Abstract
DNA has proved to be an exquisite substrate to compute at the molecular scale. However, nonlinear computations (such as amplification, comparison or restoration of signals) remain costly in term of strands and are prone to leak. Kim et al. showed how competition for an enzymatic resource could be exploited in hybrid DNA/enzyme circuits to compute a powerful nonlinear primitive: the winner-take-all (WTA) effect. Here, we first show theoretically how the nonlinearity of the WTA effect allows the robust and compact classification of four patterns with only 16 strands and three enzymes. We then generalize this WTA effect to DNA-only circuits and demonstrate similar classification capabilities with only 23 strands.
Keywords: molecular programming, strand displacement circuits, winner-take-all, pattern recognition
1. Introduction
Intense efforts have been devoted to building synthetic molecular circuits, in vitro or in vivo, in part to understand and control biological processes [1–3]. DNA, because of its programmability and biocompatibility, has emerged as the substrate of choice to build complex structures [4–11], or to compute and actuate elaborate functions [8,12–22]. In particular, strand displacement has proved an exquisite mechanism to orchestrate the hybridization of DNA strands [23–29]. In this mechanism, a strand of DNA (the output) is selectively displaced from a gate by the action of one or several strands (the inputs) [30–32]. Efforts in DNA-based molecular programming have culminated in the construction of a variety of molecular systems [33–35], including large-scale [36,37], reversible [38,39] and reprogrammable circuits [13,40], as well as circuits multiplying matrices [41] and neural networks [42]. Besides those DNA-only circuits, hybrid DNA/enzyme circuits have been reported recently. Such molecular networks—which use DNA for encoding function and signalling but enzymes for production and degradation of signals—have emerged as a general approach to construct circuits with dynamic behaviours such as oscillations or bistability [43–49].
Most logic circuits require nonlinear computations, such as the amplification, restoration or comparison of signals, which are often the most difficult parts of a circuit to implement. Here, we show how to exploit nonlinear chemical effects to naturally compute nonlinear functions in DNA circuits with a low number of strands. Kim et al. theoretically demonstrated that competition of autocatalysts for a common enzymatic resource generates a winner-take-all (WTA) effect that digitally compares concentrations by amplifying infinitesimal differences (figure 1; [50]). We generalize this effect by demonstrating theoretically WTA circuits with DNA strands only. Furthermore, we illustrate the benefit of competition with a DNA-only classifier that uses only 23 strands to recognize four patterns and their corrupted versions, and a hybrid DNA/enzyme circuit that does the same with only 16 strands. We attribute this compactness to the essential features of nonlinearity, generality and invariance of WTA circuits.
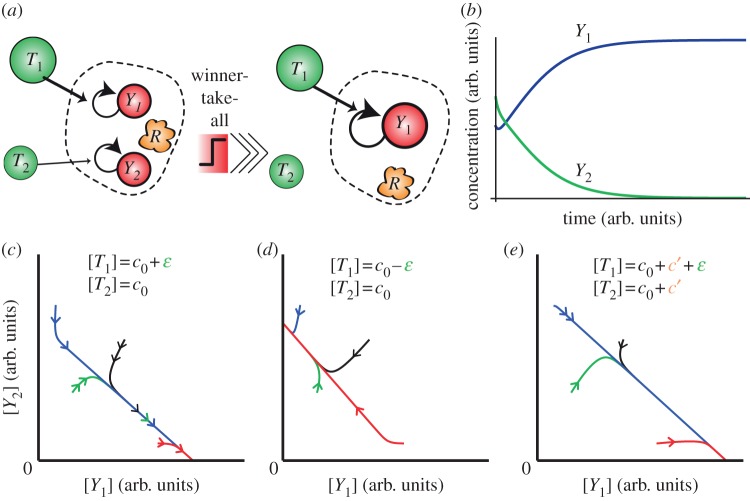
Figure 1.
WTA in enzymatic systems. (a) Several outputs Yj replicate, thanks to a specific template Tj and a common and limited enzymatic resource R. They are continuously degraded by a mechanism not shown here. The competition for the resource R leads a WTA effect. The output with the highest concentration of template evicts the others because it provides the fastest replication rate for its output (assuming all other chemical parameters are equal). (b) Schematic time-plot showing the WTA effect. Although there is initially more Y2 than Y1, the latter eventually evicts the former as [T1] > [T2]. (c) Phase plot in the space of concentrations for several trajectories with different initial conditions. The template T1 is in slight excess compared with T2. The winner, Y1, does not depend on the initial concentration of outputs, provided they are all initially present. (d) The output Y2 wins for the same initial conditions as (c), but with an excess of T2 over T1. (e) The behaviour is unchanged by a shift of all templates concentrations by a constant concentration c′.
We first describe the classification of patterns with WTA hybrid DNA/enzyme circuits. Competitive effects naturally appear in enzymatic systems, because enzymes are resources that are often shared by a great number of substrates [50–53]. Our classifiers are structurally similar to Hamming classifiers, in which a layer computes Hamming distances (the number of bits that are different) between stored and inputted patterns, and another layer selects the minimal distance [54]. Our general architecture is the following. A strand displacement layer computes several pseudo-weighted sums of the inputs. This linear layer controls a nonlinear layer that performs all nonlinear computations through a WTA circuit [55]. The nonlinear layer amplifies the maximal sum at the expense of the others, digitizing the outputs. The circuit exhibits a peculiar structure: the outputs are globally coupled, although there are no explicit connections between them.
