Abstract
The transcription factor Tbx5 is expressed in the developing heart, eyes and anterior appendages. Mutations in human TBX5 cause Holt–Oram syndrome, a condition characterized by heart and upper limb malformations. Tbx5-knockout mouse embryos have severely impaired forelimb and heart morphogenesis from the earliest stages of their development. However, zebrafish embryos with compromised tbx5 function show a complete absence of pectoral fins, while heart development is disturbed at significantly later developmental stages and eye development remains to be thoroughly analysed. We identified a novel tbx5 gene in zebrafish—tbx5b—that is co-expressed with its paralogue, tbx5a, in the developing eye and heart and hypothesized that functional redundancy could be occurring in these organs in embryos with impaired tbx5a function. We have now investigated the consequences of tbx5a and/or tbx5b downregulation in zebrafish to reveal that tbx5 genes have essential roles in the establishment of cardiac laterality, dorsoventral retina axis organization and pectoral fin development. Our data show that distinct relationships between tbx5 paralogues are required in a tissue-specific manner to ensure the proper morphogenesis of the three organs in which they are expressed. Furthermore, we uncover a novel role for tbx5 genes in the establishment of correct heart asymmetry in zebrafish embryos.
Keywords: developmental biology, zebrafish, limb, retina, heart, tbx5
2. Introduction
Tbx5 codes for a T-domain containing transcription factor that has been characterized in many vertebrate species, where it is widely expressed during the development of the heart, the eyes and the anterior set of appendages (tetrapod forelimbs and fish pectoral fins) [1–3]. Mutations in human TBX5 cause Holt–Oram syndrome (HOS; OMIM#142900), an autosomal-dominant ‘heart–hand’ condition characterized by heart and upper limb malformations [4,5]. Owing to its clinical relevance, several Tbx5 loss-of-function animal models have been developed to assess the role that this gene may play during the development of the vertebrate heart, limbs and eyes. These studies have shown that in mouse Tbx5-knockout embryos, both forelimb and heart development is severely impaired from the earliest stages of development [6–8]. Also, mis-expression of Tbx5 in the ventral retina produces altered projections of retinal ganglion cell (RGC) axons in chick embryos [9], consistent with a fundamental role for Tbx5 during eye morphogenesis.
The use of zebrafish as a model system has a series of advantages with respect to mouse, mainly their ease of embryo accessibility and the constant development of new techniques such as morpholino (MO) knock-down and transgenesis. Zebrafish pectoral fins are homologous to tetrapod forelimbs, and using genetic and transgenic techniques it has been shown that the molecular mechanisms governing the initial steps of limb/fin bud outgrowth are conserved between tetrapods and teleosts [10]. Hence, zebrafish are commonly used as a model system to study vertebrate limb development. Heart morphogenesis involves the specification and differentiation of cardiac precursors, the integration of precursors into a tube and the remodelling of the embryonic tube to create a fully functional organ [11]. Similar to limb/fin development, similarities between higher vertebrates and zebrafish heart morphogenesis have established zebrafish as a model to study cardiac development and function [12]. Finally, eye formation requires the coordination of a series of morphogenetic events and the regulated expression of several genes that are similarly conserved among vertebrate models [13].
Tbx5 function has been investigated during zebrafish development using both a MO knock-down approach and the use of a tbx5a mutant strain obtained by ENU-induced mutagenesis (heartstrings (hst) [14]). In zebrafish, and similar to amniote embryos, tbx5a is expressed in the heart, pectoral fins and dorsal retina from the earliest stages of their development. However, embryos with compromised tbx5a function show a complete absence of pectoral fins, while heart development is disturbed at a relatively late developmental stage. Defects in eye development have not been thoroughly assessed [14,15]. We identified a novel tbx5 gene in zebrafish—tbx5b—that is co-expressed with its paralogue, now referred to as tbx5a, in the developing eye and heart fields and that arose during the teleost-specific genome duplication event that took place during evolution. We hypothesized that functional redundancy of tbx5a and tbx5b in the developing heart would explain the relatively late phenotypes observed during cardiac development in fish embryos with compromised tbx5a function [16].
To test our hypothesis, we have now investigated the consequences of tbx5a and/or tbx5b downregulation during zebrafish development. Our data show that distinct relationships between tbx5 paralogues are required in a tissue-specific manner to ensure the proper morphogenesis of tissues with conspicuous expression of tbx5 genes, namely the developing heart, the retina and the pectoral fins. Finally, we also demonstrate that both tbx5 paralogues are required to direct both asymmetric events the zebrafish heart undergoes (i.e. heart tube jogging first and looping later), thus uncovering a novel and fundamental role for these genes during the establishment of cardiac left–right asymmetry.
3. Results and discussion
To understand the unique and/or redundant roles that the tbx5 paralogues have during the development of the zebrafish heart, pectoral fin and eye fields, we have used MOs against tbx5a and tbx5b to downregulate their function during embryonic development either individually or in conjunction. Briefly, we used an anti-tbx5a MO oligonucleotide for the coding sequence [15], an anti-tbx5b oligonucleotide recognizing the 5′ UTR/coding sequence boundary—tbx5b(UTR)—as well as an anti-tbx5b oligonucleotide for the exon 3/intron 4 boundary (tbx5b(SP) MO). First of all, we characterized the functionality of our MOs by generating chimeric mRNAs containing the tbx5a or tbx5b(UTR) MO-recognition sites fused to enhanced green fluorescent protein (EGFP). Injection of these RNAs (100 pg) with or without their corresponding target MO (3 ng), showed that, indeed, co-injection of our tbx5a and tbx5b(UTR) MOs caused disappearance of EGFP signal (electronic supplementary material, figure S1a–d′). In addition, and to assess tbx5b gene knock-down efficiency, we performed RT-PCR experiments from embryos that had been injected with either a control MO or a tbx5b(SP) MO. This showed that an expected 215 bp (spliced) band was obtained in control embryos in contrast to the 791 bp (unspliced) band observed after injection of 2–4 ng of tbx5b(SP) MO (electronic supplementary material, figure S1e).
