Abstract
CENP-A chromatin forms the foundation for kinetochore assembly. Replication-independent incorporation of CENP-A at centromeres depends on its chaperone HJURPScm3, and Mis18 in vertebrates and fission yeast. The recruitment of Mis18 and HJURPScm3 to centromeres is cell cycle regulated. Vertebrate Mis18 associates with Mis18BP1KNL2, which is critical for the recruitment of Mis18 and HJURPScm3. We identify two novel fission yeast Mis18-interacting proteins (Eic1 and Eic2), components of the Mis18 complex. Eic1 is essential to maintain Cnp1CENP-A at centromeres and is crucial for kinetochore integrity; Eic2 is dispensable. Eic1 also associates with Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21, components of the constitutive CCAN/Mis6/Ctf19 complex. No Mis18BP1KNL2 orthologue has been identified in fission yeast, consequently it remains unknown how the key Cnp1CENP-A loading factor Mis18 is recruited. Our findings suggest that Eic1 serves a function analogous to that of Mis18BP1KNL2, thus representing the functional counterpart of Mis18BP1KNL2 in fission yeast that connects with a module within the CCAN/Mis6/Ctf19 complex to allow the temporally regulated recruitment of the Mis18/Scm3HJURP Cnp1CENP-A loading factors. The novel interactions identified between CENP-A loading factors and the CCAN/Mis6/Ctf19 complex are likely to also contribute to CENP-A maintenance in other organisms.
Keywords: CENP-A, centromeres, epigenetics, fission yeast, Mis18
2. Introduction
Genome integrity and stability are key to the propagation of genetic information through generations. Centromeres are the specialized sites on eukaryotic chromosomes where kinetochores, which govern spindle microtubule attachment, assemble during cell division. Centromere dysfunction, and consequent kinetochore instability, can therefore cause mis-segregation of chromosomes resulting in aneuploidy and genome instability. A plethora of evidence suggests that DNA sequence is neither necessary nor sufficient for the establishment of a functional centromere [1,2], and it is generally accepted that centromere specification is epigenetically regulated in many organisms [3,4].
CENP-A, a histone H3 variant, replaces canonical H3 in specialized nucleosomes that mark active centromeres. CENP-A is both necessary and sufficient to mediate the establishment and maintenance of functional kinetochores. CENP-A associates with a distinct set of proteins that allow it to promote its own propagation to preserve centromere location [5–7]. In the fission yeast Schizosaccharomyces pombe, Cnp1CENP-A is restricted to the non-repetitive central domain of centromeres (approx. 7–10 kb; unique central core plus inverted inner-most repeats) that is flanked by heterochromatic outer repeats. Cnp1CENP-A recruitment to the central domain depends on the conserved kinetochore proteins Mis18, Mis6CENP-I/Ctf3, Sim4CENP-K/Mcm22 and Scm3HJURP [8–12]. Mis18 was originally identified as being required to maintain Cnp1CENP-A at fission yeast centromeres. Subsequently, Mis18 function was shown to be conserved at vertebrate centromeres [11,13]. Mis18 associates with the nuclear protein Mis16, which is orthologous to Saccharomyces cerevisiae Hat2 and human RbAp46/48, which are known to act as histone chaperones in many distinct histone transactions [14,15]. Mis6CENP-I/Ctf3 and Sim4CENP-K/Mcm22 reside in a larger complex consisting of approximately 16–18 centromeric proteins [16–18], most of which are conserved and collectively form the complex known as CCAN, Ctf19 and Mis6 in vertebrates, budding yeast and fission yeast, respectively [16,19,20]. The modular CCAN/Mis6/Ctf19 complex forms the main scaffold of centromere proteins that is required to recruit outer kinetochore components in order to mediate and regulate kinetochore–microtubule attachments [21]. Scm3HJURP is orthologous to the vertebrate CENP-A-specific chaperone HJURP [22]. Scm3HJURP directly associates with Cnp1CENP-A and is required for Cnp1CENP-A incorporation at centromeres [9,12].
In vertebrate cells, the assembly of new CENP-A into centromeric chromatin is uncoupled from replication and occurs in early G1, prior to S phase [23]. Consequently, following centromere replication, the level of resident CENP-A at centromeres is halved as a result of its distribution to the resulting two sister-centromeres [24]. The CENP-A loading factors Mis18α/β and Mis18BP1KNL2 and the CENP-A chaperone HJURPScm3 are transiently recruited to human centromeres in telophase and are required to replenish CENP-A in G1 [13,25,26]. Mis18 is recruited to human centromeres by Mis18BP1KNL2, which associates with centromeres via the C-terminus of the constitutive kinetochore component, CENP-CCnp3/Mif2 [27,28]. Both Mis18α and Mis18BP1KNL2 are required for the cell-cycle-regulated recruitment of HJURPScm3 prior to G1 [25,28,29]. Interestingly, in fission yeast, the CENP-C orthologue Cnp3 is not essential for viability [30] and no Mis18BP1KNL2 protein can be identified. It therefore remains unclear how Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP are recruited to centromeres to maintain Cnp1CENP-A at fission yeast centromeres. Nevertheless, as in vertebrate cells, the Cnp1CENP-A assembly factors Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP all display dynamic association with fission yeast centromeres during the cell cycle, but they are released from centromeres in mitotic prophase and re-associate in mid-anaphase [9,11–13]. This suggests that fission yeast relies on an alternative to the CENP-CCnp3/Mif2–Mis18BP1KNL2 interaction for the regulated recruitment of the Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP Cnp1CENP-A assembly factors to centromeres.
Although many kinetochore proteins have clearly been conserved between different experimental organisms over evolutionary time, it is also evident that particular proteins have been lost in some organisms and replaced by alternative pathways. For example, both Drosophila and S. cerevisiae lack the Mis18 and Mis18BP1KNL2 proteins. However, S. cerevisiae retains an Scm3HJURP orthologue, whereas Scm3HJURP function is replaced by the distinct Cal1 protein in Drosophila [31,32]. Moreover, in Arabidopsis the association dynamics of Mis18BP1KNL2 with centromeres is more similar to that of S. pombe Mis18/Scm3HJURP than human Mis18BP1KNL2/Mis18/HJURPScm3 in that it departs only briefly from centromeres during mitosis, re-associating in mid-anaphase prior to Arabidopsis CenH3CENP-A deposition in G2 [33,34]. It is well recognized that alternative solutions can evolve to mediate the same conserved process, and such alternatives can inform on parallel or other underlying pathways and mechanisms that may be more prevalent in one system relative to another.
In fission yeast, Mis16RbAp46/48/Hat2–Mis18 may promote Cnp1CENP-A assembly through a direct physical interaction detected in vitro with the Cnp1CENP-A chaperone Scm3HJURP [9,12]. However, as no direct interaction has been reported between Mis16RbAp46/48/Hat2–Mis18 and constitutive CCAN components, it remains unknown how the Mis16RbAp46/48/Hat2–Mis18 and Scm3HJURP assembly factors are recruited to the underlying constitutive kinetochore scaffold. Here, we set out to determine how fission yeast solves the problem of recruiting the Cnp1CENP-A loading machinery (Mis16RbAp46/48/Hat2/Mis18/Scm3HJURP) to centromeres without a Mis18BP1KNL2 orthologue. Through a proteomics approach, we identified two previously uncharacterized proteins as additional integral components of the Mis16RbAp46/48/Hat2-Mis18 complex, designated Eic1 and Eic2 (Eighteen Interacting Centromere proteins 1 and 2). Our analyses indicate that Eic1 is essential to maintain normal Cnp1CENP-A levels at centromeres, whereas Eic2 is dispensable. Importantly, we also demonstrate that Eic1 provides a link between Mis16RbAp46/48/Hat2-Mis18 and specific constitutive components of the kinetochore scaffold (CCAN/Mis6/Ctf19 complex components). The analyses presented identify three components within the conserved CCAN/Mis6/Ctf19 complex which we propose represent a module that is required to recruit the Cnp1CENP-A loading machinery (Mis16RbAp46/48/Hat2/Mis18/Eic1/Scm3HJURP) to fission yeast centromeres.
