Abstract
How seasonal migration originated and impacted diversification in birds remains largely unknown. Although migratory behaviour is likely to affect bird diversification, previous studies have not detected any effect. Here, we infer ancestral migratory behaviour and the effect of seasonal migration on speciation and extinction dynamics using a complete bird tree of life. Our analyses infer that sedentary behaviour is ancestral, and that migratory behaviour evolved independently multiple times during the evolutionary history of birds. Speciation of a sedentary species into two sedentary daughter species is more frequent than speciation of a migratory species into two migratory daughter species. However, migratory species often diversify by generating a sedentary daughter species in addition to the ancestral migratory one. This leads to an overall higher migratory speciation rate. Migratory species also experience lower extinction rates. Hence, although migratory species represent a minority (18.5%) of all extant birds, they have a higher net diversification rate than sedentary species. These results suggest that the evolution of seasonal migration in birds has facilitated diversification through the divergence of migratory subpopulations that become sedentary, and illustrate asymmetrical diversification as a mechanism by which diversification rates are decoupled from species richness.
Keywords: behavioural evolution, extinction rate, speciation rate, ancestral character reconstruction
1. Introduction
Seasonal migration is a behaviour shared across a wide variety of vertebrate and invertebrate taxa [1]. It is commonly considered a response to escape low food availability or harsh climatic conditions during the non-breeding season [2]. Bird migration is particularly remarkable, with journeys ranging from short distances up to 80 000 km annually [3]. The fascination for bird migration has led to a variety of studies related to orientation and navigation [4,5], disease risk [6] and climate change [7]. In comparison, few studies have investigated the origin and evolution of migratory behaviour and the impact on diversification [8].
Seasonal migration can potentially enhance speciation, because different populations within the same species may have different migratory flyways or strategies which can lead to genetic divergence between populations [9–13]. Furthermore, vagrancy, i.e. the movement of individuals outside the species’ migratory, breeding or wintering range, is likely to be more frequent in migratory than sedentary birds and increases the probability that individuals colonize new areas, leading to divergence from their ancestral populations [14,15]. Conversely, migration could reduce opportunities for speciation as it may increase gene flow between breeding populations [16], thereby reducing genetic divergence between populations [17,18]. Migrants also need to adjust their life cycle, i.e. mating and breeding, to their migratory programme [19–21], which may reduce the possibilities of shifts in phenology and thus impede sympatric speciation by divergent selection and character displacement (even if such speciation has been rarely observed [22,23]).
Seasonal migration is also assumed to affect extinction rates. During past glaciation cycles in particular, migratory species were more likely to escape changing environmental conditions and to avoid extinction than their sedentary counterparts [24]. Gradual changes in migratory distances have probably allowed migratory birds to adjust their arrival and stopover times in favourable environments during times of global climate change (e.g. when ice-sheets extended towards lower latitudes [25]). Migratory species also had a higher ability to recolonize seasonally highly productive arctic habitats following glacial retreats [26–28]. On the other hand, travelling long distances exposes individuals to high levels of stress and increases the risk of mortality [29] as well as demographic fluctuations [30,31], potentially leading to overall higher extinction rates.
Given this combination of counterbalancing influences of seasonal migration on diversification, the overall effect is unclear. The only study we know of that directly tested the effect of migratory behaviour on speciation and extinction rates in New World warblers did not detect any effect [32]. In terms of the origin and evolution of the migratory behaviour, previous comparative studies have suggested that migratory behaviour evolved repeatedly in a number of independent lineages from sedentary ancestors, for example in Old World Sylvia and Phylloscopus warblers [17] and in New World orioles [33]. The recurring evolution of migration has typically been explained by the ‘migratory threshold’ model [34–37], in which the genetic mechanisms encoding the migratory behaviour are ancestral [38], such that the ‘migratory genes’ are present in all individuals. These genes are switched on in some populations upon crossing a ‘migratory threshold’ [34–37] owing to environmental changes or when populations experience demographic booms or bottlenecks [39].
Here, we investigate the origin and evolution of migratory behaviour in birds. Furthermore, we determine how migratory behaviour has affected speciation and extinction dynamics. Specifically, we aim to understand when migratory behaviour appeared in the evolutionary history of birds and when switches between sedentary and migratory behaviour occurred. We also determine the frequency of shifts in migratory behaviour and the potential impact of such shifts on diversification. This has implications for understanding the evolution of birds and other migratory organisms.
2. Material and methods
(a). Molecular phylogenies
We downloaded the Bayesian pseudo-posterior distribution of time-calibrated bird phylogenies provided in reference [40] from the associated website (http://birdtree.org). These phylogenies include 9993 of the 10 625 extant bird species present in the taxonomy (sensu International Ornithological Committee, World Bird List Committee, 2013). From 10 000 trees sampled in the pseudo-posterior distribution, we generated a maximum clade credibility (MCC) tree, using TreeAnnotator (included in BEAST v.1.7.5 [41]). All our analyses were run on this MCC tree. Following Davis et al. [42], we ran our analyses only on trees containing at least 300 species. This resulted in analyses at the global taxonomic scale (all birds) and on the five most species-rich avian orders: Passeriformes (perching birds), Apodiformes (swifts and hummingbirds), Piciformes (woodpeckers and toucans), Psittaciformes (parrots and parakeets) and Charadriiformes (waders, gulls and auks). To test the robustness of the diversification results obtained from analyses of the MCC trees, we also randomly chose 100 trees from the posterior distribution and ran diversification analyses on each of these.
(b). Migratory categorization
We extracted seasonal latitudinal migration data (i.e. round trips synchronized with the annual cycle) for 9832 species from the distribution range maps obtained from BirdLife International and NatureServe [43]. In this worldwide dataset, each species distribution is composed by one or multiple spatial polygons. These spatial polygons are associated with characteristics of each species, such as its presence in the polygon (1, extant; 2, probably extant; 3, possibly extant; 4, possibly extinct and 5, extinct), its origin (1, native; 2, reintroduced; 3, introduced; 4, vagrant and 5, origin uncertain) and the season in which it is present in the polygon (1, resident; 2, breeding season; 3, non-breeding season; 4, passage and 5, seasonal occurrence uncertain). We considered species to be monophyletic, meaning that they belonged to only one of two categories: migratory or sedentary. Following Somveille et al. [44], we classified a species as migratory if it occurred in at least one breeding-only or one non-breeding-only polygon (seasonality 2 or 3, respectively). We hence consider migration in the broadest sense of the term, with partially migrant species considered as migrant. With these criteria, 1817 species of 9832 (18.5%) were considered migratory.
