Abstract
We have investigated the requirements for nucleosome remodeling upon transcriptional induction of the GAL1 promoter. We found that remodeling was dependent on two SAGA complex components, Gcn5 and Spt3. The involvement of the latter was surprising as its function has been suggested to be directly involved in TATA-binding protein (TBP) recruitment. We demonstrated that this novel function was in fact independent of TBP recruitment and this was further validated using a Gal4-driven synthetic promoter. Most importantly, we showed that the involvement of Spt3 in chromatin remodeling was independent of transcription, as it was also observed for a nonpromoter nucleosome located next to an activator-binding site. In an effort to explore how the Spt3 function was elicited, we found that Mot1, an ATPase of the Snf2 family that genetically interacts with Spt3, was also required for nucleosome remodeling independently of TBP recruitment. Interestingly enough, Spt3 and Mot1 were recruited on the GAL1 promoter as well as on the nonpromoter site in an interdependent manner. These findings show that the two proteins cooperate in nucleosomal transactions.
Keywords: GAL1, Mot1, nucleosome remodeling, SAGA, Spt3
Introduction
Transcription initiation in eucaryotes is accomplished through a well-orchestrated assemblage of large multisubunit complexes onto gene regulatory regions. In yeast, these complexes are recruited via interactions with DNA-binding transcriptional activators and fall into operational categories. For instance, there are those devoted in chromatin modification—a prerequisite for gene activation—typified by the Swi/Snf complex, but also those that establish physical interactions between activation domains and the basic transcriptional machinery, typified by the Mediator complex. A third class, represented by the SAGA complex, has been implicated as having both chromatin modification and adaptor functions.
The SAGA component, which is the major player in its chromatin-related functions, is Gcn5. This protein is a histone acetyltransferase (HAT) that preferentially acetylates specific lysine residues at the N-termini of H3 histone, a modification believed to assist nucleosome remodeling activities (Sterner et al, 1999; Roth et al, 2001). Other SAGA components such as Ada2 and Ada3 have been implicated in the regulation and potentiation of the Gcn5 catalytic activity (Balasubramanian et al, 2002; Sterner et al, 2002), whereas others such as Taf 68 are involved in the recognition of nucleosomal histones by Gcn5 (Grant et al, 1998). SAGA is recruited to promoters via the interaction of its Tra1 component with the activation domain of transcriptional activators (Brown et al, 2001) and its adaptor function is thought to be accomplished by components such as Spt8 and Spt3, for which a number of experiments suggest an interaction with the TATA-binding protein (TBP) (Eisenmann et al, 1992, 1994; Lee and Young, 1998; Sterner et al, 1999).
As is the case for most nonessential for integrity SAGA components, Spt3 is required for a subset of SAGA-regulated genes and in a manner largely independent of Gcn5 (Lee et al, 2000). The SPT3 gene was first identified as a suppressor of defects caused by Ty insertions into the promoter regions of certain genes (Winston et al, 1984), a scheme that led to the genetic isolation of other SPT genes encoding components of the SAGA complex as well as the yeast TBP gene (SPT15) (reviewed in Yamaguchi et al, 2001). Genetic and biochemical studies have supported a direct interaction of Spt3 with TBP (Eisenmann et al, 1992; Lee and Young, 1998; Dudley et al, 1999), and this has put forward the idea that Spt3 has a role in the recruitment of TBP at certain promoters. This idea was strengthened by the fact that Spt3 displays genetic interactions with Mot1, Not1 and TFIIA, proteins that have also been implicated in the biochemistry of TBP recruitment (Collart, 1996; Madison and Winston, 1997). Finally, this model is in concert with recent studies showing that for both GAL1 and the PHO5 promoters, Spt3 is required in vivo for TBP recruitment upon transcriptional activation (Dudley et al, 1999; Larschan and Winston, 2001; Barbaric et al, 2003).
The totality of the evidence suggests that Spt3 has a pivotal role in the function of the SAGA complex as an adaptor, perhaps equivalent to the role of Gcn5 HAT in the chromatin-related functions of this complex. We were therefore intrigued by our earlier observations, which implicated a chromatin remodeling function for Spt3 on a synthetic promoter (Syntichaki et al, 2000). In order to investigate further this possibility, we initiated studies on nucleosome transitions of the Spt3-dependent GAL1 promoter. Surprisingly, these studies revealed that Spt3 is indeed required for nucleosome remodeling and in a manner independent of TBP recruitment. These findings were further confirmed using a synthetic promoter and more importantly the involvement of Spt3 in nucleosome remodeling was also observed at a nonpromoter site (NPS) having an activator-binding DNA target. Interestingly enough, results from additional experiments gave functional significance to the genetic interaction of Spt3 with the Mot1 ATPase, as they demonstrated a TBP autonomous involvement of Mot1 in nucleosome remodeling but in a manner dependent on Spt3.