3. Results
3.1. Identification of novel proteins that associate with Mis16RbAp46/48/Hat2 or Mis18
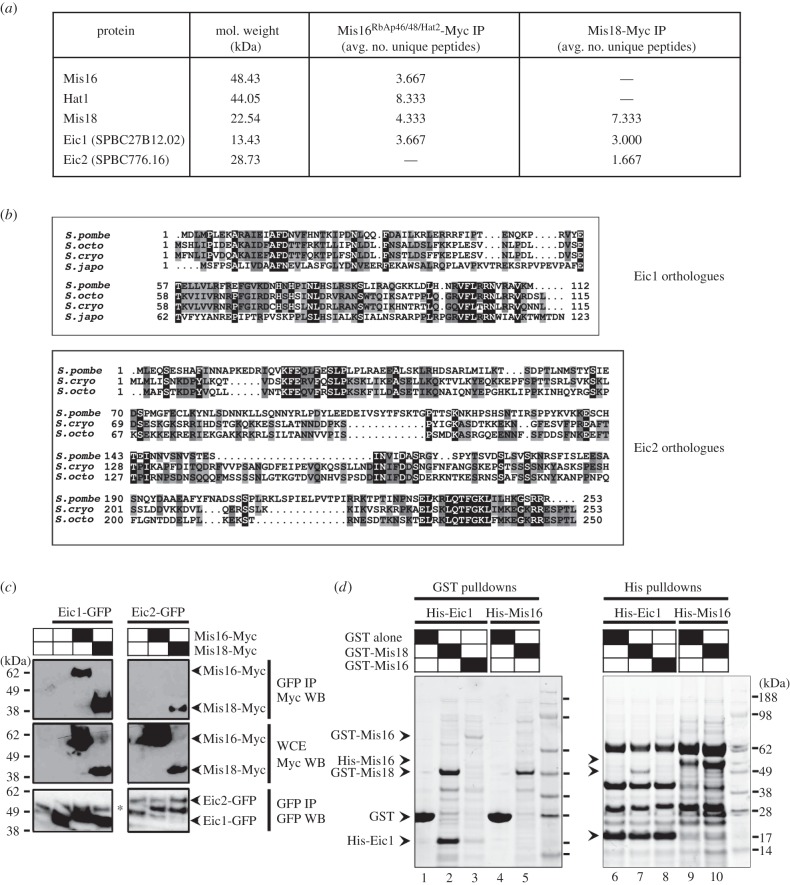
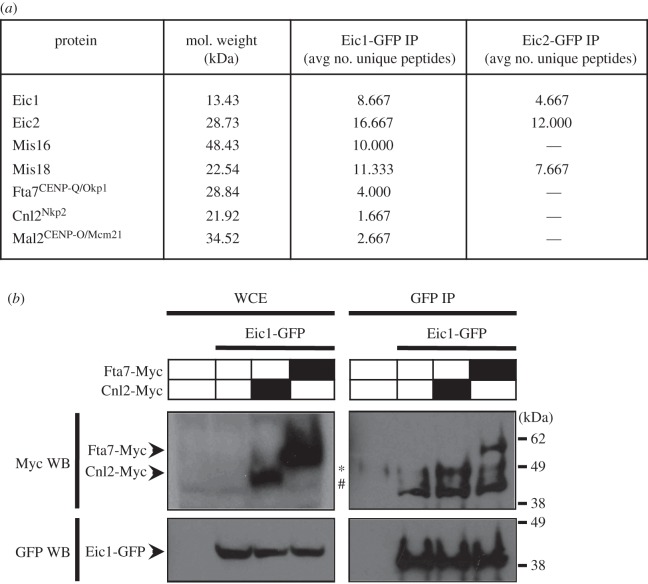
To explore the interactions of Mis16RbAp46/48/Hat2 and Mis18 with other proteins in fission yeast, Myc-tagged Mis16 and Mis18 were immunoprecipitated from extracts of cells expressing either Mis16-Myc or Mis18-Myc as fusion proteins from the endogenous genes and the captured proteins subjected to LC-MS/MS analyses. Mis18 immunoprecipitates were reproducibly found to contain two previously uncharacterized proteins, which we named Eic1 (SPBC27B12.02) and Eic2 (SPBC776.16) for Eighteen Interacting Centromere protein 1 and 2, respectively. Mis16RbAp46/48/Hat2 immunoprecipitates also reproducibly contained Eic1, along with two other known interactors: Mis18 and the histone H4 acetyltransferase Hat1 (figure 1a) [11,35]. Proteins orthologous to Eic1 were identifiable in all the sequenced genomes of the three other fission yeast species (S. octosporus, S. cryophilus and S. japonicus) by homology and synteny searches (figure 1b). However, no orthologue of Eic2 was apparent in S. japonicus (figure 1b).
Figure 1.
Eic1 and Eic2 are Mis18-interacting proteins. (a) LC-MS/MS analysis of Myc-tagged Mis16RbAp46/48/Hat2 and Mis18 immunoprecipitates from S. pombe whole cell extracts identifies two previously uncharacterized proteins, Eic1 (SPBC27B12.02) and Eic2 (SPBC776.16). Average number of unique peptides reproducibly identified from three independent experiments is shown. (b) Primary sequence alignments of Eic1 (top) and Eic2 (bottom) orthologues identified among Schizosaccharomyces species. (c) Eic1-GFP co-immunoprecipitates with both Mis16-Myc and Mis18-Myc, while Eic2-GFP only co-immunoprecipitates with Mis18-Myc. The asterisk (*) in the bottom panel denotes the IgG heavy chain. (d) Eic1 directly interacts with Mis18 and Mis16. 6xHis-Eic1 was co-expressed with GST alone, GST-Mis18 or GST-Mis16 in E. coli. Coomassie-stained SDS-PAGE gels showing reciprocal GST (lanes 1–3) and His (lanes 6–8) pulldowns from E. coli lysates are shown. Also shown are reciprocal pulldowns of 6xHis-Mis16 co-expressed with GST alone or GST-Mis18 (lanes 4–5 and 9–10).
Co-immunoprecipitation experiments verified the interactions of Eic1 and Eic2 with Mis16RbAp46/48/Hat2 and Mis18. Consistent with the LC-MS/MS results, both Mis16-Myc and Mis18-Myc were found to co-immunoprecipitate with Eic1-GFP. Also, Mis18-Myc, but not Mis16-Myc, co-immunoprecipitated with Eic2-GFP (figure 1c). This demonstrates that Mis16RbAp46/48/Hat2, Mis18, Eic1 and Eic2 associate.
To determine whether Eic1 can directly associate with Mis16RbAp46/48/Hat2 or Mis18, we co-expressed 6xHis-Eic1 with GST-Mis16 or GST-Mis18 in Escherichia coli. GST-Mis18 and 6xHis-Eic1 can clearly associate with each other in reciprocal pull down assays (figure 1d, lanes 2 and 7). 6xHis-Eic1 could also be detected in pulldowns of GST-Mis16 (figure 1d, lane 3). Similarly, a weak interaction between 6xHis-Mis16 and GST-Mis18 could be detected in vitro (figure 1d, lane 5 when compared with lanes 4 or 2).
3.2. Hat1 does not contribute to the maintenance of Cnp1CENP-A chromatin
In S. cerevisiae, Hat2 is the orthologue of Mis16 and it has previously been shown to associate with the histone H4 acetyl-transferase Hat1 [36]. Related to this, levels of acetylated histones have been shown to increase at centromeres in mis16 and mis18 mutants [11]. It is possible that altered histone acetylation at centromeres relates to the function of Mis16RbAp46/48/Hat2 and Hat1 at centromeres. We first confirmed that Mis16-Myc and Hat1-HA co-immunoprecipitate (electronic supplementary material, figure S1a) and then, to investigate the possibility that Hat1 possesses centromere-specific functions, we subjected Hat1-HA immunoprecipitates to LC/MS-MS analyses. However, we were unable to detect any Hat1-associated proteins apart from Mis16RbAp46/48/Hat2, confirming the analyses of others [35]. Moreover, Hat1-HA was not found to be enriched in the central kinetochore domain of centromeres (electronic supplementary material, figure S1b) and no loss of Cnp1CENP-A from centromeres could be detected in cells lacking Hat1 (hat1Δ; electronic supplementary material, figure S1c). Thus, it appears that Mis16RbAp46/48/Hat2 may participate in two distinct functional complexes: Mis16RbAp46/48/Hat2–Mis18–Eic1–Eic2 at centromeres (see below) and Mis16RbAp46/48/Hat2–Hat1 elsewhere in the nucleus (electronic supplementary material, figure S1d). Such a multi-functional role for Mis16RbAp46/48/Hat2 would be consistent with the known involvement of RbAp46/48 proteins in many complexes involved in histone modification and chromatin remodelling [14,15].