3.1. Knock-down of zebrafish tbx5 genes causes cardiac looping defects
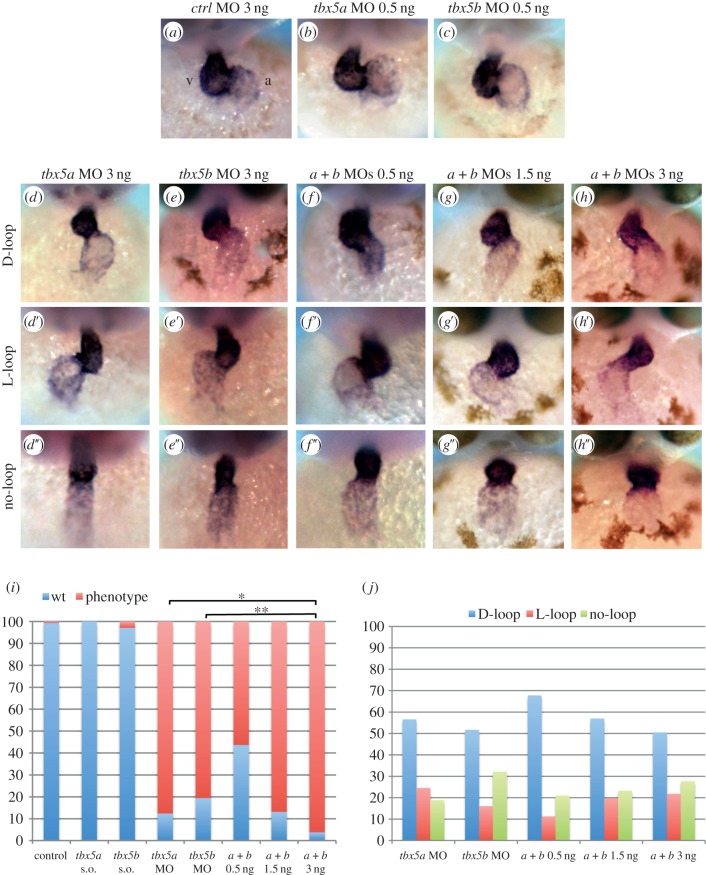
tbx5a and, as described recently, tbx5b have been implicated in cardiac looping morphogenesis [14,17]. Analyses of looping phenotypes assayed at 48 h post-fertilization (hpf) had shown that wild-type embryos had undergone complete looping (i.e. the ventricle located at the right-hand side of the embryo with the atrial and ventricular chambers sitting side by side), whereas homozygous tbx5a mutant (hst) and tbx5b morphants failed to do so, indicating that downregulation of tbx5 genes was associated with reduced looping. As tbx5b knock-down on hst mutant embryos did not increase the severity of the defects, it was argued that these paralogues do not have an overlapping function in cardiac development. In addition, looping of these hearts in the reverse orientation was never observed [17]. Similarly, we observed incomplete looping after tbx5a or tbx5b knock-down (figure 1d,e, respectively) in comparison to the characteristic looping observed in control morphants that results in the positioning of the ventricle (v) and the atrium (a) parallel to each other (figure 1a). However, in striking contrast to previous data, defects were observed not only in the degree of looping but also in the orientation of cardiac looping: tbx5 morphants can be classified into three distinguishable heart looping orientation groups (D-loop (right, normal), L-loop (left, reversed) and no-loop). After injection of a control MO, over 99% (n = 158) of the embryos displayed an S-shaped heart with the ventricle lying to the right-hand side of the embryo (D-loop; figure 1a,i). By contrast, 88% (n = 121) of the tbx5a MO-injected embryos had incomplete cardiac looping, and within these 57% showed D-loop, 24% showed L-loop and 19% showed no looping at all (figure 1d–d″,i,j). Similarly, injection of tbx5b MO also caused heart looping defects (81%, n = 108), and these embryos displayed D-looped (52%), L-looped (16%) and no-looped (34%) cardiac morphologies (figure 1e–e″,i,j).
Figure 1.
Knock-down of tbx5 genes causes cardiac looping defects. (a–c) Embryos injected with control MO or sub-optimal concentrations of tbx5a or tbx5b MOs. (d–d″) tbx5a morphant phenotypes. (e–e′) tbx5b-morphant phenotypes. (f–h″) Double knock-down of tbx5 genes (0.5 ng each MO (f–f″), 1.5 ng each MO (g–g″) and 3 ng each MO (h–h″)). (i) Quantification of the degree of looping phenotypes: wt, complete; phenotype, incomplete looping s.o., sub-otimal. (j) Quantification of the looping orientation phenotypes. A χ2 statistic has been calculated to assess significant differences between groups (**p < 0.001, *p < 0.05). Images are frontal views of 48 hpf embryos, and myl7 expression is used to highlight the developing heart.
The phenotypic similarities observed after tbx5a or tbx5b knock-down prompted us to investigate whether these genes cooperatively regulate cardiac looping in contrast to what had been previously argued. To this end, we simultaneously knocked-down their function by co-injecting sub-optimal doses of tbx5a and tbx5b MOs (0.5 ng of each MO) which, when injected alone, did not affect cardiac looping (figure 1b,c,i; n = 75 for tbx5a MO and n = 71 for tbx5b MO). This showed that over 56% (n = 110) of the double-morphants had looping defects and the three D-loop, L-loop and no-loop phenotypes were detected (68%, 11% and 21%, respectively; figure 1f–f″,i,j) suggesting that, indeed, both genes act in the same pathway and cooperate with each other to ensure the completeness and orientation of looping of the zebrafish heart. Moreover, injection of increasing concentrations of both MOs caused an increase in the percentage of phenotypes, with 87% (n = 99) and 96% (n = 105) of double-morphants displaying looping phenotypes after injection with 1.5 and 3 ng of each MO, respectively. Although, in agreement with a previous report [17], the severity of the phenotype was not enhanced by double knock-down, downregulation of both genes increased the penetrance of the phenotype (figure 1i). Similarly to single and double-morphants injected with sub-optimal doses of the tbx5a and tbx5b MOs, these double-morphant embryos also exhibited looping orientation defects since the three orientation phenotypes were detected (figure 1g–g″,h–h″,j).
3.2. tbx5 morphants exhibit cardiac tube jogging defects
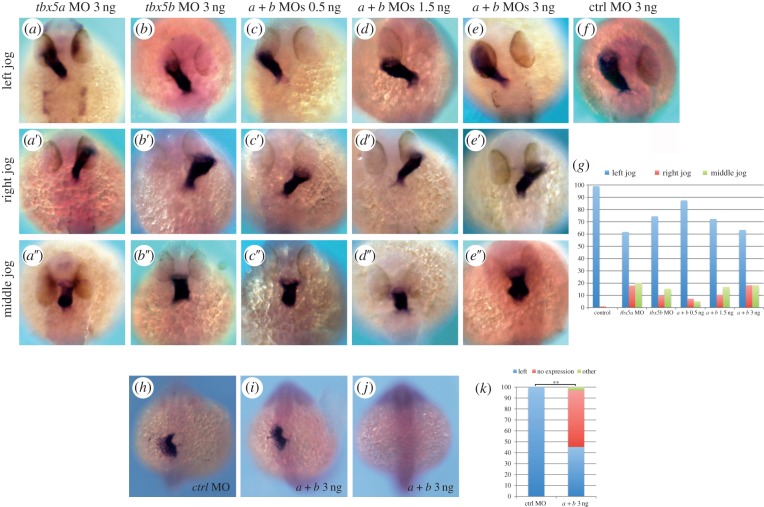
As heart looping orientation phenotypes are indicative of cardiac left–right asymmetry defects, we decided to examine whether heart jogging, the first morphologically evident break in the left–right symmetry of the zebrafish heart tube, was disrupted after knock-down of tbx5 genes. To this end, heart laterality was assessed at 26 hpf using myl7 expression to reveal positioning of the heart tube and cardiac jogging was classified as left (normal), right (reversed) or midline (no jog). After injection of 3 ng of control MO, 99% (n = 197) of the embryos developed normal left jogging (figure 2f,g). By contrast, right as well as midline jogging phenotypes were observed after tbx5a knock-down (figure 2a–a″,g; n = 183). Similarly, tbx5b morphants displayed left as well as right and middle jogging of their linear heart tubes (figure 2b–b″,g; n = 90).