Results
Spt3 is required for nucleosome remodeling at the GAL1 promoter
We investigated the possibility for the involvement of Spt3 in the Gal4-dependent remodeling of two nucleosomes positioned at the GAL1 promoter (Figure 1A, see also Lohr, 1997). We first tested the contribution of the entire SAGA complex, by eliminating the Spt20 component, which is required for its integrity (Sterner et al, 1999). As shown in Figure 1B, loss of Spt20 impaired nucleosome remodeling. We then tested the contribution of individual SAGA components such as Gcn5 and Spt3 both affecting transcriptional activation of GAL1 (Dudley et al, 1999; Papamichos-Chronakis et al, 2002). Consistent with its described role, elimination of Gcn5 blocked nucleosome remodeling (Figure 1B). Unexpectedly, the same phenotype was also exhibited when Spt3 was absent, a result that assigned a second function for Spt3, other than that involved with TBP interactions. In order to examine the autonomy of the two functions, we uncoupled TBP recruitment from chromatin remodeling using a mutation of the GAL1 TATA (Topalidou et al, 2003). On such a mutated promoter, TBP failed to be recruited and transcriptional activation was compromised (Topalidou et al, 2003), but the process of remodeling of nucleosomes −2 and −1 was both qualitatively (Figure 1B and C, top panels) and quantitatively (Figure 1B and C, bottom panels) clearly unaffected (see also Axelrod et al, 1993; Topalidou et al, 2003). We found that this remodeling was again relying on both Gcn5 and Spt3 (Figure 1C). We concluded that Spt3 functions in nucleosome remodeling at the GAL1 promoter in a manner independent of TBP recruitment.
Figure 1.
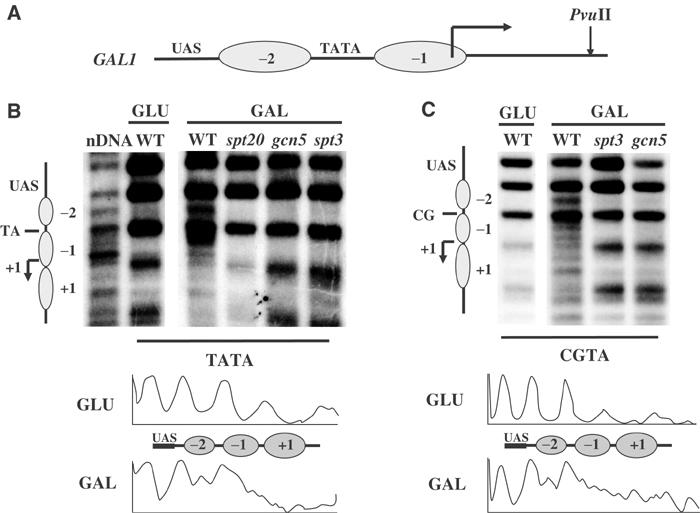
Nucleosome remodeling at the GAL1 promoter depends on Spt3 and Gcn5 but not on TBP binding. (A) Schematic representation of the GAL1 promoter indicating the Gal4-binding site (UAS), the positioned nucleosomes (−2, −1), the TATA element and the start site of transcription (arrow). The restriction endonuclease cleavage site (PvuII) used to monitor nucleosomal remodeling is also indicated. (B) Remodeling of −1 and −2 nucleosomes of the native GAL1 promoter (TATA) and (C) of a TATA less derivative (CGTA) was assayed by micrococcal nuclease sensitivity in the indicated strains grown under repressive (GLU) or inducing (GAL) conditions. nDNA is the MNase cleavage pattern for the purified GAL1 promoter. Bottom panels of (B) and (C) show the densitometry profile of the lanes corresponding to MNase pattern of the repressed (GLU) and induced (GAL) wild-type or the TATA mutated GAL1 promoter.