3.3. Eic1 and Eic2 associate with centromeres dynamically through the cell cycle
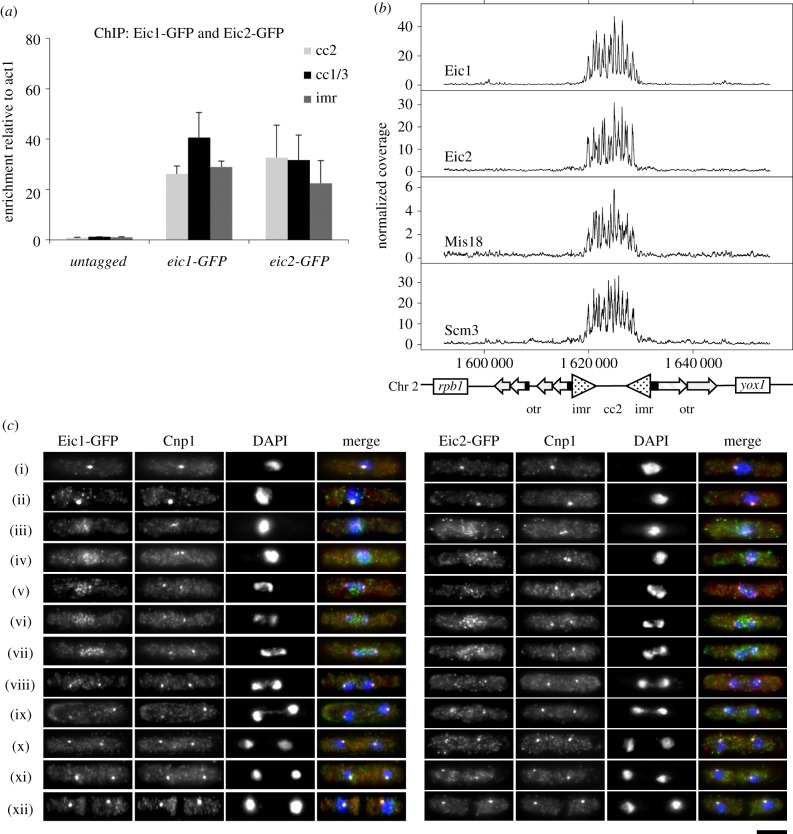
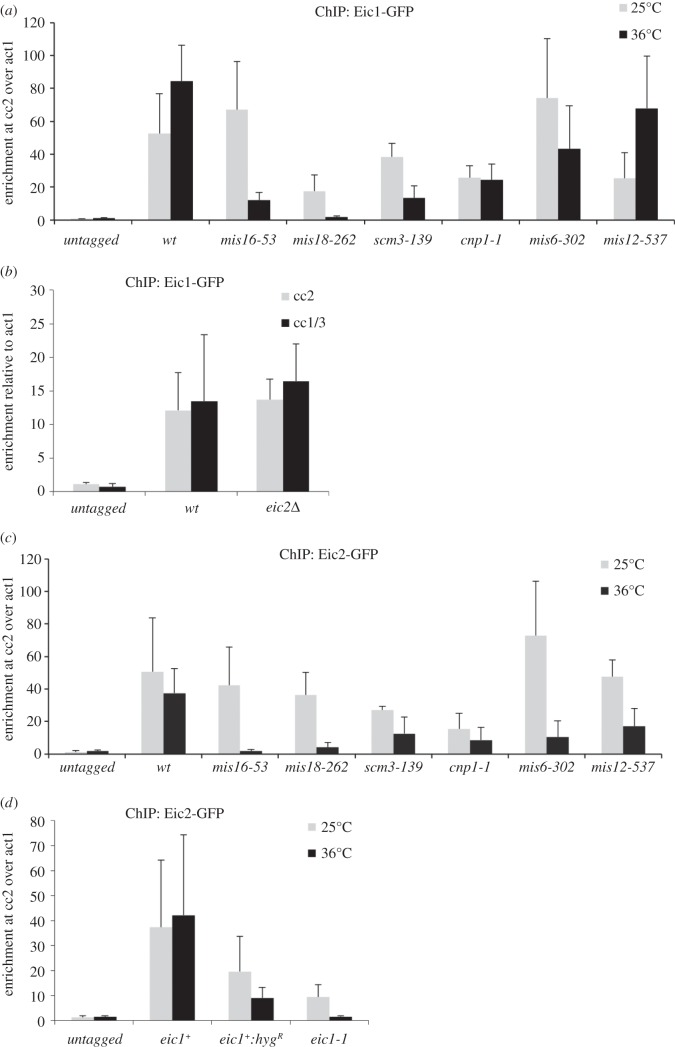
CENP-A and all known fission yeast kinetochore proteins localize specifically at centromeres and are enriched over the central domain region of centromeres. Both Eic1 and Eic2 associate with Mis18, which is known to localize to fission yeast centromeres for most of the cell cycle, apart from early prophase to mid-anaphase of mitosis [13]. To examine the localization of Eic1 and Eic2, the endogenous genes expressing Eic1 and Eic2 were fused to GFP. Quantitative chromatin immunoprecipitation (qChIP) analyses demonstrated that, as with Mis18 and other kinetochore proteins, both Eic1-GFP and Eic2-GFP are enriched over the central kinetochore domain (central core plus imr repeats) of fission yeast centromeres (figure 2a). Genome-wide ChIP-seq analyses confirmed this finding and also demonstrated that Eic1-GFP and Eic2-GFP, much like Mis18-GFP and Scm3-GFP, are undetectable at other regions of the genome including the heterochromatic outer repeats of centromeres (figure 2b and electronic supplementary material, figure S2). Immunostaining revealed that both Eic1 and Eic2 GFP-tagged proteins co-localize with Cnp1CENP-A at clustered centromeres in interphase cells (figure 2c(i),(ii)). However, as with Mis16RbAp46/48/Hat2 and Mis18 [11,13], Eic1-GFP and Eic2-GFP dissociate from centromeres in early mitosis (prometaphase), just as the centromeres (Cnp1) begin to form two separate clusters as they biorient on the spindle (figure 2c(iii),(iv)). Moreover, both Eic1-GFP and Eic2-GFP reassociate with centromeres in mid-anaphase when centromeres (Cnp1) and chromosomes (DAPI) have clearly segregated to opposite spindle poles (figure 2c(viii),(ix)). Apart from Mis16RbAp46/48/Hat2 and Mis18, the CENP-A chaperone Scm3HJURP exhibits similar temporal dissociation–reassociation at centromeres during mitosis [9,12]. Thus, our analyses identify Eic1 and Eic2 as two additional proteins that are released from centromeres in early mitosis and reloaded on centromeres in mid-anaphase. The finding that Eic1 and Eic2 exhibit similar association dynamics as Mis18 through the cell cycle is consistent with our identification of Eic1 and Eic2 being tightly associated with Mis18.
Figure 2.
Eic1 and Eic2 associate specifically with centromeres. (a) Eic1 and Eic2 bind the central domain of S. pombe centromeres. qChIP analyses showing enrichments of GFP-tagged Eic1 and Eic2 at the central cores (cc) of centromeres 1, 2 and 3 and imr repeats of centromere 1, relative to the act1 locus. Error bars represent standard deviation between at least three biological replicates. (b) Eic1 and Eic2 exhibit very similar genome-wide association profiles as Mis18 and Scm3HJURP. A comparison between the ChIP-seq profiles of GFP-tagged Eic1, Eic2, Mis18 and Scm3 across centromere 2 is presented, alongside a schematic diagram of centromere 2 (bottom). Normalized coverage represents the number of sequencing fragments obtained from anti-GFP IP normalized to that obtained from the input. (c) Eic1 and Eic2 exhibit very similar cell-cycle localization dynamics as Mis18 and Scm3HJURP. Immunofluorescence of S. pombe cells expressing GFP-tagged Eic1 or Eic2 stained with antibodies to GFP (green) and Cnp1CENP-A (red), and DAPI (blue). Both Eic1 and Eic2 dissociate from centromeres during prometaphase to mid-anaphase of mitosis ((iii)–(vii)) and subsequently reassociate. Scale bar, 5 μm.
3.4. Eic1 is required to maintain Cnp1CENP-A at centromeres
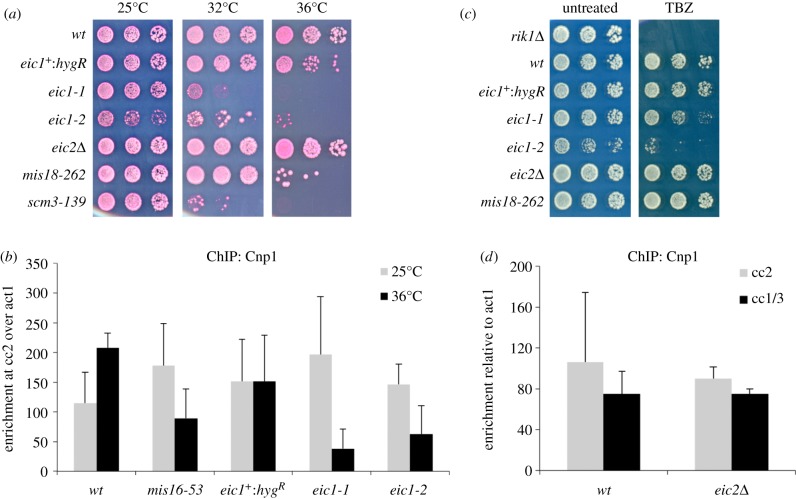
Mis18 and associated proteins have been shown to be required to maintain CENP-A at centromeres in fission yeast and vertebrates [11,13,37]. If Eic1 and Eic2 are critical for Mis18 function then they should also be required to maintain normal Cnp1CENP-A levels at centromeres. As the eic1+ gene is essential for cell viability, we generated a conditional temperature-sensitive (ts) mutant of eic1 (eic1-1:hygR, hereafter referred to as eic1-1) in which a single amino acid substitution (Phe102Ser) at a conserved residue rendered cells inviable at 36°C but with retained viability at 25°C (figure 3a). We also generated a corresponding wild-type allele of eic1 (eic1+:hygR) as a control in which, as with eic1-1, a hygromycin resistance marker was inserted within the 3′UTR of eic1 at its endogenous locus for ease of genetic manipulation (figure 3a). qChIP analyses demonstrated that Cnp1CENP-A levels were significantly diminished at centromeres in eic1-1 cells at restrictive temperature (figure 3b; electronic supplementary material, figure S3a). Cells harbouring a second ts allele of eic1 (eic1-2:hygR; two substituted residues, Lys37Glu and Tyr55His, hereafter referred to as eic1-2) also demonstrated significant loss of Cnp1CENP-A from centromeres at 36°C (figure 3a,b; electronic supplementary material, figure S3a). Furthermore, eic1-1 and eic1-2 cells displayed sensitivity to the microtubule depolymerizing drug thiabendazole (TBZ), whereas eic1+:hygR and the mis18-262 ts mutant did not confer TBZ sensitivity (figure 3c). TBZ sensitivity suggests that kinetochore–microtubule interactions are defective in eic1 mutants. Aberrant kinetochore function in eic1-1 cells was supported by cytological analyses, which revealed severe chromosome segregation defects (electronic supplementary material, figure S3b). qChIP analyses demonstrated that GFP-tagged Eic1-1 and Eic1-2 mutant proteins remained associated with centromeres even at 36°C; thus, the observed phenotypes of eic1-1 and eic1-2 cells are not a consequence of the complete absence of Eic1 protein at centromeres (electronic supplementary material, figure S3c).
Figure 3.
Eic1 is required for Cnp1CENP-A assembly, while Eic2 is dispensable. (a) ts mutations in eic1 affect cell viability, while eic2Δ cells show no defects in growth. Five-fold serial dilutions of cells spotted on YES + Phloxine B media and incubated at the indicated temperatures; dead cells stain dark pink. (b) eic1 mutants display reduced Cnp1CENP-A levels at centromeres. qChIP analyses of Cnp1CENP-A association with centromeres in the indicated strains when grown at permissive (25°C) versus restrictive temperature (36°C) for 8 h. Enrichment of cc2 DNA relative to the act1 locus is presented. (c) eic1 mutants display sensitivity to TBZ, while eic2Δ cells show no TBZ sensitivity. Five-fold serial dilutions of cells spotted on YES media (untreated) or YES media supplemented with 12.5 μg ml−1 TBZ, and incubated at 25°C. (d) eic2Δ cells display no loss of Cnp1CENP-A at centromeres. qChIP analyses of Cnp1CENP-A association with centromeres in the indicated strains when grown at 32°C. Enrichment of cc2 or cc1/3 DNA relative to the act1 locus is presented. Error bars in (b,d) represent standard deviation between at least three biological replicates.
