Abstract
Pathogenic Neisseria express type IV pili (tfp), which have been shown to play a central role in the interactions of bacteria with their environment. The regulation of piliation thus constitutes a central element in bacterial life cycle. The PilC proteins are outer membrane-associated proteins that have a key role in tfp biogenesis since PilC-null mutants appear defective for fibre expression. Moreover, tfp are also subjected to retraction, which is under the control of the PilT nucleotide-binding protein. In this work, we bring evidence that fibre retraction involves the translocation of pilin subunits to the cytoplasmic membrane. Furthermore, by engineering meningococcal strains that harbour inducible pilC genes, and with the use of meningococcus–cell interaction as a model for the sequential observation of fibre expression and retraction, we show that the PilC proteins regulate PilT-mediated fibre retraction.
Keywords: pathogenic Neisseria , PilC, PilT, retraction, type IV pili
Introduction
Type IV pili (tfp) are bacterial appendages that are widely spread among Gram-negative bacteria (Strom and Lory, 1993) and have been shown to be the key elements for the interactions of bacteria with their environment. In pathogenic Neisseria, tfp are responsible for adhesion to human host cells (Rudel et al, 1992; Nassif et al, 1994), as well as transformation competence (Rudel et al, 1995b), which are major features of the neisserial life cycle. Current models consider that the pilus is assembled from pilin subunits that are located in the cytoplasmic membrane, in a process that might be energised by the PilF component, a putative nucleotide-binding protein (Freitag et al, 1995; Merz and Forest, 2002). Once assembled, the fibre can be extruded to the bacterial surface through an outer membrane secretin named PilQ (Wolfgang et al, 2000).
Pilus retraction is responsible for twitching motility, and has been shown to play a central role in the interactions of pathogenic Neisseria with human cells (Pujol et al, 1999; Merz et al, 2000). It requires de novo protein synthesis and relies on the presence of the PilT protein (Merz et al, 2000). Neisserial PilT protein was found in the cytosol as well as associated with the cytosolic membrane (Brossay et al, 1994). Forces up to 110 pN are generated by the retraction of the fibre, and the rate of PilT-mediated retraction is under the control of the presence of ATP (Maier et al, 2002). PilT proteins from other species were shown to have ATPase activity (Okamoto and Ohmori, 2002) and to form oligomers (Herdendorf et al, 2002). They are remote members of the AAA superfamily of proteins, a ubiquitous group of proteins whose members have the ability to display nucleotide-dependant conformational switches that may apply tension to bound proteins (Vale, 2000; Merz and Forest, 2002). They thus act as multimeric mechanoenzymes that are able to disassemble stable protein–protein complexes.
Among the components of the neisserial tfp machinery, the PilC proteins play a crucial but still enigmatic role (Nassif et al, 1994; Rudel et al, 1995a; Wolfgang et al, 2000). They are associated with the outer membrane but can also be recovered from purified pili (Rudel et al, 1995c; Rahman et al, 1997). PilC-null strains show impairment in pilus expression and are not competent for transformation. However, the mechanism by which the PilC proteins promote piliation remains unknown. They act as pilus tip adhesins in Neisseria gonorrhoeae (gonococcus) (Rudel et al, 1995b), and the two pilC loci harboured by this species are functionally interchangeable. On the other hand, in Neisseria meningitidis (meningococcus), only PilC1 is equivalent to the gonococcal PilC proteins and promotes adhesion. PilC2, which is independently expressed from PilC1, fails to promote adhesion despite identical functions in piliation and transformation competence (Nassif et al, 1994; Morand et al, 2001). The abolition of PilT in a PilC-null background restores piliation (Wolfgang et al, 1998b). This suggests that both components have antagonistic effects on tfp biogenesis, and it was hypothesised that the role of the PilC proteins in pilus biogenesis is to stabilise nascent fibres before translocation through the outer membrane (Wolfgang et al, 2000), by acting either as initiator of extrusion or as a chaperone for tfp formation (Wolfgang et al, 1998b).
The apparent piliation status of a bacterial population corresponds to the sum of fibre extension and retraction events on every single cell. Most bacteria express more than one pilus fibre, and the coordination of all pilus sites within a single bacterium is not fully understood (Long et al, 2001). The concomitance of pilus elongation and retraction within a bacterial population can easily be observed as bacterial ‘twitching' in liquid growth. In order to get insights into tfp retraction, one needs a model in which retraction can be dissociated from fibre extension for a whole bacterial population. Adhesion of N. meningitidis to human cells is a major tfp-related phenotype in which both PilC and PilT have been shown to have crucial functions, and which typically involves two distinct successive steps. The first step corresponds to the localised adhesion of bacterial microcolonies onto host cells. This is pilus mediated and characterised by the upregulation of the meningococcal PilC1 protein (Taha et al, 1998). Following this stage, pili are retracted and bacteria subsequently appear non-piliated, forming a monolayer covering the apical surface of the cells (Pujol et al, 1997, 1999). Given that pilus retraction is observed for the whole adhering bacterial population, meningococcal interaction with host cells represents a unique synchronised physiological model for studying tfp retraction.
In this work, using the interaction of meningococci with human cells as a model for sequential tfp extension and retraction, we bring evidence that the tfp biogenesis role of the PilC proteins is to prevent the PilT-induced ‘pilus melting' into the cytoplasmic membrane.
Results
Meningococcal piliation correlates with the level of PilC in a PilT+ background
To investigate the mechanism by which PilC promotes piliation in pathogenic Neisseria, we engineered C2SC1WEa, a meningococcal strain harbouring an IPTG-inducible pilC1 gene (pilC1ind) (Materials and methods). To alleviate for interferences due to the wild-type pilC2 variant that is simultaneously expressed in the Nm2C4.3 (wild type) strain, the pilC2 locus was inactivated. As expected, the expression of PilC1 was dependent upon IPTG concentration, whereas the amount of PilE and PilT remained unchanged in all conditions of induction (Figure 1A). Transcriptional analysis using real-time PCR showed that the transcriptional level of pilC1ind could be tuned up to twice that of pilC1wt with 1 mM IPTG (Figure 1B). The piliation status of this PilC1ind/PilTwt strain was assessed by using immunofluorescence staining of pilin, and correlated with IPTG concentration (Figure 2, panel PilC1ind/PilTwt). Thus, as long as pilin and PilT expressions are unchanged, the level of piliation correlates with that of pilC1 transcription. Identical results were obtained when the meningococcal pilC2 gene was induced instead of pilC1 (data not shown), confirming previous data showing that both meningococcal PilC variants are functionally identical for tfp biogenesis (Morand et al, 2001).
Figure 1.