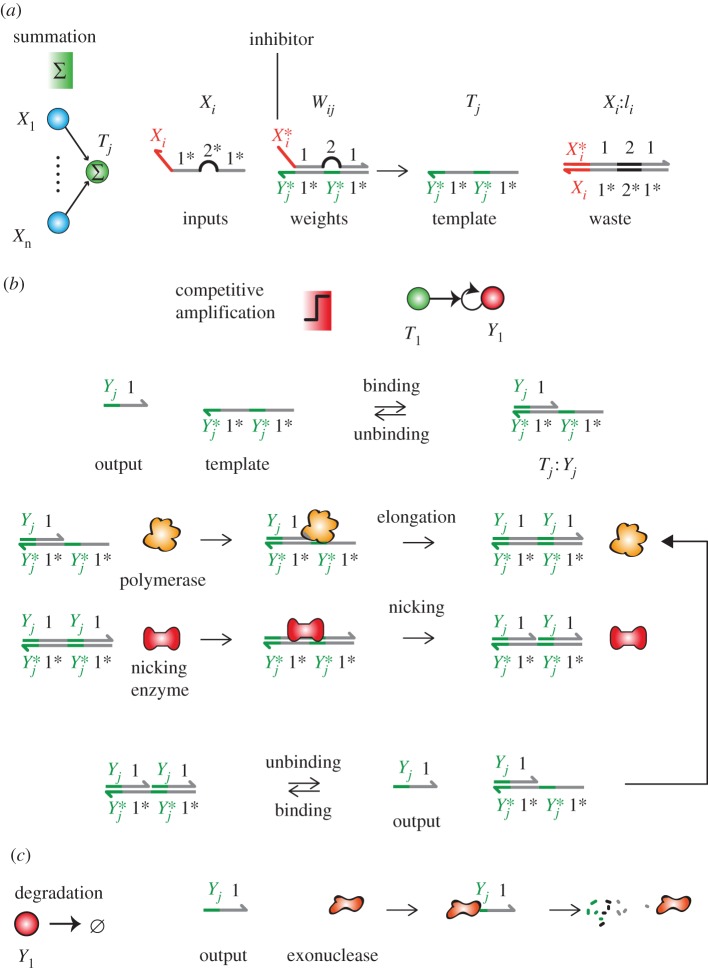
The chemical reactions used for the computations are shown in figure 2. The chemistry is based on the enzymatic toolbox of Montagne et al. [45]: protected single-stranded DNA templates encode the topology of a circuit, directing the enzymatic production of transient signal oligonucleotides. The input strands Xi represent digital variables: their initial concentrations [Xi](0) can be either 0 (FALSE), c0 (TRUE) or 0.5 c0 (ambiguous input), the latter modelling corruption of an input. The summation operation is based on the mass action kinetics of strand displacement. An input Xi displaces an inhibiting strand Ii from a weight complex Wij = Ii : Tj to yield an activated template Tj:
| 1.1 |
Figure 2.
Chemical operation of hybrid DNA/enzyme circuits. By a slight abuse of notation, Xi refers either to the output Xi or its specific domain. The number-labelled domains are global, shared by all outputs. The letter-labelled domains are specific to an input or output. (a) Strand displacement performs pseudo-weighted-sums of the inputs concentrations. An input Xi displaces an inhibitor Ii from a weight complex Wij = Ii : Tj to give an active template Tj. (b) Amplification. A template Tj catalyses the replication of Yj through a mechanism mediated by a polymerase and a nicking enzyme. Binding of the output Yj to the template triggers its elongation by the polymerase and nicking by a nicking enzyme. Spontaneous melting releases the outputs. (c) Degradation. Outputs are continuously degraded by an exonuclease enzyme which hydrolyses single-stranded DNA.
The mass action constant of strand displacement kf is 106 M−1 s−1, which gives a half-time of displacement of approximately 10 s for Xi and Wij in the 100 nM range [56]. Because mass action kinetics are linear in each reactant, the concentration of released template Tj varies approximately linearly with Xi, thus mimicking a weighted sum. This can be easily seen when the input strands are in large excess (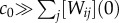 ), and the level of inputs is either 0 or c0 (no ambiguous inputs). After completion of displacement, the amount of active template [Tj](∞) is [Tj](∞) = ∑i min([Wij](0), [Xi](0)). Because [Xi](0) is either 0 or c0, [Tj](∞) will simplify to the weighted sum
), and the level of inputs is either 0 or c0 (no ambiguous inputs). After completion of displacement, the amount of active template [Tj](∞) is [Tj](∞) = ∑i min([Wij](0), [Xi](0)). Because [Xi](0) is either 0 or c0, [Tj](∞) will simplify to the weighted sum
The enzymatic layer uses the dual-repeat templates Tj produced by the linear layer to replicate outputs Yj [45]. This replication proceeds as follows: the template Tj (reversibly) binds to Yj to form a primer–template substrate, which triggers the elongation of Yj by the polymerase. A nicking enzyme recognizes this duplex and cuts one of its strands to yield two bound output strands Yj, which spontaneously unbind from the template (the inhibitor strand is chemically protected from being nicked). The temperature and lengths of domains are chosen so that outputs—but not inhibitors—spontaneously unbind from the templates on the experimental timescale. As a rule of thumb, eight nucleotides for domain 1 and 4 nucleotides for the specific domain of Yj should be appropriate and provide enough room to accommodate four different sequences of outputs. Inputs and templates are chemically protected from degradation, for example by phosphorothioate backbone modifications [45].
The net reactions for Yj are as follows. For the reversible binding, we have
| 1.2 |
The binding of a template Tj to its output Yj is designed to be reversible, with a fast equilibration (less than or equal to 1 s) and a large dissociation constant (Kb = 1 μM). We thus have 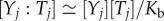 . Kinetic and thermodynamic constants are taken to be identical for all templates, outputs and inputs. Regarding the enzymatic steps, we have
. Kinetic and thermodynamic constants are taken to be identical for all templates, outputs and inputs. Regarding the enzymatic steps, we have
| 1.3 |
for the replication and decay. The polymerase has a Michaelis–Menten constant Km = 40 nM. At full speed, the polymerase replicates outputs at a rate of Vmax = 10 nM s−1. The output strands are degraded by an exonuclease, assuming a first-order degradation with a rate of kr = 0.01 s−1. We assume nicking is much faster than polymerization, which becomes the kinetically limiting step. The production of DNA is therefore approximated by a competitive Michaelis–Menten model for the polymerase. We assume that the degradation of DNA is first order. This yields equation (1.4), governing the evolution of the concentration of output strands
| 1.4 |
Competition is reflected by the presence in the denominator of all outputs Yi that use the polymerase [52]. We base our discussion and simulations on biochemical constants found in the literature [45].
The WTA effect is a nonlinear kinetic effect that amplifies selectively one output to the detriment of the others. It spontaneously emerges when several outputs compete for the polymerase (figure 1). The higher the level of an output, the higher the fraction of polymerase it sequesters. This nonlinear feedback reinforces the output with the quickest replication rate and gradually evicts other outputs—leaving only one output at the steady state [50,53]. From a perspective of signal processing, the WTA effect is a perfect nonlinear primitive to digitize signals. We use WTA effects to compare the replication rates of species, amplify the fastest and suppress the others, thereby instantiating the response of the circuit.
