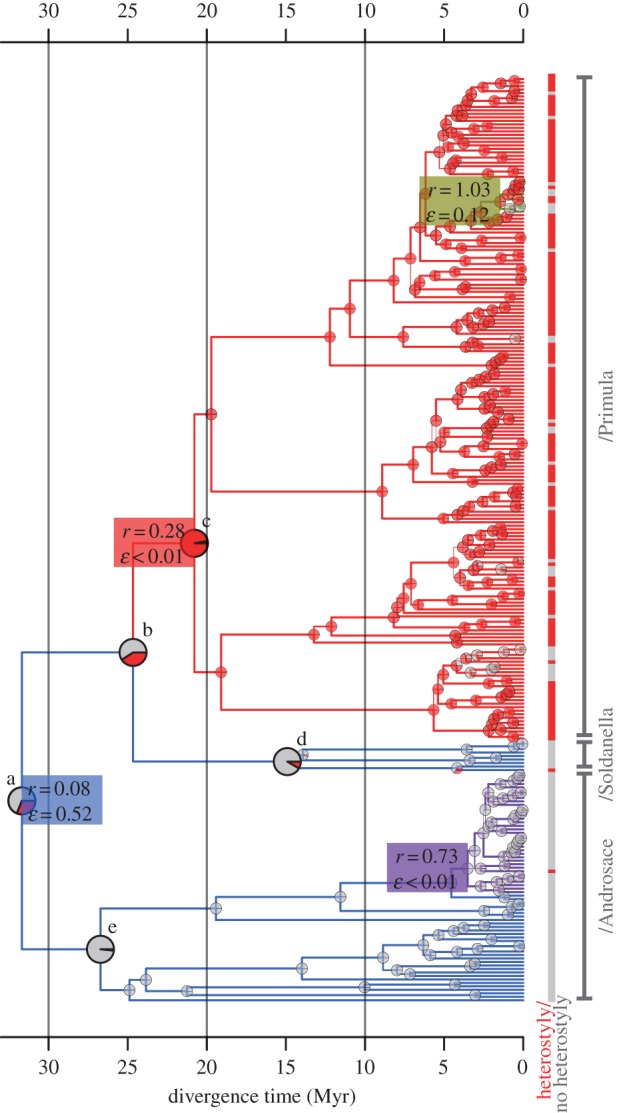
Maximum clade credibility (MCC) chronogram of Primulaceae with median node heights based on the uncorrelated lognormal (UCLN) dating analysis. Pie charts at internal nodes indicate maximum-likelihood estimates of marginal ancestral states as the proportion of likelihood associated with presence (red) and absence (grey) of heterostyly (under scheme 2; electronic supplementary material, text S1), magnified for labelled nodes of interest a–e (for values and statistical support, see the electronic supplementary material, table S1). Tip states are indicated in the same colour scheme. Branch colours designate tree partitions inferred to evolve at four significantly different rates by MEDUSA, with maximum-likelihood estimates of net-diversification rate (r) and extinction fraction (ɛ) for each tree partition. The rightmost vertical bar indicates the three major clades. See the electronic supplementary material, table S1 for statistical support of ancestral character states. Branch thickness is proportional to posterior probability (for values, see the electronic supplementary material, figure S1).
Bayesian and maximum-likelihood character-state estimates indicate that heterostyly arose early in the history of Primulaceae (electronic supplementary material, table S1) and imply that the most recent common ancestor of /Primula was heterostylous [47]. The single origin of heterostyly along the stem lineage of /Primula (figure 2, branch b–c) is inferred to have been followed by several, recent losses within deeply nested lineages. Alternative scenarios of earlier evolution of heterostyly with additional losses along the stem lineages of /Androsace and/or /Soldanella find no support by Bayes factors (electronic supplementary material, table S1). Heterostyly appears to have evolved independently in Androsace vitaliana (/Androsace) and Hottonia palustris (/Soldanella; figure 2), but in forms different from that of /Primula (electronic supplementary material, text S1).
If heterostyly is associated with higher rates of species accumulation over time, we would expect: (i) differential rates of speciation and/or extinction between heterostylous and non-heterostylous lineages across the phylogeny, and (ii) higher net-diversification rates in the heterostylous clade /Primula (82% of species heterostylous) compared with the /Androsace + /Soldanella grade (1% of species heterostylous). Bayesian and maximum-likelihood estimates under the binary-state speciation and extinction (BiSSE) model [50] indicate that heterostyly is associated with greatly reduced extinction rates, whereas speciation rates do not significantly differ between heterostylous and non-heterostylous lineages (table 1 and figure 3a; electronic supplementary material, table S3). The apparent impact of heterostyly on extinction rate translates to a 2.3- to 4.4-fold higher net-diversification rate, depending on specific analytical settings (table 1; electronic supplementary material, table S3). This result did not change qualitatively when we accounted for polyploidy (which is sometimes associated with loss of heterostyly [35,51]), by either excluding polyploids from analyses, or by assigning them to distinct states under a multi-state speciation and extinction model (MuSSE [52]; details and caveats of this approach are discussed in the electronic supplementary material, text S1).
Table 1.
Best-fitting models of speciation and extinction rates using BayesRate (BR) and BiSSE for datasets of longer (family-level) and shorter (/Primula level) time-scales. (Median rates of speciation (λ) and extinction (μ) of heterostylous (subscript 1) and non-heterostylous (subscript 0) species and difference in their net-diversification rates (positive values indicate higher diversification rates for heterostylous species), estimated ± interquartile range across 100 trees, for the best-fitting diversification models considered; electronic supplementary material, tables S2 and S3 list full results. Support for models with a free parameter indicates support for effects of heterostyly on that parameter. Model fit: BayesRate, comparison of marginal log likelihoods using Bayes factors (BF) indicates very strong support over the second-best model; BiSSE, median maximum-likelihood estimates ± interquartile range across 100 trees and Akaike information criteria (AIC)-weights (BiSSE) indicate the two models together representing 0.99 probability that these are the best among the set of candidate models. Note that although likelihoods based on different datasets cannot be directly compared, their parametrizations can.)