In contrast to eic1 mutants, cells lacking the eic2+ gene (eic2Δ) displayed no loss of Cnp1CENP-A from centromeres (figure 3d). One possibility is that Eic2 is not required to maintain Cnp1CENP-A at centromeres but is required to establish Cnp1CENP-A on naive centromeric DNA. To test this, a plasmid bearing centromeric DNA that efficiently establishes Cnp1CENP-A chromatin and functional kinetochores in wild-type cells (pH-cc2) [38] was transformed into eic2Δ cells. No defect in the establishment of Cnp1CENP-A chromatin or functional kinetochores was observed (electronic supplementary material, figure S4). Furthermore, eic2Δ cells were not sensitive to TBZ (figure 3c). We conclude that Eic1 is required for Cnp1CENP-A maintenance and kinetochore integrity, whereas Eic2 is dispensable.
3.5. The recruitment of Cnp1CENP-A assembly factors is reduced at centromeres in eic1 and eic2 mutants
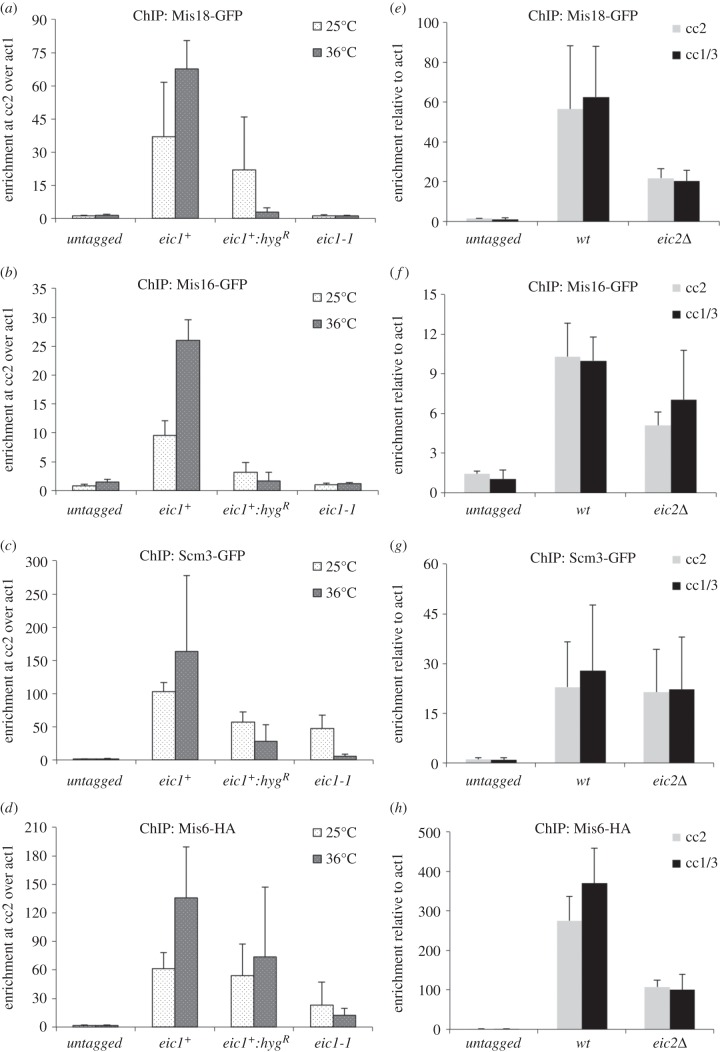
Previous studies have shown that defective Mis16RbAp46/48/Hat2 or Mis18 function affects the localization of the Cnp1CENP-A loading factors Scm3HJURP and Mis6CENP-I/Ctf3 at centromeres [9,11,12]. Vertebrate Mis18α/β and Mis18BP1KNL2 have also been shown to be required for the recruitment of the CENP-A chaperone HJURPScm3 to centromeres, and consequently the incorporation of CENP-A at centromeres [13,25]. We find that the levels of GFP-tagged Mis16RbAp46/48/Hat2 and Mis18 associated with fission yeast centromeres are greatly reduced in the eic1-1 mutant even at 25°C (figure 4a,b), consistent with the observed synthetic genetic interaction between eic1-1 and the GFP-tagged alleles of mis16 and mis18 (electronic supplementary material, figure S5). Additionally, we find that Scm3HJURP association with centromeres is also dependent on Eic1 function (figure 4c). Interestingly, we find Mis6CENP-I/Ctf3 association with centromeres to be only partially dependent on Eic1 (figure 4d), as intermediate levels of Mis6CENP-I/Ctf3 are retained at centromeres even when Eic1 function is compromised. Thus Eic1, in conjunction with its interacting partners Mis16RbAp46/48/Hat2 and Mis18, is essential to maintain normal levels of Cnp1CENP-A as well as Cnp1CENP-A loading factors on centromeres.
Figure 4.
Eic1 and Eic2 promote normal levels of association of Cnp1CENP-A assembly factors with centromeres. (a,b) Mis18-GFP and Mis16-GFP association with centromeres is entirely dependent on Eic1, and (e,f) partly dependent on Eic2. (c) Scm3-GFP association with centromeres is dependent on Eic1, and (g) largely independent of Eic2. (d) Mis6-HA association with centromeres is partly dependent on Eic1, and (h) Eic2. qChIP analyses of (a,e) Mis18-GFP, (b,f) Mis16-GFP, (c,g) Scm3-GFP and (d,h) Mis6-HA association with centromeres in the indicated strains when grown at permissive (25°C) versus restrictive temperature (36°C) for 8 h (a–d); or at 32°C (e–h). Enrichment of cc2 DNA relative to the act1 locus is presented in (a–d). Enrichment of cc2 or cc1/3 DNA relative to the act1 locus is presented in (e–h). Error bars represent standard deviation between at least three biological replicates.
Surprisingly, eic2Δ cells exhibited reduced levels of Mis18, Mis16RbAp46/48/Hat2 and Mis6CENP-I/Ctf3 at centromeres, whereas Scm3HJURP remained unaffected (figure 4e–h). As normal levels of Cnp1CENP-A are retained at centromeres and no defect in chromosome segregation was detectable in eic2Δ cells (figure 3c,d and electronic supplementary material, figure S4), the observed reduction of Mis18, Mis16RbAp46/48/Hat2 and Mis6CENP-I/Ctf3 at centromeres must not be sufficient to significantly affect Cnp1CENP-A maintenance.
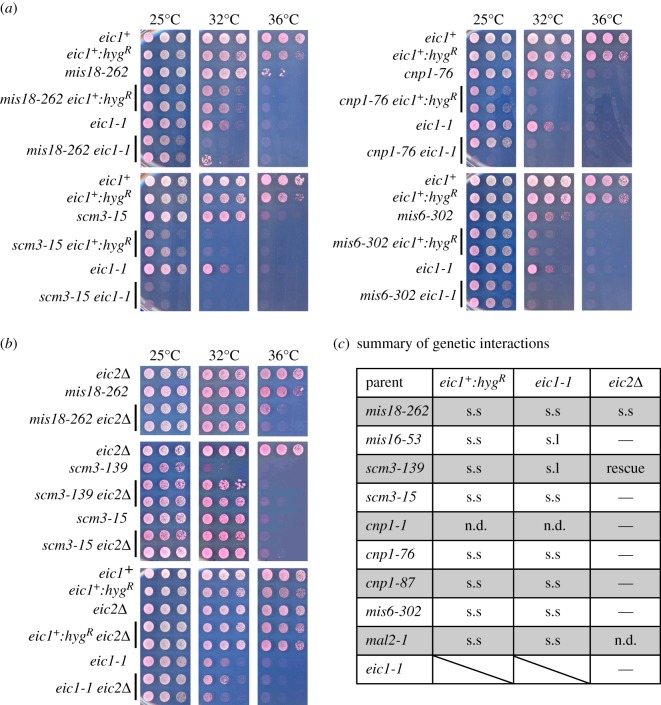
Epistasis analyses (double mutant combinations) frequently reveal positive and negative genetic interactions between mutants and consequently inform on the functional niche of specific proteins. The eic1-1 mutation exhibited significantly reduced growth when combined with ts mutations in mis18, scm3, cnp1 and mis6 (figure 5a and electronic supplementary material, figure S6a; summarized in figure 5c). We failed to generate eic1-1 mis16-53 and eic1-1 scm3-139 double mutant strains, suggesting that eic1-1 has a synthetic lethal interaction with both mis16-53 and scm3-139. Surprisingly, such synthetic interactions could also be observed for the eic1+:hygR allele, which must compromise eic1 function in sensitized genetic backgrounds and therefore be considered a hypomorphic allele of eic1 (figure 5 and electronic supplementary material, figure S6a). These results, in conjunction with our ChIP analyses of Cnp1CENP-A assembly factors (figure 4a–d), clearly demonstrate that Eic1 makes important contributions to the proper localization of Cnp1CENP-A assembly factors and Cnp1CENP-A itself.
Figure 5.