Spt3 functions in chromatin remodeling independently of promoter context
GAL1 is a complex promoter and its transcriptional activation requires, in addition to Gal4, derepression from a Mig1–Ssn6–Tup1-mediated repression (Johnston et al, 1994; Papamichos-Chronakis et al, 2002). In order to generalize the above observations, we utilized a simplified synthetic promoter based on PHO5 and in which a Gcn4 response element was inserted in the linker region between nucleosomes −1 and −2 (Figure 2A, see also Topalidou et al, 2003). Gal4-dependent transcriptional activation of this promoter was achieved through a chimeric protein bearing the Gcn4 DNA-binding domain fused to the Gal4 activation domain and expressed under the translational control system of native Gcn4 (Thireos et al, 1984). In this way, we could monitor Gal4-dependent transcriptional activation through amino-acid limitation, thus avoiding the galactose-induced signaling. As shown in Figure 2B, amino-acid limitation resulted in a marked increase in transcription through this promoter, which was highly dependent on Spt3 but not on Gcn5. This dependency was not an indirect consequence of the prohibition of activator recruitment as this was unaffected by the Spt3 lesion (Figure 2C, left panel). Examination of the requirement for nucleosome remodeling revealed that this process also required Spt3 but not Gcn5 (Figure 2C, right panel). Finally, the Spt3 involvement in nucleosome remodeling was independent of TBP recruitment as it was also evident on a promoter mutated for the TATA element (Figure 2C, right panel). We concluded that the TBP-independent involvement of Spt3 in chromatin remodeling does not depend on the promoter context.
Figure 2.
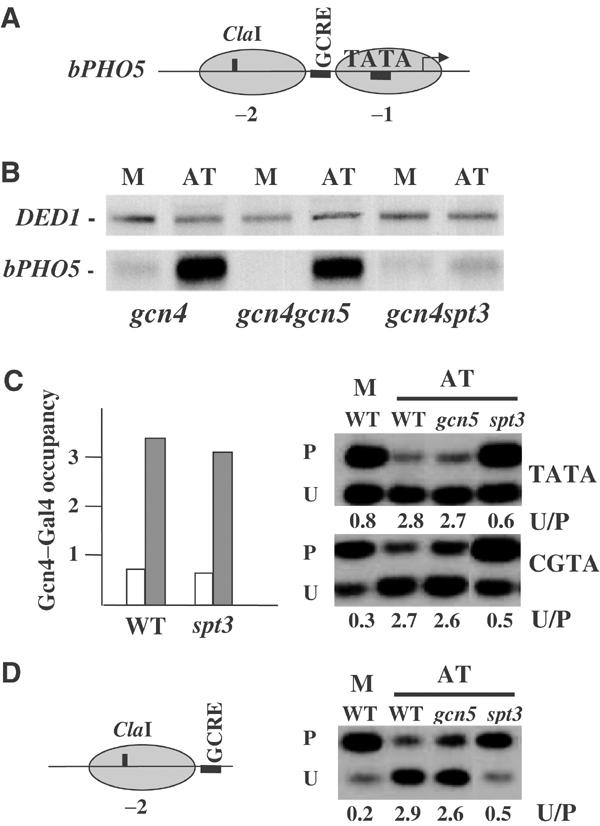
Spt3 is involved in nucleosome remodeling independently of the promoter context. (A) Schematic representation of the bPHO5 synthetic promoter indicating the positioned nucleosomes (−2, −1), the Gcn4-binding site (GCRE), the TATA element, the start site of transcription (arrow) and the ClaI cleavage site used for monitoring nucleosome remodeling. (B) bPHO5–HIS3 mRNA (bPHO5) isolated from the indicated strains expressing the Gcn4–Gal4 chimeric activator and grown under repressive (M) or inducing (AT) conditions. DED1 mRNA was used as a loading control. (C) Left: occupancy of the bPHO5 promoter by the Gcn4–Gal4 hybrid activator in gcn4 (WT) and gcn4spt3 (spt3) strains growing either under repressive (open bars) or inducing (gray bars) conditions. This was determined by ChIP and quantitative PCR and numbers reflect the fold enrichment of the tested DNA relative to a region of PHO5 ORF after correction for the ratios of amplification achieved using input DNA. Right: remodeling of the −2 nucleosome at the bPHO5 synthetic promoter (TATA) and its mutated derivative (CGTA). Remodeling was assayed by accessibility to restriction enzyme ClaI in the gcn4 (WT), gcn4gcn5 (gcn5) and gcn4spt3 (spt3) strains expressing the chimeric activator and growing under repressive (M) or inducing (AT) conditions. P is the protected DNA, whereas U is a fragment that results by ClaI cleavage and secondary digestions. Also shown is the quantification of these experiments expressed as the ratio of unprotected to protected (U/P) DNA fragments. (D) Left: schematic representation of the NPS indicating the −2 nucleosome, the Gcn4-binding site (GCRE) and the ClaI cleavage site used for monitoring nucleosome remodeling. Right: remodeling of the −2 nucleosome at the NPS. Remodeling was assayed and quantified as described in (C).