(c). Diversification analyses
To test the effect of migratory behaviour on speciation and extinction rates, we performed character-dependent diversification analyses [45]. Specifically, we used the cladogenetic state speciation and extinction model (ClaSSE [46], implemented in the R package diversitree 0.6-1 and 0.7-6 [47]). This model extends the binary state speciation and extinction model (BiSSE [48,49]) by allowing characters to evolve not only along phylogenetic branches (anagenetically), but also at speciation (cladogenetic) events. Hence, speciation is characterized by four parameters: two parameters correspond to ‘symmetrical’ speciation (here denoted λMMM and λSSS), meaning that the two daughter species resulting from the speciation event inherit the ancestral character (here, λMMM captures migratory species (M) diversifying into two migratory species (M and M) and λSSS captures sedentary species (S) diversifying into two sedentary species (S and S)); the two other parameters correspond to ‘asymmetrical’ speciation (here denoted λMSM and λSSM), meaning that there is a character change at speciation. Here, λMSM captures migratory species (M) diversifying into a sedentary (S) and a migratory (M) daughter species, whereas λSSM captures sedentary species (S) diversifying into a sedentary (S) and a migratory (M) daughter species. We disregarded the possibility that a sedentary species diversifies into two migratory daughter species (λSMM), and alternatively that a migratory species diversifies into two sedentary daughter species (λMSS); we considered these events to be implausible and rare in nature. This was confirmed by additional tests in which λSMM and λMSS were estimated close to 0 (results not shown). In addition to the parameters characterizing speciation, two parameters characterized extinction (μmigr and μsed, corresponding to the extinction rates of migratory and sedentary species, respectively), and two parameters characterized anagenetic character change (qmigr−sed and qsed−migr, corresponding to transitions from migratory to sedentary behaviour and from sedentary to migratory behaviour, respectively).
We considered sixteen diversification scenarios. Eight of these 16 models accounted for asymmetrical speciation, and eight did not (i.e. λSSM = 0 and λMSM = 0). Among each set of eight models, four had equal transition rates (qsed−migr = qmigr−sed) and four had different transition rates (qsed−migr≠qmigr−sed). In each of these four categories, we considered the four following scenarios: (i) diversification rates independent of migratory behaviour (λSSS = λMMM and μsed = μmigr), (ii) migratory behaviour affects symmetrical speciation but not extinction rates (λSSS≠λMMM and μsed = μmigr), (iii) migratory behaviour affects extinction but not symmetrical speciation rates (λSSS = λMMM and μsed≠μmigr), and (iv) migratory behaviour affects both symmetrical speciation and extinction rates (λSSS ≠ λMMM and μsed≠μmigr).
In the eight models without asymmetrical speciation, the overall migratory (or sedentary) speciation rate is given by the corresponding symmetrical speciation rate (i.e. λmigr = λMMM and λsed = λSSS). In the eight models with asymmetrical speciation, the overall migratory (or sedentary) speciation rate is given by the sum of the symmetrical and asymmetrical speciation rates (λmigr = λMMM + λMSM and λsed = λSSS + λSSM).
We fitted the 16 different diversification scenarios to each of the MCC trees and to 100 randomly chosen phylogenies, while accounting for incomplete taxon sampling [49]. We further computed the likelihood and Akaike information criterion (AIC) corresponding to each scenario. We checked support for the selected model—the model with the lowest AIC—against all models nested within it using the likelihood ratio test (LRT). If the model with lowest AIC was supported by LRT, it was considered the best. If it was not supported by LRT (p > 0.05), the simpler model—with less parameters—was considered the best. Net diversification rates (r = λ − μ) were computed from speciation and extinction rate estimates. To examine the confidence around parameter estimates, we ran Markov chain Monte Carlo (MCMC) analyses on the consensus tree, using the best-selected model for each group. We used an exponential prior 1/(2r) to be as conservative as possible [45] and started the chain with the parameters obtained by maximum-likelihood. We applied a burn-in of 500 steps and ran 20 000 steps of MCMC for each phylogeny. The likelihood and the parameters estimates were very stable along the chain.
(d). Ancestral character reconstruction
Migratory states were reconstructed by maximum-likelihood and mapped onto the nodes and tips of the consensus tree using diversitree. Ancestral reconstruction is not implemented in ClaSSE (diversitree version 0.9–6 [47]); we therefore used the ancestral reconstruction algorithm associated with BiSSE, using the best-fitting model without asymmetrical speciation.
3. Results
In the global-scale phylogeny as well as the order-level phylogenies, the best-fitting model revealed a significant association between migratory behaviour and speciation rates, extinction rates, or both (figures 1 and 2; electronic supplementary material, tables S1 and S2).
Figure 1.
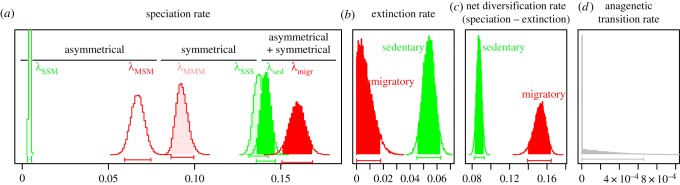
Migration promotes bird diversification. In comparison with sedentary birds (green), migratory birds (red) have low symmetrical and high asymmetrical speciation rates (a), low extinction rates (b) and high net diversification rates (c). Anagenetic transition rates are very low (d). Posterior distributions not different between migratory and sedentary species are shown in grey. Posterior distributions estimates are from MCMC analyses on all birds, using the best-fitting model.
Figure 2.