Because the autocatalysis is first order (only one Yi is required to catalyse the production of a new Yi), the WTA circuits have an important property of invariance: the winning output Yj depends only on the kinetic parameters and the steady-state concentration of templates, and not on the initial concentrations of outputs [53,57]. (The situation would be different for higher-order autocatalysis [50].) Any large excess of a slowly replicating output will eventually be overcome by a tiny fraction of another output replicating faster. Because all kinetic parameters are assumed equal, the winner is determined solely by the concentration of activated templates. Thus, the winner is unchanged by a shift of all templates concentrations by a common constant (figure 1e). This invariance a priori allows all the weights to be positive, and surmounts a limitation of molecular circuits, which cannot easily handle negative values because concentrations are always positive. Molecular engineers often overcome this limitation with a dual-rail representation: the positive and negative parts of a signal are carried by two distinct species [37]. Unfortunately, the dual-rail representation complicates the synthesis and debugging of logic circuits because it doubles their size [37,42]. Our circuits do not require negative values, because we constrain the weights to be positive in the selection algorithm.
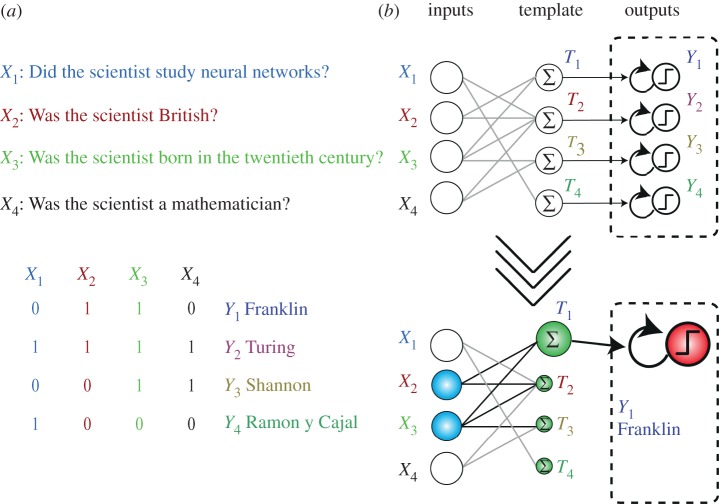
The general structure of the classification circuit is shown in figure 3. It takes as input the answers to four questions about a scientist and returns as output the corresponding scientist (following the game proposed by Qian et al. [42]). Input strands X1 … X4 encode the answers of a human player to the questions. The circuit's answer is given by the surviving output strand Yj at the steady state.
Figure 3.
General structure of the classification circuit. The circuit takes as input four questions about a scientist and returns an output strand that corresponds to the scientist. (a) Questions associated with the scientists. (b) Organization of the circuit. Inputs Xi lead to the production of templates Tj, which in turn catalyse the competitive amplification of outputs Yj in a WTA layer.
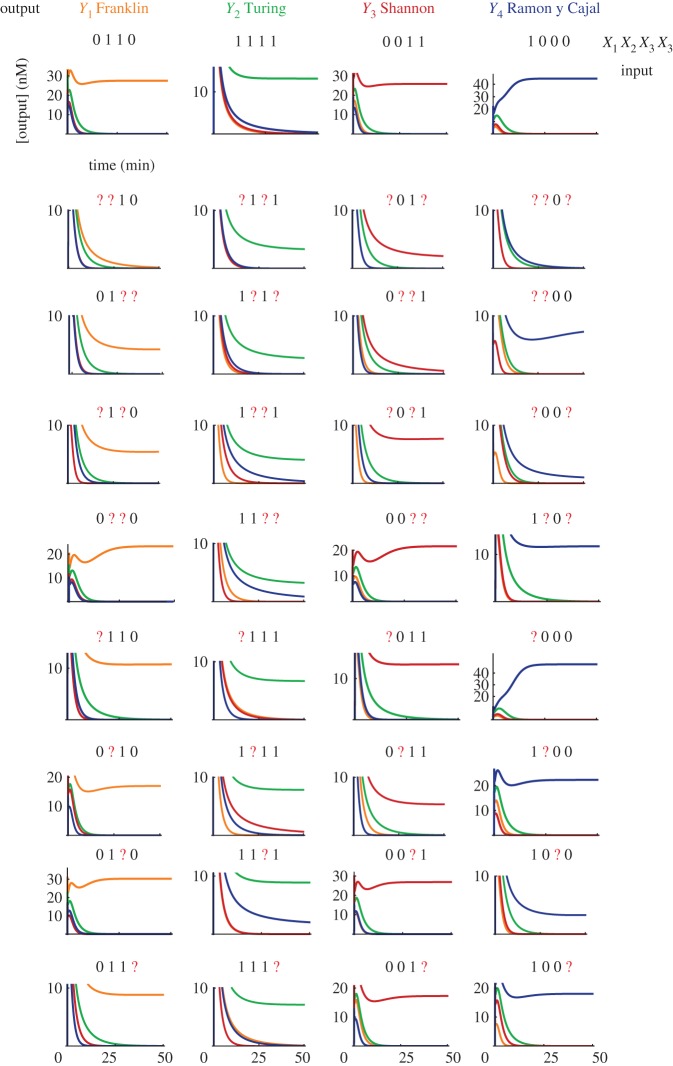
We found the weights using a randomized search based on the perceptron algorithm (see the electronic supplementary material). For the simulations in figure 4, the concentration of c0 is 300 nM, and the concentration of the weights is (in nM):
 |
1.5 |
Figure 4.
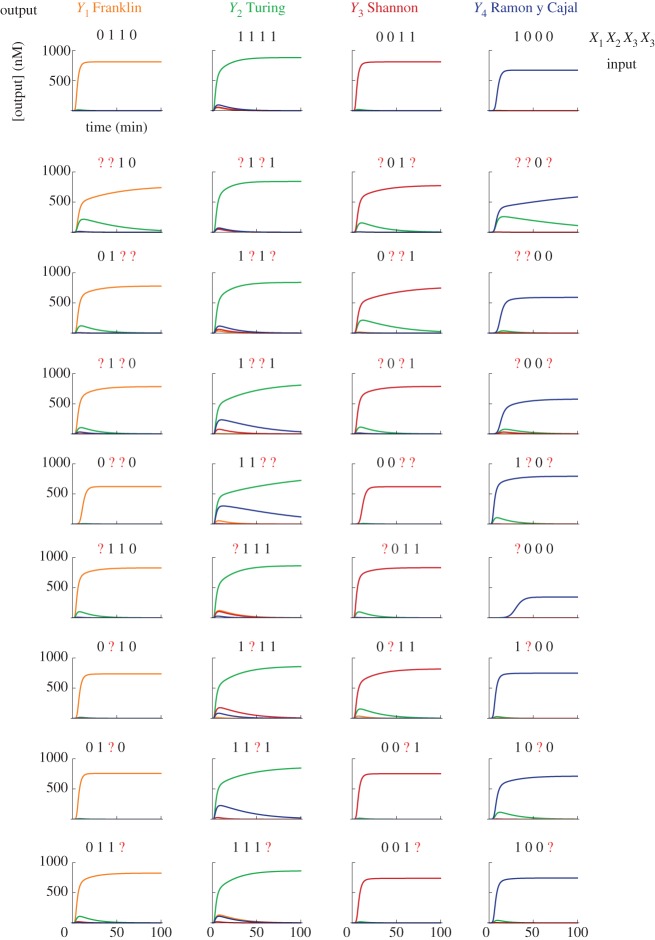
Simulation of pattern classification with the hybrid DNA/enzyme circuit. The plots show the temporal evolution of free outputs Yj in response to the injection of the four correct patterns and 32 corrupted versions of those. Injected patterns are indicated on top of each plot. A question mark (?) indicates a corrupted input, whose concentration is 0.5 c0 rather than 0 or c0. Patterns are injected at t = 0. We set a constant level of undegradable outputs at ε = 10 pM, which serves to initiate amplification.