Spt3 functions in nucleosome remodeling at a nonpromoter site
Based on the fact that SAGA can be recruited by Gal4 even at nonpromoter regions containing the cognate DNA target (Bhaumik and Green, 2001), we transferred the DNA region occupied by nucleosome −2 along with the Gcn4-binding site on a yeast episomal vector (Figure 2D, left, see also Topalidou and Thireos, 2003). As expected, high amounts of the Gcn4–Gal4 hybrid protein could recruit SAGA on this site (see below) but not TBP (data not shown). When growth was shifted to amino-acid limitation conditions, the nucleosome organized by this DNA was remodeled in a manner dependent on Spt3 but not on Gcn5 (Figure 2D, right). This result was important as it uncoupled completely Spt3's participation in nucleosome remodeling not only from TBP recruitment but also from other steps in transcription initiation.
Mot1 is required for Gal4-directed nucleosome remodeling
Given the above results, the obvious question was how Spt3 could mediate chromatin remodeling. We approached this question by examining factors that have been previously shown to interact genetically with Spt3 and are required for GAL1 transcription. One of these is the TBP interacting protein Mot1 that belongs to the Snf2 family of ATPases (Auble et al, 1994; Collart, 1996; Madison and Winston, 1997; Prelich, 1997). Strains mutated for the MOT1 gene proved to have some interesting properties. Firstly, in the absence of a functional Mot1, both transcriptional activation and nucleosome remodeling at the GAL1 promoter could not be accomplished (Figure 3A, top and 3B) and this was not due to a failure of Gal4 recruitment (Figure 3A, bottom). Secondly, the positive function of Mot1 on nucleosome remodeling involved its ATPase domain, as a point mutation that affects this function (Dasgupta et al, 2002) exhibited the same phenotypes (Figure 3B). Finally, Mot1 was also required for nucleosome remodeling at the NPS as assayed both by restriction endonuclease sensitivity and micrococcal nuclease digestions (Figure 3C), and this was not the indirect result of activator recruitment (Figure 3D). Interestingly enough, Mot1 dependence was activator independent as transcriptional activation through the bPHO5 promoter directed by the Gcn4 activation domain was also compromised in a mot1-1 strain (data not shown). We concluded that Mot1, a protein that, similarly to Spt3, has been connected with TBP recruitment (Andrau et al, 2002; Geisberg et al, 2002), is involved in Gal4-guided nucleosome remodeling in a TBP recruitment autonomous manner.
Figure 3.
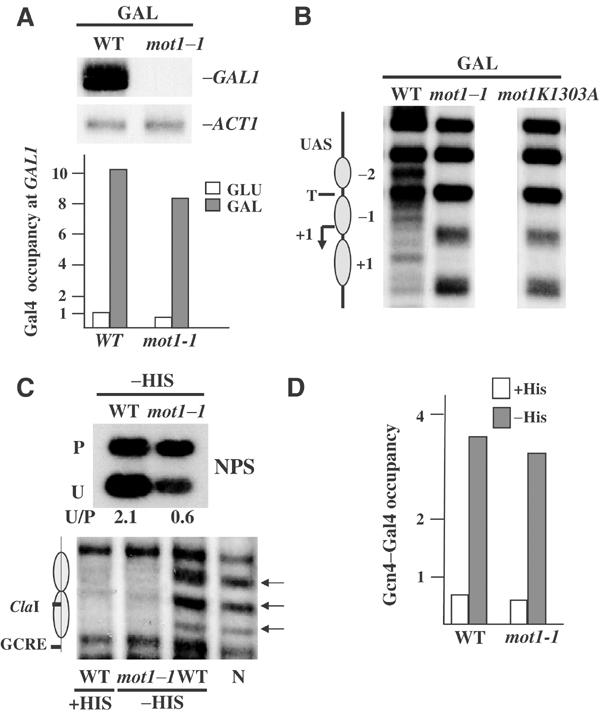
Mot1 is required for Gal4-mediated transcription and nucleosome remodeling. (A) Top: Northern blot analysis of GAL1 mRNA isolated from wild-type (WT) and mot1-1 strains grown under inducing conditions (GAL). ACT1 mRNA was used as a loading control. Bottom: GAL1 promoter occupancy by Gal4 monitored by ChIP in the indicated strains grown either in glucose (GLU) or galactose (GAL). Numbers in the x-axis are as described in the legend of Figure 2C. (B) Nucleosome remodeling at the GAL1 promoter as assayed by MNase sensitivity. Wild-type (WT), mot1-1 and mot1-1 expressing the ATPase-deficient form of MOT1 (mot1K1303A) strains were grown under inducing conditions (GAL). (C) Remodeling of the nucleosome at the NPS as assayed by either accessibility to ClaI (top), quantified as in Figure 2C, or to MNase (bottom) in gcn4 (WT) and gcn4mot1-1 (mot1-1) strains expressing the chimeric activator and growing under normal (+His) or histidine-depleted conditions (−His). Also shown is the pattern of MNase digestion of naked DNA (N). Arrows indicate sites that become hypersensitive to MNase following induction. (D) Nonpromoter GCRE site (NPS) occupancy by the Gcn4–Gal4 hybrid activator determined by ChIP in gcn4 (WT) and gcn4mot1-1 (mot1-1) strains growing under normal (+His) or histidine-depleted conditions (−His). Numbers in the x-axis are as described in the legend of Figure 2C.