| model specification |
model fit |
parametrization |
Net effect | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| framework | speciation | extinction | log likelihood | ΔAIC | AIC-weight | BF | λ0 | λ1 | μ0 | μ1 | (λ1 − μ1) − (λ0 − μ0) |
| family-level BR | constrained | free | −663.06 | — | — | 23.2 | 0.77 ± 0.31 | 0.77 ± 0.31 | 0.68 ± 0.30 | 0.37 ± 0.23 | 0.29 ± 0.14 |
| family-level BiSSE | free | free | −698 ± 84 | 0.00 | 0.56 | — | 0.43 ± 0.14 | 0.32 ± 0.11 | 0.36 ± 0.14 | 0 ± 0 | 0.24 ± 0.10 |
| constrained | free | −699 ± 84 | 0.48 | 0.44 | — | 0.33 ± 0.11 | 0.33 ± 0.11 | 0.24 ± 0.10 | 0 ± 0 | 0.24 ± 0.10 | |
| /Primula-level BiSSE | free | constrained | −486 ± 69 | 0.00 | 0.72 | — | 0.64 ± 0.26 | 0.19 ± 0.08 | 0 ± 0 | 0 ± 0 | −0.43 ± 0.23 |
| free | free | −486 ± 69 | 2.00 | 0.27 | — | 0.64 ± 0.26 | 0.19 ± 0.08 | 0 ± 0 | 0 ± 0 | −0.44 ± 0.22 | |
Figure 3.
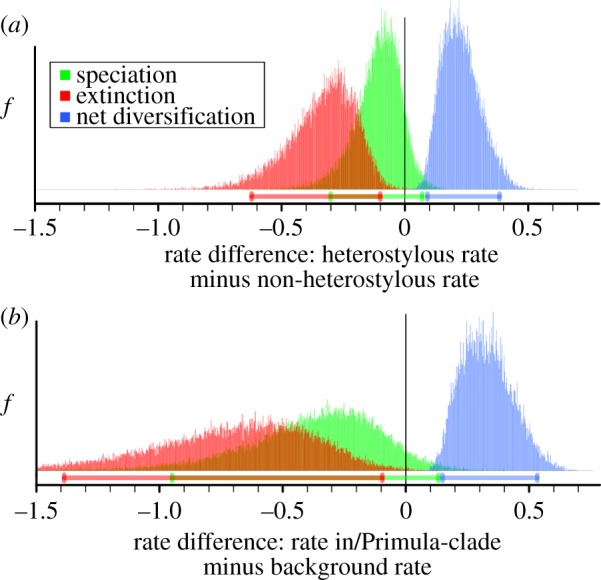
Posterior densities of differences in rates of speciation, extinction and net diversification between heterostylous and non-heterostylous lineages. (a) BiSSE (comparing rates of heterostylous and non-heterostylous lineages) and (b) BayesRate (comparing rates in mostly heterostylous /Primula with background rates in /Soldanella + /Androsace) analyses both identify an association between presence of heterostyly and lower rates of extinction, resulting in higher net diversification. Ninety-five per cent intervals of highest posterior density are indicated below densities not including zero indicate significant differences between compared rates. Based on UCLN dating analysis; electronic supplementary material, tables S2 and S3 list full results.
The BiSSE results were corroborated by analyses that locate shifts in diversification rate independent of the phylogenetic distribution of heterostyly, allowing us to assess whether factors other than heterostyly might be important to diversification in this group. Analyses using MEDUSA identified three shifts in diversification rate (figure 2; electronic supplementary material, table S2; [3]). Notably a 2.2- to 3.5-fold increase in diversification rate was detected along the same branch where heterostyly was inferred to have evolved (see root and b–c, figure 2). This shift was robust to phylogenetic uncertainty, being recovered in each of the 100 trees sampled from the posterior distribution of phylogeny estimates. Topology-based analysis using SymmeTree [53] also recovered a diversification-rate shift along the same branch (albeit marginally significant: p = 0.06; electronic supplementary material, table S2). Two other, deeply nested shifts identified with the MEDUSA analyses represented increases in net diversification that involved relatively few species and were not detected by SymmeTree (electronic supplementary material, table S2).
The implied shift in diversification rate along the stem lineage of /Primula was further corroborated by BayesRate [54] analyses, which reveal that the shift is owing to lower extinction rates (but similar speciation rates) in the heterostylous clade (/Primula) relative to the non-heterostylous grade (/Soldanella + /Androsace), leading to higher net-diversification rates in the heterostylous clade (figure 3 and table 1; electronic supplementary material, figure S2 and table S3). The net effects of extinction-rate differences owing to the evolution of heterostyly in /Primula were further confirmed using cross-validation predictive diversity densities (CVPDD) [55]. These results indicate that /Primula contains significantly more species than would be expected if the group had diversified under the background (non-heterostylous) diversification rate of /Soldanella and /Androsace (electronic supplementary material, figure S3). These results collectively provide compelling support both for a significant decrease in extinction rates associated with the evolution of heterostyly in Primulaceae (prediction (i)) and an increase in net-diversification rate along the branch where heterostyly evolved (prediction (ii)).
Additional BiSSE analyses investigating the loss of heterostyly in more recent times, i.e. including only heterostylous and non-heterostylous species of /Primula, indicate that, within /Primula, the loss of heterostyly is associated with increased net-diversification rates owing to a significantly higher speciation rate in non-heterostylous lineages (table 1; electronic supplementary material, table S3). In sharp contrast to the effects of heterostyly across the family as a whole, extinction rates of heterostylous and non-heterostylous lineages within /Primula are not significantly different (table 1; electronic supplementary material, table S3).