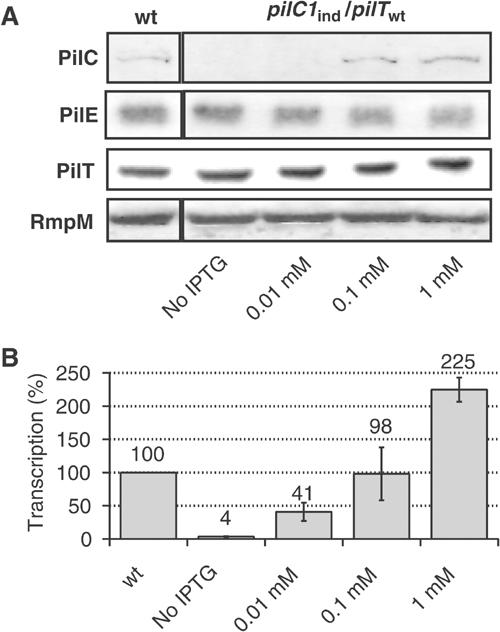
Phenotypes associated with the induction of pilC1. Strains NmC2SC1WEa (PilC1ind/PilTwt) and Nm2C4.3 (wild type), as reference, were grown on GC-agar plates supplemented with IPTG (concentration up to 1 mM). (A) Western blot analysis. Total bacterial extracts were analysed for the presence of PilC, pilin (PilE), PilT and RmpM (extracts calibration). Semiquantitative analysis of Western blots showed an approximate two-fold overexpression of PilC1ind with 1 mM IPTG, compared with PilC1wt. (B) Comparison of the transcriptional level of pilC1ind (NmC2SC1WEa) for IPTG concentration up to 1 mM, with that of pilC1wt (Nm2C4.3, 100% reference). The transcription of pilC1ind is expressed as a percentage of that of pilC1wt. Data are mean±standard error of the mean, resulting from at least six measurements.
Figure 2.
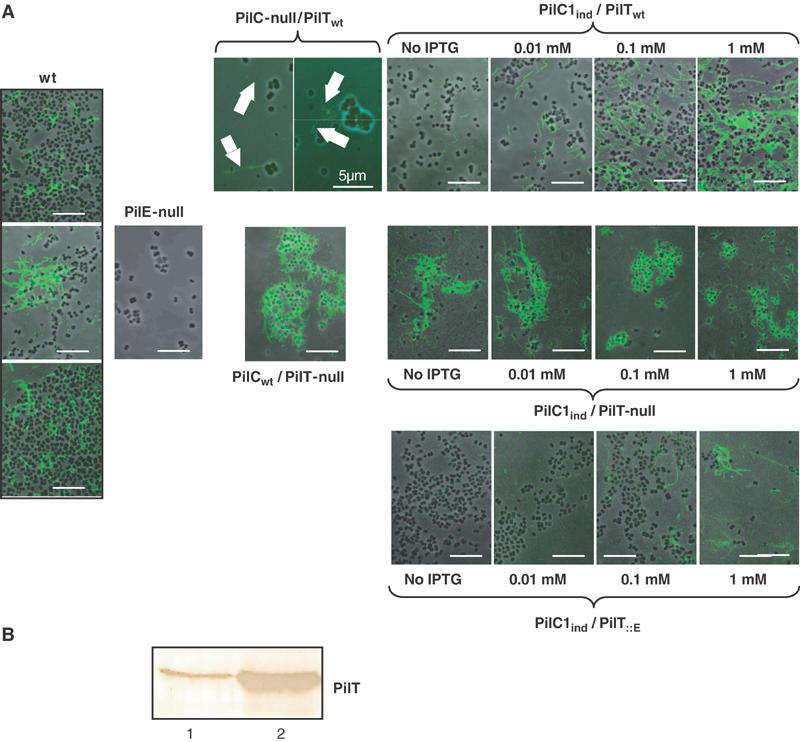
Piliation phenotypes in relation with the balance between PilC1 and PilT. (A) Strains Nm2C4.3 (wild type), PilE-null, PilC-null/PilTwt, PilCwt/PilT-null, PilC1ind/PilTwt (NmC2SC1WEa), PilC1ind/PilT-null (NmC1ITSE) and PilC1ind/PilT∷E (NmC1IESK) were grown on GC-agar plates supplemented with up to 1 mM IPTG. Piliation was assessed by immunofluorescence staining of pilin. Bacteria appear in phase contrast and pilin in green; the arrows show pili. Unless stated, the scale bar is 10 μm. In a PilT+ background, piliation correlates with the expression of PilC1 (panel PilC1ind/PilTwt), whereas abolition of PilT-driven retraction leads to hyperpiliation, independent of the amount of PilC1 (panel PilC1ind/PilT-null). For each level of pilC1 induction, increased expression of PilT leads to decreased piliation (panels PilC1ind/PilTwt and PilC1ind/PilT∷E). (B) Immunolabelling of PilT: Similar amounts of total protein extracts of wild-type (Nm2C4.3, lane 1) and PilC1ind/PilT∷E (NmC1IESK, lane 2) strains were used for Western blotting. Expression of the pilT∷E construct leads to increased levels of PilT, compared to the wild type.
It should be pointed out that pili were observed in noninduced samples (Figure 2, panel PilC1ind/PilTwt). This could be attributed to promoter leakage, as previously described with other genes (Long et al, 2001). However, some pili were surprisingly also visible on the surface of virtually all PilC-null/PilTwt bacteria. These fibres were of low amount and appeared very thin, suggesting that they were not forming large bundles like those of the wild type. They could only be observed using immunofluorescence, but could not be reliably detected by electron microscopy. Thus, the level of piliation is regulated by the expression of the PilC proteins, although these molecules are dispensable for tfp biogenesis.
PilT and PilC coordinately regulate fibre retraction
As mentioned above, the abolition of PilT in a PilC-null background restores piliation (Wolfgang et al, 1998b). This prompted us to assess the role of PilC in a PilT-null background. We engineered NmC1ITSE, a PilT-null derivative of the above-described strain that harbours pilC1ind (Materials and methods). In contrast with the PilTwt isogenic background (Figure 2, panel PilC1ind/PilTwt), the high level of piliation in a PilT-null background remained unchanged regardless of the induction of pilC1 (Figure 2, panel PilC1ind/PilT-null). Thus, abolition of retraction by loss of function of PilT shows that pilus assembly and extrusion to the bacterial surface take place independent of the amount of PilC. This result does not support the hypothesis that PilC proteins act as chaperones for fibre assembly, but suggests that PilC stabilises the pilus once it is formed and extruded to the bacterial surface, probably by counteracting PilT-mediated retraction.
In such a model, enhanced retraction for defined levels of PilC would result in decreased piliation. In order to test this hypothesis, we engineered NmC1IESK (Materials and methods), a pilC1ind strain that expresses pilT under the control of a pilE promoter (pilT∷E), thus leading to increased levels of PilT compared to the wild type (Figure 2B). Piliation of this pilC1ind/pilT∷E strain was assessed by immunofluorescence for different levels of induction of pilC1. For each condition of induction, hyperexpression of PilT resulted in decreased piliation, compared to the isogenic pilC1ind/pilTwt strain (Figure 2). These results support the hypothesis that the balance between the transcription of pilC and pilT controls the level of piliation by regulating pilus retraction.