Interdependent recruitment of Spt3 and Mot1
The similar phenotypes of mot1-1 and spt3Δ prompt us to investigate any dependencies in the recruitment of the corresponding proteins on the tested chromatin templates. We first investigated whether Mot1 occupied these regions. For the GAL1 promoter, we observed that under inducing conditions Mot1 was indeed recruited and this was dependent on both the presence of Gcn5 and Spt3 (Figure 4A). These dependencies were not due to an indirect effect on the amount or the recruitment potential of Mot1, as its reported occupancy (Andrau et al, 2002) at the SAGA-independent PGK1 promoter (Lee et al, 2000) was unchanged in either gcn5 or spt3 strains (Figure 4A). In addition, we should note that in both strains wild-type levels of Gal4 are recruited on GAL1 (Dudley et al, 1999). Interestingly enough, Mot1 was also recruited at the NPS in the absence of any bound TBP but this was only dependent on Spt3 (Figure 4B). Neither Mot1 occupancy on PGK1 promoter (Figure 4B, left) nor Gcn4–Gal4 recruitment at the GCRE site (Figure 2C, left) was dependent on Spt3, two facts arguing again against indirect effects resulting from the absence of this SAGA component. Surprisingly, Spt3 (and presumably SAGA) could not be assembled on both tested sites in the absence of Mot1 (Figure 4C and D). It should be noted that in mot1-1 strains, Gal4 recruitment on GAL1 as well as that of Gcn4–Gal4 at the nonpromoter GCRE were unaffected (Figure 3A, bottom and 3D). In addition, Mot1 was not required for the integrity of SAGA as in a mot1-1 strain, SAGA was assembled properly on promoters requiring its function (Bhaumik and Green, 2002) but not requiring Mot1 (Andrau et al, 2002) such as that of ADH1 (Figure 4C and D). This observed interdependent recruitment indicated a tight coordination for the assembly and chromatin remodeling function of Mot1 and Spt3 on Gal4-regulated promoters.
Figure 4.
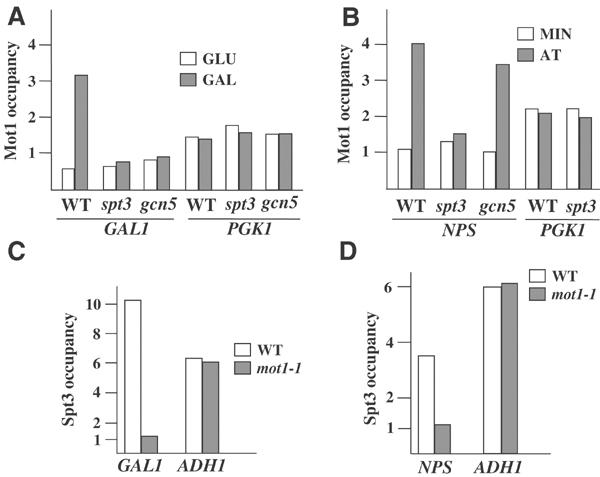
Interdependent recruitment of Spt3 and Mot1 on the GAL1 promoter and the NPS. (A) Occupancy of the GAL1 and PGK1 promoters by Mot1. Chromatin immunoprecipitation was conducted using the indicated strains expressing Myc-tagged Mot1 and growing under repressive (GLU) or inducing (GAL) conditions. (B) Occupancy of the NPS and PGK1 promoter by Mot1. ChIP was performed in gcn4 (WT), gcn4gcn5 (gcn5) and gcn4spt3 (spt3) strains expressing the chimeric activator and Myc-tagged Mot1 and growing under repressive (MIN) or inducing (AT) conditions. (C) Occupancy of GAL1 and ADH1 promoters by Spt3. ChIP was performed in the indicated strains grown in galactose and containing the Myc-tagged version of Spt3. (D) Occupancy of the NPS and ADH1 promoter by Spt3. ChIP was performed in gcn4 (WT) and gcn4mot1-1 (mot1-1) strains expressing the chimeric activator and the Myc-tagged Spt3 and grown in histidine-depleted media. In all experiments numbers in the x-axis are as described in the legend of Figure 2C.