The sum of a column j in W gives the maximal amount of template Tj produced when all inputs are present in large excess. Simulations of a hybrid DNA/enzyme circuit that classifies 36 patterns (four correct and 32 corrupted) are shown in figure 4. The patterns are the same as electronic supplementary material, fig. S15 of Qian et al. [42]. We model the corruption of an input by injecting an ambiguous concentration of 0.5 c0 rather than 0 or c0.
For large and random patterns, an algorithmic search for the weights is probably not needed. Indeed, for Hamming classifiers, the pattern matrix (or its transpose, depending on how it is defined) always provides correct weights [54]. In our case, the pattern matrix should be an appropriate weight matrix for large and random patterns. Indeed, consider a set of patterns Xj with N bits, each bit of each pattern being chosen independently and randomly to be 0 or 1. We have
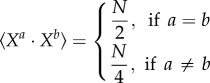 |
where the brackets denote the average value of the dot product. Thus, for two distinct patterns Xa and Xb, the average of (Xa · Xa − Xa · Xb) is on the order of N, whereas its standard deviation is on the order of 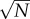 . For a large number of bits N, it is extremely unlikely that there will exist a spurious pattern Xb such that (Xa · Xa − Xa · Xb) < 0 and the weight matrix
. For a large number of bits N, it is extremely unlikely that there will exist a spurious pattern Xb such that (Xa · Xa − Xa · Xb) < 0 and the weight matrix 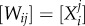 will provide satisfying weights with a high probability. Note that for small patterns such as those used here, this approach will not necessarily yield correct weights. For example, the pattern 1111 would win for all presented inputs if we used the pattern matrix as a weight matrix.
will provide satisfying weights with a high probability. Note that for small patterns such as those used here, this approach will not necessarily yield correct weights. For example, the pattern 1111 would win for all presented inputs if we used the pattern matrix as a weight matrix.
Note that the steady-state level of the winning output varies according to the pattern presented. This occurs because the concentration of template released also varies between patterns. In order to obtain a truly digital behaviour, a set of threshold/amplifying gates could be added downstream to normalize the concentration of outputs [36,37].
The classification is robust to corruption of patterns; it classifies without errors all the presented correct and corrupted patterns (similar to Qian et al. [42]). The classification is quick; it takes about 5–15 min for the winning output to reach 50 per cent of its steady state. For comparison, the half-time for similarly complex computations with a DNA-only circuit ranges from 30 min to 10 h [34,37,42].
An interesting metric for a DNA circuit is the number of strands required for its construction. This gives a proxy for the practical complexity of the synthesis and preparation. Note that similar but distinct metrics, such as the number of DNA complexes or the total number of base pairs, also inform on the experimental complexity of DNA circuits. According to the strand count, our hybrid circuit is remarkably compact: it classifies four patterns and 32 corrupted versions with only 16 strands (four inputs, four templates, four outputs and four inhibitors). As a point of comparison, we can compare our circuit with some propositions [50] or demonstrations [42] of Hopfield associative memories (which are however recurrent classification circuits). The memory of Qian et al. [42]—which recognizes and completes four-bit patterns—is built from 71 strands (112 strands with the reporters) [42]. The proposal of Kim et al. for the construction of an associative memory with a hybrid DNA/enzyme circuit would require 28 strands [50].
Three factors contribute to the compactness of our circuit based on enzymatic competition. First, DNA strands serve only to encode the circuit, whereas enzymes are in charge of the digitization machinery. In DNA-only circuits, a DNA machinery specific to each gate is in charge of digitization [34,36,37]. Second, competitive inhibition between n outputs usually requires O(n2) connections between them. We do not need those mutual connections, because the WTA effect is a general and global effect: the variation of one output immediately affects the replication rate of all others. The polymerase acts as a means to communicate information between outputs. Similar use of subtle physical effects to implement WTA effects in electronic circuits has been demonstrated [58]. Finally, the invariance property of the WTA effect dispenses with the use of dual-rail logic, which roughly doubles the size of logic circuits. More generally, our circuits highlight how neural networks are a promising paradigm for molecular computation, given their capacity to encode a larger number of functions with fewer species than Boolean circuits [42].
However, there are important differences between the recurrent circuits of Qian et al. or Kim et al., and the circuits presented here. First, our classification circuit is feed-forward (it associates an output to a set of inputs), whereas the circuits of Qian et al. and Kim et al. are recurrent (they complete their inputs so as to converge to a stable pattern). Thus, if we require some information about the bits of the classified patterns (turning our classifier into a memory), a look-up table is needed, which will increase the size of the circuit. Conversely, if we need to interpret the result of recurrent circuits (turning the memory into a classifier), then a look-up table is equally needed. Second, the storage capacity of Hopfield memories is limited, growing sublinearly with the number of bits in the patterns [59]. The feed-forward circuits presented here could potentially store many more patterns, because they are structurally similar to Hamming classifiers (which use positive or negative values for weights and inputs) and whose capacity grows exponentially with the number of bits in the patterns [60]. Third, the distributive architecture of recurrent networks confers them robustness to deletion of strands. By contrast, in our feed-forward circuit, omission of an output in the preparation of the circuit will prevent this output from competing, thus effectively erasing the corresponding pattern from the classifier.
The competition leading to a WTA effect is not limited to enzymes and applies equally to amplification mechanisms based on DNA only. It is straightforward to enforce competition in synthetic DNA circuits, because strands with common domains can easily be designed, thanks to the modularity of Watson–Crick base pairing.