Figure 2.
tbx5 morphants exhibit cardiac jogging and lefty2 expression defects. (a–a″) tbx5a morphant phenotypes. (b–b″) tbx5b morphants. (c–e″) Left, right and middle jog phenotypes obtained by co-injection of tbx5a and tbx5b MOs at 0.5 ng (c–c″), 1.5 ng (d–d″) or 3 ng (e–e″) of each MO. (f) Control (ctrl) morphant. (g) Quantification of the phenotypes. (h–j) lefty2 expression in control (h) and double-morphant (i,j) 22-somite stage embryos. (k) Quantification of the phenotypes. A χ2 test has been used to assess significant differences between groups (**p < 0.001). All images are dorsal views with anterior to the top, and myl7 expression is used to highlight the developing heart tube in a–f.
Once again, the phenotypic similarities between tbx5a and tbx5b morphants suggested these genes cooperatively regulate heart jogging. To confirm this, we injected sub-optimal doses of tbx5a and tbx5b MOs which showed that, in agreement with both genes cooperating to ensure the normal left-sided jog of the heart tube, 12.5% (n = 96) of these morphant embryos had a defective middle or right jog of the heart (figure 2c–c″,g). Moreover, and similar to the heart looping defects discussed above, injection of increasing concentrations of both MOs increased the percentage of phenotypes observed, with nearly 30% (n = 94) and 40% (n = 71) of double-morphant embryos showing defective heart jogging after injection of 1.5 and 3 ng of each MO, respectively (figure 2d–e″,g).
The cardiac phenotypes caused by tbx5a and/or tbx5b knock-down (namely cardiac jogging and looping orientation defects) demonstrate that tbx5 genes are required to direct both asymmetric events the zebrafish heart undergoes [18]. First, and during the process referred to as cardiac jogging, the cardiac cone, formed at the embryonic midline, is converted into a linear tube by the repositioning of the atrial cells to the anterior and left of the ventricular cells due to their higher migration rates [19,20]. Second, and during the conserved process of cardiac looping, the ventricle is positioned to the right of the atrium. These two processes are defective in tbx5a and/or tbx5b morphants. Interestingly, known left–right cardiac determinants such as Bmp4 have been isolated in screens aimed to find Tbx5-induced genes [21], and bioinformatic approaches have highlighted the presence of Tbx5-binding sites in the vicinity of the lefty2 locus (N. Mercader 2013, personal communication), another well-known left–right asymmetry determination factor. To address whether tbx5 genes may be regulating lefty2 expression in developing embryos, we assessed the expression of this gene after tbx5 genes knock-down. In support of tbx5 genes being upstream of lefty2 expression, over half of the tbx5a and tbx5b double-morphants (52%, n = 106) showed no expression of lefty2, whereas 100% of control MO injected embryos (n = 43) had the characteristic left-sided expression of lefty2 at the 22-somite stage (figure 2h–k). These experiments show that tbx5 genes are upstream of lefty2 expression, and hence the cardiac laterality phenotypes observed in tbx5 morphants may be explained by this relationship between tbx5 and the asymmetric gene lefty2. However, how bilaterally expressed genes such as tbx5 can specifically regulate a left-side specific gene like lefty2 remains unclear. Nevertheless, one can hypothesize that lefty2 will only become activated in the left-hand side of the cardiac cone where tbx5 acts with a co-activator that is only present in the left side of the developing embryo. Conversely, a repressor only found in the right side of the embryo may be inhibiting the activation of lefty2 by tbx5 genes in the right side of the cardiac cone.
The reasons for the discrepancies between our results, implicating the tbx5 genes in the asymmetry events the zebrafish heart undergoes (namely cardiac jogging first and looping later), and those of others [17] are unclear. One possibility is that we have used a MO against tbx5a to downregulate its function, whereas Parrie et al. [17] used the tbx5a mutant line hst to analyse the effects of this gene loss-of-function. The hst mutation introduces a premature STOP codon at residue 316 of the tbx5a open reading frame, which leaves the mutated protein with intact N-terminal and T-box (DNA binding) domains and part of the C-terminal domain. It is therefore possible that the hst mutation behaves as an hypomorphic allele with regard to the left–right asymmetric development of the heart. In agreement with this hypothesis, most of the TBX5 mutations causing a clear HOS phenotype lie upstream of the predicted hst mutation. To test whether, indeed, the hst mutation behaves as an hypomorphic allele with regards to cardiac development, we developed an assay to assess whether the laterality phenotype of tbx5a morphants could be rescued by introducing specific MO-insensitive forms of the tbx5a mRNA: (i) a full-length tbx5a mRNA that should be able to rescue the tbx5a MO-mediated phenotype, (ii) a tbx5a mRNA that is identical to that produced in hst mutant embryos and (iii) a severely truncated version of tbx5a that we have engineered by introducing a premature STOP codon within the T-box domain (figure 3a). Notably, both the full-length and the hst-like forms of tbx5a were able to partially rescue the cardiac jogging phenotype when co-injected with the tbx5a MO (figure 3b; n = 65 and n = 168, respectively). Similarly, a full-length tbx5b form was able to rescue the jogging phenotype of tbx5b morphants when co-injected with our tbx5b MO (figure 3b; n = 117). By contrast, the severely truncated form of tbx5a was not able to rescue the laterality phenotype (figure 3b, n = 79). Overall, these data demonstrate not only the specificity of the cardiac phenotypes caused by MO-mediated knock-down of tbx5a and/or tbx5b, but also that the hst mutation behaves as a hypomorphic allele with regard to cardiac laterality. We have ourselves analysed heart tube jogging in hst mutants (n = 38) and all of them displayed a normal left-jog as visualized by myl7 expression in 26 hpf embryos (figure 3c,d). These results underline the need for caution when using the hst mutation as a tbx5a loss-of-function allele.
Figure 3.
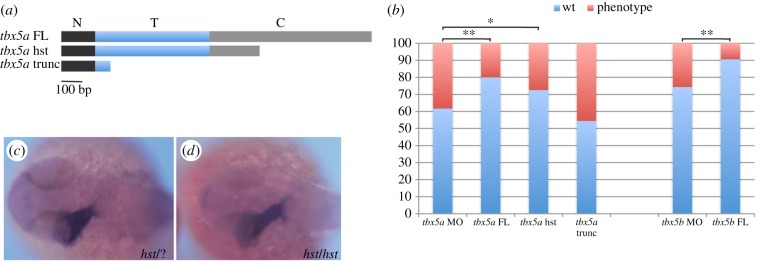
The hst mutation is a hypomorphic allele with regards to cardiac laterality. (a) tbx5a variants generated to perform the rescue experiments: a tbx5a full-length (tbx5a FL) version includes the whole N-terminal (N, black rectangle), T- (T, blue rectangle) and C-terminal (C, grey rectangle) domains; a heartstrings version (tbx5a hst) containing the whole N-terminal domain and T-domain and a truncated C-terminal domain; and a tbx5a severely truncated version (tbx5a trunc) that contains the whole N-terminal domain and a truncated T-domain. (b) Quantification of the rescue experiments (wt, left jog; phenotype, right and middle jog). A χ2 statistic has been calculated to assess significant differences between groups (**p < 0.01, *p < 0.05). (c,d) Dorsal views with anterior towards the left of 26 hpf +/+ or +/hst (c) and hst mutant (d) embryos showing normal leftward jogging of the embryonic heart tube highlighted by myl7 expression.