3. Discussion
(a). Phylogenetic evidence for effects of heterostyly on diversification
In this study, we demonstrate that the evolution of heterostyly increased the rate of species accumulation over time in the primrose family. We find evidence for an association between the evolution of heterostyly and a decrease in extinction rate, but not with an elevated speciation rate (figure 3 and table 1; electronic supplementary material, figure S2 and table S3). Although the accuracy of absolute extinction rates estimated from phylogenies is contentious, especially when these phylogenies are incomplete or when diversification rates vary among lineages [56], our results are based on strong agreement among numerous likelihood-based methods (figure 3; electronic supplementary material, figure S2). Our analyses include methods that accommodate the potentially confounding effects of trait evolution and diversification rates (BiSSE and MuSSE analyses: figures 2 and 3; electronic supplementary material, figure S2, and tables S2 and S3), methods that rely on differences in relative rather than absolute extinction rates and methods that do not require extinction-rate estimates (CVPDD analysis; electronic supplementary material, figure S3). Furthermore, our results are robust to various forms of phylogenetic uncertainty, including uncertainty in divergence-time estimates (by methods that average over dated trees and those that leverage topological information to identify diversification-rate shifts), and to uncertainty in the choice of relaxed-clock models (electronic supplementary material, tables S1–S3 and figures S1–S3). These considerations collectively provide compelling evidence for a correlation between the evolution of heterostyly and decreased extinction, rather than increased speciation.
For multiple reasons, we believe it is unlikely that the inferred correlation between heterostyly and increased diversification rate in primroses is, in fact, attributable to other traits. First, analyses that estimate the location of diversification-rate shifts independent of character histories detected a diversification-rate shift along the /Primula stem lineage, the branch where heterostyly was independently inferred to have evolved (figure 2 and table 1; electronic supplementary material, table S1). The failure of these trait-independent methods to detect diversification-rate shifts towards the tips of the phylogeny associated with more recent losses or gains of heterostyly probably reflects a simple lack of power, given the relatively small number of species in these recent subclades. For instance, heterostylous A. vitaliana shows comparatively high genetic and morphological diversity among its subspecies [57]. This diversity could represent an early stage of rapid accumulation of high incipient species diversity that is not yet culminated in high species diversification rates.
Second, /Primula, which contains nearly all the heterostylous Primulaceae species, has no well-defined synapomorphies or otherwise obvious intrinsic attributes that might provide alternative explanations for the differential diversification rates that we attribute to heterostyly. There are no floral morphological or growth-form differences among major clades of Primulaceae (cf. Androsace section Pseudoprimula; electronic supplementary material, text S1). In addition, /Primula species with and without heterostyly display similar overall life history and morphological variation [58]. Thus, we consider it unlikely that a species’ membership of /Primula or presence of heterostyly is consistently correlated with other specific traits that could intrinsically drive species diversification and as such confound the possible effects of heterostyly.
Third, heterostylous and non-heterostylous clades of Primulaceae both have their centres of species diversity in the physiographically extremely heterogeneous eastern Himalayan region [46,59]. As the heterostylous and non-heterostylous clades have similar percentages of species occurring in the eastern Himalaya (electronic supplementary material, text S1), this region probably provided both heterostylous and non-heterostylous lineages comparable ecological opportunities for rapid, extrinsically driven species diversification [60]. Given this similarity in geographical distribution of heterostylous and non-heterostylous species, ecological opportunities alone cannot explain the differential rates of diversification detected in Primulaceae.
Finally, polyploidy, which is sometimes associated with loss of heterostyly (e.g. in Primula section Aleuritia [35,51,61,62]) is well established as a major force in angiosperm evolution [63] that potentially impacts diversification rates [64]. However, analyses accounting for polyploidy yielded results that were congruent with analyses agnostic to ploidy level (electronic supplementary material, text S1 and table S3).
All these considerations lead us to conclude that the evolution of heterostyly in Primulaceae 35–15 Myr ago (node c in figure 2; electronic supplementary material, figure S1) resulted in increased species accumulation via decreased extinction, rather than increased speciation. In addition, repeated losses of heterostyly throughout /Primula (generally less than 2–3 Myr ago; figure 2; electronic supplementary material, figure S1) are associated with elevated speciation and net-diversification rates (table 1; electronic supplementary material, table S3).
(b). Heterostyly and mechanisms of increased diversification
The obligate outcrossing enforced via heteromorphic self-incompatibility may convey long-term genetic advantages for survival of heterostylous lineages, while a release from obligate outcrossing via loss of heterostyly may promote shorter term success of self-compatible lineages in /Primula. In contrast to obligately outcrossing heterostylous taxa, non-heterostylous taxa across Primulaceae clades are self-compatible (with no known exceptions among investigated species; electronic supplementary material, text S1), implying higher selfing rates, although some species may be partially outcrossing through other mechanisms [65]. Losses of heterostyly within /Primula are thought to arise via crossing-over in the heterostyly supergene, giving rise to mutants (termed homostyles) that combine male and female aspects of both floral morphs in the same flower, which are thus self-fertile [35,38,65]. Generally, lower levels of inbreeding in outcrossing versus selfing populations lead to larger effective population sizes, where fixation of slightly deleterious alleles via drift is decreased [66], and higher genetic diversity is maintained [28,29]. Higher genetic diversity, in turn, may increase the probability of adaptation to changing environmental conditions [67], thus mitigating extinction risks, with positive outcomes on long-term evolutionary survival [30,68–70]. These positive population-genetic effects may be important in the physiographically and ecologically heterogeneous alpine/arctic habitats favoured by primroses [13,71,72]. Outcrossed, heterostylous species are more likely to include individuals with genetic combinations that are adaptive across a variety of rapidly changing ecological niches, promoting survival and adaptation following environmental change [67]. By contrast, inbreeding, non-heterostylous species might be more prone to extinction in the face of environmental change [67,68]. The long-term positive effect of heterostyly on diversification suggests that population-genetic advantages of heterostyly outweigh extinction risks associated with dependence on potentially scarce or unreliable pollinators in alpine/arctic habitats where primroses occur.