Analysis of genetic interactions between eic1 or eic2 mutants and mutations in Cnp1CENP-A or Cnp1CENP-A assembly factors. (a) eic1+:hygR and eic1-1 cells display reduced growth when combined with mutations in mis18, scm3, cnp1 or mis6. (b) eic2Δ cells display genetic interactions when combined with mis18-262 and scm3-139, but not eic1-1. Five-fold serial dilutions of cells spotted on YES + Phloxine B media and incubated at the indicated temperatures; dead cells stain dark pink. (c) A tabular summary of genetic interactions analysed in this study. s.s, synthetic sick/reduced growth; s.l, synthetic lethal; n.d., not determined; —, no interaction detected.
eic2Δ showed no combinatorial effects when combined with cnp1, mis6 or mis16 ts mutants (electronic supplementary material, figure S6b; summarized in figure 5c). However, eic2Δ displayed a negative interaction with mis18-262, notably reducing growth at 36°C relative to mis18-262 alone (figure 5b). By contrast, eic2Δ partially suppressed the temperature sensitivity of the scm3-139 (Leu73Phe) but not the scm3-15 (Ser281Leu) mutant (figure 5b; summarized in figure 5c). The negative interaction of Eic2 with Mis18 confirms that Eic2 contributes to the function of the Mis18 complex. As the protein levels as well as association of Scm3HJURP with centromeres remain unaffected in eic2Δ cells (figure 4c; electronic supplementary material, figure S6c), the mechanism of partial suppression of temperature sensitivity observed in eic2Δ scm3-139 cells remains unclear. eic2Δ showed no combinatorial effects when combined with eic1-1 (figure 5b; summarized in figure 5c), suggesting that Eic2 may not influence Eic1 function even though Eic1 and Eic2 physically associate.
3.6. Eic1 and Eic2 association with centromeres is dependent on Cnp1CENP-A loading factors
Previous analyses in fission yeast have shown that Mis16RbAp46/48/Hat2 and Mis18 are required for the localization of the Cnp1CENP-A-specific chaperone Scm3HJURP and the CCAN protein Mis6CENP-I/Ctf3 to centromeres [9,11,12]. Moreover, the localization of Mis18 at centromeres is unaffected by mutations in Cnp1CENP-A, Mis6CENP-I/Ctf3 or Scm3HJURP, whereas Scm3HJURP is dependent on functional Mis6CENP-I/Ctf3, Mis16RbAp46/48/Hat2 and Mis18 for its centromeric localization [9,11,12]. Such analyses suggest that Mis18, and probably Mis16RbAp46/48/Hat2, function to mediate the recruitment of the Cnp1CENP-A assembly factors Mis6CENP-I/Ctf3 and Scm3HJURP to centromeres and thus the incorporation of Cnp1CENP-A itself. To further dissect the role of Eic1 and Eic2, and to determine whether they act together with Mis16RbAp46/48/Hat2 and Mis18, we used qChIP to examine the dependencies of Eic1 and Eic2 for centromere localization in strains expressing various ts kinetochore proteins. Eic1 association with centromeres was found to be entirely dependent on functional Mis18, as indicated by undetectable levels of Eic1 at centromeres in mis18-262 cells grown at 36°C (figure 6a and electronic supplementary material, figure S7a). Lower Eic1 levels were also detected on centromeric central cores in mis16-53 and scm3-139 mutants at restrictive temperature; however, they were largely unchanged in cells expressing mutant Cnp1CENP-A, Mis6CENP-I/Ctf3 and Mis12 proteins (figure 6a and electronic supplementary material, figure S7a). Centromeric Eic1 levels also remained unaffected in eic2Δ cells (figure 6b). Thus, the localization of Eic1 at kinetochores is mainly dependent on its partner proteins Mis16RbAp46/48/Hat2 and Mis18, but not Eic2.
Figure 6.
Eic1 and Eic2 depend on distinct Cnp1CENP-A assembly factors for their association with centromeres. (a,b) Eic1 association with centromeres is dependent on Mis18, Mis16RbAp46/48/Hat2 and Scm3HJURP, but is largely independent of Cnp1CENP-A, Mis6CENP-I/Ctf3, Mis12 and Eic2. qChIP analyses of Eic1-GFP association with centromeres in the indicated strains when grown at permissive (25°C) versus restrictive temperature (36°C) for 8 h (a), or when grown at 32°C (b). (c,d) Eic2 association with centromeres is dependent on Mis18, Mis16RbAp46/48/Hat2, Scm3HJURP, Cnp1CENP-A, Mis6CENP-I/Ctf3, Mis12 and Eic1. qChIP analyses of Eic2-GFP association with centromeres in the indicated strains when grown at permissive (25°C) versus restrictive temperature (36°C) for 8 h. Enrichment of cc2 DNA relative to the act1 locus is presented in (a,c,d). Enrichment of cc2 or cc1/3 DNA relative to the act1 locus is presented in (b). Error bars represent standard deviation between at least three biological replicates.
The level of Eic2 at centromeres was found to be greatly dependent on functional Mis16RbAp46/48/Hat2, Mis18 and Eic1, and partially dependent on functional Scm3HJURP, Cnp1CENP-A and Mis6CENP-I/Ctf3 (figure 6c,d and electronic supplementary material, figure S7b). Unexpectedly, Eic2 levels at centromeres were found to be reduced in mis12-537 (a mutation in an outer kinetochore component) cells at restrictive temperature (figure 6c and electronic supplementary material, figure S7b). These analyses suggest that Eic2 is also recruited to centromeres by other mechanisms that are independent of its association with Mis18.
3.7. Eic1 associates with CCAN/Mis6/Ctf19 complex components that influence its recruitment to centromeres
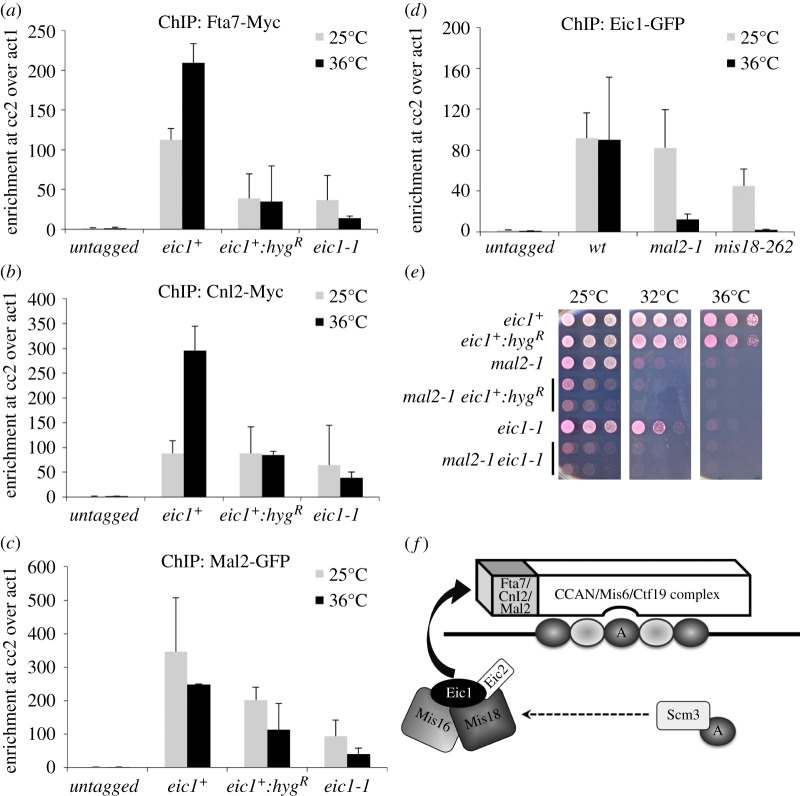
To further investigate the functional niche of Eic1 and Eic2, immunoprecipitates of both GFP-tagged proteins were subjected to LC-MS/MS analyses to identify associated factors. Only Eic1 and Mis18 were detected in Eic2-GFP immunoprecipitates (figure 7a). However, three CCAN/Mis6/Ctf19 complex components were found to associate with Eic1-GFP: Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21 (figure 7a). Co-immunoprecipitation of Myc-tagged Fta7CENP-Q/Okp1 or Cnl2Nkp2 with Eic1-GFP verified that these CCAN/Mis6/Ctf19 complex components associate with Eic1 (figure 7b). ChIP analyses revealed that the association of Myc-tagged Fta7CENP-Q/Okp1, Myc-tagged Cnl2Nkp2 and GFP-tagged Mal2CENP-O/Mcm21 with centromeres is only partially dependent on functional Eic1 (figure 8a–c), as these proteins are detectable to a significant degree at centromeres in eic1-1 cells even at non-permissive temperature. This is consistent with the finding that centromeric association of the CCAN component Mis6CENP-I/Ctf3 also only partly depends on Eic1 function (figure 4d).
Figure 7.
Eic1 interacts with three essential subunits of the CCAN/Mis6/Ctf19 complex. (a) LC-MS/MS analysis of GFP-tagged Eic1 or Eic2 immunoprecipitates from S. pombe whole cell extracts identifies Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21 as Eic1-interacting proteins. Average number of unique peptides reproducibly identified from three independent experiments is shown. (b) Eic1-GFP co-immunoprecipitates with Fta7-Myc and Cnl2-Myc. In the top panel, the asterisk (*) denotes the IgG heavy chain, and the hash tag (#) denotes a non-specific band.
Figure 8.
The CCAN components Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21 influence Eic1 association with centromeres. (a) Fta7-Myc, (b) Cnl2-Myc and (c) Mal2-GFP association with centromeres is partly dependent on Eic1. (d) Eic1-GFP association with centromeres is greatly dependent on Mal2. qChIP analyses of (a) Fta7-Myc, (b) Cnl2-Myc, (c) Mal2-GFP and (d) Eic1-GFP association with centromeres in the indicated strains when grown at permissive (25°C) versus restrictive temperature (36°C) for 8 h. Enrichment of cc2 DNA relative to the act1 locus is presented. Error bars represent standard deviation between at least three biological replicates. (e) eic1-1 displays a severe negative genetic interaction when combined with mal2-1. Five-fold serial dilutions of cells spotted on YES + Phloxine B media and incubated at the indicated temperatures; dead cells stain dark pink. (f) A model for Cnp1CENP-A maintenance at S. pombe centromeres mediated by Eic1. The CCAN components Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21 that are constitutively bound to centromeres together form a module that recruits Eic1, and thereby regulates the temporal association of Eic1, Mis16RbAp46/48/Hat2, Mis18 and Eic2 with centromeres. Once bound, Mis16RbAp46/48/Hat2/Mis18 then likely recruit the Cnp1CENP-A-specific chaperone Scm3HJURP to centromeres (the dashed arrow indicates that only an in vitro association between these proteins has been demonstrated), and thus ensures replenishment of Cnp1CENP-A (‘A’ in closed oval) at centromeres in every cell cycle.