3.3. tbx5 genes are essential for correct dorsoventral retina regionalization
Tbx5 genes are also conspicuously expressed in the dorsal retina, a feature conserved among vertebrates [1–3,22]. However, the consequences of Tbx5 loss-of-function in this domain have led to controversial results. Tbx5 gene function in the developing retina has been examined in developing chick embryos by mis-expressing this gene in the ventral domain, where it is not normally expressed. This caused dorsalization of the ventral retina as determined by upregulation of dorsal markers and downregulation of ventral ones, as well as altered projections of RGCs [9]. tbx5a knock-down in zebrafish led to downregulation of dorsal retina markers, while ventral markers did not seem to be affected and ganglion cell projections were not analysed [23]. We hypothesized that as tbx5a and tbx5b are co-expressed in the dorsal domain of the developing retina, functional redundancy may explain the controversial results found between different models. This prompted us to determine the consequences of the downregulation of tbx5 genes in this territory.
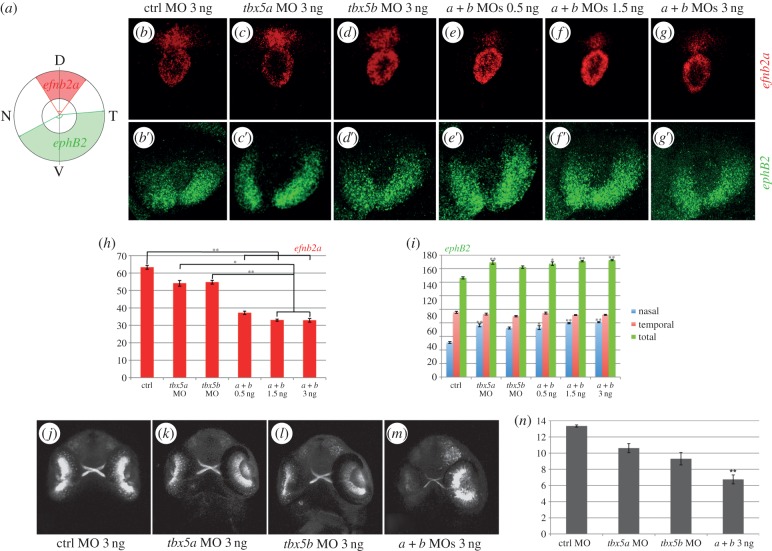
We analysed the expression of the dorsally expressed ephrin, efnb2a, and the ventrally expressed ephrin receptor, ephB2, because restricted ephrinB/ephB expression along the dorsoventral axis has been shown to play a key role in retinotectal topographic map formation [24,25]. To quantify the extent of efnb2a expression, we measured the angle of expression of this gene by setting a ‘hinge’ in the centre of the lens (figure 4a). In control embryos, the efnb2a expression domain was measured to form an average angle of 63° (figure 4b,h). Knock-down of either tbx5a or tbx5b caused a reduction of the efnb2a angle of expression leading to an average angle of 54°, although this decrease was not found to be statistically significant (figure 4c,d,h). To investigate whether both paralogues function in conjunction to determine the extent of dorsal efnb2a expression, we co-injected sub-optimal doses of tbx5a and tbx5b MOs. Remarkably, the efnb2a expression domain was greatly reduced to an average angle of 37°, a statistically significant 41% reduction compared with control embryos (figure 4e,h). Injection of increasing concentrations of both MOs caused slightly more severe effects than those observed after co-injection of sub-optimal doses of tbx5a and tbx5b MOs (figure 4f,g,h). Notably, further statistical analyses showed that significant differences are found between the injection of single tbx5a and tbx5b MOs with respect to their co-injection at these higher doses (1.5 and 3 ng each; figure 4h), suggesting that tbx5 paralogues act redundantly in the dorsal retina to ensure efnb2a expression in this territory. To better map the decrease of efnb2a expression observed in tbx5a- and/or tbx5b-morphant retinas, we divided the angles obtained into two, a dorso-nasal angle and a dorso-temporal angle, by setting the dorsal-most point as the point lying dorsal to the ventrally located choroid fissure (D in figure 4a). This showed that both dorso-nasal and dorso-temporal borders of expression similarly decreased accordingly to the total angle of efnb2a expression measured (electronic supplementary material, figure S2a–c).
Figure 4.
tbx5 genes are required for dorsoventral retina organization. (a) Schematics of our quantification method. Expression of efn2a and ephB2 in control (ctrl) embryos (b–b′), tbx5a morphants (c–c′) and tbx5b morphants (d–d′). (e–g′) Expression of efn2a and ephB2 in embryos co-injected with different concentrations of both tbx5a and tbx5b MOs. (h–i) Quantification of the results obtained for the expression of efn2a (h) and ephB2 (i). (j–m) Retinal projections of 48 hpf ath5:GFP embryos injected with control, tbx5a, tbx5b or tbx5a and tbx5b MO. (n) Optic nerve diameter quantifications. Data are represented as the mean ± s.e. A Kruskal–Wallis test was used to determine statistical differences among experimental groups (*p < 0.05, **p < 0.001). D, dorsal; N, nasal; T, temporal; V, ventral.
To ascertain whether this decrease in efnb2a expression domain was concomitant with a deregulation of ephB expression in the ventral retina, we measured the extent of ephB2 expression. In control embryos, the expression domain of ephB2 formed an average angle of 146° (figure 4b′,i). tbx5a and tbx5b downregulation caused an increase of this angle to an average of 169° and 162°, respectively (figure 4c′–d′,i). Moreover, injection of sub-optimal doses of tbx5a and tbx5b MOs similarly caused an increase in the expression extent of the ventral marker ephB2 (to an average of 167°, figure 4e′,i). Again, injection of increasing concentrations of both MOs caused similar effects to those observed after co-injection of sub-optimal doses of tbx5a and tbx5b MOs (figure 4f′–g′,i). In contrast to the results obtained for the bilaterally symmetrical loss of expression of the dorsal marker efnb2a, statistically significant expansions of the ephB2 expression domain towards its ventro-nasal border were largely enough to explain the increase in the global domain of ephB2 expression in the different conditions (figure 4i).
Interestingly, the homeodomain transcription factor meis1 has been implicated in the establishment of the proper retinotectal map of the developing zebrafish. meis1 knock-down causes a decrease of dorsal efnb2a expression and an increase of ventral ephB2, which is also associated with downregulation of dorsal tbx5a expression [26]. We show that downregulation of tbx5a is enough to explain the defects in dorsoventral-restricted expression of the ephrinB/ephB and therefore propose a model by which meis1 acts upstream of tbx5 genes expression to ensure the correct dorsoventral expression of efnb2a and ephB2 in the developing retina.
Notably, dorsal efnb2a gene expression is not completely abolished after tbx5a and tbx5b knock-down, suggesting that other factors are acting with the tbx5 paralogues to maintain dorsal retina identity. Strikingly, T-box genes have been shown to cooperatively interact in many developmental processes [27,28]. Regarding dorsal retina identity, several related T-box genes are co-expressed with tbx5 in this domain, namely the other three genes that, together with tbx5, form the Tbx2 subfamily of T-box genes, i.e. tbx2, tbx3 and tbx4 [29–31]. Functional redundancy between these genes may therefore explain the lack of complete downregulation of dorsal retina markers.