Pilus retraction during meningococcus–cell interactions requires downregulation of PilC1
To further address the involvement of PilC in pilus retraction, we took advantage of the induction of pilus retraction occurring in N. meningitidis upon contact with human cells. As mentioned above, meningococcal adhesion involves two steps in which bacteria successively undergo intense tfp expression and fibre retraction. Figure 3 illustrates these steps (panel A, 1H and 8H), as well as the requirement of PilT-driven fibre retraction for the loss of piliation (panel B) (Pujol et al, 1999). We used this model to synchronise pilus retraction among a bacterial population, and to monitor the transcription of three components known to act as modulators of pilus biogenesis (i.e. pilT, pilC1 and pilF) in the course of adhesion of a wild-type meningococcal strain onto human umbilical vein endothelial cells (HUVECs). Figure 4 shows the transcriptional level of each gene during localised (1H) and intimate (8H) adhesion, measured by real-time quantitative PCR analysis (Materials and methods). A chromosomal kanamycin resistance aphA3 cassette was used as reference, and the ‘induction index' was calculated for each gene considered at every time point, using non-cell-associated bacteria grown in an identical infection medium as calibrator. For the three genes, transcription of the calibrator (bacteria grown in the absence of cells) appeared to be stable throughout the experiment (data not shown). The transcription of pilF remained stable throughout adhesion, thus appearing independent of the variation of piliation in this model. The transcription of pilT was moderately affected by the interaction of the bacteria with the cells. On the other hand, the transcription of pilC1 was upregulated during the initial localised adhesion, when bacteria are heavily piliated, and subsequently reduced by two-fold during the intimate adhesion, when pili are retracted. Despite repeated washings of the infected cell monolayer, residual re-infection during the course of adhesion is probably responsible for the slight upregulation of pilC1 that can be observed after 8 h, compared to bacteria grown in infection medium alone. The initial upregulation of PilC1 has previously been shown to be essential for an efficient meningococcus–cell interaction (Taha et al, 1998). On the other hand, the subsequent downregulation of pilC1 transcription is associated with the PilT-driven loss of pili.
Figure 3.
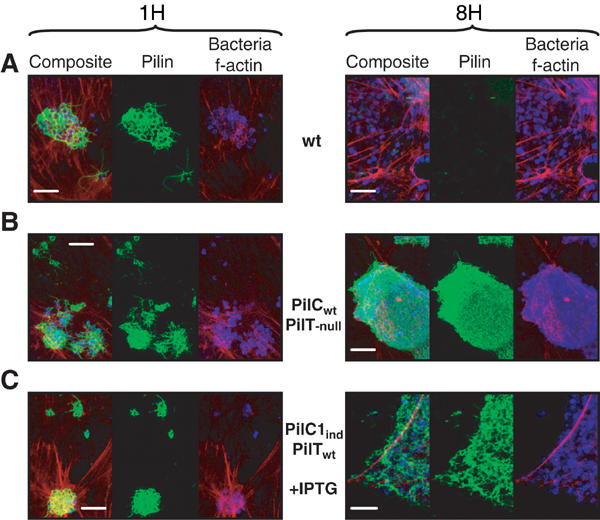
Piliation status of meningococci during adhesion. The wild-type (Nm2C4.3, panel A), PilCwt/PilT-null (panel B) and PilC1ind/PilTwt (NmC2SC1WEa, panel C) strains were allowed to adhere onto HUVECs for 1 h (localised adhesion) or 8 h (intimate adhesion). Bacteria appear blue, f-actin red and pili green. The scale bar is 10 μm. (A) Adhesion of the wild type is characterised by the loss of pili at intimate adhesion. (B) Tfp retraction relies on the presence of PilT: The PilT-null strain remains piliated at a time corresponding to intimate adhesion for the wild type. (C) Constant induction of pilC1 with IPTG in the PilC1ind/PilTwt strain prevents fibre retraction at late adhesion times.
Figure 4.
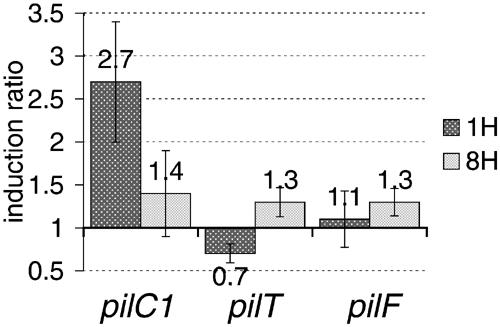
Transcriptional measurements for pilC1, pilT and pilF during adhesion. Relative transcription levels of pilC1, pilT and pilF were measured at localised (1H) and intimate (8H) adhesion stages. The induction ratio compares the transcription of genes in cell-associated bacteria with that of meningococci grown in the absence of human cells (infection medium alone). The transcription of pilC2 was not monitored since it was previously shown that it is not affected by adhesion (Taha et al, 1998). Data represent mean±standard error of the mean, resulting from at least six independent measurements.
These data strengthen the model that PilC proteins prevent pilus retraction. To confirm this hypothesis, we performed adhesion onto human cells using the above-described pilC1ind/pilTwt strain (NmC2SC1WEa). The use of 1 mM IPTG allowed a pilC1ind transcription level that was twice that of the wild type, thus being close to the overtranscription of pilC1 during initial localised adhesion of wild-type N. meningitidis (Figure 4). The piliation status of cell-associated bacteria was assessed under sustained induction, at times corresponding to localised and intimate adhesion steps. Unlike the wild type that exhibited a dramatic loss of pili in the onset of intimate adhesion (Figure 3A), the adhering IPTG-induced bacteria expressed a highly piliated phenotype at all time points (Figure 3C). Thus, the persistence of pilC1 expression in adhering bacteria leads to the persistence of abundant tfp expression and prevents pilus retraction. These results suggest that the lack of downregulation of pilC1 transcription during meningococcus–cell interaction prevents PilT-induced pilus retraction, and argue that PilC is involved in the regulation of piliation by refraining PilT-driven retraction.
Pilus retraction is associated with the relocalisation of mature pilin subunits along the cytoplasmic membrane
The above results strongly suggest that the pilus biogenesis role of PilC is the repression of PilT-driven fibre retraction. Since little is known about the molecular mechanisms involved in retraction, we sought to determine the location of pilin subunits during the course of meningococcus–cell interaction in adhesion. Using the wild type, as well as the PilT-null mutant as negative control for tfp retraction, we performed pilin gold-labelling experiments on ultra-thin sections of resin-embedded infected cell monolayers (Figure 5). During localised adhesion (1H), both wild-type and PilCwt/PilT-null strains displayed similar pilin distribution patterns. The pilin-bound gold beads were seen in all bacterial compartments, as well as outside the bacterial cell as parts of tfp fibres. This observation correlates with the high level of piliation of both wild-type and PilCwt/PilT-null strains at this stage of adhesion (Figure 3). On the other hand, at a time corresponding to intimate adhesion in the wild type (8H), both strains featured dramatically different patterns. In the wild type, pilin-bound gold beads were predominantly associated with the bacterial cytoplasmic membrane, whereas the PilT-null mutant did not display significant modification of pilin distribution compared to early time point. The relative bead diameter and membrane thickness made it difficult to conclude whether pilin subunits were within the membrane or simply attached to it. However, structural models suggest that pilin monomers are not soluble in aqueous media and that the amino-terminal end of the protein is very hydrophobic, which is consistent with the presence of the pilin subunits within the cytoplasmic membrane (Finlay et al, 1986; Parge et al, 1995; Merz and Forest, 2002).