Interestingly, Eic1-GFP association with centromeres was found to greatly depend on functional Mal2CENP-O/Mcm21 (figure 8d), although it appeared to be largely independent of functional Mis6CENP-I/Ctf3 (figure 6a and electronic supplementary material, figure S7a). Additionally, the mal2-1 ts mutant exhibited a more severe growth defect than mis6-302 when combined with eic1-1 (figures 5a and 8e). Together, these results suggest that the CCAN components Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21 may act in concert to recruit Eic1 to centromeres via their physical association with Eic1.
4. Discussion
In this study, we have identified and characterized two novel Mis18-interacting proteins, Eic1 and Eic2, in fission yeast. Eic1 is a small protein of approximately 13.4 kDa that is well conserved within Schizosaccharomyces species (figure 1b). The absence of an Eic2 orthologue in S. japonicus suggests that the primary sequence of Eic2 may have rapidly diverged over evolutionary time so that it is now undetectable within the S. japonicus genome; alternatively, other proteins may undertake the function of Eic2 in S. japonicus. The lack of any specific domains within Eic1 and Eic2 hinders the detection of similar or related proteins in other organisms, and thus orthologues of Eic1 and Eic2 remain unidentified outside of the Schizosaccharomyces clade.
The human Mis18 complex (consisting of hMis18α/β and Mis18BP1KNL2) primes CENP-A assembly by associating with centromeres specifically in telophase, subsequently recruiting the CENP-A-specific chaperone HJURPScm3 to mediate CENP-A deposition in G1 [13,25,26,28,29]. The phosphorylation of Mis18BP1KNL2 by CDK1/2 has been shown to regulate the recruitment of Mis18BP1KNL2 and timing of CENP-A assembly during the cell cycle [39]. In fission yeast, the known Cnp1CENP-A loading factors, Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP, also exhibit a very specific cell-cycle-regulated localization to centromeres in that they dissociate before metaphase and re-associate in mid-anaphase [9,11–13]. Our analyses demonstrate that both Eic1 and Eic2 are centromere-specific proteins with cell-cycle dynamics that are very similar to that of Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP (figure 2 and electronic supplementary material, figure S2). The re-association of these five factors in anaphase–telophase is presumably required to allow the subsequent replication-independent deposition of Cnp1CENP-A in G2 phase of the cell cycle [40,41]. Recent analysis of Arabidopsis Mis18BP1KNL2 suggests that it conforms to the S. pombe, rather than the vertebrate, pattern of CENP-A loading factor recruitment to centromeres during the cell cycle [33].
Our analyses show that functional Eic1 is required for the association of its interacting partners Mis16RbAp46/48/Hat2 and Mis18, as well as Scm3HJURP with centromeres (figure 4a–c). The eic1+:hygR and eic1-1 alleles also display strong negative interactions when combined with mutant Cnp1CENP-A or mutant Cnp1CENP-A assembly factors, Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP (figure 5a; summarized in figure 5c). Such interactions are consistent with Eic1 being required to promote Cnp1CENP-A assembly by mediating the association of the Cnp1CENP-A chaperone Scm3HJURP as well as Mis16RbAp46/48/Hat2 and Mis18 with centromeres. Eic1 is required to recruit Mis18 to centromeres, and functional Mis18 has been shown to affect the localization of the CCAN/Mis6/Ctf19 complex component Mis6CENP-I/Ctf3 to centromeres [11]. It was thus surprising that our analyses only detected a partial reduction in Mis6CENP-I/Ctf3 levels at centromeres in eic1 mutant cells (figure 4d). Nevertheless, consistent with this finding, our analyses show that the association of other CCAN components with centromeres is only partly dependent on Eic1 function (Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21; figure 8a–c). The CCAN proteins Mis6CENP-I/Ctf3, Sim4CENP-K/Mcm22, Mis15CENP-N/Chl4 and Mis17CENP-U/Ame1 have been shown to facilitate Cnp1CENP-A assembly [8,10,11]. Thus, although functional Eic1 is essential for the recruitment of the key Cnp1CENP-A assembly factors Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP to centromeres, it is partly dispensable for maintaining constitutive CCAN components at centromeres.
Eic2, the second Mis18-interacting protein that we identified, contributes to Mis18 function (figure 5b) as it promotes normal levels of Mis18, Mis16RbAp46/48/Hat2 and Mis6CENP-I/Ctf3 association with centromeres (figure 4e,f,h). However, Eic2 is dispensable for maintaining Cnp1CENP-A and Scm3HJURP association with centromeres (figures 3d and 4g). It was therefore unexpected that loss of Eic2 (eic2Δ) resulted in the partial rescue of scm3-139 temperature sensitivity (figure 5b; summarized in figure 5c). Given that the levels of Scm3HJURP protein and its association with centromeres remain unaffected in eic2Δ cells (figure 4g; electronic supplementary material, figure S6c), we speculate that Scm3HJURP–Mis18 interactions are perhaps stabilized in the absence of Eic2. Regardless of these changes, the levels of loading factors remaining at centromeres in eic2Δ cells are sufficient to maintain normal Cnp1CENP-A levels. The fact that the localization and function of Mis18, but not Eic1 or Cnp1CENP-A (figures 3d, 4e, 5b and 6b), is compromised in eic2Δ cells suggests that Eic1 and Eic2 function independently. Eic2 may act to bolster Mis18 function under particular conditions of stress.
Eic1 association with centromeres is entirely dependent on functional Mis18, but only partly dependent on functional Mis16RbAp46/48/Hat2 and Scm3HJURP. Moreover, Eic1 levels at centromeres remain largely unaffected when Cnp1CENP-A, Mis6CENP-I/Ctf3 or Mis12 function is compromised (figure 6a and electronic supplementary material, figure S7a). These and other observations (figure 4a–d) suggest that Eic1 acts in concert with Mis16RbAp46/48/Hat2 and Mis18 to promote recruitment of the Cnp1CENP-A-specific chaperone Scm3HJURP to centromeres and thereby facilitates Cnp1CENP-A assembly. Thus, although no obvious protein similarity is evident between Eic1 and Mis18BP1KNL2, we propose that Eic1 serves as the functional counterpart of Mis18BP1KNL2 [13, 33,37].
The maintenance of epigenetically specified centromeres on chromosomes requires a feedback mechanism where, once established, constitutive centromere proteins themselves recruit the CENP-A assembly factors and thereby ensure their preservation. In vertebrates, the CENP-A assembly factors HJURPScm3/Mis18/Mis18BP1KNL2 are recruited by the interaction of Mis18BP1KNL2 with CENP-CCnp3/Mif2 [27,28]. Our analyses suggest that Eic1 performs the equivalent function to Mis18BP1KNL2 by association with the Fta7CENP-Q/Okp1, Cnl2Nkp2 and Mal2CENP-O/Mcm21 subunits of the constitutive CCAN/Mis6/Ctf19 complex (figure 7). In support of this view, we find that Eic1 recruitment to centromeres is particularly dependent on functional Mal2CENP-O/Mcm21, and eic1 mal2 double mutants are severely compromised (figure 8d,e). By contrast, Eic1 association with centromeres is only partly dependent on the CCAN component Mis6CENP-I/Ctf3 (figure 6a and electronic supplementary material, figure S7a). Mcm21Mal2/CENP-O and Okp1Fta7/CENP-Q along with Ctf19Fta2/CENP-P and Ame1Mis17/CENP-U are known to form COMA, a biochemically distinct subcomplex within the larger Ctf19 complex in S. cerevisiae [19,42], much like the stable CENP-O/P/Q/U subcomplex described in vertebrate cells [43]. We propose that Mal2CENP-O/Mcm21, Fta7CENP-Q/Okp1 and Cnl2Nkp2 form an analogous module within the CCAN/Mis6/Ctf19 complex that ensures the propagation of Cnp1CENP-A chromatin and kinetochores by recruiting Eic1 and consequently the Cnp1CENP-A assembly factors Mis16RbAp46/48/Hat2, Mis18 and Scm3HJURP to fission yeast centromeres.
Undoubtedly, the analyses of centromere–kinetochore architecture in distinct model systems have provided insights into the function of particular components. The lack of specific components in one organism reveals its reliance on alternative pathways. For example, Drosophila lack the kinetochore protein CENP-TCnp20/Cnn1, and consequently their kinetochores are completely reliant on the KMN (Knl1–Mis12–Ndc80) pathway for attachment to microtubules [4,18]. In vertebrates, Mis18BP1KNL2 directly associates with CENP-CCnp3/Mif2, allowing the maintenance and propagation of centromeric chromatin through the recruitment of Mis18, HJURPScm3 and thus CENP-A [27,28]. Remarkably, the CENP-ACID assembly pathway appears to be completely rewired during evolution of Drosophila because the main CENP-A loading factors HJURPScm3, Mis18 and Mis18BP1KNL2 have been lost and replaced by the Cal1 protein, which directly associates with CENP-CCnp3/Mif2 [31,32]. In S. cerevisiae, where centromere specification is driven by the recognition of specific DNA elements by DNA-binding proteins [15], a self-propagation mechanism is no longer necessary and consequently proteins equivalent to Mis18/Mis18BP1KNL2, Cal1 or Eic1 are absent. Interestingly, fission yeast Cnp3CENP-C/Mif2 is not essential indicating that it is not required for the propagation of Cnp1CENP-A chromatin. Eic1 appears to perform an equivalent role to Mis18BP1KNL2 but associates with the essential constitutive CCAN components Mal2CENP-O/Mcm21, Fta7CENP-Q/Okp1 and Cnl2Nkp2, rather than Cnp3CENP-C/Mif2. Thus, it is possible that the highly conserved CCAN components CENP-OMal2/Mcm21 and CENP-QFta7/Okp1 also function in other organisms to recruit CENP-A assembly factors; they may even have an equivalent underlying function that is redundant with CENP-CCnp3/Mif2 at vertebrate centromeres.