Finally, to assess whether altered efnb2a and ephB2 expression in tbx5a and/or tbx5b morphants altered the normal formation of the retinotectal map, we injected our MOs into one-cell stage ath5:gfp embryos that express the gfp transgene in RGCs under the regulation of the ath5 promoter, the zebrafish homologue of the Drosophila atonal gene [32]. By 48 hpf, RGCs have extended their axons to form the optic nerve that crosses the ventral midline to form the optic chiasm and project dorsally to the contralateral optic tectum (figure 4j). Injection of tbx5a and/or tbx5b MOs did not cause observable pathfinding errors (figure 4k–m). However, these experiments showed that the optic nerve of double-morphants was considerably thinner than that of control MO-injected siblings (figure 4m). Moreover, and in agreement with both tbx5 genes acting redundantly to ensure proper optic nerve formation, double-morphant embryos showed a significantly thinner optic nerve (figure 4n). It is noteworthy that space cadet mutants, fish that carry a mutation in the retinoblastoma gene rb1, exhibit thinner optic nerves than wild-type siblings [33] and that it has been shown that Tbx2 (closely related to Tbx5) molecularly interacts with Rb1 [34]. It is therefore tempting to speculate that, likewise, Tbx5 may be interacting with Rb1 to regulate the normal formation of the optic nerve in zebrafish embryos.
Altogether, our data show that knock-down of tbx5 genes causes an alteration of dorsoventral ephrinb/ephB expression in the retina and the formation of a thinner optic nerve.
3.4. tbx5b knock-down causes a delay in pectoral fin growth
Although we had not previously observed tbx5b expression in developing pectoral fins [16], others have recently described it in the pectoral fin bud mesenchyme of 36 hpf embryos [17]. In agreement with tbx5b playing a role during zebrafish pectoral fin morphogenesis, tbx5b morphants had smaller pectoral fins when compared with control embryos at 3 dpf (figure 5a,i; [17]), which is reminiscent of the phenotypes observed upon subtle downregulation of tbx5a function [14,15] or downregulation of tbx5 target genes [35]. To get further insight into where in the limb developmental pathway tbx5b function is required, we used a series of markers to assess the state of the two tissues required for and involved in the process of fin outgrowth: the fin mesenchyme and the overlying fin ectoderm. Briefly, bi-directional fibroblast growth factor (FGF) signals emanate from and are received by both tissues, creating a positive feedback loop that is required to sustain pectoral fin outgrowth [10].
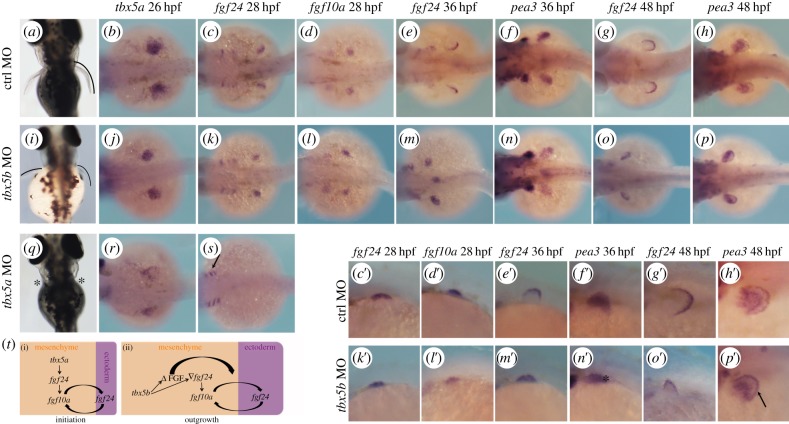
Figure 5.
tbx5b knock-down causes a delay in pectoral fin growth. (a,i,q) Pectoral fin morphology at 3 dpf. Dorsal views are shown with anterior to the top. (b–h) Expression of the developing pectoral fin markers in control (ctrl) MO-injected embryos. (c′–h′) Higher magnifications of (c–h). (j–p) Pectoral fin markers expression in tbx5b-morphant embryos. (k′–p′) Higher magnifications of (k–p). (r,s) tbx5a morphants. (t) Model for the differential requirements for the tbx5 genes during pectoral fin development. b–h, j–p,r,s are dorsal views with anterior to the left. c′–h′,k′–p′ are lateral views with anterior to the left.
In control embryos, compacted pectoral fin territories can be observed by means of tbx5a expression at 26 hpf (figure 5b) and expression of the tbx5a target gene, fgf24, is activated in this territory, namely the pectoral fin mesenchyme (figure 5c–c′). By contrast, tbx5a morphants failed to compact the tbx5a-labelled mesenchymal tissue (figure 5r) and fgf24 expression was never activated in the mesenchyme although its expression was readily detected in other tissues such as the branchial arches (figure 5s, arrow). Similar to control embryos, tbx5b morphants displayed compacted expression of tbx5a and fgf24 expression was activated in this territory (figure 5j,k–k′). fgf24 expressed in the pectoral fin bud mesenchyme is required to activate the expression of another FGF, namely fgf10a [36]. Activation of fgf10a expression is observed in both control and tbx5b morphants (figure 5d–d′,l–l′), indicating that the fin bud outgrowth initiation programme is properly established in tbx5b-compromised embryos. However, in contrast to the similarities observed between control and tbx5b morphants regarding mesenchymal FGF expression, it was striking to note that at 36 hpf, when fgf24 expression has been downregulated in this tissue and activated in the overlying fin ectoderm in control embryos, this did not occur after tbx5b depletion (figure 5e–e′,m–m′). To determine whether, indeed, the only tissue with active FGF signalling was the fin bud mesenchyme and not the overlying ectoderm of tbx5b morphants, we used pea3 expression, a direct read-out of cellular exposure to FGF, as a marker [37]. In control embryos, pea3 expression is detected in both mesenchymal and ectodermal compartments of the fin bud (figure 5f–f′), whereas only mesenchymal pea3 was detected after tbx5b knock-down (figure 5n–n′; the asterisk labels lack of ectodermal expression). Interestingly, ectodermal fgf24 was observed in 48 hpf tbx5b-morphant fins (figure 5o–o′) and these fins resembled control 36 hpf ones (figure 5e′,o′). In agreement with FGF signalling being active in both mesenchymal and ectodermal tissues of tbx5b-depleted fins, pea3 expression was now evident in these two tissues (figure 5p–p′; the arrow marks expression in the tbx5b-morphant ectoderm), similarly to what is found in control MO-injected embryos (figure 5h–h′).