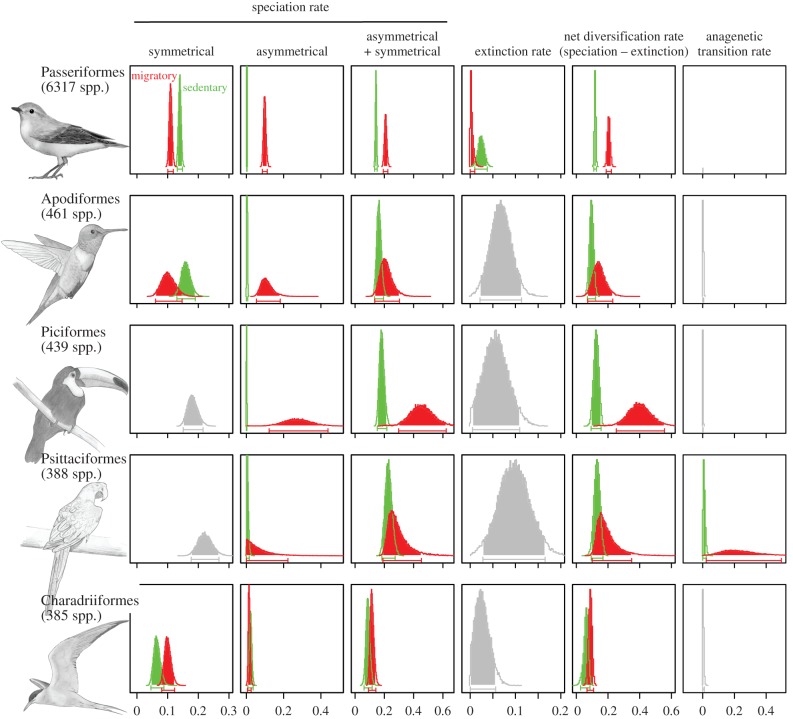
Migration promotes diversification in all species-rich orders. Posterior distributions of migratory (red) and sedentary (green) speciation—symmetrical and asymmetrical—, extinction, net diversification and anagenetic transition rates, computed using the best-fitting model. In the majority of groups, in comparison with sedentary birds, migratory birds have higher speciation rates and lower—or equal—extinction rates, resulting in higher net diversification rates. Anagenetic transition rates are very low. Posterior distributions not different between migratory and sedentary species are shown in grey. Posterior distributions estimates are from MCMC analyses on all birds, using the best-fitting model.
(a). Migration and diversification of all birds
One-fifth (1817/9832 = 18.5%) of all birds are migratory. Accounting for cladogenetic character changes greatly improved the fit of the models (ΔAIC = 736 and p < 0.05 between the overall best-fitting model and the best non-cladogenetic model; electronic supplementary material, table S1). The best-fitting model was a model with migratory behaviour affecting speciation rates (λSSS≠λMMM) and extinction rates (μsed≠μmigr); models with differential anagenetic transitions rates between migratory to sedentary behaviour (qsed−migr≠qmigr−sed) were not supported (figure 1). Symmetrical speciation rates were higher for sedentary than migratory species (λSSS = 0.141 ± 0.009 Myr−1 and λMMM = 0.091 ± 0.005 Myr−1), whereas asymmetrical speciation rates were much higher for migratory than sedentary species (λMSM = 0.067 ± 0.006 Myr−1 and λSSM = 0.005 ± 3.88 × 10−04 Myr−1) species. This resulted in an overall higher migratory speciation rate (λMMM + λMSM = 0.158 ± 0.005 Myr−1) compared with the overall sedentary speciation rate (λSSS + λSSM = 0.146 ± 0.004 Myr−1). Extinction rates were lower in migratory than sedentary species (μmigr = 0.007 ± 0.005 Myr−1 and μsed = 0.056 ± 0.01 Myr−1). Net diversification rates were thus higher for migratory versus sedentary species (net migratory diversification rate: 0.151 ± 0.01 Myr−1; net sedentary diversification rate: 0.09 ± 0.005 Myr−1).
While models excluding character change at cladogenetic events supported high anagenetic transitions from migratory to sedentary behaviour (qmigr−sed = 0.084 ± 0.008 Myr−1), anagenetic changes were estimated to be rare in models including cladogenetic character changes (qsed−migr ∼ qmigr−sed ∼ 0). In the latter, anagenetic character changes were ‘replaced’ by cladogenetic changes (λMSM = 0.067 ± 0.006 Myr−1). Hence, transition rates were better explained by cladogenetic than anagenetic changes, with frequent speciations of migratory species into a migratory and a sedentary daughter species.
(b). Migration and diversification of major bird orders
The proportion of migratory species is heterogeneously distributed in the five most species-rich orders (14% in Passeriformes, 9% in Apodiformes, 3% in Piciformes, 4% in Psittaciformes and 67% in Charadriiformes). In agreement with results obtained at the global scale, we found higher net diversification rates for migratory species in all five taxonomic groups (figure 2 and electronic supplementary material, table S2). This trend, however, was significant only for Passeriformes and Piciformes (electronic supplementary material, figure S1). Higher migratory net diversification rates were mainly explained by higher speciation rates for migratory than sedentary species (figure 2 and electronic supplementary material, table S2). Asymmetrical speciation rates were higher for migratory than sedentary species in all orders, except in Charadriiformes for which asymmetrical speciation rates were higher for sedentary species (figure 2 and electronic supplementary material, table S2). Symmetrical speciation rates were higher for sedentary species in Passeriformes and Apodiformes, and higher for migratory species in Charadriiformes. In Passeriformes, higher migratory net diversification rates were also due to low migratory extinction rates (figure 2 and electronic supplementary material, table S2). The only order in which we found anagenetic transitions to be important was Psittaciformes, in which anagenetic transitions from a migratory to a sedentary behaviour seemed to occur frequently (figure 2 and electronic supplementary material, table S2).