In conclusion, our analysis has identified Eic1 as a factor that connects Mis16RbAp46/48/Hat2 and Mis18, which are required for Cnp1CENP-A incorporation, with constitutive kinetochore components within the CCAN/Mis6/Ctf19 complex. We propose that Eic1 serves two major functions: (i) priming Cnp1CENP-A assembly in concert with Mis16RbAp46/48/Hat2 and Mis18 and (ii) promoting kinetochore integrity in conjunction with the CCAN/Mis6/Ctf19 complex. The interdependency relationships among Eic1, Mis16RbAp46/48/Hat2 and Mis18, along with their similar dynamic localization to centromeres, indicate that Eic1, Mis16RbAp46/48/Hat2 and Mis18 form a complex that is temporarily released from centromeres during mitosis and can associate with centromeres independently of Cnp1CENP-A. The novel physical interaction via Eic1 that we have uncovered between constitutively bound CCAN components and Mis18 is likely to be fundamental for the temporal regulation of Eic1–Mis16RbAp46/48/Hat2–Mis18 association with centromeres, and thereby the cell-cycle-dependent assembly of Cnp1CENP-A chromatin. The identification of Eic1 and Eic2 adds to the repertoire of known cell-cycle-regulated centromeric proteins involved in maintaining CENP-A at centromeres and provides a good example of how distinctly different proteins can contribute to a conserved cellular process in diverse organisms.
5. Material and methods
5.1. Yeast strains and standard techniques
Standard methods were used for fission yeast growth, genetics and manipulation [44]. Five-fold serial dilutions of the indicated strains were spotted onto YES media containing Phloxine B for growth assays, or DMSO or 12.5 μg ml−1 TBZ for TBZ sensitivity assays. Gene deletions, tagging and centromeric plasmid transformations were carried out by either the lithium acetate transformation method or electroporation. The eic1+:hygR allele was generated by integrating a hygromycin resistance marker within the 3′UTR of eic1+ at its endogenous locus. The eic1-1 and eic1-2 alleles were generated likewise, but derived by error-prone PCR using the GeneMorph II random mutagenesis kit (Agilent Technologies). The epitope-tagged alleles of Eic1, Eic2, Hat1, Fta7 and Cnl2 were generated at their respective endogenous loci by integrating an in-frame GFP, 3HA or 13myc cassette at their respective C termini [45].
5.2. Immunoaffinity purification and mass spectrometry
For Mis16 and Mis18 pulldowns, 5 g of pulverized S. pombe cells expressing Myc-tagged Mis16 or Mis18 were used for immunoprecipitation with anti-Myc antibody 9E10 (Covance) coupled to Protein G Dynabeads (Life Technologies), alongside an untagged control. For Eic1 and Eic2 pulldowns, 5 g of pulverized S. pombe cells expressing GFP-tagged Eic1 or Eic2 were used for immunoprecipitation with anti-GFP antibody A11122 (Life Technologies) coupled to Protein G Dynabeads, alongside an untagged control. After washes, Dynabeads with immunoprecipitated material were subjected to on-bead tryptic digestion, following which the samples were treated as described [9]. The average number of unique peptides corresponding to proteins that were reproducibly enriched in the epitope-tagged samples but consistently absent in the untagged controls, over three independent biological replicates, is presented.
5.3. Multiple sequence alignment
Orthologues of Eic1 and Eic2 among the Schizosaccharomyces species were identified by BLAST, PSI-BLAST or synteny searches against the Schizosaccharomyces group database available at the Broad Institute Schizosaccharomyces Comparative Genome Project [46]. Primary sequences of Eic1 or Eic2 orthologues were aligned using ClustalW and T-Coffee.
5.4. Co-immunoprecipitations and western analyses
For co-immunoprecipitation experiments, 2 g of pulverized S. pombe cells expressing the indicated epitope-tagged proteins were used for immunoprecipitation using anti-GFP antibody A11122 (Life Technologies), anti-Myc antibody 9E10 (Covance), anti-Myc antibody 9B11 (Cell Signaling) or anti-HA antibody 12CA5 (Roche) coupled to Protein G Dynabeads (Life Technologies). The same antibodies were also used for western analyses as indicated. Anti-Bip1 was used as a loading control where indicated.
5.5. Recombinant protein co-expression and binding assays
Codon-optimized ORFs of Eic1, Mis16 and Mis18 were synthesized by GeneArt (Life Technologies), for expression in E. coli. These were then sub-cloned into pGEX-6P-1 (GST) (GE Healthcare, gift from J. Welburn) or pEC(K)-3CHis (His) (gift from A. A. Jeyaprakash) expression vectors as indicated, and co-transformed into the BL21-Gold-pLysS E. coli strain (Agilent Technologies). Co-transformants were grown in SuperBroth supplemented with carbenicillin and kanamycin as appropriate, and the indicated proteins co-expressed by induction with 0.3 mM IPTG. GST pulldowns were done using glutathione agarose (Sigma). His pulldowns were done using Ni-NTA agarose beads (Qiagen). Samples were resolved on NuPAGE Bis-Tris gels (Life Technologies) and stained using InstantBlue (Expedeon).
5.6. Quantitative chromatin immunoprecipitation
The indicated S. pombe strains were grown in YES media at 32°C. If appropriate, cells were shifted to restrictive temperature (36°C) for 8 h, or continued to grow for the same length of time at permissive temperature (25°C) before fixation. For ChIPs on centromeric plasmids, cells harbouring pH-cc2 [38,47] were grown in PMG media minus adenine, minus uracil, at 32°C. To confirm that plasmids were behaving episomally and had not integrated, a plasmid stability test was performed at the time of fixation. Cells (100–1000) were plated onto YES supplemented with 1/10th adenine and allowed to form colonies. Samples exhibiting no integrations were used for ChIP.
ChIP was performed as described [9], or with the following modifications. Cells were fixed with 1% formaldehyde (Sigma) for 20 min at room temperature and lysed using a bead beater (Biospec Products). Cell lysates were sonicated in a Bioruptor (Diagenode) (15 min, 30 s On and 30 s Off at ‘High’ (200 W) position). For Cnp1CENP-A ChIPs, anti-Cnp1CENP-A antiserum was used with Protein G agarose beads (Roche). For all other ChIPs, Protein G Dynabeads (Life Technologies) were used along with anti-GFP A11122 (Life Technologies), anti-HA 12CA5 (Roche) or anti-Myc 9B11 (Cell Signaling), as appropriate. Immunoprecipitated DNA was recovered using the Chelex-100 resin (BioRad) [48]. ChIPs were analysed by real-time PCR using Lightcycler 480 SYBR Green (Roche) with primers specific to the central cores of centromere 2 (cc2) or centromeres 1/3 (cc1/3), the innermost repeats of centromere 1 (imr) or act1. All ChIP enrichments were calculated as % DNA immunoprecipitated at the locus of interest relative to the corresponding input samples, and normalized to % DNA immunoprecipitated at the act1 locus. Histograms represent data averaged over at least three biological replicates. Error bars represent standard deviations from at least three biological replicates.
5.7. ChIP-seq analysis
Schizosaccharomyces pombe strains expressing the indicated GFP-tagged proteins were grown in YES media at 32°C to 1.25 × 109 cells at a density of 1 × 107 cells ml−1. Cells were fixed for 15 min in 1% formaldehyde (Sigma) and lysed with 0.4 mg ml−1 Zymolyase 100 T (AMS Biotechnology Europe) in PEMS for 1 h at 36°C. Cell lysates were sonicated in a Bioruptor (Diagenode) (20 min, 30 s On and 30 s Off at ‘High’ (200 W) position) and immunoprecipitated overnight using anti-GFP antibody A11122 (Life Technologies) and Protein G Dynabeads (Life Technologies). The samples were washed and cross-links reversed using 1% SDS for 4 h at 65°C. Immunoprecipitated DNA was recovered using a Qiagen PCR purification kit, and centromeric enrichment was verified by qPCR using Lightcycler 480 SYBR Green (Roche). Illumina libraries were prepared following the TruSeq Nano DNA kit (Illumina) guidelines using NEXTflex (Bio Scientific) adapters with internal barcodes. Multiplexed libraries were 100 bp paired-end sequenced on an Illumina HiSeq2000 (Ark Genomics, Edinburgh, UK). ChIP-seq data were mapped onto the S. pombe genome assembly EF2 (Ensemble) using Bowtie2. Coverage calculations and peak calling were done using MACS and PeakSplitter.
5.8. Cytology
Immunolocalization and microscopy were performed as described [9]. If appropriate, cells were shifted to restrictive temperature (36°C) for 8 h before fixation. Cells were fixed for 7–10 min with 3.7% formaldehyde (Sigma). Fixation of cells for tubulin staining used formaldehyde and 0.06% glutaraldehyde. Antibodies used were anti-GFP A11122 (1 : 200) (Life Technologies), TAT1 anti-tubulin (1 : 15) (gift from K. Gull) or anti-Cnp1CENP-A antiserum (1 : 1000). Alexa Fluor 594- and 488-coupled secondary antibodies were used at 1 : 1000 (Life Technologies).