Taken together, we show that pectoral fin development has a different requirement for each of the tbx5 paralogues: tbx5a function is required for the earliest steps of initiation of fin outgrowth, whereas tbx5b functions later to ensure properly timed and sustained fin outgrowth. Both these requirements are linked by the connection between tbx5 genes and the downstream regulation of FGF signalling. Briefly, during the initiation of pectoral fin outgrowth (figure 5t(i)), tbx5a expressed prior to overt fin outgrowth is required to initiate fgf24 signalling in the pectoral fin mesenchyme, and hence in the absence of tbx5a the limb initiation programme is never established and pectoral fins fail to form (figure 5q, asterisks; [14,15]). Later, once the limb initiation programme has commenced and FGF signalling is active in the pectoral fin mesenchyme, tbx5b is required for the maintenance of pectoral fin growth. tbx5b morphants have a delay in pectoral fin growth: these embryos fail to downregulate fgf24 expression in the fin bud mesenchyme and activate this gene expression in the overlaying ectoderm at 36 hpf. Nevertheless, 12 h later, fgf24 expression is no longer detected in the mesenchyme and becomes clearly observed in the ectodermal tissue. Strikingly, 48 hpf tbx5b-morphant pectoral fins resemble younger (36 hpf) control fins, suggesting that tbx5b is required to setup a certain threshold of FGF activity in the mesenchyme that is necessary to (i) signal to the overlying ectoderm and activate FGF signalling in this tissue and (ii) downregulate fgf24 expression in the mesenchyme (figure 5t(ii)). Thus, owing to this delay in FGF signalling activation in the ectoderm, tbx5b-deficient pectoral fins appear smaller than control fins (figure 5a,i). Given the critical requirement of tbx5a to establish the pectoral fin bud outgrowth initiation programme, it is not clear whether tbx5a may function, similarly to tbx5b, during these later stages of fin outgrowth. It is tempting to speculate that this is indeed the case, because, as mentioned before, the subtle downregulation of tbx5a function or its target genes is reminiscent of the tbx5b loss-of-function shown here.
3.5. Distinct tissue-specific requirements for tbx5a and tbx5b
Our characterization of the consequences of tbx5a and/or tbx5b knock-down in the three tissues where these genes are expressed demonstrates the existence of differential requirements for these paralogues in each tissue and distinct functional inter-relationships between the tbx5 genes.
Firstly, cardiac looping is affected in both single tbx5a and tbx5b morphants, and downregulation of both tbx5 genes does not increase the severity of the looping phenotype, indicating the essential function each of these genes plays to achieve the complete looping of the zebrafish heart. Moreover, we show that both genes act in the same pathway and cooperate with each other to ensure looping morphogenesis, because co-injection of sub-optimal concentrations of tbx5a and tbx5b MOs similarly causes looping phenotypes and increasing concentrations of both MOs caused an increase in the percentage of phenotypes (figure 1i). With regard to heart asymmetric development, we also show that both paralogues are essential for normal leftward heart tube jogging and consequent dextral looping to occur, because tbx5a and tbx5b morphants show jogging and looping orientation defects and co-inhibition of both genes does not result in either more severe or higher phenotypic penetrance (figures 1j and 2g). Secondly, a synergistic effect of tbx5a and tbx5b is necessary for proper efnb2a expression in the dorsal retina. efnb2a expression is not significantly affected in tbx5a or tbx5b morphants, whereas it is decreased by 50% in double tbx5a and tbx5b morphants, suggesting these two genes act together to guarantee the proper extent of efnb2a expression in this domain. Further, the optic nerves of double-morphants appear thinner than those of control siblings (figure 4). Finally, it is noteworthy that still another relationship between tbx5 genes is found regarding fin bud development, where tbx5a and tbx5b are differentially required to ensure the proper initiation of outgrowth first and maintenance of fin growth later, respectively (figure 5).
The tight regulation of Tbx5 gene dosage has been shown to be fundamental for many developmental processes to take place normally, because both the subtle upregulation and downregulation of its function has been shown to cause developmental defects [38–41]. Our data demonstrate that, moreover, tbx5 gene(s) dosage needs to be strongly controlled in a tissue-dependent manner to ensure the proper morphogenesis of the distinct tissues where this gene is most prominently expressed.
4. Material and methods
4.1. Animal welfare
The local ethics committee approved animal studies and all procedures conformed to the essential ethical rules and the current applicable legislation (Council Directive 86/609/EEC; Law 5/1995/GC; Order 214/1997/GC; Law 1201/2005/SG). Adult fish are kept in a designated fish facility with a designated manager and welfare officers. When animals need to be euthanized, an overdose of tricaine methane sulfonate (MS222, 200–300 mg l−1) by prolonged immersion was used, which is a well-established humane method.
4.2. Animal maintenance
Adult zebrafish were bred under standard conditions and embryos obtained by natural spawning and incubated at 28.5°C in E3 medium [42]. They were further staged and fixed at specific time-points as described by Kimmel et al. [43]. Wild-type and ath5:GFP [32] fish were used in this study.
4.3. Morpholino oligonucleotides
MO oligonucleotides (Gene Tools LLC) were dissolved to 1 mM and 0.5–3 ng injected into one-cell stage embryos. MOs were co-injected with an anti-p53 MO (7.5 ng) to avoid off-target effects caused by toxicity, and all experiments were performed with at least three independent replicates. The MOs used were: a control MO; an anti-tbx5a MO for the coding sequence [15]; an anti-tbx5b MO against the 5′UTR/coding sequence boundary—tbx5b(UTR) MO—5′ GGATTCGCCATATTCCCGTCTGAGT 3′; and an anti-tbx5b oligonucleotide for the exon 3/intron 4 boundary—tbx5b(SP) MO—5′ TTAAAAAACTAGGCACTCACCGGCC 3′. To test the knock-down efficiency of the tbx5b(SP) MO, RT-PCR was performed using whole-embryo RNA from 24 hpf embryos that had been injected with either control or tbx5b(SP) MO. RNA was isolated using Trizol reagent (Invitrogen) and a reverse transcription reaction with SuperScript II RNase H—reverse transcriptase (Invitrogen) was then performed to generate cDNA following the manufacturer's instructions. The PCR was performed using the primers zftbx5b_ex3fwd 5′ AGTATGGAGGGAATTAAAGTTTA 3′ (sequence present in the third exon of the tbx5b gene) and zftbx5b_ex4rev 5′ CATTTGTTATCTGCAAACTTATAC 3′ (present in the fourth exon of the tbx5b gene) to detect spliced and un-spliced tbx5b transcripts. As equivalent phenotypes were obtained with both tbx5b MOs, for most of the experiments results with the tbx5b(UTR) MO are shown, unless otherwise indicated.
4.4. Morpholino functionality and specificity
To assess for the functionality of the MOs used, we generated chimeric mRNAs in which the tbx5a or tbx5b(UTR) MO recognition sites (underlined) were fused to EGFP by PCR amplification using a plasmid containing EGFP-polyA as template. The following primers were used: tbx5aMO_EGFP_fwd, 5′ ATGGCGGACAGTGAAGACACCTTTCGGGTGAGCAAGGGCGAGGAGC 3′; tbx5bMO_EGFP_fwd, 5′ ACTCAGACGGGAATATGGCGAATCCAGTGAGCAAGGGCGAGGAGC 3′; in conjunction with the reverse primer: FP_SV40rev 5′ AAGCTTGATGAGTTTGGACAAACCAC 3′. The resulting products were cloned into the pGEM T-easy vector (Promega) and further transferred into the pCS2+ vector to obtain full-length mRNAs. The mMessage Machine kit (Ambion) was used to obtain full-length mRNAs according to the manufacturer's protocol. One hundred picograms of mRNA with or without the corresponding MO (3 ng) was injected into one-cell stage embryos and the presence of GFP expression assessed at 24 hpf.