Figure 5 shows how to exploit competition for a fuel strand in order to perform the WTA effect in DNA-only systems. First, inputs displace outputs from weights strands (similar to the way inputs release templates in figure 4).
| 1.6 |
Figure 5.
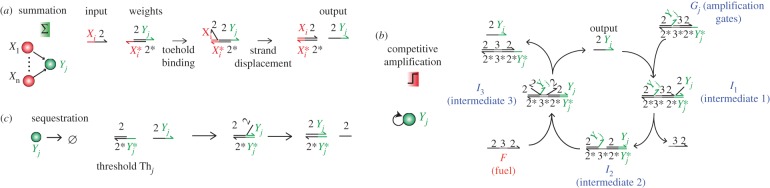
Pattern recognition with DNA-only circuits. (a) Summation. The concentrations of inputs are summed by translation gates. (b) Competitive amplification. Outputs are amplified by a mechanism based on toehold exchange. During amplification, outputs compete for a limited and global fuel. (c) Sequestration. Outputs Yj are also irreversibly sequestered by threshold gates Thj. When there is just enough fuel to saturate one but not all thresholds, amplification becomes competitive. It is expected that the output (or outputs) with the highest initial concentration (or concentrations) survive(s).
The rest of the circuit is based on the strand-exchange mechanism of Zhang et al. [31]. The autocatalytic outputs compete for a common fuel and are continuously sequestered by threshold gates [42].
| 1.7 |
and
| 1.8 |
Those processes replace the continuous production–degradation of DNA in hybrid circuits. The crucial point is that all outputs cannot survive, because there is not enough fuel to saturate all the respective thresholds, [Thj](0) < [Fuel](0) < ∑[Thi](0). The fuel becomes a limiting resource for the replication of outputs. Intuitively, this shortage of fuel should lead to a competition between outputs that nonlinearly amplifies small differences, leading to a WTA effect. Contrary to the hybrid circuits, the winner of this competition should depend on initial concentrations, because exhaustion of fuel will stop amplification before the circuit has had enough time to ‘forget’ its initial state. Because the concentrations of gates and thresholds are initially identical for all outputs, we expect that the initial concentration of outputs will determine the surviving output after exhaustion of fuel. The DNA-only circuits are compact for reasons similar to the hybrid circuits (no dual-rail, nonlinear and global nature of competition). They require the synthesis and mixing of 23 strands, which should allow their fast preparation. An experimental implementation of this scheme should be careful of potential cross-interactions between components. For example, the toehold of the weights may initiate an invasion of the amplification or threshold gates through a four-way branch migration (the speed of which is greatly reduced in a magnesium buffer [61]).
We used visual DNA strand displacement (Visual DSD) [62–64] to simulate this circuit and verify numerically the existence of a WTA effect (code in the electronic supplementary material). Inspired by model-checking in computer science, Visual DSD automatically compiles and simulates strand displacement reactions from a domain-level definition of strands. We used the standard kinetic parameters and default compilation of Visual DSD. All toehold domains have identical lengths and binding constants.
The response of the circuit to the 36 patterns is computed in deterministic mode, without leaks simulated (figure 6). After a transient period of about 15 min, the output corresponding to the presented pattern emerges as the winner for the majority of patterns. The final concentration of the winning output is a small fraction of the total amount of output produced; the vast majority of outputs are sequestered by their threshold gates. The final level of the winning output varies by a factor of 10, a larger ratio than for the hybrid circuit. For some patterns, all the outputs decay (such as ??10, 0??1, ??0?), or a winner exists but with a very low level (?00?). Note that while we have found numerically conditions in which a WTA effect occurs in DNA-only circuits, it is not guaranteed that a unique winner will exist whenever competition for fuel occurs. The competition condition ([Thj](0) < [Fuel](0) < ∑[Thi](0)) does not exclude cases where a winner may fail to emerge, or more than one winner may exist. Thorough analytical studies are needed to better understand the regimes that lead to it.
Figure 6.
Visual DSD simulations of pattern classification with a DNA-only circuit (deterministic simulations, default compilation, no leak). The plots show the temporal response for free outputs Yj to the same patterns as figure 4. We use the concentration for the weights complexes given by the matrix in equation (1.5). [Fuel](0) = [Amplifying gate](0) = 1700 nM, [Threshold gates](0) = 680 nM. For the input concentration, we use c0 = 250 nM. The inputs are injected at t = 0.
Strand displacement is prone to leak, which is thought to arise mainly from blunt-end displacement (displacement of a strand from a duplex without the assistance of a toehold) [32,65,66]. Leak is especially problematic for first-order autocatalytic circuits because they amplify exponentially even a miniscule leak (contrary to second-order amplification which occurs only above a threshold, in the presence of first-order degradation) [31]. Yet, we expect our classification circuit to be robust to leak because of the invariance of the WTA effect. A small, uniform leak of outputs will shift their levels by a common concentration, which should not change the winner. Note that there are limits to the invariance of the DNA-only circuit, because shifting the level of all outputs by a very large concentration will annihilate the WTA effect, because all thresholds will be saturated.
Fully simulating all possible leak reactions is prohibitive for deterministic simulations, so we simulated the circuit with the just-in-time mode (which performs stochastic simulations and compiles reactions on-the-fly) in order to evaluate the robustness to leaks [62,67]. Figure 7 shows the effect of increased leaking rates on the circuit. For the standard rate of leak of 1 M−1 s−1 [56,66], the classification is robust, and only the correct winner persists. For a stronger leak rate of 10 M−1 s−1, the correct winner initially dominates, but after a dozen of minutes, the levels of all outputs drift upwards. This is not unexpected as strong leakage should accelerate the convergence of such single-use circuit towards a thermodynamic equilibrium, in which the concentration of all free outputs is non null (see figure 7 right). Similar drifts in DNA circuits have been observed experimentally before [23,31,34,37]. The robustness of WTA circuits to moderate leakage, which we probed at the stochastic level for technical reasons, is a property that we expect to be true at the deterministic level.
Figure 7.
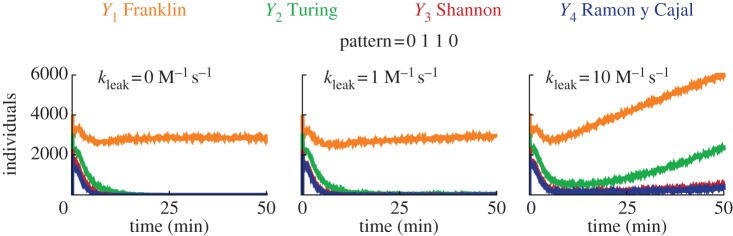
Stochastic simulations of the effect of leak on the classification (Visual DSD). The plots show the free concentration of outputs in response to the injection of pattern 0110, for various kinetic rates of leak. We selected the just-in-time mode, the directive scale 100 and default compilation. The concentrations for the circuit are identical to figure 6.