5.9. Centromeric plasmid selection system and stability
pH-cc2 (H denotes an otr heterochromatic element and cc denotes central domain DNA) carries ura4+ and sup3-5 (suppressor of ade6-704) selection systems [38,47]. Cells without ura4+ cannot grow on minus-uracil plates, while ade6-704 cells do not grow without adenine and form red colonies on 1/10th adenine plates. The sup3-5-tRNA gene suppresses a premature stop in ade6-704, allowing growth on minus-adenine plates. Cells containing pH-cc2 form a high percentage of white or sectored colonies on 1/10th adenine indicator plates, demonstrating their relative mitotic stability in wild-type cells. In cells lacking Clr4, however, their mitotic stability is lost due to a lack of heterochromatin-dependent centromeric cohesion. To confirm that plasmids were behaving episomally and had not integrated, a plasmid stability test was performed at the time of fixation for ChIP. Cells (100–1000) were plated onto YES supplemented with 1/10th adenine and allowed to form colonies. Samples exhibiting no integrations were used for ChIP.
Note added in proof
A concurrent study has identified Eic1 and Eic2 as Mis19 and Mis20, respectively, in association with Mis18 [49].
Supplementary Material
Acknowledgements
We are grateful to F. de Lima Alves for mass spectrometry support, and A. A. Jeyaprakash for advice on recombinant protein co-expression and pulldowns. We also thank L. Sanchez-Pulido for attempting to bioinformatically find Eic1 and Eic2 orthologues. Additionally, we thank M. Yanagida and U. Fleig for strains; A. A. Jeyaprakash and J. Welburn for E. coli expression vectors; and A. Pidoux, E. Choi, S. Catania and members of the Allshire lab for advice, discussion and reagents. We also thank A. Marston and A. Pidoux for critical comments on the manuscript. L.S. performed experiments. N.R.T.T. performed ChIP-seq analyses. J.R. provided resources for mass spectrometry. L.S. and R.C.A. jointly conceived and designed the study, and wrote the manuscript.
Data accessibility
ChIP-seq data have been submitted to the Gene Expression Omnibus under accession no. GSE 54685.
Funding statement
L.S. was supported by an EC FP7 Marie Curie International Incoming Fellowship (PIIF-GA-2010-275280) and an EMBO Long Term Fellowship (ALTF 1491-2010). The Darwin Trust and a Principal's Career Development scholarship supported N.R.T.T. The Wellcome Trust supported the work of R.C.A. (095021 and 065061) and J.R. (084229) along with funding from the European Commission Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082) to R.C.A. The Wellcome Trust Centre for Cell Biology (092076) and mass spectrometry instrumentation (091020) are supported by funding from the Wellcome Trust. R.C.A. is a Wellcome Trust Principal Research Fellow.
References
- 1.Scott KC, Sullivan BA. 2013. Neocentromeres: a place for everything and everything in its place. Trends Genet. 30, 66–74. (doi:10.1016/j.tig.2013.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stimpson KM, Matheny JE, Sullivan BA. 2012. Dicentric chromosomes: unique models to study centromere function and inactivation. Chromosome Res. 20, 595–605. (doi:10.1007/s10577-012-9302-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomez-Rodriguez M, Jansen LE. 2013. Basic properties of epigenetic systems: lessons from the centromere. Curr. Opin. Genet. Dev. 23, 219–227. (doi:10.1016/j.gde.2012.11.002) [DOI] [PubMed] [Google Scholar]
- 4.Catania S, Allshire RC. 2014. Anarchic centromeres: deciphering order from apparent chaos. Curr. Opin. Cell Biol. 26, 41–50. (doi:10.1016/j.ceb.2013.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller S, Almouzni G. 2013. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochim. Biophys. Acta 1839, 241–250. (doi:10.1016/j.bbagrm.2013.11.008) [DOI] [PubMed] [Google Scholar]
- 6.Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. 2011. Drosophila CENH3 is sufficient for centromere formation. Science 334, 686–690. (doi:10.1126/science.1206880) [DOI] [PubMed] [Google Scholar]
- 7.Fachinetti D, et al. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066. (doi:10.1038/ncb2805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Chen ES, Yanagida M. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288, 2215–2219. (doi:10.1126/science.288.5474.2215) [DOI] [PubMed] [Google Scholar]
- 9.Pidoux AL, et al. 2009. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311. (doi:10.1016/j.molcel.2009.01.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pidoux AL, Richardson W, Allshire RC. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161, 295–307. (doi:10.1083/jcb.200212110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118, 715–729. (doi:10.1016/j.cell.2004.09.002) [DOI] [PubMed] [Google Scholar]
- 12.Williams JS, Hayashi T, Yanagida M, Russell P. 2009. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33, 287–298. (doi:10.1016/j.molcel.2009.01.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell 12, 17–30. (doi:10.1016/j.devcel.2006.11.002) [DOI] [PubMed] [Google Scholar]
- 14.Loyola A, Almouzni G. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677, 3–11. (doi:10.1016/j.bbaexp.2003.09.012) [DOI] [PubMed] [Google Scholar]
- 15.Allshire RC, Karpen GH. 2008. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet. 9, 923–937. (doi:10.1038/nrg2466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, McLeod I, Anderson S, Yates JR, III, He X. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24, 2919–2930. (doi:10.1038/sj.emboj.7600762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiroiwa Y, Hayashi T, Fujita Y, Villar-Briones A, Ikai N, Takeda K, Ebe M, Yanagida M. 2011. Mis17 is a regulatory module of the Mis6-Mal2-Sim4 centromere complex that is required for the recruitment of CenH3/CENP-A in fission yeast. PLoS ONE 6, e17761 (doi:10.1371/journal.pone.0017761) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14, 604–613. (doi:10.1038/ncb2493) [DOI] [PubMed] [Google Scholar]
- 19.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, III, Chan CS, Drubin DG, Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. (doi:10.1016/S0092-8674(02)00973-X) [DOI] [PubMed] [Google Scholar]
- 20.Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469. (doi:10.1038/ncb1397) [DOI] [PubMed] [Google Scholar]
- 21.Santaguida S, Musacchio A. 2009. The life and miracles of kinetochores. EMBO J. 28, 2511–2531. (doi:10.1038/emboj.2009.173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. 2009. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137, 1173–1174. (doi:10.1016/j.cell.2009.06.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen LET, Black BE, Foltz DR, Cleveland DW. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805. (doi:10.1083/jcb.200701066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunleavy EM, Almouzni G, Karpen GH. 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2, 146–157. (doi:10.4161/nucl.2.2.15211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. 2009. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484. (doi:10.1016/j.cell.2009.02.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. 2009. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497. (doi:10.1016/j.cell.2009.02.040) [DOI] [PubMed] [Google Scholar]
- 27.Dambacher S, et al. 2012. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3, 101–110. (doi:10.4161/nucl.18955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moree B, Meyer CB, Fuller CJ, Straight AF. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194, 855–871. (doi:10.1083/jcb.201106079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. 2011. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194, 229–243. (doi:10.1083/jcb.201012017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka K, Chang HL, Kagami A, Watanabe Y. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343. (doi:10.1016/j.devcel.2009.08.004) [DOI] [PubMed] [Google Scholar]
- 31.Phansalkar R, Lapierre P, Mellone BG. 2012. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 20, 493–504. (doi:10.1007/s10577-012-9299-7) [DOI] [PubMed] [Google Scholar]
- 32.Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. 2014. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 204, 313–329. (doi:10.1083/jcb.201305036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, Sandmann M, Demidov D, Schubert V, Schubert I. 2013. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 25, 3389–3404. (doi:10.1105/tpc.113.114736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lermontova I, Rutten T, Schubert I. 2011. Deposition, turnover, and release of CENH3 at Arabidopsis centromeres. Chromosoma 120, 633–640. (doi:10.1007/s00412-011-0338-5) [DOI] [PubMed] [Google Scholar]
- 35.Tong K, Keller T, Hoffman CS, Annunziato AT. 2012. Schizosaccharomyces pombe Hat1 (Kat1) is associated with Mis16 and is required for telomeric silencing. Eukaryot. Cell 11, 1095–1103. (doi:10.1128/EC.00123-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parthun MR. 2007. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 26, 5319–5328. (doi:10.1038/sj.onc.1210602) [DOI] [PubMed] [Google Scholar]
- 37.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. 2007. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176, 757–763. (doi:10.1083/jcb.200701065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folco HD, Pidoux AL, Urano T, Allshire RC. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319, 94–97. (doi:10.1126/science.1150944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE. 2012. Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 22, 52–63. (doi:10.1016/j.devcel.2011.10.014) [DOI] [PubMed] [Google Scholar]
- 40.Lando D, et al. 2012. Quantitative single-molecule microscopy reveals that CENP-ACnp1 deposition occurs during G2 in fission yeast. Open Biol. 2, 120078 (doi:10.1098/rsob.120078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. 2008. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol. Biol. Cell 19, 682–690. (doi:10.1091/mbc.E07-05-0504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Wulf P, McAinsh AD, Sorger PK. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17, 2902–2921. (doi:10.1101/gad.1144403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hori T, Okada M, Maenaka K, Fukagawa T. 2008. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell 19, 843–854. (doi:10.1091/mbc.E07-06-0556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 45.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. (doi:10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- 46.Rhind N, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332, 930–936. (doi:10.1126/science.1203357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baum M, Ngan VK, Clarke L. 1994. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol. Biol. Cell 5, 747–761. (doi:10.1091/mbc.5.7.747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185. (doi:10.1038/nprot.2006.27) [DOI] [PubMed] [Google Scholar]
- 49.Hayashi T, Ebe M, Nagao K, Kokubu A, Sajiki K, Yanagida M. In press. Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP-A through interacting with NMD factors and the SWI/SNF complex. Genes Cells (doi:10.1111/gtc.12152) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data have been submitted to the Gene Expression Omnibus under accession no. GSE 54685.