However, even though fixation of slightly deleterious mutations and depauperate genetic diversity in self-compatible lineages may decrease their fitness and overall adaptive potential, both effects require time to build up, thus possibly delaying the onset of higher extinction after the loss of heterostyly [30,66,68,69]. By contrast, loss of heterostyly may rapidly and positively influence speciation rates, as found in /Primula (table 1). Self-compatibility immediately increases the likelihood of allopatric speciation, because self-compatible lineages are more likely to establish new populations following (long-distance) dispersal than self-incompatible lineages [43]. Furthermore, adaptation may proceed more quickly in selfing than in outcrossing species, when it occurs via selection from standing genetic variation before variation becomes depleted, mainly because fixation times are decreased in selfers [66,67]. Finally, when loss of heterostyly is associated with polyploidy (discussed above), the resulting, immediate reproductive isolation between cytotypes can lead to rapid speciation [51,61,62]. Indeed, the clade within /Primula for which a shift to higher net-diversification rates was detected by MEDUSA (brown clade, figure 2) mainly comprises non-heterostylous, polyploid species, suggesting a link between loss of heterostyly, polyploidy and immediate increases in diversification.
In summary, while non-heterostylous lineages may originate often and sometimes diversify faster than heterostylous relatives over the short term, these lineages tend to ‘live fast and die young’. The immediate advantages of selfing are offset by its longer term genetic disadvantages, in line with an ‘ephemeral speciation model’ [73], where self-compatible lineages may sometimes diversify rapidly (see [58] for a /Primula-specific discussion), but rarely persist over longer evolutionary time-scales.
This interpretation is supported by the observation that across angiosperms as a whole, net-diversification rates of families with mechanisms for self-sterility are higher than those of families lacking them [32]. In Solanaceae, where self-incompatibility systems are under multi-allelic, gametophytic control (rather than di-allelic, sporophytic control as in Primulaceae), the negative effects of selfing were linked with high extinction and low net-diversification rates in self-compatible lineages over long time-scales [31]. In evening primroses (Oenothera, Onagraceae), lack of outcrossing, owing to a special chromosomal arrangement preventing recombination, was associated with increased diversification rates over shorter time-scales [33]. A similar pattern of short-term success and long-term demise has been suggested for polyploid lineages that originate frequently, but rarely persist into deep evolutionary time [64]. These results from disparate plant groups and traits suggest that net time-dependent effects of a trait on the evolutionary dynamics of diversification may be a much more general phenomenon in angiosperm evolution than currently recognized.
The higher species diversity associated with heterostyly seen in Primulaceae is not evident in all of the 28 plant families that include heterostylous species [35,74,75]. Clade-specific patterns could be owing to differences in floral morphology, ecology and habitat associated with heterostyly [34], or dependence on geographical settings (e.g. physiographic heterogeneity; see above). It is also possible that the mitigating effects of heterostyly on extinction rates remain undetectable until enough time has passed for closely related non-heterostylous lineages to experience higher extinction rates, and hence for differential species numbers to accumulate [76]. Detailed phylogenetic and diversification-rate analyses with adequate taxon sampling are still lacking for other heterostylous groups, but are needed to evaluate the macro-evolutionary effects of heterostyly on a clade-by-clade basis over a range of evolutionary time-scales [77,78].
While evolutionary innovations in plant reproductive systems have been widely predicted as key drivers of diversification in flowering plants, their impacts have rarely been quantified. Our results illustrate the complexity of possible diversification trajectories across heterostylous angiosperms, with both early gains and later losses of heterostyly prompting increased net diversification. Heterostyly is one of many floral innovations that were repeatedly gained and lost in angiosperms [16,17]. Many such innovations, and especially those affecting reproductive ecology and mating systems, are likely to have had similarly complex effects on patterns of species diversification [26]. In order to more fully understand the evolutionary dynamics and extraordinary evolutionary success of angiosperms, detailed analyses of a wider range of floral traits at varying temporal and geographical scales will be required [79,80].
4. Material and methods
Details of the analyses are described in the electronic supplementary material, text S1.
We sampled 265 out of ca 730 species of Primulaceae s.str. (i.e. Primulaceae subfamily Primuloideae sensu APGIII) representing all genera and sections (electronic supplementary material, table S4). To minimize bias owing to incomplete taxon sampling, the sampling intensity was held proportional to the size of genera and sections and to the distribution of heterostyly (i.e. the fraction of taxa with heterostyly among sampled and extant taxa was approximately equal across clades; electronic supplementary material, text S1 and table S4). Phylogenetic relationships were inferred from DNA sequences of coding (matK) and non-coding (trnL, trnL-trnF, rpl16) plastid loci, which were generated de novo (210 sequences) or downloaded from GenBank (820 sequences; electronic supplementary material, table S6). Divergence times and phylogenetic relationships were jointly estimated using BEAST v. 1.6.2 [81], under the uncorrelated lognormal (UCLN) and uncorrelated exponential (UCEXP) relaxed-clock branch-rate models. Chronograms were calibrated based on fossil seeds of Primula riosiae from the Miocene [82] and the Primulaceae crown age inferred in an independent analysis of a six-locus, 21-taxon plastid DNA sequence dataset for Ericales (electronic supplementary material, table S5), which was in turn calibrated with the fossil taxa Eurya and Saurauia from the Upper Cretaceous [83].
The distribution of heterostyly among sampled species was scored from taxonomic literature (electronic supplementary material, table S6). We employed four character-coding schemes (reflecting different interpretations of atypical heterostyly in A. vitaliana and H. palustris and conflicting evidence for breeding systems in 10 other sampled species) to assess the effects of character-state polymorphism. Character history was estimated using Bayesian and maximum-likelihood methods implemented in BayesTraits v. 1.0 [84] and the R-package diversitree [52], respectively. The latter accommodates the potential effects of character states on diversification rates when estimating ancestral states. Statistical support for heterostyly as the ancestral state was evaluated for five key nodes based on Bayes factors, for which marginal likelihoods were inferred using the harmonic-mean estimator implemented in BayesTraits.