Figure 5.
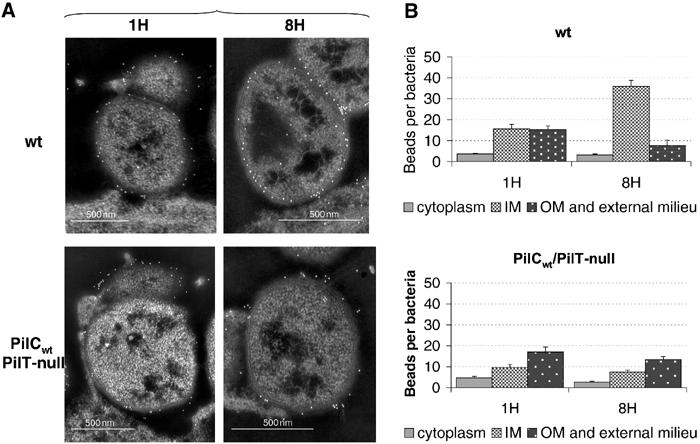
Pilin gold labelling on ultra-thin sections of adhering meningococci. Wild-type (Nm2C4.3) or PilCwt/PilT-null strains were allowed to adhere to HUVECs for 1 h (1H) to 8 h (8H). (A) In the wild type (upper panel), no gold beads are seen in the external milieu or associated with the outer membrane at intimate adhesion (8H), whereas numerous beads are observed on the surface of the bacterial cell or along pili at early time point (1H). In the absence of PilT-mediated retraction (lower panel), similar pilin distribution patterns are observed for the PilCwt/PilT-null strain throughout adhesion (1H and 8H). (B) Bead counts for each bacterial compartment at both time points for the wild-type and the PilCwt/PilT-null strains. The distribution of beads in the bacterial compartments is dramatically altered when tfp retraction occurs (wild type), but is stable in the absence of PilT-mediated retraction (PilCwt/PilT-null). Data represent mean±standard error of the mean.
The absence of noticeable changes in the pattern of pilin distribution during the course of adhesion of a PilT-null mutant is consistent with the lack of fibre retraction and the hyperpiliated phenotype observed in this strain (Figure 3B). Western blot experiments performed on cell-associated bacteria at the intimate adhesion stage showed that both adhering wild-type and PilT-null meningococci featured mature pilin patterns, although pilin in the PilT-null strain consistently showed a slightly altered electrophoretic mobility (Figure 6). The absence of detectable prepilin for both strains suggests that the detection of pilin along the cytoplasmic membrane during retraction is not due to the accumulation of prepilin. Furthermore, unlike gonococcus, N. meningitidis expresses very low amounts of the truncated soluble form of pilin, of which no trace could be detected. Similarly, no other putative degradation peptide could be recognised in cell-associated bacteria.
Figure 6.
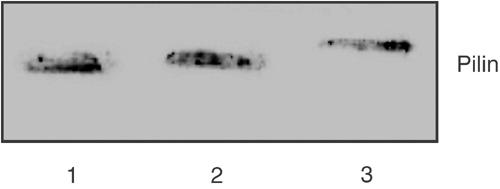
Pilin immunoblotting at intimate adhesion time. At intimate adhesion time (8H), HUVEC monolayers infected with wild-type or PilCwt/PilT-null bacteria were scrapped and similar amounts of bacterial total protein extracts were probed for pilin by Western blot. Lane 1, wild type (Nm2C4.3); lane 2, PilCwt/PilT-null; lane 3, PilD-null strain (bacterial extract as control for pilin size, in which prepilin is not cleaved into mature pilin). Both wild-type and PilCwt/PilT-null strains show mature pilin patterns. Neither prepilin nor S-pilin could be seen.
Besides relocalisation of pilin subunits to the inner membrane in the course of retraction, we sought to investigate if pilus retraction indeed involves pilin translocation from surface-expressed pili to an intrabacterial compartment. This was addressed by performing a de-biotinylation protection experiment on living bacteria (Figure 7). Together with the wild type, the meningococcal PilCwt/PilT-null, PilC-null/PilTwt and PilE-null mutants were also tested. Liquid-grown bacteria were submitted to surface biotinylation, using a labelling derivative that could subsequently be cleaved off by washing bacteria under reducing conditions (Materials and methods). We then performed Western blot analysis on total bacterial extracts, probing with streptavidin in the search of proteins that would be protected from surface biotin cleavage. In order to confirm that such bands would correspond to pilin, the membrane was subsequently re-probed with an anti-pilin antibody. The retraction-proficient strains (i.e. wild type and PilC-null/PilTwt) displayed a specific band of biotinylated pilin that was absent in the PilE-null as well as in the retraction-deficient PilCwt/PilT-null strain, although the total amount of pilin in the latter was similar to those of the wild type and the PilC-null/PilTwt. The specific presence in retraction-proficient strains, but not in the PilT-null strain, of a form of pilin that is resistant to surface biotin cleavage suggests that retraction involves the translocation of surface-exposed pilin subunits to an intracellular bacterial compartment that is inaccessible to biotin removal.
Figure 7.
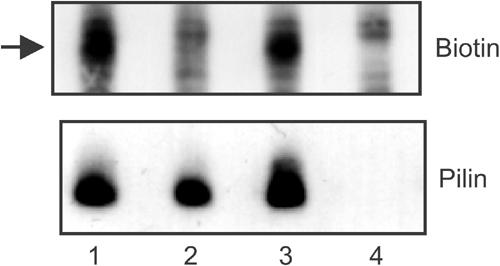
De-biotinylation protection experiment on living bacteria. Bacteria were allowed to grow, and twitch in relevant case, in the presence of a biotinylation agent that could subsequently be cleaved off the bacterial surface under reducing conditions. Lane 1, wild type (Nm2C4.3); lane 2, PilCwt/PilT-null; lane 3, PilC-null/PilTwt; lane 4, PilE-null. Upper panel: Whole bacterial extracts were used in Western blots with HRP-coupled streptavidin in the search of biotinylated proteins that would be protected from surface de-biotinylation. The arrow indicates de-biotinylation-protected pilin, which is only observed in retraction-proficient strains. Lower panel: Biotinylated bands corresponding to pilin were identified by subsequent re-probing of the membrane with an anti-pilin antibody.