To assess for the specificity of the MOs used and functionality of tbx5a variants, mRNAs to perform rescue experiments were generated. The following primers were used (mis-matched nucleotides are shown in small letters): tbx5a_fwd, 5′ ATGGCcGAttcaGAgGAtACgTTcaGGCTCCAAAACTCTCCCAGTG 3′ in conjunction with FLrev 5′ TTAGCTGGCTTCATTCCAGTC 3′, hstQ316rev 5′ CTaTGTGTGTCCGTGGTAGGAGC 3′ or T-boxtruncrev 5′ CTATGCTTTGGTGATGATCATCTCTG 3′ were used to generate full-length, heartstrings or truncated variants of tbx5a, respectively. tbx5b_fwd 5′ ATGGCcAAcCCAATGTTCGAATCTCTACGG 3′ and FLtbx5b_rev 5′ TCAACTCCCCCCACACCAGTTG 3′ were used to generate full-length tbx5b. Resulting products were cloned into the pGEM T-easy vector (Promega) and further transferred into the pCS2+ vector to obtain mRNAs. Eighty picograms of mRNA with the corresponding MO (3 ng) were co-injected into one-cell stage embryos and heart laterality assessed at 26 hpf by means of myl7 expression.
4.5. Whole mount in situ hybridizations
The antisense RNA probes used were: myl7 [44], lefty2 (kindly provided by N. Mercader), efnb2a (kindly provided by J. Terriente), ephB2 (kindly provided by R. Dorsky), tbx5a [16], fgf24 [36], fgf10a (obtained by RT-PCR on RNA from 24 hpf with the primers fwd 5′ ATGGAAAGTGACTAAGGGTGC 3′ and rev 5′ CTACACGATAGGAATGGGGAG 3′) and pea3 (obtained by RT-PCR with the primers fwd 5′ AGAAAGAGCCGCAGAGTCCC 3′ rev 5′ TCCTGTTTGACCATCATATGGG 3′).
Chromogenic whole mount in situ hybridizations were carried out as described by Albalat et al. [16]. Embryos were observed in an OLYMPUS MVX10 macroscope and photographed with the OLYMPUS CellD software. Fluorescent whole mount in situs were carried out as described by Brend et al. [45]. Embryos were embedded in 1% low melting agarose (Sigma) dissolved in PBS and observed in a Leica SP2 confocal microscope. Acquired images are projections of z-stacks.
4.6. Quantification of retinal phenotypes
The extent of efnb2a and ephB2 expression domain was quantified by setting an imaginary hinge in the centre of the lens. The total angle of expression was sub-divided into nasal versus temporal by considering the choroid fissure as the ventral-most point. The Kruskal–Wallis test was used to assess statistical differences among experimental conditions.
4.7. Immunofluorescence
ath5:GFP 48 hpf embryos were fixed in 4% paraformaldehyde (PFA) at 4°C, washed with PBST (0.5% Triton), digested with 10 mg ml−1 proteinase K for 40 min and post-fixed in 4% PFA for 20 min. After PBST washes, embryos were blocked with 1% BSA and an anti-GFP antibody (Invitrogen, 1 : 600) was subsequently left overnight at 4°C. The antibody was washed out with 1% BSA washes before adding the secondary antibody (anti-rabbit Alexa488 1 : 200, Molecular Probes) and left overnight at 4°C. Secondary antibody washes were performed with PBST. The acquired images are projections of z-stacks taken with a Leica SP2 confocal microscope.
Supplementary Material
Acknowledgements
We thank N. Mercader, J. Terriente and R. Dorsky for providing reagents and J. Giblin, N. Mercader, M. Logan, E. Nöel, D. Norris and C.M. and E.M-B's laboratories for fruitful discussions. We are indebted to Amayra Hernandez-Vega, Maria Marsal, Carla Prat and Marta Munar for experimental help and to Florencia Cavodeassi for kindly providing the ath5:GFP transgenic line. The authors declare that they have no conflict of interest.
Funding statement
Research was supported by a Spanish Ministerio de Economía y competitividad grant no. (BFU2011-24772). C.M. held a Ramón y Cajal contract and A.P-R. was supported by a ‘La Caixa’ Fellowship.
References
- 1.Begemann G, Ingham PW. 2000. Developmental regulation of Tbx5 in zebrafish embryogenesis. Mech. Dev. 90, 299–304. (doi:10.1016/S0925-4773(99)00246-4) [DOI] [PubMed] [Google Scholar]
- 2.Chapman DL, et al. 1996. Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev. Dyn. 206, 379–390. (doi:10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F) [DOI] [PubMed] [Google Scholar]
- 3.Gibson-Brown JJ, IAgulnik S, Silver LM, Papaioannou VE. 1998. Expression of T-box genes Tbx1–Tbx5 during chick organogenesis. Mech. Dev. 74, 165–169. (doi:10.1016/S0925-4773(98)00056-2) [DOI] [PubMed] [Google Scholar]
- 4.Basson CT, et al. 1997. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt–Oram syndrome. Nat. Genet. 15, 30–35. (doi:10.1038/ng0197-30) [DOI] [PubMed] [Google Scholar]
- 5.Li QY, et al. 1997. Holt–Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat. Genet. 15, 21–29. (doi:10.1038/ng0197-21) [DOI] [PubMed] [Google Scholar]
- 6.Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. 2003. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130, 623–633. (doi:10.1242/dev.00191) [DOI] [PubMed] [Google Scholar]
- 7.Bruneau BG, et al. 2001. A murine model of Holt–Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106, 709–721. (doi:10.1016/S0092-8674(01)00493-7) [DOI] [PubMed] [Google Scholar]
- 8.Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MP. 2003. Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130, 2741–2751. (doi:10.1242/dev.00473) [DOI] [PubMed] [Google Scholar]
- 9.Koshiba-Takeuchi K, et al. 2000. Tbx5 and the retinotectum projection. Science 287, 134–137. (doi:10.1126/science.287.5450.134) [DOI] [PubMed] [Google Scholar]
- 10.Mercader N. 2007. Early steps of paired fin development in zebrafish compared with tetrapod limb development. Dev. Growth Differ. 49, 421–437. (doi:10.1111/j.1440-169X.2007.00942.x) [DOI] [PubMed] [Google Scholar]
- 11.Glickman NS, Yelon D. 2002. Cardiac development in zebrafish: coordination of form and function. Semin. Cell Dev. Biol. 13, 507–513. (doi:10.1016/S1084952102001040) [DOI] [PubMed] [Google Scholar]
- 12.Bakkers J. 2011. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 91, 279–288. (doi:10.1093/cvr/cvr098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow RL, Lang RA. 2001. Early eye development in vertebrates. Annu. Rev. Cell Dev. Biol. 17, 255–296. (doi:10.1146/annurev.cellbio.17.1.255) [DOI] [PubMed] [Google Scholar]
- 14.Garrity DM, Childs S, Fishman MC. 2002. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development 129, 4635–4645. [DOI] [PubMed] [Google Scholar]
- 15.Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. 2002. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature 417, 754–758. (doi:10.1038/nature00814) [DOI] [PubMed] [Google Scholar]
- 16.Albalat R, Baquero M, Minguillon C. 2010. Identification and characterisation of the developmental expression pattern of tbx5b, a novel tbx5 gene in zebrafish. Gene Expr. Patterns 10, 24–30. (doi:10.1016/j.gep.2009.11.003) [DOI] [PubMed] [Google Scholar]