Diversification-rate analyses were performed on chronograms estimated under both the UCLN and UCEXP models to accommodate for uncertainty in the choice of relaxed-clock model (Bayes-factor comparisons indicate that both models provide a similar fit to the data). We used methods that explicitly assess the correlation between heterostyly and diversification rate, and we also used methods that agnostically identify the location of diversification-rate shifts (i.e. independent of the reproductive trait data; electronic supplementary material, text S1). These approaches are complementary and collectively provide a rigorous evaluation of the effects of heterostyly on diversification rates.
We estimated the effects of heterostyly on rates of speciation and extinction under the BiSSE model [50,52]. The BiSSE model includes independent diversification parameters (speciation and extinction) for both states of the binary trait (heterostyly and non-heterostyly), as well as parameters for the rate of change between the two states. Accordingly, the BiSSE model potentially permits us to distinguish between alternate scenarios for the prevalence of a given state among extant species (i.e. the state either promotes rates of diversification and/or there is a bias in transitions towards that state). However, the ability to perform reliable inference under this parameter-rich model may be compromised when one state greatly dominates among observed tips, or when trees have few taxa [85], conditions that do not apply in our analyses. We variously constrained the six parameters of the full BiSSE model to specify nested diversification models, for which the relative fit to the data was assessed using Akaike information criteria-weights. For each diversification model, we accommodated phylogenetic uncertainty by averaging inferences over a sample of 100 trees drawn from the marginal posterior probability distributions estimated under each of the relaxed-clock models. Specifically, we performed maximum-likelihood estimation under the following BiSSE models: (i) state-specific speciation and extinction rates; (ii) state-specific speciation rates, but state-independent extinction rates; (iii) state-independent speciation rates, but state-specific extinction rates; and (iv) state-independent speciation and extinction rates. We also performed Bayesian inference under the full BiSSE model using Markov chain Monte Carlo to estimate marginal posterior probability densities for the diversification rate parameters.
To assess possible impacts of heterostyly on diversification rates across different time-scales, we performed BiSSE analyses on a subset of the tree, comprising a more recent temporal window, and compared results to those obtained on the whole tree. To this end, we performed the full set of BiSSE analyses (as described above for the whole tree) on /Primula (i.e. after excluding the 75 species of /Androsace and /Soldanella). To provide a preliminary assessment of possible confounding effects owing to polyploidy, BiSSE analyses at the family and /Primula level were also performed on datasets that did not contain polyploid species. In addition, we performed MuSSE analyses (implemented in diversitree) that assigned polyploid species to separate states. Issues related to incomplete knowledge of character states and limits of the information in the data are discussed in the electronic supplementary material, text S1.
We performed a separate series of analyses to identify the location of diversification-rate shifts independent of the phylogenetic distribution of heterostyly. This included maximum-likelihood analyses using MEDUSA [3], applied to the maximum clade credibility (MCC) trees and averaged over 100 trees sampled from the posterior distribution of trees, and using SymmeTree v. 1.1 applied to the majority-rule consensus tree [53]. MEDUSA analyses did not employ any correction for incomplete taxon sampling, because it was not possible to assign unsampled taxa to extant clades (electronic supplementary material, text S1).
Subsequently, we tested the significance of detected shifts in diversification rate using Bayesian methods. BayesRate v. 1.3.41 [54] was used to test for differences in speciation and extinction rates between the largely heterostylous /Primula (82% of species are heterostylous) and the nearly exclusively non-heterostylous grade comprising /Soldanella and /Androsace. This method accommodates uncertainty in the inferred phylogeny, divergence times, speciation and extinction-rate parameters, and incorporated missing species by specifying the proportion of species sampled. The cross-validated posterior diversity densities approach [55] implemented in R was also used to test shifts in diversification. This approach does not require inference of extinction rates, which are notoriously difficult to estimate [56]. BEAST was used to estimate the background diversification rate based on /Soldanella and /Androsace; the timing for the origin of heterostyly was based on stem and crown ages of /Primula.
Acknowledgements
We thank Daniele Silvestro for tailoring BayesRate code, Michael Nowak, Richard Carter and Barbara Keller for helpful discussions, the associate editor and two anonymous reviewers for their insightful comments, and Bert de Vos for photos in figure 1.
Data accessibility
DNA sequences: GenBank accessions are listed in the electronic supplementary material, tables S5 and S6; phylogenetic data: TreeBase accession number TB2:S14824.
Funding statement
We thank the University of Zürich, the G. & A. Claraz Foundation and the Austrian Science Fund (FWF, P16104) for financial support.