Together with transmission electron microscopy (TEM) data showing pilin accumulation along the cytoplasmic membrane, these results suggest that PilT-driven fibre retraction involves the translocation of fibre-forming pilin subunits to the cytoplasmic membrane. Interestingly, the PilC-null strain exhibits comparable amounts of protected biotinylated pilin to the wild type. Since these bacteria are poorly piliated, this observation supports the hypothesis that, in the absence of PilC, surface-expressed pili are immediately retracted.
Discussion
In this work, we show that the PilC proteins are involved in tfp biogenesis by regulating PilT-driven fibre retraction, and we provide with a set of direct evidence suggesting that tfp retraction involves the redistribution of pilin subunits to the cytoplasmic membrane.
The regulation of piliation is a central element in the physiology of pathogenic Neisseria. Previous reports have shown that the level of piliation is under the control of pilE transcription level (Long et al, 2001). Besides, the amount of PilT, as well as the availability of ATP, is a limiting factor for retraction (Maier et al, 2002). The PilC proteins are associated with the outer membrane. They can also be recovered from purified pili and, at least in N. gonorrhoeae, act as pilus tip-located adhesins. Their major role in tfp-related functions is exemplified by their requirement for transformation competence, and by the fact that all but the PilC2 meningococcal variants mediate adhesion to cells. In this regard, the regions specifically involved in adhesion have been localised within the amino-terminal part of the adhesion-promoting variants (Morand et al, 2001). Our data show that the PilC proteins have a key role in the regulation of piliation by preventing pilus retraction. This relies on three sets of evidence. First, efficient tfp assembly and expression to the bacterial surface is achieved regardless of the level of PilC, provided that retraction is abolished. Second, in an adhesion model where pilus extension and retraction are sequentially observed, the downregulation of PilC is required to observe pilus retraction. Third, abolition of PilC leads to the detection of pilin that is protected from de-biotinylation. These findings are consistent with the hypothesis that PilC-null strains show a defect in piliation because they express PilT (Wolfgang et al, 1998b), and with previous reports stating that PilC proteins are essential neither for fibre formation nor for fibre surface localisation (Wolfgang et al, 2000). Cell-associated transcriptional analysis constitutes a model in which enhanced piliation and retraction are concomitant for a large number of bacteria. In the absence of cells, growing bacteria are not ‘on phase', and pilus extension is observed along with fibre retraction. This phenomenon is clearly observed when liquid-grown bacteria form twitching bacterial clumps.
Our data show that retraction can be regulated by a variation of two-fold in pilC transcription. Although one cannot exclude that the slight increase of pilT transcription during the course of adhesion to human cells also participates in the balance between PilC and PilT, it seems that the downregulation of pilC transcription is required to allow efficient PilT-driven retraction. The ratio of two-fold in pilC transcription observed during meningococcus–host cell interaction is consistent with the previously reported measurement of activity of transcriptional pilC gene fusions using β-galactosidase (Taha et al, 1998). This two-fold variation is further supported by the use of the IPTG-inducible pilC1 gene.
The presence of tfp on PilC-null strains was described in earlier reports (Rudel et al, 1992, 1995a), but knowledge of the genetic background for such mutants was incomplete. Other reports, based on TEM examination to assess piliation, suggested that PilC-null neisserial strains exhibited an absolute lack of tfp (Nassif et al, 1994), unless fibre expression was restored by concomitant abolition of PilT (Wolfgang et al, 1998b). In this work, pilC1 and/or pilC2 expressions were abolished by deletion and insertion of antibiotic resistance cassettes, so that no incidental frameshifting in transcription or transduction could be responsible for the expression of low amounts of protein. TEM examination, which is highly valuable for qualitative analysis of pili, could not reliably detect tfp fibres in the PilC-null strain. It is likely that tfp detection was possible because a polyclonal antibody raised against the whole hydrophilic moiety of the pilin variant expressed by the strain was used.
The observation that pilin subunits are associated with the inner membrane after pilus retraction is concordant with a ‘melting' model of tfp retraction, in which the pilus would be disassembled within the cytoplasmic membrane by ‘dilution' of the subunits into the inner membrane bi-layers (Merz and Forest, 2002). This model is strengthened by the correlation of retraction with a form of pilin that is protected from surface de-biotinylation. This form of pilin is not observed in a PilT-null strain but is present in a PilC-null strain, in comparable amounts to that observed in the wild type. Since PilC-null bacteria appear poorly piliated, these results suggest that lack of PilC allows fibre retraction as soon as pili are formed, and support the role of PilC as counteracting PilT-mediated retraction. Although little is known about the fate of pilin subunits that are retrieved from fibres, the accumulation of pilin within the cytoplasmic membrane might also suggest that this bacterial compartment acts as a reservoir for the assembly of novel pilus fibres. Metabolic protein synthesis and energetic models suggest that, in the absence of pilin recycling, tfp biogenesis would require most of the protein synthesis capacity of the bacterial cell (Merz and Forest, 2002). Furthermore, unlike gonococcus, N. meningitidis expresses very low amounts of the truncated soluble form of pilin (Marceau and Nassif, 1999), of which no trace could be detected in Western blot experiments performed on bacteria that have undergone pilus retraction, and no other putative degradation peptide could be recognised. Although one cannot exclude that N. meningitidis might degrade pilin issued from fibre retraction so efficiently that we could not capture this event using conventional tools, we favour the hypothesis that fibre-derived pilin subunits that are targeted to the inner membrane during retraction could be, at least partially, recycled into new fibres.
Taken together, our results help to elucidate some steps of tfp biogenesis and suggest a model presented in Figure 8. Tfp retraction is intimately linked to the anabolic phases of biogenesis, that is, fibre assembly and translocation through the outer membrane secretin PilQ. The amount of PilE available for assembly appears to be an essential limiting factor (Long et al, 2001), but little is known about the role of other components of the tfp machinery in the regulation of fibre assembly and expression at the bacterial surface. Since it is required for tfp expression and also belongs to the AAA family of proteins (Merz and Forest, 2002), models suggest that PilF would be the anabolic counterpart of PilT. However, the steady transcription of pilF at all adhesion steps might indicate that fibre assembly and extrusion are controlled independent of fibre retraction. The absence of prepilin accumulation in any conditions of adhesion or of PilC expression suggests that, in physiological conditions, pilin processing by the prepilin peptidase PilD is not a limiting factor for tfp biogenesis. Since increased levels of PilC refrain PilT-driven retraction, and since pilT transcription is moderately altered in the course of cell-contact retraction, it is tempting to hypothesise that, as long as enough pilin subunits are provided for assembly, the regulation of piliation mostly relies on the tuning of retraction, through the regulation of pilC transcription. In this model, each tfp fibre that is expressed on the cell surface is bound to be retracted, provided that ATP and functional PilT are available at the proper site along the cytoplasmic membrane. This model would be concordant with previous reports showing that the action of PilT can be observed on fibres that are expressed on the cell surface (i.e. in a PilC-null mutant), as well as on fibres that have not yet been extruded through the outer membrane (i.e. in a PilQ-null strain) (Wolfgang et al, 2000).