(c). Evolution of seasonal migration
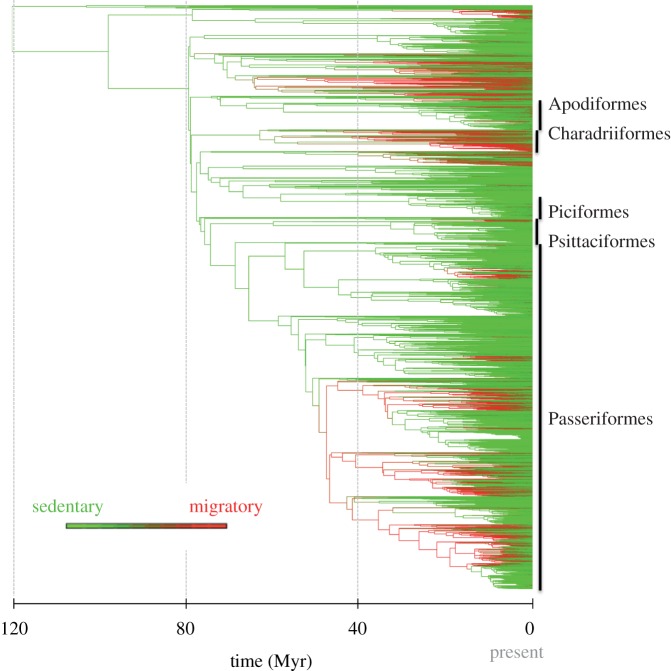
According to our ancestral character state reconstruction analyses, migration appeared around 80 Ma and reappeared independently several times during the evolutionary history of birds (figure 3). All lineages in the ancient part of the phylogeny (from 120 to 80 Ma) were inferred to be sedentary. Accordingly, many ancient nodes in the bird tree, in particular, the crown ancestors of the five major orders, were reconstructed as sedentary (probability 0.89 for Passeriformes, 0.92 for Apodiformes, 0.94 for Piciformes, 0.88 for Psittaciformes and 0.60 for Charadriiformes).
Figure 3.
Ancestral bird lineages are reconstructed as sedentary (green), with repeated apparitions of the migratory behaviour (red). The ancestral state reconstruction is based on the best-fitting BiSSE model and is plotted on the dated consensus tree. The colour of a given branch is scaled from completely green (inferred probability of being migratory equals 0) to completely red (inferred probability of being migratory equals 1), based on the mean probability of the two adjacent nodes.
4. Discussion
(a). Seasonal migration as a major driver of bird diversification
Few studies have explicitly investigated the impact of migration on diversification in birds [17,32]. Our analyses suggest that migration has played an important role in the diversification of birds. Both at the global scale (in the phylogeny of all birds) and across the five major bird orders, migratory behaviour is overall associated with high speciation rates and low extinction rates, resulting in high net diversification rates.
These results contrast with previous findings by Winger et al. [32], who reported no relationship between migratory behaviour and diversification rates in New World warblers. New World warblers are a relatively small group (less than 150 species), below the 300 species threshold recommended by Davis et al. [42] to avoid type II error rates. Hence, Winger et al. [32] may simply have lacked the statistical power to detect the effect of migration on diversification [42]. Moreover, New World warblers have a specific genus—Setophaga—which experienced a well-documented explosive radiation owing to competition for food [50]. Such particularities may blur the association between diversification and migratory behaviour. The higher net diversification rate we detected in migratory compared with sedentary species is consistent with previous studies that identified a positive correlation between annual dispersal and richness [51,52]. The authors of the later studies suggested that migratory clades may have larger geographical extents which could potentially increase a clade-wide carrying capacity [51–53].
Despite a higher overall speciation rate in migratory than sedentary species, speciation with no character change (symmetrical speciation) is estimated to be more frequent in sedentary than in migratory species in the whole phylogeny, in the Passeriformes and in the Apodiformes. In other words, the divergence of a migratory species into two migratory daughter species tends to be less frequent than the divergence of a sedentary species into two sedentary daughter species. This finding is consistent with the predictions of Helbig [17] and Claramunt et al. [18] that genetic differentiation is reduced in species with high dispersal capacity, such as migratory birds. The annual mix of individuals from different breeding populations on shared wintering sites may also tend to homogenize gene pools within a species [18,54]. The rare occurrence of events when migratory species diverge into two migratory species may also be linked to the difficulty of changing migratory trajectories, which has been widely illustrated in the literature. For example, Oenanthe oenanthe conserved its ancestral migration route from Alaska to West Africa instead of choosing a shorter route to Central America [55].
The main reason why speciation rates are overall higher in migratory birds is that the divergence of migratory species into a migratory and a sedentary daughter species is more frequent than the divergence of sedentary species into a migratory and a sedentary daughter species. These asymmetrical speciation events are almost as frequent as the symmetrical speciation events in migratory bird species. These results hold at the global scale as well as in four of the five most speciose bird orders, and suggest that the exceptional dispersal capacities of migratory species may also promote the colonization of new areas where small founder colonies are well adapted, turn sedentary and diverge from their ancestral migratory species. Such settling down of migratory populations has, for example, been observed in Turdus pilaris, with populations establishing in Greenland in few generations [56,57].
The Charadriiformes (waders, gulls and auks) stand out in terms of their speciation patterns. The overall speciation rate is also higher in migratory than sedentary species in this group. However, the higher migratory speciation rate is linked to a higher symmetrical migratory speciation rate, whereas the asymmetrical speciation rate is lower for migratory than sedentary species. This atypical speciation pattern in Charadriiformes may, in part, be explained by the particular ecology of the group, in which species are good dispersers and are highly associated with aquatic environments [43,58]. High dispersal capacities associated with the possibility of taking advantage of rich seasonal marine resources available at high latitudes may explain why few species settle down and turn sedentary in this group.
Migration tends to decrease extinction rates in birds as a whole and in the Passeriformes. Migratory species may be less restricted in space and have the capacity to escape and find suitable habitat, if climate is changing locally or shifting spatially. For example, migration may have avoided the extirpation of populations during major climate changes, such as glaciation events linked to Milankovitch cycles [59]. Conversely, sedentary species may be constrained by space, as is the case for insular species, and may be limited to altitudinal displacement to cope with climate and/or habitat changes [60]. We did not detect a significant association between migration and extinction rates in four of five of the most species-rich taxonomic orders (Apodiformes, Piciformes, Psittaciformes and Charadriiformes). It could be that there is indeed no effect of migration on extinction rates in these groups. However, there is also a possibility that the lack of significant effect arises from low statistical power. Davis et al. [42] warned against a potential lack of statistical power in trait-dependent analyses when the distribution of character states is highly unbalanced, such that one of the character states is present in less than 10% of the species. This was the case in Apodiformes, Piciformes and Psittaciformes, in which less than 10% of the species are migratory. In the Charadriiformes, both the distribution of character states and the size of the phylogeny complied with the recommendations of Davis et al. [42]. Still, no effect on extinction was detected in this group. Extinction is notoriously difficult to estimate from molecular phylogenies, in particular, when rate heterogeneity across lineages is not accounted for [61–64]. It is thus difficult to conclude whether migration indeed had no effect on extinction in these four groups, or if we lacked power to detect the effect.