Discussion
In this report, we have revealed a novel role involving nucleosome remodeling for two transcriptional regulators, Spt3 and Mot1, both implicated in taking part, either positively or negatively, in the mechanisms of TBP recruitment (Auble et al, 1994, 1997; Dudley et al, 1999; Larschan and Winston, 2001; Andrau et al, 2002; Bhaumik and Green, 2002; Barbaric et al, 2003). Our results demonstrate unequivocally that their function in nucleosome remodeling is independent of the stable recruitment of TBP, as it was evident in both tested promoters in the absence of a TATA element. Most importantly, both proteins were required for the remodeling of a nucleosome located just next to an activator landing site where not only TBP cannot be assembled but also transcription initiation is precluded.
Elegant genetic studies have identified allele-specific interactions between spt3 and spt15 mutations (Madison and Winston, 1997; Larschan and Winston, 2001). These data, combined with the evidence for interaction between Spt3 and TBP (Eisenmann et al, 1992), led to the proposal that this SAGA component is involved in the adaptor functions of the complex that support the recruitment of TBP (Larschan and Winston, 2001; Bhaumik and Green, 2002; Barbaric et al, 2003). Now we show that Spt3 has a function in nucleosome remodeling, which is independent of TBP recruitment. These two functions could be completely separable or alternatively they might be detached but highly coordinated. Finally, we cannot exclude the possibility that both phenotypes could reflect a single but yet elusive function in which one reaction permits both nucleosome remodeling and the recruitment of TBP.
It is well established that another SAGA component involved in nucleosome remodeling is the Gcn5 HAT. Whole genome studies have shown that among the SAGA-dependent genes, the requirements for Gcn5 or Spt3 are distinct and only a small proportion requires both proteins for transcriptional activation (Lee et al, 2000). Indeed, both activation and remodeling of the synthetic promoter and its derivatives were Gcn5 independent, a fact demonstrating that the involvement of Spt3 in nucleosome remodeling is not mediated by this HAT. On the other hand, nucleosome remodeling at the GAL1 promoter was dependent on both Gcn5 and Spt3. It is possible that these requirements reflect the complexity of the transcriptional activation of this promoter that necessitates relief from Ssn6/Tup1-mediated repression. This repression, being mediated by HDACs, could explain the requirement for an opposing activity such as the Gcn5 HAT on the native GAL1 promoter.
If Gcn5 does not mediate Spt3 function then how is this elicited? A clue that could offer mechanistic explanations on the role of Spt3 in nucleosome remodeling was offered by our findings for a similar function performed by the Mot1 ATPase. Mot1 was initially identified as a negative regulator of transcription involved in the mechanics of TBP recruitment (Auble et al, 1994). Mot1 was found to bind in vitro to either free or DNA bound TBP and to effect an ATP-dependent dissociation of TBP from its target DNA (Auble et al, 1994, 1997). Although this enzymatic property can explain its repressive function, it is insufficient to provide reasoning for the recently discovered positive function of Mot1 in transcription (Andrau et al, 2002; Dasgupta et al, 2002). Indeed, whole genome studies revealed that approximately 7% of the yeast genes do require Mot1 for transcription and this was positively correlated with TBP recruitment (Andrau et al, 2002; Geisberg et al, 2002). This correlation is far from demonstrating a direct involvement of Mot1 in TBP recruitment, but it is in concert with an indirect effect as suggested by our findings. Thus, the positive role of Mot1 in transcription and specifically that of its ATPase activity can be attributed to its role in nucleosome remodeling, which in turn allows for the stable recruitment of TBP. Mechanistically, this role can be direct and analogous to that of Snf2, a known player in nucleosome remodeling that belongs to the same ATPase family.