Figure 8.
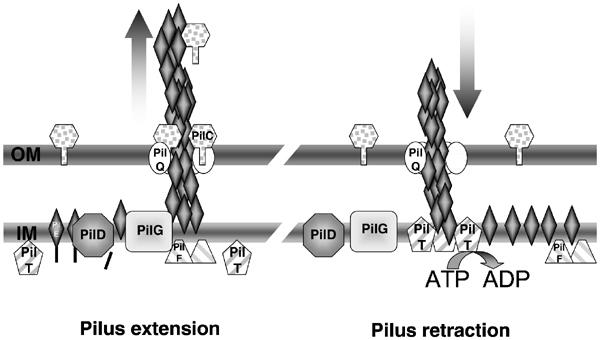
Model for the expression and retraction of neisserial tfp. Pilus extension: Tfp is formed in/along the cytoplasmic membrane (IM) by the assembly of mature pilin subunits that are stalled into the membrane, after processing by the prepilin peptidase PilD. The growing fibre is subsequently translocated to the bacterial surface through the outer membrane (OM) secretin PilQ. By homology with other machineries, the neisserial PilF is thought to have nucleotide-binding activities (Fernandez and Berenguer, 2000; Sandkvist, 2001). PilT-driven retraction is prevented as long as sufficient PilC proteins, which might act as a clip on the pilus, are expressed. Pilus retraction: Reduced amounts of PilC cannot prevent the action of PilT. Mature pilin subunits are thought to be translocated from the base of the fibre to the cytoplasmic membrane in an ATP-dependant process, thus leading to fibre retraction and accumulation of mature pilin along this membrane. Although not formally demonstrated for the neisserial machinery, reports in other species show that PilT is an ATPase and can form oligomers (Herdendorf et al, 2002; Okamoto and Ohmori, 2002). This model surmises that, when the PilT component is absent, fibres are normally assembled and extruded to the bacterial surface (situation in the left panel), but are not subjected to retraction. This results in the accumulation of tfp on the cell surface and bacteria appear hyperpiliated whatever the expression of PilC. The abolition of PilC in a PilTwt background allows increased retraction rates (situation in the right panel), and fibres are immediately retracted after expression on the bacterial surface, thus leading to poor piliation.
Hypothesis can also be drawn on how the PilC components, which are associated with the outer membrane, can prevent the retraction of the fibre. Given that PilC can be found associated with tfp, the association of PilC with the fibre may act as a clip, causing sterical constraints and refraining fibre retraction. In this case, the association of PilC with the pilus should be reversible since retraction should not be totally inhibited, and retraction may not be abolished for all fibres on a single bacterium at the same time. However, one cannot rule out the presence of another component through which PilC could exert its effect on retraction. A possible candidate could be the channel-forming secretin PilQ, which could be functionally regulated by the presence of PilC. This hypothesis would be supported by previous reports showing that, in the absence of PilQ, PilC is shed in the external milieu (Drake et al, 1997).
Materials and methods
Bacterial strains and media
Nm2C4.3 is a derivative of 8013, a serogroup C, class 1 strain that produces the SB pilin variant (Nassif et al, 1993). This strain is piliated and adherent to human cells, Opa−, Opc−, PilC1+ and PilC2+. Neisseria were grown at 37°C in a 5% CO2 atmosphere on GC medium (Difco) containing Kellog's supplement (Kellogg et al, 1968). For selection of meningococci, kanamycin (Km) was used at 100 μg/ml, erythromycin (Em) at 2 μg/ml, tetracyclin (Tet) at 2 μg/ml and spectinomycin (Sp) at 50 μg/ml. The PilCwt/PilT-null and PilE-null mutants were previously described (Pujol et al, 1999), as well as the PilC-null strain (Morand et al, 2001). All meningococcal strains used in this work express the SB pilin variant. Quantitative RT–PCR analyses of meningococcal adhesion onto cells were performed on a derivative of the serogroup A Z5463 strain that belongs to the same epidemics as Z2491 (Sarkari et al, 1994), which carries an intergenic chromosomal insertion of an aphA3 kanamycin resistance cassette.
Construction of strains harbouring an inducible pilC gene
All enzymes were supplied by New England Biolabs. The region upstream of pilC1 and the 5′ part of the gene were simultaneously cloned into pUC19, inserting a NotI restriction site upstream of the Shine-Dalgarno sequence of pilC1. Two fragments were amplified from Nm2C4.3 using oligonucleotides C1-152-US (5′-GCCGTCTGAACCTGCCCGACGGTATCCCGCG AAGC-3′) and C1R-14-Not (5′-GCGGCCGCGGTCAAACCCCCCGCCCTTCG-3′), and C1F-SD-Not (5′-GCGGCCGCCCAAAAGGAAATACGATGAAT-3′) and CR328-BsiW1 (Morand et al, 2001). The pHSX-ermC-lacIOP plasmid (gift from HS Seifert) (Seifert, 1997) was restricted with NotI, to release a 3.1 kb fragment containing ermC, lacIq, and the tandem lac operator promoter sequences tacOP and UV5OP. This cassette was cloned into the NotI site upstream of pilC1. The resulting plasmid of correct orientation was cut with BsiWI and used for transformation into a pilC1+/ΔpilC2∷aadA derivative of Nm2C4.3. The resulting NmC2SC1WEa transformant harbouring pilC1ind was selected with Sp and Em. Induction of PilC was observed in liquid as well as agar-grown bacteria.
Similar procedure was used for strain NmC1ITSE, harbouring pilC1ind and inactivated pilT, but the pHSX-ermC-lacIOP plasmid was replaced by its tetracyclin-resistant counterpart, pHSX-tet-lacIOP (also gift from HS Seifert). Inactivation of pilT was subsequently carried out with insertion of a pilT∷ermAM cassette (Pujol et al, 1999), by transformation in the presence of IPTG and selection for Tet and Em.
For NmC1IESK, a translational fusion of the pilE promoter to pilT gene was introduced in the genome of NmC2SC1WEa by transformation, and selection for Campbell-type integration with Km, Sp and Em. Briefly, a PCR product was generated using primers PilE5′T (5′-TAAGCCTGAGGCATTTCCCCTTTCAATTAGG AGTAATTTTATGCAGATTACCGACTTACTCG-3′) and SS6Eco (5′-GATCCCCACCGGAATTGCG-3′), and plasmid p11.2.7 (Wolfgang et al, 1998a) as template. After complete Bsu36 and partial EcoR1 restriction, the largest resulting fragment was cloned into the Bsu36–EcoRI-restricted pPilE plasmid (Wolfgang et al, 2000). PilE expression in NmC1IESK was identical to that of the parental strain and wild type.