It may seem counterintuitive that despite a higher migratory net diversification rate, there are many more sedentary than migratory extant bird species. In fact, although migration is an important factor promoting diversification, migratory species contribute substantially to the generation of sedentary species. On average, every time a sedentary species diversifies into two sedentary daughters species, a migratory species generates only one migratory daughter species and one sedentary species. This results in a decoupling between diversification rates and species richness.
(b). Accounting for cladogenetic transitions
Character-dependent analyses are receiving a lot of attention in the literature [45,65–67]. A large majority of these analyses have been carried out with models accounting for potential anagenetic changes, but not for cladogenetic changes (e.g. BiSSE or multiple state speciation and extinction [65–67]). In our analyses, allowing for cladogenetic transitions significantly improved the fit of the models, and dramatically changed the results. In BiSSE-type models, not accounting for cladogenetic character changes, frequent anagenetic transition rates from migratory to sedentary species were estimated; however, the ClaSSE analyses revealed that these changes are, in fact, tightly coupled with the speciation process. Allowing for speciation and character changes provided an interesting result that would have been completely missed with BiSSE-type analyses: the settlement and subsequent divergence of migrant populations into sedentary species [56,68] seem to be a frequent mode of speciation in migratory birds. In future studies, more systematically accounting for cladogenetic character changes, using ClaSSE [46] or BiSSE-ness [69], could lead to similar surprises.
Interestingly, we can understand the parameters estimated with BiSSE-type models in the light of those estimated in models incorporating cladogenetic character changes. The rate of anagenetic transition from migratory to sedentary behaviour in BiSSE-type models (qmigr−sed = 0.084 ± 0.008 Myr−1) was very similar to the rate of migratory asymmetric speciation (λMSM = 0.067 ± 0.006 Myr−1). This correspondence between parameters estimated with BiSSE-type versus ClaSSE-type models suggests that at least in our study, and in the absence of a character reconstruction implementation associated with ClaSSE-type models, using BiSSE-type models is reasonable: estimated diversification rates associated with the migratory and sedentary behaviour are similar, as well as transition rates from migratory to sedentary behaviour, the only change being that the shifts occur along branches instead of during speciation events.
(c). Transition rates and the evolution of the migratory behaviour
The extent to which migratory behaviour is a conserved trait is a controversial question [70,71]. It has been argued that because migratory behaviour can emerge or disappear within a few generations in a given population, it is highly labile [17,35,37]. However, recent studies have found that migratory traits are phylogenetically structured in New World warblers [32], suggesting that shifts in migratory strategies may be less frequent than previously thought. The very low anagenetic transition rates we found when asymmetrical speciation is accounted for also support that birds as a whole rarely shift between sedentary and migratory behaviour, or vice versa. Rather, sedentary species often evolve through divergence of a migratory ancestor into a sedentary and a migratory daughter species. Once migration is expressed in a lineage, it tends to be maintained during evolution.
Our ancestral state reconstruction results, which suggest that sedentary behaviour is ancestral for all birds, are in line with previous findings for other groups, such as the Phylloscopidae [17]. This does not imply that the genes involved in migratory behaviour are not ancestral. It has previously been argued that, because seasonal migration appeared repeatedly, and because the genetic structure involved in seasonal migration is complex, the genes required for migration were present in the common ancestors of birds [17,37,70–72]. Our results confirm that the expression of migratory behaviour has been triggered multiple times during the evolutionary history of birds, supporting the hypothesis that the genetic structure involved in migration is present in all birds. The behaviour is however activated only in some lineages, maybe as a result of genetic or environmental changes [17,34,37]. Other more direct sources of evidence would, however, be necessary to further support this hypothesis, which would imply a better understanding of the genetic basis for the dispersal behaviour and potentially the ancestral reconstruction of the implicated genes.
(d). Potential biases
The results presented in this study rely on several important assumptions, such as: (i) the correctness of the phylogeny, (ii) the homogeneity of rates across lineages within migratory or sedentary species, and (iii) the constancy of diversification rates through time.
— (i) Diversification analyses directly rely on the topological and dating quality of the phylogeny used. The phylogeny we used is the most up-to-date nearly complete phylogeny of birds [40]. This tree is controversial, because not all species have molecular sequences [73], yet it currently represents the best tree for such global-scale analyses. We accounted for some of the uncertainty in phylogenetic reconstruction by running our analyses on several trees from the posterior distribution, and found consistent results.
— (ii) Migratory (and sedentary) lineages may have heterogeneous diversification rates for many reasons, including differences in habitat [67], body size, niche size, diet [66] or sexual selective pressures such as colour polymorphism [65]. We ran the various models at the order level to reduce the bias introduced by this heterogeneity in our global-scale analyses. There are probably heterogeneities within orders as well, yet the consistency of our results at the global scale and across orders suggest that these results are robust to rate variations across lineages. Heterogeneity in rates may, however, to some extent explain the lack of evidence for the effect of migratory behaviour on extinction rates, as discussed above.
— (iii) Diversification rates may vary through time for a variety of reasons including variations in the abiotic and biotic environment [64,74]. In particular, there is considerable evidence for slowdowns in the diversification rates of small- to medium-sized clades of birds, which has often been interpreted as evidence that diversity-dependent processes are at play [74–77]. Diversity dependence is unlikely to operate at the large taxonomic scales considered here. Jetz et al. [40] instead suggested that diversification is accelerating towards the present for birds as a whole. We ran diversity-dependent analyses [78] on the whole phylogeny and on the five most speciose orders, confirming that current bird diversity is far from a potential ‘carrying capacity’ (results not shown). Although diversity-dependent processes are unlikely to bias our results, other processes responsible for time-variation in diversification rates might. Future analyses testing the effect of migration on diversification while allowing for time-variation in rates would be useful to test the robustness of our results to such potential time variations.