References
- 1.Mayr E. 1982. The growth of biological thought: diversity, evolution, and inheritance. Cambridge, MA: Harvard University Press. [Google Scholar]
- 2.Benton MJ, Emerson BC. 2007. How did life become so diverse? The dynamics of diversification according to the fossil record and molecular phylogenetics. Palaeontology 50, 23–40. ( 10.1111/j.1475-4983.2006.00612.x) [DOI] [Google Scholar]
- 3.Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13 410–13 414. ( 10.1073/pnas.0811087106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiz-Palacios O, Schneider H, Heinrichs J, Savolainen V. 2011. Diversification of land plants: insights from a family-level phylogenetic analysis. BMC Evol. Biol. 11, 341 ( 10.1186/1471-2148-11-341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegmann BM, et al. 2011. Episodic radiations in the fly tree of life. Proc. Natl Acad. Sci. USA 108, 5690–5695. ( 10.1073/pnas.1012675108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schirrmeister BE, de Vos JM, Antonelli A, Bagheri HC. 2013. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the great oxidation event. Proc. Natl Acad. Sci. USA 110, 1791–1796. ( 10.1073/pnas.1209927110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scotland RW, Sanderson MJ. 2004. The significance of few versus many in the tree of life. Science 303, 643 ( 10.1126/science.1091483) [DOI] [PubMed] [Google Scholar]
- 8.Rabosky DL, Slater GJ, Alfaro ME. 2012. Clade age and species richness are decoupled across the eukaryotic tree of life. PLoS Biol. 10, e1001381 ( 10.1371/journal.pbio.1001381.t002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheffers BR, Joppa LN, Pimm SL, Laurance WF. 2012. What we know and don‘t know about Earth's missing biodiversity. Trends Ecol. Evol. 27, 501–510. ( 10.1016/j.tree.2012.05.008) [DOI] [PubMed] [Google Scholar]
- 10.Crepet WL, Niklas KJ. 2009. Darwin's second ‘abominable mystery’: why are there so many angiosperm species? Am. J. Bot. 96, 366–381. ( 10.3732/ajb.0800126) [DOI] [PubMed] [Google Scholar]
- 11.Darwin C. 1903. More letters of Charles Darwin: a record of his work in a series of hitherto unpublished letters . London, UK: John Murray. [Google Scholar]
- 12.Crepet WL. 2000. Progress in understanding angiosperm history, success, and relationships: Darwin's abominably ‘perplexing phenomenon’. Proc. Natl Acad. Sci. USA 97, 12 939–12 941. ( 10.1073/pnas.97.24.12939) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vamosi JC, Vamosi SM. 2011. Factors influencing diversification in angiosperms: at the crossroads of intrinsic and extrinsic traits. Am. J. Bot. 98, 460–471. ( 10.3732/ajb.1000311) [DOI] [PubMed] [Google Scholar]
- 14.Sanderson MJ, Donoghue MJ. 1994. Shifts in diversification rate with the origin of angiosperms. Science 264, 1590–1593. ( 10.1126/science.264.5165.1590) [DOI] [PubMed] [Google Scholar]
- 15.Smith SA, Beaulieu JM, Stamatakis A, Donoghue MJ. 2011. Understanding angiosperm diversification using small and large phylogenetic trees. Am. J. Bot. 98, 404–414. ( 10.3732/ajb.1000481) [DOI] [PubMed] [Google Scholar]
- 16.Endress PK. 2011. Changing views of flower evolution and new questions. In Flowers on the tree of life (eds Wanntorp L, de Craene LR.), pp. 120–141. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 17.Endress PK. 2011. Evolutionary diversification of the flowers in angiosperms. Am. J. Bot. 98, 370–396. ( 10.3732/ajb.1000299) [DOI] [PubMed] [Google Scholar]
- 18.Kay KM, Sargent RD. 2009. The role of animal pollination in plant speciation: integrating ecology, geography, and genetics. Annu. Rev. Ecol. Evol. Syst. 40, 637–656. ( 10.1146/annurev.ecolsys.110308.120310) [DOI] [Google Scholar]
- 19.van der Niet T, Johnson SD. 2012. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends Ecol. Evol. 27, 353–361. ( 10.1016/j.tree.2012.02.002) [DOI] [PubMed] [Google Scholar]
- 20.Burger WC. 1981. Why are there so many kinds of flowering plants? Bioscience 31, 577–581. [Google Scholar]
- 21.Stebbins GL. 1981. Why are there so many species of flowering plants? Bioscience 31, 573–577. ( 10.2307/1308219) [DOI] [Google Scholar]
- 22.Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Annu. Rev. Ecol. Evol. Syst. 35, 375–403. ( 10.1146/annurev.ecolsys.34.011802.132347) [DOI] [Google Scholar]
- 23.Grant V. 1949. Pollination systems as isolating mechanisms in angiosperms. Evolution 3, 82–97. ( 10.2307/2405454) [DOI] [PubMed] [Google Scholar]
- 24.Sargent RD. 2004. Floral symmetry affects speciation rates in angiosperms. Proc. R. Soc. Lond. B 271, 603–608. ( 10.1098/rspb.2003.2644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodges SA, Arnold ML. 1995. Spurring plant diversification: are floral nectar spurs a key innovation? Proc. R. Soc. Lond. B 262, 343–348. ( 10.1098/rspb.1995.0215) [DOI] [Google Scholar]
- 26.Fernandez-Mazuecos M, Blanco-Pastor JL, Gomez JM, Vargas P. 2013. Corolla morphology influences diversification rates in bifid toadflaxes (Linaria sect. Versicolores). Ann. Bot. 112, 1705–1722. ( 10.1093/aob/mct214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stebbins GL. 1970. Adaptive radiation of reproductive characteristics in angiosperms, I: pollination mechanisms. Annu. Rev. Ecol. Syst. 1, 307–326. ( 10.1146/annurev.es.01.110170.001515) [DOI] [Google Scholar]
- 28.Lloyd DG. 1980. Demographic factors and mating patterns in angiosperms. In Botanical monographs 15: demography and evolution in plant populations (ed. Solbrig OT.), pp. 67–88. Oxford, UK: Blackwell Scientific Publications. [Google Scholar]
- 29.Hamrick JL, Godt M. 1996. Effects of life history traits on genetic diversity in plant species. Phil. Trans. R. Soc. Lond. B 351, 1291–1298. ( 10.1098/rstb.1996.0112) [DOI] [Google Scholar]