Proteins and immunoblotting
Immunoblottings were performed as previously described (Marceau et al, 1995). Pilin was detected with the 5C5 monoclonal antibody (Marceau et al, 1995), PilT with a polyclonal antibody raised in rabbit against the whole molecule, the RmpM protein with a specific monoclonal antibody and PilC with a polyclonal antibody (Morand et al, 2001).
Cell culture and adhesion assays
Adhesion assays were performed on glass coverslips in 24-well plates, at an MOI of 10. Incubation was carried out up to 8 h and the medium was replaced every hour to minimise monolayer re-infection. HUVECs (PromoCell, Heidelberg, Germany) were used between passages 1 and 8 and grown in Endo-SFM (Life Technologies, Grand Island), with 10% FCS, 1 ng/ml β-FGF (Boehringer-Mannhein, Meylan, France), 2 mM L-glutamine (Life Technologies, Grand Island), 0.5 UI/ml heparin (Sigma, Saint Louis) and 1.25 μg/ml endothelial cell growth supplement (Sigma, Saint Louis). Cells were seeded at 5 × 104 cells/cm2 and grown at 37°C in a humidified incubator under 5% CO2 for 2–3 days. The day before infection, culture medium was replaced by Endo-SFM supplemented with 10% FCS. For LR-White embedding, the same protocol was used but HUVECs were grown in 0.4-μm-pore-sized Transwell-COL wells (Corning Costar Corp., Cambridge, MA). 1 mM IPTG was maintained throughout adhesion with NmC2SC1WEa. Detection of pilin at intimate adhesion stage was performed on total extracts of PBS-washed infected cell monolayers.
RNA extraction and real-time PCR assays
Bacteria were either scrapped with HUVECs, harvested from infection media or from GC-agar plates supplemented with IPTG. Care was taken to avoid bacterial lysis by preventing overgrowth, and bacteria were checked by immunofluorescence to be piliated and to conserve propensity to form clumps and twitch (for the wild type). In all, 400 μl of bacterial suspension, 700 μl of prewarmed Trizol (Life Technologies, Gaithersburg, MD) and 50 μl of 150–212 μm glass beads were vortexed. The mix was placed on dry ice and then thawed, before the addition of 200 μl chloroform. After centrifugation, the aqueous phase was re-extracted once with Trizol, and RNA was precipitated with isopropanol. RNA pellet was washed with 75% ethanol and re-dissolved in RNase-free TE buffer. DNA was removed by addition of DNAse I (Takara Shuzo, Otsu, Shiga, Japan), subsequently inactivated at 70°C for 10 min. The complete removal of DNA was checked by the absence of signal in a PCR using the RNA sample as template.
Reverse transcription was performed using oligonucleotides complementary to the 3′ end of the genes and the SuperscriptTM II reverse transcriptase (Life Technologies, Gaithersburg, MD). A validation experiment was run in order to ensure that efficiencies of amplification for target and reference were approximately equal. Real-time PCR was run on an ABI PRISM 7700 (Applied Biosystems, Foster City, CA) with SYBR-Green PCR Master Mix, according to the manufacturer's instructions. The relative quantitation of gene transcription was performed by the comparative threshold cycle (Ct) method, according to the manufacturer's instructions (User Bulletin #2, Applied Biosystems, Foster City, CA). Briefly, fluorescence is proportional to the amount of DNA during the exponential phase of PCR, and Ct is defined as the number of cycles necessary for the fluorescence, which is measured at every PCR cycle, to pass a predefined threshold. The relative amount of target DNA for each gene, which is normalised using the aphA3 gene as an internal reference, is given by 2(Ctgene_CtaphA3. For the transcription analysis of the pilC1ind gene (Figure 1B), results are expressed as a percentage of the transcription of pilC1wt in identical conditions (calibrator): 100 × 2(CtpilC1wt_CtaphA3wt)/2(CtpilC1ind_CtaphA3ind). For the comparison of adhering bacteria to bacteria grown in infection media (Figure 4), the calibrator is the bacteria grown in infection medium, and the level of gene transcription is calculated as a ratio: 2(Ctgene_CtaphA3)infectionmedia/2(Ctgene_CtaphA3)adheringbacteria. Data result from at least six measurements, on at least two distinct RNA extractions.
Immunofluorescence labelling
Piliation of whole bacteria was assessed by immunofluorescence using a polyclonal anti-pilin antibody raised against the whole hydrophilic carboxy-terminal part of the SB pilin variant of Nm2C4.3. Bacterial suspensions were laid on glass slides and fixed with methanol. Samples were saturated with PBS–0.2% gelatin, and all reagents were diluted in PBS–0.2% gelatin. A488-labelled donkey anti-rabbit secondary antibody was used for fluorescence labelling (Jackson Immunoresearch, West Grove, PA).
Immunofluorescence on infected human cells was performed as previously described (Morand et al, 2001). Pili were labelled using the anti-pili 20D9 monoclonal antibody (Pujol et al, 1997), f-actin using Alexa568–Phalloidin (Molecular Probes, Eugene, OR), and bacteria using a previously described polyclonal antibody that does not recognise the SB pilin variant (Pron et al, 1997). Secondary antibodies directed against rabbit or mouse immunoglobulins were coupled with Cy5 or Alexa488, respectively (Jackson Immunoresearch, West Grove, PA).
TEM embedding of cell monolayers
For pilin gold-labelling experiments, infected human cells grown on Transwell-COL were embedded in LR-White medium resin (Sigma, St Louis, MO). After three washes in cacodylate buffer (0.1 M sodium cacodylate, 5 mM CaCl2, 5 mM MgCl2, pH 7.2), cells were fixed with 2% paraformaldehyde and 0.1% glutaraldehyde in cacodylate buffer for 1 h at 4°C. Fixed cells were washed with cacodylate buffer, and remaining aldehyde activity was quenched in the same buffer containing 50 mM NH4Cl. The membranes were cut out of the Transwell, embedded in 2% low melting point agarose in cacodylate buffer, and were successively washed in water and Michaelis buffer (pH 6.0), before the addition of 1% uranyl-acetate in Michaelis buffer for 30 min at room temperature. After progressive dehydration of the sample in increasing ethanol concentration up to 70%, LR-White resin embedding was performed. The resin was allowed to polymerise for at least 4 days at 45°C before ultra-thin sectioning and labelling.
Gold-labelling experiments
Labelling was performed on ultra-thin sections of resin-embedded infected cell monolayers, which were laid on Formwar-coated nickel grids. Pilin was labelled using a polyclonal antibody that was raised against the whole hydrophilic moiety of the SB pilin variant. Secondary labelling was performed with protein A-coupled 10 nm gold beads (Department of Cell Biology, University School of Medicine, Utrecht, Netherlands). Prior to labelling, ultra-thin sections were saturated with blocking solution (Aurion, Wageningen, Netherlands). After labelling, preparations were successively counterstained with 2% uranyl-acetate, 50 mM lead citrate, and extensively rinsed in distilled water. TEM was performed on a JEOL JEM-100CX-II apparatus. Bead counts were performed on randomly selected ultra-thin bacterial sections, provided that bacterial membranes were clearly distinguishable. Beads located in the cytoplasm and more than 50 nm away from the cytoplasmic membrane were counted as ‘cytoplasm'; beads located along the outer membrane, as well as those outside the bacterial cells but closer than 200 nm from the membrane were counted as ‘OM and external milieu'; beads associated with the cytoplasmic membrane were counted as ‘IM'. Some beads in the external milieu were not counted when they were part of pili but could not be assigned to a defined bacterium.