- 17.Parrie LE, Renfrew EM, Wal AV, Mueller RL, Garrity DM. 2013. Zebrafish tbx5 paralogs demonstrate independent essential requirements in cardiac and pectoral fin development. Dev. Dyn. 242, 485–502. (doi:10.1002/dvdy.23953) [DOI] [PubMed] [Google Scholar]
- 18.Chen JN, van Eeden FJ, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. 1997. Left–right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124, 4373–4382. [DOI] [PubMed] [Google Scholar]
- 19.Veerkamp J, Rudolph F, Cseresnyes Z, Priller F, Otten C, Renz M, Schaefer L, Abdelilah-Seyfried S. 2013. Unilateral dampening of Bmp activity by nodal generates cardiac left–right asymmetry. Dev. Cell 24, 660–667. (doi:10.1016/j.devcel.2013.01.026) [DOI] [PubMed] [Google Scholar]
- 20.Lenhart KF, Holtzman NG, Williams JR, Burdine RD. 2013. Integration of nodal and BMP signals in the heart requires FoxH1 to create left–right differences in cell migration rates that direct cardiac asymmetry. PLoS Genet. 9, e1003109 (doi:10.1371/journal.pgen.1003109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plageman TF, Jr, Yutzey KE. 2006. Microarray analysis of Tbx5-induced genes expressed in the developing heart. Dev. Dyn. 235, 2868–2880. (doi:10.1002/dvdy.20923) [DOI] [PubMed] [Google Scholar]
- 22.Gruenauer-Kloevekorn C, Reichel MB, Duncker GI, Froster UG. 2005. Molecular genetic and ocular findings in patients with Holt-Oram syndrome. Ophthalmic Genet. 26, 1–8. (doi:10.1080/13816810590918073) [DOI] [PubMed] [Google Scholar]
- 23.French CR, Erickson T, French DV, Pilgrim DB, Waskiewicz AJ. 2009. Gdf6a is required for the initiation of dorsal-ventral retinal patterning and lens development. Dev. Biol. 333, 37–47. (doi:10.1016/j.ydbio.2009.06.018) [DOI] [PubMed] [Google Scholar]
- 24.Mann F, Ray S, Harris W, Holt C. 2002. Topographic mapping in dorsoventral axis of the Xenopus retinotectal system depends on signaling through ephrin-B ligands. Neuron 35, 461–473. (doi:10.1016/S0896-6273(02)00786-9) [DOI] [PubMed] [Google Scholar]
- 25.Hindges R, McLaughlin T, Genoud N, Henkemeyer M, O'Leary D. 2002. EphB forward signaling controls directional branch extension and arborization required for dorsal–ventral retinotopic mapping. Neuron 35, 475–487. (doi:10.1016/S0896-6273(02)00799-7) [DOI] [PubMed] [Google Scholar]
- 26.Erickson T, French CR, Waskiewicz AJ. 2010. Meis1 specifies positional information in the retina and tectum to organize the zebrafish visual system. Neural Dev. 5, 22 (doi:10.1186/1749-8104-5-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greulich F, Rudat C, Kispert A. 2011. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 91, 212–222. (doi:10.1093/cvr/cvr112) [DOI] [PubMed] [Google Scholar]
- 28.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. 2005. T-box genes in vertebrate development. Annu. Rev. Genet. 39, 219–239. (doi:10.1146/annurev.genet.39.073003.105925) [DOI] [PubMed] [Google Scholar]
- 29.Behesti H, Holt JK, Sowden JC. 2006. The level of BMP4 signaling is critical for the regulation of distinct T-box gene expression domains and growth along the dorso-ventral axis of the optic cup. BMC Dev. Biol. 6, 62 (doi:10.1186/1471-213X-6-62) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross JM, Dowling JE. 2005. Tbx2b is essential for neuronal differentiation along the dorsal/ventral axis of the zebrafish retina. Proc. Natl Acad. Sci. USA 102, 4371–4376. (doi:10.1073/pnas.0501061102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse-Bend R, Rosenthal J, Quist TS, Veien ES, Fuhrmann S, Dorsky RI, Chien CB. 2012. Extraocular ectoderm triggers dorsal retinal fate during optic vesicle evagination in zebrafish. Dev. Biol. 371, 57–65. (doi:10.1016/j.ydbio.2012.08.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masai I, et al. 2003. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development 130, 2479–2494. (doi:10.1242/dev.00465) [DOI] [PubMed] [Google Scholar]
- 33.Gyda M, Wolman M, Lorent K, Granato M. 2012. The tumor suppressor gene retinoblastoma-1 is required for retinotectal development and visual function in zebrafish. PLoS Genet. 8, e1003106 (doi:10.1371/journal.pgen.1003106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vance KW, Shaw HM, Rodriguez M, Ott S, Goding CR. 2010. The retinoblastoma protein modulates Tbx2 functional specificity. Mol. Biol. Cell 21, 2770–2779. (doi:10.1091/mbc.E09-12-1029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey SA, Logan MP. 2006. sall4 acts downstream of tbx5 and is required for pectoral fin outgrowth. Development 133, 1165–1173. (doi:10.1242/dev.02259) [DOI] [PubMed] [Google Scholar]
- 36.Fischer S, Draper BW, Neumann CJ. 2003. The zebrafish fgf24 mutant identifies an additional level of Fgf signaling involved in vertebrate forelimb initiation. Development 130, 3515–3524. (doi:10.1242/dev.00537) [DOI] [PubMed] [Google Scholar]
- 37.Raible F, Brand M. 2001. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech. Dev. 107, 105–117. (doi:10.1016/S0925-4773(01)00456-7) [DOI] [PubMed] [Google Scholar]
- 38.Chiavacci E, Dolfi L, Verduci L, Meghini F, Gestri G, Evangelista AM, Wilson SW, Cremisi F, Pitto L. 2012. MicroRNA 218 mediates the effects of Tbx5a over-expression on zebrafish heart development. PLoS ONE 7, e50536 (doi:10.1371/journal.pone.0050536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDermott DA, Hatcher CJ, Basson CT. 2008. Atrial fibrillation and other clinical manifestations of altered TBX5 dosage in typical Holt–Oram syndrome. Circ. Res. 103, e96 (doi:10.1161/CIRCRESAHA.108.181834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori AD, et al. 2006. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 297, 566–586. (doi:10.1016/j.ydbio.2006.05.023) [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi JK, et al. 2011. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2, 187 (doi:10.1038/ncomms1187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westerfield M. 2000. A guide for the laboratory use of zebrafish (Danio rerio), 4th edn Eugene, OR: University of Oregon Press. [Google Scholar]
- 43.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. (doi:10.1002/aja.1002030302) [DOI] [PubMed] [Google Scholar]
- 44.Yelon D, Horne SA, Stainier DY. 1999. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23–37. (doi:10.1006/dbio.1999.9406) [DOI] [PubMed] [Google Scholar]
- 45.Brend T, Holley SA. 2009. Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J. Vis. Exp. 25, e1229 (doi:10.3791/1229) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.