- 30.Takebayashi N, Morrell PL. 2001. Is self-fertilization an evolutionary dead end? Revisiting an old hypothesis with genetic theories and a macroevolutionary approach. Am. J. Bot. 88, 1143–1150. ( 10.2307/3558325) [DOI] [PubMed] [Google Scholar]
- 31.Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igić B. 2010. Species selection maintains self-incompatibility. Science 330, 493–495. ( 10.1126/science.1194513) [DOI] [PubMed] [Google Scholar]
- 32.Ferrer MM, Good SV. 2012. Self-sterility in flowering plants: preventing self-fertilization increases family diversification rates. Ann. Bot. 110, 535–553. ( 10.1093/aob/mcs124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson MTJ, FitzJohn RG, Smith SD, Rausher MD, Otto SP. 2011. Loss of sexual recombination and segregation is associated with increased diversification in evening primroses. Evolution 65, 3230–3240. ( 10.1111/j.1558-5646.2011.01378.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett SCH. 1992. Evolution and function of heterostyly. Monogr. Theor. Appl. Genet. 15, 85–127. [Google Scholar]
- 35.Naiki A. 2012. Heterostyly and the possibility of its breakdown by polyploidization. Plant Spec. Biol. 27, 3–29. ( 10.1111/j.1442-1984.2011.00363.x) [DOI] [Google Scholar]
- 36.Darwin C. 1877. The different forms of flowers on plants of the same species. London, UK: John Murray. [Google Scholar]
- 37.Ferrero V, Arroyo J, Castro S, Navarro L. 2012. Unusual heterostyly: style dimorphism and self-incompatibility are not tightly associated in Lithodora and Glandora (Boraginaceae). Ann. Bot. 109, 655–665. ( 10.1093/aob/mcr222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett S, Shore JS. 2008. New insights on heterostyly: comparative biology, ecology and genetics. In Self-incompatibility in flowering plants: evolution, diversity and mechanisms (ed. Franklin-Tong VE.), pp. 3–32. Berlin, Germany: Springer. [Google Scholar]
- 39.Lewis D, Jones DA. 1992. The genetics of heterostyly. In Evolution and function of heterostyly (ed. Barrett SCH.), pp. 129–150. Berlin, Germany: Springer. [Google Scholar]
- 40.Keller B, Thomson JD, Conti E. In press. Heterostyly promotes disassortative pollination and reduces sexual interference in Darwin's primroses evidence from experimental studies. Funct. Ecol. ( 10.1111/1365-2435.12274) [DOI] [Google Scholar]
- 41.Armbruster WS, Muchhala N. 2008. Associations between floral specialization and species diversity: cause, effect, or correlation? Evol. Ecol. 23, 159–179. ( 10.1007/s10682-008-9259-z) [DOI] [Google Scholar]
- 42.Keller B, de Vos JM, Conti E. 2012. Decrease of sexual organ reciprocity between heterostylous primrose species, with possible functional and evolutionary implications. Ann. Bot. 110, 1233–1244. ( 10.1093/aob/mcs199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker HG. 1955. Self-compatibility and establishment after ‘long-distance'dispersal. Evolution 9, 347–349. ( 10.2307/2405656) [DOI] [Google Scholar]
- 44.Källersjö M, Bergqvist G, Anderberg AA. 2000. Generic realignment in primuloid families of the Ericales s.l.: a phylogenetic analysis based on DNA sequences from three chloroplast genes and morphology. Am. J. Bot. 87, 1325–1341. ( 10.2307/2656725) [DOI] [PubMed] [Google Scholar]
- 45.Mast AR, Kelso S, Richards AJ, Lang DJ, Feller DM, Conti E. 2001. Phylogenetic relationships in Primula L. and related genera (Primulaceae) based on noncoding chloroplast DNA. Int. J. Plant Sci. 162, 1381–1400. ( 10.1086/323444) [DOI] [Google Scholar]
- 46.Schneeweiss GM, Schönswetter P, Kelso S, Niklfeld H. 2004. Complex biogeographic patterns in Androsace (Primulaceae) and related genera: evidence from phylogenetic analyses of nuclear internal transcribed spacer and plastid trnL-F sequences. Syst. Biol. 53, 856–876. ( 10.1080/10635150490522566) [DOI] [PubMed] [Google Scholar]
- 47.Mast AR, Kelso S, Conti E. 2006. Are any primroses (Primula) primitively monomorphic? New Phytol. 171, 605–616. ( 10.1111/j.1469-8137.2006.01700.x) [DOI] [PubMed] [Google Scholar]
- 48.Yan HF, He CH, Peng CI, Hu C-M, Hao G. 2010. Circumscription of Primula subgenus Auganthus (Primulaceae) based on chloroplast DNA sequences. J. Syst. Evol. 48, 123–132. ( 10.1111/j.1759-6831.2010.00068.x) [DOI] [Google Scholar]
- 49.Boucher FC, Thuiller W, Roquet C, Douzet R, Aubert S, Alvarez N, Lavergne S. 2012. Reconstructing the origins of high-alpine niches and cushion life form in the genus Androsace s.l. (Primulaceae). Evolution 66, 1255–1268. ( 10.1111/j.1558-5646.2011.01483.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 51.Guggisberg A, Mansion G, Kelso S, Conti E. 2006. Evolution of biogeographic patterns, ploidy levels, and breeding systems in a diploid–polyploid species complex of Primula. New Phytol. 171, 617–632. ( 10.1111/j.1469-8137.2006.01722.x) [DOI] [PubMed] [Google Scholar]
- 52.FitzJohn RG. 2012. diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092. ( 10.1111/j.2041-210X.2012.00234.x) [DOI] [Google Scholar]
- 53.Chan KMA, Moore BR. 2005. SymmeTree: whole-tree analysis of differential diversification rates. Bioinformatics 21, 1709–1710. ( 10.1093/bioinformatics/bti175) [DOI] [PubMed] [Google Scholar]
- 54.Silvestro D, Schnitzler J, Zizka G. 2011. A Bayesian framework to estimate diversification rates and their variation through time and space. BMC Evol. Biol. 11, 311 ( 10.1186/1471-2148-11-311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore BR, Donoghue MJ. 2009. A Bayesian approach for evaluating the impact of historical events on rates of diversification. Proc. Natl Acad. Sci. USA 106, 4307–4312. ( 10.1073/pnas.0807230106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pyron RA, Burbrink FT. 2013. Phylogenetic estimates of speciation and extinction rates for testing ecological and evolutionary hypotheses. Trends Ecol. Evol. 28, 729–736. ( 10.1016/j.tree.2013.09.007) [DOI] [PubMed] [Google Scholar]