5. Conclusion
Our results provide, to our knowledge, a first insight into how seasonal migration may have impacted and shaped the history of avian diversification. Remarkably, seasonal migration promotes speciation by increasing the probability that migratory species form new sedentary species. At the same time, a migratory species rarely diversifies into two migratory species. Hence, although seasonal migration enhances speciation and reduces extinction, diversification rates and species richness are decoupled, with the overwhelming majority of extant birds—sedentary species—diversifying slower than migratory species.
Acknowledgements
K.A.J. acknowledges support from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme (FP7/2007–2013) under REA grant agreement no. PIEF-GA-2011-300924. Conceived and designed the experiments: J.R., F.J., H.M. Performed the experiments: J.R. Analysed the data: J.R. Contributed reagents/materials/analysis tools: J.R. Wrote the paper: J.R., F.J., K.A.J., F.L.C., H.M. We thank Albert Phillimore and one anonymous reviewer for advice that significantly improved the manuscript.
Funding statement
Funding was provided by the Chaire Modélisation Mathématique et Biodiversité of Véolia Environnement-Ecole Polytechnique-Muséum National d'Histoire Naturelle-Fondation X, the French National Center for Scientific Research (CNRS), and grant ECOEVOBIO-CHEX2011 from the French National Research Agency (A.N.R.) awarded to H.M.
References
- 1.Dingle H, Drake VA. 2007. What is migration? Bioscience 57, 113–121. ( 10.1641/B570206) [DOI] [Google Scholar]
- 2.Wiener P, Tuljapurkar S. 1994. Migration in variable environments: exploring life history evolution using structured population models. J. Theor. Biol. 166, 75–90. ( 10.1006/jtbi.1994.1006) [DOI] [PubMed] [Google Scholar]
- 3.Egevang C, Stenhousec IJ, Phillips RA, Petersen A, Fox JW, Silk JRD. 2010. Tracking of Arctic terns Sterna paradisaea reveals longest animal migration. Proc. Natl Acad. Sci. USA 107, 2078–2081. ( 10.1073/pnas.0909493107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alerstam T. 2006. Conflicting evidence about long-distance animal navigation. Science 291, 300–303. ( 10.1126/science.291.5502.300) [DOI] [PubMed] [Google Scholar]
- 5.Tøttrup AP, et al. 2012. The annual cycle of a trans-equatorial Eurasian–African passerine migrant: different spatio-temporal strategies for autumn and spring migration. Proc. R. Soc. B 279, 1008–1016. ( 10.1098/rspb.2011.1323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altizer S, Bartel R, Han BA. 2011. Animal migration and infectious disease risk. Science 331, 296–302. ( 10.1126/science.1194694) [DOI] [PubMed] [Google Scholar]
- 7.Hurlbert AH, Liang Z. 2012. Spatiotemporal variation in avian migration phenology: citizen science reveals effects of climate change. PLoS ONE 7, e31662 ( 10.1371/journal.pone.0031662) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg R, Marra PP. 2005. Birds of two worlds: the ecology and evolution of migration. Washington, DC: Smithsonian Institution. [Google Scholar]
- 9.Bearhop S, et al. 2005. Assortative mating as a mechanism for rapid evolution of a migratory divide. Science 310, 502–504. ( 10.1126/science.1115661) [DOI] [PubMed] [Google Scholar]
- 10.Trierweiler C, et al. 2014. Migratory connectivity and population-specific migration routes in a long-distance migratory bird. Proc. R. Soc. B 281, 20132897 ( 10.1098/rspb.2013.2897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmore KE, Fox JW, Irwin DE. 2012. Dramatic intraspecific differences in migratory routes, stopover sites and wintering areas, revealed using light-level geolocators. Proc. R. Soc. B 279, 4582–4589. ( 10.1098/rspb.2012.1229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Tris J, Bensch S, Carbonell R, Helbig AJ, Telleria JL. 2004. Historical diversification of migration patterns in passerine birds. Evolution 58, 1819–1832. ( 10.1554/03-731) [DOI] [PubMed] [Google Scholar]
- 13.Prochazka P, et al. 2011. Low genetic differentiation among reed warbler Acrocephalus scirpaceus populations across Europe . J. Avian Biol. 42, 103–113. ( 10.1111/j.1600-048X.2010.05161.x) [DOI] [Google Scholar]
- 14.Joseph L, Wilkie T, Alpers D. 2003. Independent evolution of migration on the South American landscape in a long-distance temperate-tropical migratory bird, Swainson's flycatcher Myiarchus swainsoni. J. Biogeogr. 30, 925–937. ( 10.1046/j.1365-2699.2003.00841.x) [DOI] [Google Scholar]
- 15.Outlaw DC, Voelker G, Milá B, Girman DJ. 2003. Evolution of long-distance migration in and historical biogeography of Catharus thrushes: a molecular phylogenetic approach. The Auk 120, 299–310. ( 10.1642/0004-8038(2003)120[0299:EOLMIA]2.0.CO;2) [DOI] [Google Scholar]
- 16.Dawideit BA, Phillimore AB, Laube I, Leisler B, Bohning-Gaese K. 2009. Ecomorphological predictors of natal dispersal distances in birds. J. Anim. Ecol. 78, 388–395. ( 10.1111/j.1365-2656.2008.01504.x) [DOI] [PubMed] [Google Scholar]
- 17.Helbig AJ. 2003. Evolution of bird migration: a phylogenetic and biogeographic perspective. In Avian migration (eds Berthold P, Gwinner E, Sonnenschein E.), pp. 3–20. Berlin, Germany: Springer. [Google Scholar]
- 18.Claramunt S, Derryberry EP, Remsen JV, Jr, Brumfield RT. 2012. High dispersal ability inhibits speciation in a continental radiation of passerine birds. Proc. R. Soc. B 279, 1567–1574. ( 10.1098/rspb.2011.1922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwinner E. 1996. Circannual clocks in avian reproduction and migration. Ibis 138, 47–63. ( 10.1111/j.1474-919X.1996.tb04312.x) [DOI] [Google Scholar]