We have presented two implementations of the WTA effect in synthetic DNA systems. While they both use autocatalysis and competition for a resource, their properties are different. The hybrid DNA/enzyme circuit used competition for an enzyme, whereas the DNA-only circuit used competition for a fuel. The winning output of the hybrid DNA/enzyme circuit was determined by the concentration of template released and was independent of the initial concentration of outputs. In the DNA-only circuits, the winning output was determined by the initial concentration of output Yi released (all gates have equal concentrations). Those differences in design illustrate the flexibility of the WTA effect and, in principle, any limiting and shared resource is appropriate to enforce competition [50,53].
The precise implementation of the WTA effect may be dictated by criteria of performance, robustness or ease of design. DNA-only circuits are entirely synthetic, which facilitates their debugging and increases their modularity. Cascading WTA modules appears easier with DNA-only circuits than hybrid DNA/enzyme circuits, because the competitive effects can be localized to various subnetworks using different fuel strands (whereas competition for enzymes would lead to system-wide coupling). On the other hand, hybrid DNA/enzyme designs may prove more suitable for actuating circuits that respond dynamically to change in their input patterns, given that they continuously degrade and produce DNA strands (instead of simply releasing and sequestering them).
2. Conclusion
We have sought to exploit low-level physical effects inherent to biochemistry in order to compute nonlinear functions. We have shown that a powerful computational primitive (WTA effect) is naturally implemented in DNA circuits when replicating strands are forced to compete for a limited resource. We identify three features of the WTA effect (invariance, nonlinearity and compactness) which allow robust nonlinear computations from a reduced set of strands. We believe that such low-level routines may considerably widen the repertoire of DNA logic circuits, while facilitating their preparation. Last, our work further highlights how competition for a limited resource may provoke the emergence of complex dynamics, which has often been noted in studies on the origin of life, and searches for early replicating peptides or nucleic acids [57,68–70].
Acknowledgements
We thank Jongmin Kim, Lulu Qian, David Soloveichik, Andrew Turberfield, Erik Winfree and David Zhang for discussions, as well as Andrew Phillips for help with Visual DSD. A preliminary version of this manuscript appeared as an extended abstract in Genot et al. [71].
References
- 1.Benenson Y. 2009. Biocomputers: from test tubes to live cells. Mol. Biosyst. 5, 675–685 10.1039/b902484k (doi:10.1039/b902484k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nandagopal N, Elowitz MB. 2011. Synthetic biology: integrated gene circuits. Science 333, 1244–1248 10.1126/science.1207084 (doi:10.1126/science.1207084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hockenberry AJ, Jewett MC. 2012. Synthetic in vitro circuits. Curr. Opin. Chem. Biol. 16, 253–259 10.1016/j.cbpa.2012.05.179 (doi:10.1016/j.cbpa.2012.05.179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothemund PWK. 2006. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 10.1038/nature04586 (doi:10.1038/nature04586) [DOI] [PubMed] [Google Scholar]
- 5.Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. 2009. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 10.1038/nature08016 (doi:10.1038/nature08016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen ES, et al. 2009. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 459, 73–75 10.1038/nature07971 (doi:10.1038/nature07971) [DOI] [PubMed] [Google Scholar]
- 7.Ackermann D, Schmidt TL, Hannam JS, Purohit CS, Heckel A, Famulok M. 2010. A double-stranded DNA rotaxane. Nat. Nanotechnol. 5, 436–442 10.1038/nnano.2010.65 (doi:10.1038/nnano.2010.65) [DOI] [PubMed] [Google Scholar]
- 8.Han DR, Pal S, Liu Y, Yan H. 2010. Folding and cutting DNA into reconfigurable topological nanostructures. Nat. Nanotechnol. 5, 712–717 10.1038/nnano.2010.193 (doi:10.1038/nnano.2010.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han DR, Pal S, Nangreave J, Deng ZT, Liu Y, Yan H. 2011. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 10.1126/science.1202998 (doi:10.1126/science.1202998) [DOI] [PubMed] [Google Scholar]
- 10.Ke YG, Ong LL, Shih WM, Yin P. 2012. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 10.1126/science.1227268 (doi:10.1126/science.1227268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arbona JM, Aime JP, Elezgaray J. 2013. Cooperativity in the annealing of DNA origamis. J. Chem. Phys. 138, 015105. 10.1063/1.4773405 (doi:10.1063/1.4773405) [DOI] [PubMed] [Google Scholar]
- 12.Wilner OI, Weizmann Y, Gill R, Lioubashevski O, Freeman R, Willner I. 2009. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotechnol. 4, 249–254 10.1038/nnano.2009.50 (doi:10.1038/nnano.2009.50) [DOI] [PubMed] [Google Scholar]
- 13.Elbaz J, Lioubashevski O, Wang FA, Remacle F, Levine RD, Willner I. 2010. DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechnol. 5, 417–422 10.1038/nnano.2010.88 (doi:10.1038/nnano.2010.88) [DOI] [PubMed] [Google Scholar]
- 14.Elbaz J, Wang FA, Remacle F, Willner I. 2012. pH-programmable DNA logic arrays powered by modular DNAzyme libraries. Nano Lett. 12, 6049–6054 10.1021/nl300051g (doi:10.1021/nl300051g) [DOI] [PubMed] [Google Scholar]
- 15.Lund K, et al. 2010. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 10.1038/nature09012 (doi:10.1038/nature09012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickham SFJ, Endo M, Katsuda Y, Hidaka K, Bath J, Sugiyama H, Turberfield AJ. 2011. Direct observation of stepwise movement of a synthetic molecular transporter. Nat. Nanotechnol. 6, 166–169 10.1038/nnano.2010.284 (doi:10.1038/nnano.2010.284) [DOI] [PubMed] [Google Scholar]