Irrespective as to whether Mot1 involvement in chromatin remodeling is direct, such a function could mediate the observed role of Spt3 in nucleosome remodeling. Support for such mediation comes from the reported genetically deduced interaction of the two proteins, and our findings showing the requirement of Spt3 for the stable recruitment of Mot1. The fact though, that Mot1 was also required for recruitment of SAGA, excludes a simple model based on sequential dependencies for recruitments and suggests a functional coordination on chromatin remodeling that determines the stability of the recruitments. Given the fact that upon remodeling the DNA contacts with nucleosomal histones are lost (Boeger et al, 2003; Reinke and Horz, 2003), an interesting speculation is that following an initial Spt3-dependent Mot1-driven nucleosome remodeling, SAGA could be stabilized through contacts of the now free DNA with its histone fold containing TAF II components (Selleck et al, 2001). In turn, stabilization of Mot1 and thus maintenance of the remodeled state could be secured through its interaction with SAGA, a fact supported by relevant immunoprecipitation experiments (our own unpublished observations).
Materials and methods
Yeast strains and media
The gcn4Δ, gcn5Δ, gcn4Δgcn5Δ, spt3Δ and spt3Δgcn4Δ strains were obtained by appropriate gene disruptions of a GAL2 S288C parental strain with the genotype MATa trp1-1, leu2-3, ura3-52 as described in Georgakopoulos et al (1995). The FY1291 strain (MATa, leu2Δ1, ura3-52, arg4-12, lys2-173R2, trp1Δ63, spt20Δ::ARG4) was provided by F. Winston. The JD215B strain (Mata, leu2-3,112, ura3-52, trp1-1, his4-519, can1-101 mot1-1) was rendered gcn4Δ using kanamycin selection. Wild-type, gcn5Δ or spt3Δ strains bearing the mutated GAL1 CGTA element were derived by two step replacements of their endogenous loci as described previously (Topalidou et al, 2003). Mot1 and Spt3 were tagged at the C terminus with 9 Myc epitopes, as described in Topalidou and Thireos (2003). Repressed growth conditions were either rich medium (YPD) or minimal medium supplemented with the required amino acids. High levels of Gcn4-dependent transcription were achieved either by adding 3-AT to the medium at a concentration of 10 mM and thus eliciting histidine limitation or by histidine depletion of histidine auxotrophic strains. GAL1 induction was achieved by growing cells to mid log phase in rich media containing 2% raffinose and then shifted to YPG (2% galactose) for 30 min.
Plasmid constructions
The synthetic reporter bPHO5 and mononucleosomal construct NPS have been described in Topalidou and Thireos (2003). The bPHO5–HIS3 reporter was derived by replacing the LacZ ORF of bPHO5 with a truncated HIS3 gene. The Gcn4–Gal4 hybrid was constructed by replacing the activation domain of a genomic clone of Gcn4 (amino acids 54–168) with the activation domain of Gal4 (amino acids 768–881). The TATA box mutations within the synthetic reporter or the GAL1 gene were obtained by site-directed mutagenesis as described in Topalidou and Thireos (2003). The plasmid expressing the Mot1 ATPase mutant (mot1–K1303A) was a gift from D Auble (Dasgupta et al, 2002).
Gene expression analysis and in vivo nucleosome remodeling assays
Total yeast RNA was isolated from yeast cells grown to an OD550 ∼0.6–0.8 by the hot phenol method as described in Topalidou et al (2003). Remodeling of nucleosome −2 was assayed by the restriction enzyme accessibility assay as described in Topalidou and Thireos (2003). Accessibility was monitored following secondary digestions with EcoRI/HindIII or PvuII/HindIII (for the mononucleosomal construct) and probing with the EcoRI–HindIII PHO5 fragment. Micrococcal nuclease assays were performed using nystatin-permeabilized spheroplasts (Topalidou et al, 2003). Following MNase digestions, the nucleosomal structure of the GAL1 promoter was analyzed by the indirect end labeling method, using PvuII for secondary digestion and the BsaI–PvuII fragment of GAL1 as a probe. Similarly, the nucleosomal structure of the region upstream of the nonpromoter GCRE was analyzed by indirect end labeling using SspI for secondary digestion and a PCR fragment encompassing positions 1360–1700 of pRS316.