- 20.Berthold P, Gwinner E, Sonnenschein E. 2003. Avian migration. Berlin, Germany: Springer. [Google Scholar]
- 21.Teplitsky C, Mouawad NG, Balbontín J, de Lope F, Møller AP. 2011. Quantitative genetics of migration syndromes: a study of two barn swallow populations. J. Evol. Biol. 24, 2025–2038. ( 10.1111/j.1420-9101.2011.02342.x) [DOI] [PubMed] [Google Scholar]
- 22.Monteiro LR, Furness RW. 1998. Speciation through temporal segregation of Madeiran storm petrel (Oceanodroma castro) populations in Azores? Phil. Trans. R. Soc. Lond. B 353, 945–953. ( 10.1098/rstb.1998.0259) [DOI] [Google Scholar]
- 23.Bolton M, et al. 2008. Monteiro's storm-petrel Oceanodroma monteiroi: a new species from the Azores. Ibis 150, 717–727. ( 10.1111/j.1474-919X.2008.00854.x) [DOI] [Google Scholar]
- 24.Taberlet P, Fumagalli L, Wust-Saucy A-G, Cosson J-F. 1998. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 7, 453–464. ( 10.1046/j.1365-294x.1998.00289.x) [DOI] [PubMed] [Google Scholar]
- 25.Tøttrup AP, et al. 2012. Drought in Africa caused delayed arrival of European songbirds. Science 338, 130 ( 10.1126/science.1227548) [DOI] [PubMed] [Google Scholar]
- 26.Lindström Å, Agrell J. 1999. Global change and possible effects on the migration and reproduction of arctic-breeding waders. Ecol. Bull. 47, 145–159. [Google Scholar]
- 27.Mila B, Smith T, Wayne R. 2006. Postglacial population expansion drives the evolution of long-distance avian migration in a songbird. Evolution 60, 2403–2409. ( 10.1111/j.0014-3820.2006.tb01875.x) [DOI] [PubMed] [Google Scholar]
- 28.Ruegg KC, Hijmans RJ, Moritz C. 2006. Climate change and the origin of migratory pathways in the Swainson's thrush, Catharus ustulatus. J. Biogeogr. 33, 1172–1182. ( 10.1111/j.1365-2699.2006.01517.x) [DOI] [Google Scholar]
- 29.Perez-Tris J, Telleria JL. 2002. Regional variation in seasonality affects migratory behaviour and life-history traits of two Mediterranean passerines. Acta Oecol. 23, 13–21. ( 10.1016/S1146-609X(01)01129-8) [DOI] [Google Scholar]
- 30.Sanderson FJ, Donald PF, Pain DJ, Burfield IJ, van Bommel FPJ. 2006. Long-term population declines in Afro-Palearctic migrant birds. Biol. Conserv. 131, 93–105. ( 10.1016/j.biocon.2006.02.008) [DOI] [Google Scholar]
- 31.Møller AP, Rubolini D, Lehikoinen E. 2008. Populations of migratory bird species that did not show a phenological response to climate change are declining. Proc. Natl Acad. Sci. USA 105, 16 195–16 200. ( 10.1073/pnas.0803825105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winger BM, Lovette IJ, Winkler DW. 2012. Ancestry and evolution of seasonal migration in the Parulidae. Proc. R. Soc. B 279, 610–618. ( 10.1098/rspb.2011.1045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo B, Omland KE, Johnson KP. 2007. Ancestral state reconstruction of migration: multistate analysis reveals rapid changes in New World orioles (Icterus sp.). The Auk 124, 410–419. ( 10.1642/0004-8038(2007)124[410:ASROMM]2.0.CO;2) [DOI] [Google Scholar]
- 34.Pulido F, Berthold P, van Noordwijk AJ. 1996. Frequency of migrants and migratory activity are genetically correlated in a bird population: evolutionary implications. Proc. Natl Acad. Sci. USA 93, 14 642–14 647. ( 10.1073/pnas.93.25.14642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulido F. 2007. The genetics and evolution of avian migration. BioScience 57, 165–174. ( 10.1641/B570211) [DOI] [Google Scholar]
- 36.Berthold P. 1996. Control of bird migration, p. 355 London, UK: Chapman & Hall. [Google Scholar]
- 37.Berthold P. 1999. A comprehensive theory for the evolution, control and adaptability of avian migration. Ostrich 70, 1–11. ( 10.1080/00306525.1999.9639744) [DOI] [Google Scholar]
- 38.Salewski V, Bruderer B. 2007. The evolution of bird migration: a synthesis. Naturwissenschaften 94, 268–279. ( 10.1007/s00114-006-0186-y) [DOI] [PubMed] [Google Scholar]
- 39.Able KP, Belthoff JR. 1998. Rapid ‘‘evolution’’ of migratory behaviour in the introduced house finch of eastern North America. Proc. R. Soc. Lond. B 265, 2063–2071. ( 10.1098/rspb.1998.0541) [DOI] [Google Scholar]
- 40.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491, 444–448. ( 10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 41.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 ( 10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis MP, Midford PE, Maddison W. 2013. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evol. Biol. 13, 38 ( 10.1186/1471-2148-13-38) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birdlife International, NatureServe. 2011. Bird species distribution maps of the world. BirdLife International, Cambridge, United Kingdom and NatureServe, Arlington, United States. Available upon request at: http://www.birdlife.org/datazone/ (Accessed 2013 June).
- 44.Somveille M, Manica A, Butchart SHM, Rodrigues ASL. 2013. Mapping global diversity patterns for migratory birds. PLoS ONE 8, e70907 ( 10.1371/journal.pone.0070907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzjohn R. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092. ( 10.1111/j.2041-210X.2012.00234.x) [DOI] [Google Scholar]
- 46.Goldberg EE, Igic B. 2012. Tempo and mode in plant breeding system evolution. Evolution 66, 3701–3709. ( 10.1111/j.1558-5646.2012.01730.x) [DOI] [PubMed] [Google Scholar]
- 47.Fitzjohn R. 2013. diversitree: comparative phylogenetic analyses of diversification. Available at http://www.cran.r-project.org/web/packages/diversitree.