- 17.Modi S, Swetha MG, Goswami D, Gupta GD, Mayor S, Krishnan Y. 2009. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 4, 325–330 10.1038/nnano.2009.83 (doi:10.1038/nnano.2009.83) [DOI] [PubMed] [Google Scholar]
- 18.Duose DY, Schweller RM, Hittelman WN, Diehl MR. 2010. Multiplexed and reiterative fluorescence labeling via DNA circuitry. Bioconjug. Chem. 21, 2327–2331 10.1021/bc100348q (doi:10.1021/bc100348q) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duose DY, Schweller RM, Zimak J, Rogers AR, Hittelman WN, Diehl MR. 2012. Configuring robust DNA strand displacement reactions for in situ molecular analyses. Nucleic Acids Res. 40, 3289–3298 10.1093/nar/gkr1209 (doi:10.1093/nar/gkr1209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKee ML, Milnes PJ, Bath J, Stulz E, Turberfield AJ, O'Reilly RK. 2010. Multistep DNA-templated reactions for the synthesis of functional sequence controlled oligomers. Angew. Chem. Int. Ed. 49, 7948–7951 10.1002/anie.201002721 (doi:10.1002/anie.201002721) [DOI] [PubMed] [Google Scholar]
- 21.Chandran H, Gopalkrishnan N, Yurke B, Reif J. 2012. Meta-DNA: synthetic biology via DNA nanostructures and hybridization reactions. J. R. Soc. Interface 9, 1637–1653 10.1098/rsif.2011.0819 (doi:10.1098/rsif.2011.0819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweller RM, Zimak J, Duose DY, Qutub AA, Hittelman WN, Diehl MR. 2012. Multiplexed in situ immunofluorescence using dynamic DNA complexes. Angew. Chem. Int. Ed. 51, 9292–9296 10.1002/anie.201204304 (doi:10.1002/anie.201204304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin P, Choi HMT, Calvert CR, Pierce NA. 2008. Programming biomolecular self-assembly pathways. Nature 451, 318–322 10.1038/nature06451 (doi:10.1038/nature06451) [DOI] [PubMed] [Google Scholar]
- 24.Elbaz J, Moshe M, Willner I. 2009. Coherent activation of DNA tweezers: a ‘SET–RESET’ logic system. Angew. Chem. Int. Ed. 48, 3834–3837 10.1002/anie.200805819 (doi:10.1002/anie.200805819) [DOI] [PubMed] [Google Scholar]
- 25.Zhang DY, Seelig G. 2011. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 10.1038/nchem.957 (doi:10.1038/nchem.957) [DOI] [PubMed] [Google Scholar]
- 26.Zhang DY. 2011. Cooperative hybridization of oligonucleotides. J. Am. Chem. Soc. 133, 1077–1086 10.1021/ja109089q (doi:10.1021/ja109089q) [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. 2011. A DNA tile actuator with eleven discrete states. Angew. Chem. Int. Ed. 50, 3983–3987 10.1002/anie.201007642 (doi:10.1002/anie.201007642) [DOI] [PubMed] [Google Scholar]
- 28.Xing YZ, Yang ZQ, Liu DS. 2011. A responsive hidden toehold to enable controllable DNA strand displacement reactions. Angew. Chem. Int. Ed. 50, 11 934–11 936 10.1002/anie.201105923 (doi:10.1002/anie.201105923) [DOI] [PubMed] [Google Scholar]
- 29.Genot AJ, Zhang DY, Bath J, Turberfield AJ. 2011. Remote toehold: a mechanism for flexible control of DNA hybridization kinetics. J. Am. Chem. Soc. 133, 2177–2182 10.1021/ja1073239 (doi:10.1021/ja1073239) [DOI] [PubMed] [Google Scholar]
- 30.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. 2000. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 10.1038/35020524 (doi:10.1038/35020524) [DOI] [PubMed] [Google Scholar]
- 31.Zhang DY, Turberfield AJ, Yurke B, Winfree E. 2007. Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 10.1126/science.1148532 (doi:10.1126/science.1148532) [DOI] [PubMed] [Google Scholar]
- 32.Soloveichik D, Seelig G, Winfree E. 2010. DNA as a universal substrate for chemical kinetics. Proc. Natl Acad. Sci. USA 107, 5393–5398 10.1073/pnas.0909380107 (doi:10.1073/pnas.0909380107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojanovic MN, Stefanovic D. 2003. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 21, 1069–1074 10.1038/nbt862 (doi:10.1038/nbt862) [DOI] [PubMed] [Google Scholar]
- 34.Seelig G, Soloveichik D, Zhang DY, Winfree E. 2006. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 10.1126/science.1132493 (doi:10.1126/science.1132493) [DOI] [PubMed] [Google Scholar]
- 35.Ran T, Kaplan S, Shapiro E. 2009. Molecular implementation of simple logic programs. Nat. Nanotechnol. 4, 642–648 10.1038/nnano.2009.203 (doi:10.1038/nnano.2009.203) [DOI] [PubMed] [Google Scholar]
- 36.Qian L, Winfree E. 2011. A simple DNA gate motif for synthesizing large-scale circuits. J. R. Soc. Interface 8, 1281–1297 10.1098/rsif.2010.0729 (doi:10.1098/rsif.2010.0729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian L, Winfree E. 2011. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 10.1126/science.1200520 (doi:10.1126/science.1200520) [DOI] [PubMed] [Google Scholar]
- 38.Genot AJ, Bath J, Turberfield AJ. 2011. Reversible logic circuits made of DNA. J. Am. Chem. Soc. 133, 20 080–20 083 10.1021/ja208497p (doi:10.1021/ja208497p) [DOI] [PubMed] [Google Scholar]
- 39.Orbach R, Remacle F, Levine RD, Itamar W. 2012. Logic reversibility and thermodynamic irreversibility demonstrated by DNAzyme-based Toffoli and Fredkin logic gates. Proc. Natl Acad. Sci. USA 109, 21 228–21 233 10.1073/pnas.1219672110 (doi:10.1073/pnas.1219672110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei RJ, Matamoros E, Liu MH, Stefanovic D, Stojanovic MN. 2010. Training a molecular automaton to play a game. Nat. Nanotechnol. 5, 773–777 [DOI] [PubMed] [Google Scholar]
- 41.Genot AJ, Bath J, Turberfield A. 2013. Combinatorial displacement of DNA strands: application to matrix multiplication and weighted sums. Angew. Chem. Int. Ed. 52, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 42.Qian L, Winfree E, Bruck J. 2011. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 10.1038/nature10262 (doi:10.1038/nature10262) [DOI] [PubMed] [Google Scholar]
- 43.Kim J, White KS, Winfree E. 2006. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2, article 68 10.1038/msb4100099 (doi:10.1038/msb4100099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takinoue M, Kiga D, Shohda KI, Suyama A. 2008. Experiments and simulation models of a basic computation element of an autonomous molecular computing system. Phys. Rev. E 78, 041921 (doi:10.1103/PhysRevE.78.041921) [DOI] [PubMed] [Google Scholar]