- 57.Dixon CJ, Schönswetter P, Vargas P, Ertl S, Schneeweiss GM. 2009. Bayesian hypothesis testing supports long-distance Pleistocene migrations in a European high mountain plant (Androsace vitaliana, Primulaceae). Mol. Phylogenet. Evol. 53, 580–591. ( 10.1016/j.ympev.2009.07.016) [DOI] [PubMed] [Google Scholar]
- 58.de Vos JM, Wüest RO, Conti E. 2014. Small and ugly? Phylogenetic analyses of the ‘selfing syndrome’ reveal complex evolutionary fates of monomorphic primrose flowers. Evolution 68, 1042–1057. [DOI] [PubMed] [Google Scholar]
- 59.Richards AJ. 2003. Primula, 2nd edn Portland, OR: Timber Press. [Google Scholar]
- 60.Qiu Y-X, Fu C-X, Comes HP. 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of quaternary climate and environmental change in the world's most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. ( 10.1016/j.ympev.2011.01.012) [DOI] [PubMed] [Google Scholar]
- 61.Guggisberg A, Mansion G, Conti E. 2009. Disentangling reticulate evolution in an arctic-alpine polyploid complex. Syst. Biol. 58, 55–73. ( 10.1093/sysbio/syp010) [DOI] [PubMed] [Google Scholar]
- 62.Theodoridis S, Randin C, Broennimann O, Patsiou T, Conti E. 2013. Divergent and narrower climatic niches characterize polyploid species of European primroses in Primula sect. Aleuritia. J. Biogeogr. 40, 1278–1289. ( 10.1111/jbi.12085) [DOI] [Google Scholar]
- 63.Soltis DE, et al. 2009. Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348. ( 10.3732/ajb.0800079) [DOI] [PubMed] [Google Scholar]
- 64.Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, Rieseberg LH, Otto SP. 2011. Recently formed polyploid plants diversify at lower rates. Science 333, 1257 ( 10.5061/dryad.6hf21) [DOI] [PubMed] [Google Scholar]
- 65.Vos JM, Keller B, Isham ST, Kelso S, Conti E. 2012. Reproductive implications of herkogamy in homostylous primroses: variation during anthesis and reproductive assurance in alpine environments. Funct. Ecol. 26, 854–865. ( 10.1111/j.1365-2435.2012.02016.x) [DOI] [Google Scholar]
- 66.Wright SI, Ness RW, Foxe JP, Barrett SC. 2008. Genomic consequences of outcrossing and selfing in plants. Int. J. Plant Sci. 169, 105–118. ( 10.1086/523366) [DOI] [Google Scholar]
- 67.Glémin S, Ronfort J. 2013. Adaptation and maladaptation in selfing and outcrossing species: new mutations versus standing variation. Evolution 67, 225–240. ( 10.1111/j.1558-5646.2012.01778.x) [DOI] [PubMed] [Google Scholar]
- 68.Frankham R. 2005. Genetics and extinction. Biol. Conserv. 126, 131–140. ( 10.1016/j.biocon.2005.05.002) [DOI] [Google Scholar]
- 69.Igić B, Lande R, Kohn JR. 2008. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 169, 93–104. ( 10.1086/523362) [DOI] [Google Scholar]
- 70.Escobar JS, Cenci A, Bolognini J, Haudry A, Laurent S, David J, Glémin S. 2010. An integrative test of the dead-end hypothesis of selfing evolution in Triticeae (Poaceae). Evolution 64, 2855–2872. ( 10.1111/j.1558-5646.2010.01045.x) [DOI] [PubMed] [Google Scholar]
- 71.Benton MJ. 2009. The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science 323, 728–732. ( 10.1126/science.1157719) [DOI] [PubMed] [Google Scholar]
- 72.Vamosi JC, Vamosi SM. 2010. Key innovations within a geographical context in flowering plants: towards resolving Darwin's abominable mystery. Ecol. Lett. 13, 1270–1279. ( 10.1111/j.1461-0248.2010.01521.x) [DOI] [PubMed] [Google Scholar]
- 73.Rosenblum EB, et al. 2012. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol. 39, 255–261. ( 10.1007/s11692-012-9171-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferrero V, Rojas D, Vale A, Navarro L. 2012. Delving into the loss of heterostyly in Rubiaceae: is there a similar trend in tropical and non-tropical climate zones? Perspect. Plant Ecol. Evol. Syst. 14, 161–167. ( 10.1016/j.ppees.2011.11.005) [DOI] [Google Scholar]
- 75.Tippery NP, Les DH. 2011. Phylogenetic relationships and morphological evolution in Nymphoides (Menyanthaceae). Syst. Bot. 36, 1101–1113. ( 10.1600/036364411X605092) [DOI] [Google Scholar]
- 76.Klak C, Reeves G, Hedderson T. 2004. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature 427, 63–65. ( 10.1038/nature02243) [DOI] [PubMed] [Google Scholar]
- 77.Cohen JI. 2010. ‘A case to which no parallel exists’: the influence of Darwin's different forms of flowers. Am. J. Bot. 97, 701–716. ( 10.3732/ajb.0900395) [DOI] [PubMed] [Google Scholar]
- 78.Wake DB, Wake MH, Specht CD. 2011. Homoplasy: from detecting pattern to determining process and mechanism of evolution. Science 331, 1032–1035. ( 10.1126/science.1188545) [DOI] [PubMed] [Google Scholar]
- 79.Moore BR, Donoghue MJ. 2007. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am. Nat. 170(Suppl. 2), S28–S55. ( 10.1086/519460) [DOI] [PubMed] [Google Scholar]
- 80.Drummond CS, Eastwood RJ, Miotto STS, Hughes CE. 2012. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Syst. Biol. 61, 443–460. ( 10.1093/sysbio/syr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Czaja A. 2003. Paläokarpologische Untersuchungen von Taphozönosen des Unter- und Mittelmiozäns aus dem Braunkohlentagebau Berzdorf/Oberlausitz (Sachsen). Palaeontogr. Abteilung B 265, 1–148. [Google Scholar]
- 83.Knobloch E, Mai DH. 1986. Monographie der Früchte und Samen in der Kreide von Mitteleuropa. Rozpravy ústredního ústavu geologickenho, Praha 47, 1–219. [Google Scholar]
- 84.Pagel M, Meade A. 2006. BayesTraits. See: http://www.evolution.rdg.ac.uk/BayesTraits.html.
- 85.Davis MP, Midford PE, Maddison W. 2013. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol. Biol. 13, 38 ( 10.1186/1471-2148-13-38) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
DNA sequences: GenBank accessions are listed in the electronic supplementary material, tables S5 and S6; phylogenetic data: TreeBase accession number TB2:S14824.