Chromatin immunoprecipitation
Chromatin was immunoprecipitated as described in Topalidou et al (2003) using either anti-Myc antisera (Santa Cruz) or an antibody against Gal4 C-terminal activation domain (Abcam). The recovered DNA was subjected to quantitative real-time PCR analysis. GAL1 promoter was analyzed using primers encompassing the Gal4 UAS and nucleosome −2 (primer coordinates −169 and −370). A primer corresponding to the GCRE element along with a primer specific to the region of nucleosome −2 (5′ CATTGGTAATCTCGAAT 3′) were used for PCR amplification of the mononucleosomal reporter DNA. The coordinates of the primers amplifying the ADH1 promoter were −220 and −9 and those of PGK1 were −310 and −69 relative to the start codon. Quantification reflected the fold enrichment of the tested DNA relative to a region of PHO5 ORF (primer coordinates +1017 and +1220) after correction for the ratios of amplification achieved using input DNA. Each chromatin IP was repeated at least three times and the variation between experiments was ±8–10%.
Acknowledgments
We thank David Auble for the MOT1 ATPase mutant and Martin Collart for the mot1-1 strain. This work was supported by PENED 2002 (GT) and RTN (DT) grants. IT and MPC are graduate students at Department of Biology, University of Crete.
References
- Andrau JC, Van Oevelen CJ, Van Teeffelen HA, Weil PA, Holstege FC, Timmers HT (2002) Mot1p is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J 21: 5173–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S (1994) Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev 8: 1920–1934 [DOI] [PubMed] [Google Scholar]
- Auble DT, Wang D, Post KW, Hahn S (1997) Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol 17: 4842–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod JD, Reagan MS, Majors J (1993) GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev 7: 857–869 [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S (2002) Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem 277: 7989–7995 [DOI] [PubMed] [Google Scholar]
- Barbaric S, Reinke H, Horz W (2003) Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol 23: 3468–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15: 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol Cell Biol 22: 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger H, Griesenbeck J, Strattan JS, Kornberg RD (2003) Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell 11: 1587–1598 [DOI] [PubMed] [Google Scholar]
- Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337 [DOI] [PubMed] [Google Scholar]
- Collart MA (1996) The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol 16: 6668–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT (2002) Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci USA 99: 2666–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F (1999) The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev 13: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F (1992) SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6: 1319–1331 [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F (1994) The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics 137: 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Kuras L, Struhl K (2002) Mot1 associates with transcriptionally active promoters and inhibits association of NC2 in Saccharomyces cerevisiae. Mol Cell Biol 22: 8122–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos T, Gounalaki N, Thireos G (1995) Genetic evidence for the interaction of the yeast transcriptional co-activator proteins GCN5 and ADA2. Mol Gen Genet 246: 723–728 [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR III, Workman JL (1998) A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94: 45–53 [DOI] [PubMed] [Google Scholar]
- Johnston M, Flick JS, Pexton T (1994) Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol 14: 3834–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Winston F (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15: 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704 [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA (1998) Regulation of gene expression by TBP-associated proteins. Genes Dev 12: 1398–1408 [DOI] [PubMed] [Google Scholar]
- Lohr D (1997) Nucleosome transactions on the promoters of the yeast GAL and PHO genes. J Biol Chem 272: 26795–26798 [DOI] [PubMed] [Google Scholar]
- Madison JM, Winston F (1997) Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol 17: 287–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D (2002) Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell 9: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Prelich G (1997) Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2alpha homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol 17: 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD (2001) Histone acetyltransferases. Annu Rev Biochem 70: 81–120 [DOI] [PubMed] [Google Scholar]
- Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S (2001) A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol 8: 695–700 [DOI] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL (1999) Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol 19: 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL (2002) The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J Biol Chem 277: 8178–8186 [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G (2000) The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404: 414–417 [DOI] [PubMed] [Google Scholar]
- Thireos G, Penn MD, Greer H (1984) 5′ untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci USA 81: 5096–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Papamichos-Chronakis M, Thireos G (2003) Post-TATA binding protein recruitment clearance of Gcn5-dependent histone acetylation within promoter nucleosomes. Mol Cell Biol 23: 7809–7817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Thireos G (2003) Gcn4 occupancy of open reading frame regions results in the recruitment of chromatin-modifying complexes but not the mediator complex. EMBO Rep 4: 872–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Durbin KJ, Fink GR (1984) The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39: 675–682 [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Narita T, Inukai N, Wada T, Handa H (2001) SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J Biochem (Tokyo) 129: 185–191 [DOI] [PubMed] [Google Scholar]