- 45.Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. 2011. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, article 466 10.1038/msb.2010.120 (doi:10.1038/msb.2010.120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, Winfree E. 2011. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, article 465 10.1038/msb.2010.119 (doi:10.1038/msb.2010.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ayukawa S, Takinoue M, Kiga D. 2011. RTRACS: a modularized RNA-dependent RNA transcription system with high programmability. Acc. Chem. Res. 44, 1369–1379 10.1021/ar200128b (doi:10.1021/ar200128b) [DOI] [PubMed] [Google Scholar]
- 48.Padirac A, Fujii T, Rondelez Y. 2012. PNAS plus: bottom-up construction of in vitro switchable memories. Proc. Natl Acad. Sci. USA 109, E3212–E3220 10.1073/pnas.1212069109 (doi:10.1073/pnas.1212069109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rondelez Y, Fujii T. 2013. Predator–prey molecular ecosystems. ACS Nano 7, 27–37 [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Hopfield JJ, Winfree E. 2004. Neural network computation by in vitro transcriptional circuits. In Advances in neural information processing systems, vol. 17 (eds Saul LK, Weiss Y, Bottou L.), pp. 681–688 Cambridge, MA: MIT Press [Google Scholar]
- 51.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 10.1016/j.cell.2011.07.014 (doi:10.1016/j.cell.2011.07.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rondelez Y. 2012. Competition for catalytic resources alters biological network dynamics. Phys. Rev. Lett. 108, 018102. 10.1103/PhysRevLett.108.018102 (doi:10.1103/PhysRevLett.108.018102) [DOI] [PubMed] [Google Scholar]
- 53.Genot AJ, Fujii T, Rondelez Y. 2012. Computing with competition in biochemical networks. Phys. Rev. Lett. 109, 208102. 10.1103/PhysRevLett.109.208102 (doi:10.1103/PhysRevLett.109.208102) [DOI] [PubMed] [Google Scholar]
- 54.Lippmann R, Gold B, Malpass M. 1987. A comparison of Hamming and Hopfield neural nets for pattern classification. Technical report 769 Cambridge, MA: MIT Press [Google Scholar]
- 55.Maass W. 2000. On the computational power of winner-take-all. Neural Comput. 12, 2519–2535 10.1162/089976600300014827 (doi:10.1162/089976600300014827) [DOI] [PubMed] [Google Scholar]
- 56.Yurke B, Mills AP. 2003. Using DNA to power nanostructures. Genet. Prog. Evol. Mach. 4, 111–122 10.1023/A:1023928811651 (doi:10.1023/A:1023928811651) [DOI] [Google Scholar]
- 57.Szathmáry E. 2006. The origin of replicators and reproducers. Phil. Trans. R. Soc. B 361, 1761–1776 10.1098/rstb.2006.1912 (doi:10.1098/rstb.2006.1912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazzaro J, Ryckebusch S, Mahowald MA, Mead CA. 1988. Winner-take-all networks of O(N) complexity. Technical report CALTECH–CS–TR–21–88. Pasadena, CA: California Institute of Technology
- 59.McEliece RJ, Posner EC, Rodemich ER, Venkatesh SS. 1987. The capacity of the Hopfield associative memory. IEEE Trans. Inf. Theory 33, 461–482 10.1109/TIT.1987.1057328 (doi:10.1109/TIT.1987.1057328) [DOI] [Google Scholar]
- 60.Hassoun M, Watta P. 1996. The hamming associative memory and its relation to the exponential capacity DAM. In Proc. IEEE Int. Conf. Neural Networks, vol. 1, ICNN-96, June 3–6, pp. 583–587 Washington, DC [Google Scholar]
- 61.Panyutin IG, Hsieh P. 1994. The kinetics of spontaneous DNA branch migration. Proc. Natl Acad. Sci. USA 91, 2021–2025 10.1073/pnas.91.6.2021 (doi:10.1073/pnas.91.6.2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lakin MR, Youssef S, Polo F, Emmott S, Phillips A. 2011. Visual DSD: a design and analysis tool for DNA strand displacement systems. Bioinformatics 27, 3211–3213 10.1093/bioinformatics/btr543 (doi:10.1093/bioinformatics/btr543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lakin MR, Youssef S, Cardelli L, Phillips A. 2012. Abstractions for DNA circuit design. J. R. Soc. Interface 9, 470–486 10.1098/rsif.2011.0343 (doi:10.1098/rsif.2011.0343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lakin MR, Parker D, Cardelli L, Kwiatkowska M, Phillips A. 2012. Design and analysis of DNA strand displacement devices using probabilistic model checking. J. R. Soc. Interface 9, 1470–1485 10.1098/rsif.2011.0800 (doi:10.1098/rsif.2011.0800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reynaldo LP, Vologodskii AV, Neri BP, Lyamichev VI. 2000. The kinetics of oligonucleotide replacements. J. Mol. Biol. 297, 511–520 10.1006/jmbi.2000.3573 (doi:10.1006/jmbi.2000.3573) [DOI] [PubMed] [Google Scholar]
- 66.Zhang DY, Winfree E. 2009. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17 303–17 314 10.1021/ja906987s (doi:10.1021/ja906987s) [DOI] [PubMed] [Google Scholar]
- 67.Pauleve L, Youssef S, Lakin MR, Phillips A. 2010. A generic abstract machine for stochastic process calculi. In CMSB 2010: Proc. 8th Int. Conf. Computational Methods in Systems Biology, Trento, Italy, pp. 43–54 New York, NY: ACM [Google Scholar]
- 68.Lee DH, Severin K, Yokobayashi Y, Ghadiri MR. 1997. Emergence of symbiosis in peptide self-replication through a hypercyclic network. Nature 390, 591–594 10.1038/37569 (doi:10.1038/37569) [DOI] [PubMed] [Google Scholar]
- 69.Voytek SB, Joyce GF. 2009. Niche partitioning in the coevolution of 2 distinct RNA enzymes. Proc. Natl Acad. Sci. USA 106, 7780–7785 10.1073/pnas.0903397106 (doi:10.1073/pnas.0903397106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vaidya N, Manapat ML, Chen IA, Xulvi-Brunet R, Hayden EJ, Lehman N. 2012. Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77 10.1038/nature11549 (doi:10.1038/nature11549) [DOI] [PubMed] [Google Scholar]
- 71.Genot A, Fujii T, Rondelez Y. 2012. Molecular computations with competitive neural networks that exploit linear and nonlinear kinetics. In Turing-100. The Alan Turing centenary (ed. Voronkov A.), pp. 113–117 EasyChair Proceedings in Computing (EPiC), vol. 10. EasyChair: Manchester, UK. See http://www.easychair.org/publications/?page=171364546 Manchester [Google Scholar]