- 48.Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol. 56, 701–710. ( 10.1080/10635150701607033) [DOI] [PubMed] [Google Scholar]
- 49.FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611. ( 10.1093/sysbio/syp067) [DOI] [PubMed] [Google Scholar]
- 50.Lovette IJ, Bermingham E. 1999. Explosive speciation in the New World Dendroica warblers. Proc. R. Soc. Lond. B 266, 1629–1636. ( 10.1098/rspb.1999.0825) [DOI] [Google Scholar]
- 51.Owens IPF, Bennett PM, Harvey PH. 1999. Species richness among birds: body size, life history, sexual selection or ecology? Proc. R. Soc. Lond. B 266, 933–939. ( 10.1098/rspb.1999.0726) [DOI] [Google Scholar]
- 52.Phillimore AB, Freckleton RP, Orme CDL, Owens IPF. 2006. Ecology predicts large-scale patterns of phylogenetic diversification in birds. Am. Nat. 168, 220–229. ( 10.1086/505763) [DOI] [PubMed] [Google Scholar]
- 53.Price TD. 2008. Speciation in birds. Greenwood Village, CO: Roberts & Company Publishers. [Google Scholar]
- 54.Winker K. 2000. Migration and speciation. Nature 404, 36 ( 10.1038/35003651) [DOI] [PubMed] [Google Scholar]
- 55.Bairlein F, et al. 2012. Cross-hemisphere migration of a 25 g songbird. Biol. Lett. 8, 505–507. ( 10.1098/rsbl.2011.1223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salomonsen F. 1950. The immigration and breeding of the fieldfare (Turdus pilaris L.) in Greenland. Proc. Int. Ornithol. Congr. 10, 515–526. [Google Scholar]
- 57.Gill FB. 1995. Ornithology, 2nd edn New York, NY: WH. Freeman and Co. [Google Scholar]
- 58.Hoyo JD, Elliott A, Sargatal J, Cabot J. 1992. Handbook of the birds of the world, 3rd edn Barcelona, Spain: Lynx Edicions. [Google Scholar]
- 59.Jansson R. 2003. Global patterns in endemism explained by past climatic change. Proc. R. Soc. Lond. B 270, 583–590. ( 10.1098/rspb.2002.2283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cahill AE, et al. 2013. How does climate change cause extinction? Proc. R. Soc. B 280, 20121890 ( 10.1098/rspb.2012.1890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabosky DL. 2010. Extinctions should not be estimated from molecular phylogenies. Evolution 64, 1816–1824. ( 10.1111/j.1558-5646.2009.00926.x) [DOI] [PubMed] [Google Scholar]
- 62.Quental TB, Marshall CR. 2010. Diversity dynamics: molecular phylogenies need the fossil record. Trends Ecol. Evol. 25, 434–441. ( 10.1016/j.tree.2010.05.002) [DOI] [PubMed] [Google Scholar]
- 63.Morlon H, Parsons TL, Plotkin JB. 2011. Reconciling molecular phylogenies with the fossil record. Proc. Natl Acad. Sci. USA 108, 16 327–16 332. ( 10.1073/pnas.1102543108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morlon H. 2014. Phylogenetic approaches for studying diversification. Ecol. Lett. 17, 508–525. ( 10.1111/ele.12251) [DOI] [PubMed] [Google Scholar]
- 65.Hugall AF, Stuart-Fox D. 2012. Accelerated speciation in colour-polymorphic birds. Nature 485, 631–634. ( 10.1038/nature11050) [DOI] [PubMed] [Google Scholar]
- 66.Price SA, Hopkins SSB, Smith KK, Roth VL. 2012. Tempo of trophic evolution and its impact on mammalian diversification. Proc. Natl Acad. Sci. USA 109, 7008–7012. ( 10.1073/pnas.1117133109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rolland J, Condamine FL, Jiguet F, Morlon H. 2014. Faster speciation and reduced extinction in the tropics contribute to the mammalian latitudinal diversity gradient. PLoS Biol. 12, e1001775 ( 10.1371/journal.pbio.1001775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasner CA, Yeh P, Eggert LS, Hunt KE, Woodruff DS, Price TD. 2004. Genetic and morphological evolution following a founder event in the dark-eyed junco, Junco hyemalis thurberi. Mol. Ecol. 13, 671–681. ( 10.1046/j.1365-294X.2004.02104.x) [DOI] [PubMed] [Google Scholar]
- 69.Magnuson-Ford K, Otto SP. 2012. Linking the investigations of character evolution and species diversification. Am. Nat. 180, 225–245. ( 10.1086/666649) [DOI] [PubMed] [Google Scholar]
- 70.Rappole JH. 2003. An integrative framework for understanding the origin and evolution of avian migration . J. Avian Biol. 34, 124–128. ( 10.1034/j.1600-048X.2003.03170.x) [DOI] [Google Scholar]
- 71.Zink RM. 2002. Towards a framework for understanding the evolution of avian migration. J. Avian Biol. 33, 433–436. ( 10.1034/j.1600-048X.2002.03081.x) [DOI] [Google Scholar]
- 72.Dingle H. 1996. The biology of life on the move. New York, NY: Oxford University Press. [Google Scholar]
- 73.Gewin V. In press ‘Tree of life’ constructed for all living bird species. Nature. ( 10.1038/nature.2012.11712) [DOI] [Google Scholar]
- 74.Moen DS, Morlon H. 2014. Why does diversification slow down? Trends Ecol. Evol. 29, 190–197. ( 10.1016/j.tree.2014.01.010) [DOI] [PubMed] [Google Scholar]
- 75.Phillimore AB, Price TD. 2008. Density-dependent cladogenesis in birds. PLoS Biol. 6, 483–489. ( 10.1371/journal.pbio.0060071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morlon H, Potts MD, Plotkin JB. 2010. Inferring the dynamics of diversification: a coalescent approach. PLoS Biol. 8, e1000493 ( 10.1371/journal.pbio.1000493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McPeek MA. 2008. The ecological dynamics of clade diversification and community assembly. Am. Nat. 172, E270–E284. ( 10.1086/593137) [DOI] [PubMed] [Google Scholar]
- 78.Leventhal GE. 2013. expoTree: Calculate density dependent likelihood of a phylogenetic tree. Available at http://www.cran.r-project.org/web/packages/expoTree.