De-biotinylation protection experiment
Agar-grown bacteria were resuspended in minimal twitching media (PBS, 1 g/l D-glucose), and allowed to grow freely for 15 min. Sulpho-NHS-SS-Biotin (Pierce, www.piercenet.com) was then added (final concentration 1 g/l) and growth was prolonged for 15 min. Bacterial viability could be assessed (for the wild type) by clump formation and twitching motility. Labelling was stopped with 20 mM glycine and cold PBS washings. De-biotinylation was performed for 20 min in PBS–MESNA (2-mercaptoethanesulphonic acid, 60 g/l, Sigma, St Louis, MO). After three PBS washings, total protein extracts were used in nonreducing SDS–PAGE. After transfer onto PVDF, samples were first probed with horse-radish peroxydase-coupled streptavidin, and subsequently with the 5C5 anti-pilin antibody. Biotinylation of the periplasmic FBP (ferric ion-binding protein) was similar and of very low level for the four tested strains, suggesting no differences in labelling dynamics between these strains.
Acknowledgments
We acknowledge HS Seifert for kindly providing with the IPTG-inducible constructs, and AP Pugsley and D Heuer for stimulating discussions. We also thank C Fréhel and E Pellegrini for their valuable help in TEM and gold labelling. PCM was funded by FRM, CANAM, Fondation BAYER and Max-Planck-Institut für Infektionsbiologie. The laboratory of XN is supported by INSERM and Université Paris V-René Descartes.
References
- Brossay L, Paradis G, Fox R, Koomey M, Hebert J (1994) Identification, localization, and distribution of the PilT protein in Neisseria gonorrhoeae. Infect Immun 62: 2302–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake SL, Sandstedt SA, Koomey M (1997) PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol 23: 657–668 [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Berenguer J (2000) Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol Rev 24: 21–44 [DOI] [PubMed] [Google Scholar]
- Finlay BB, Pasloske BL, Paranchych W (1986) Expression of the Pseudomonas aeruginosa PAK pilin gene in Escherichia coli. J Bacteriol 165: 625–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag NE, Seifert HS, Koomey M (1995) Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol 16: 575–586 [DOI] [PubMed] [Google Scholar]
- Herdendorf TJ, McCaslin DR, Forest KT (2002) Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J Bacteriol 184: 6465–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DS Jr, Cohen IR, Norins LC, Schroeter AL, Reising G (1968) Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol 96: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CD, Hayes SF, van Putten JP, Harvey HA, Apicella MA, Seifert HS (2001) Modulation of gonococcal piliation by regulatable transcription of pilE. J Bacteriol 183: 1600–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Potter L, So M, Seifert HS, Sheetz MP (2002) Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci USA 99: 16012–16017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Beretti JL, Nassif X (1995) High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol Microbiol 17: 855–863 [DOI] [PubMed] [Google Scholar]
- Marceau M, Nassif X (1999) Role of glycosylation at Ser63 in production of soluble pilin in pathogenic Neisseria. J Bacteriol 181: 656–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz AJ, Forest KT (2002) Bacterial surface motility: slime trails, grappling hooks and nozzles. Curr Biol 12: R297–R303 [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP (2000) Pilus retraction powers bacterial twitching motility. Nature 407: 98–102 [DOI] [PubMed] [Google Scholar]
- Morand PC, Tattevin P, Eugene E, Beretti JL, Nassif X (2001) The adhesive property of the type IV pilus-associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N-terminal part of the molecule. Mol Microbiol 40: 846–856 [DOI] [PubMed] [Google Scholar]
- Nassif X, Beretti JL, Lowy J, Stenberg P, O'Gaora P, Pfeifer J, Normark S, So M (1994) Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc Natl Acad Sci USA 91: 3769–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif X, Lowy J, Stenberg P, O'Gaora P, Ganji A, So M (1993) Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohmori M (2002) The cyanobacterial PilT protein responsible for cell motility and transformation hydrolyzes ATP. Plant Cell Physiol 43: 1127–1136 [DOI] [PubMed] [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA (1995) Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature 378: 32–38 [DOI] [PubMed] [Google Scholar]
- Pron B, Taha MK, Rambaud C, Fournet JC, Pattey N, Monnet JP, Musilek M, Beretti JL, Nassif X (1997) Interaction of Neisseria meningitidis with the components of the blood–brain barrier correlates with an increased expression of PilC. J Infect Dis 176: 1285–1292 [DOI] [PubMed] [Google Scholar]
- Pujol C, Eugene E, de Saint, Martin, Nassif X (1997) Interaction of Neisseria meningitidis with a polarized monolayer of epithelial cells. Infect Immun 65: 4836–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C, Eugene E, Marceau M, Nassif X (1999) The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc Natl Acad Sci USA 96: 4017–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Källström H, Normark S, Jonsson AB (1997) PilC of pathogenic Neisseria is associated with the bacterial surface. Mol Microbiol 25: 11–25 [DOI] [PubMed] [Google Scholar]
- Rudel T, Boxberger HJ, Meyer TF (1995a) Pilus biogenesis and epithelial cell adherence of Neisseria gonorrhoeae pilC double knock-out mutants. Mol Microbiol 17: 1057–1071 [DOI] [PubMed] [Google Scholar]
- Rudel T, Facius D, Barten R, Scheuerpflug I, Nonnenmacher E, Meyer TF (1995b) Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA 92: 7986–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Scheurerpflug I, Meyer TF (1995c) Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature 373: 357–359 [DOI] [PubMed] [Google Scholar]
- Rudel T, van PJ, Gibbs CP, Haas R, Meyer TF (1992) Interaction of two variable proteins (PilE and PilC) required for pilus-mediated adherence of Neisseria gonorrhoeae to human epithelial cells. Mol Microbiol 6: 3439–3450 [DOI] [PubMed] [Google Scholar]
- Sandkvist M (2001) Biology of type II secretion. Mol Microbiol 40: 271–283 [DOI] [PubMed] [Google Scholar]
- Sarkari J, Pandit N, Moxon ER, Achtman M (1994) Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol Microbiol 13: 207–217 [DOI] [PubMed] [Google Scholar]
- Seifert HS (1997) Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene 188: 215–220 [DOI] [PubMed] [Google Scholar]
- Strom MS, Lory S (1993) Structure–function and biogenesis of the type IV pili. Annu Rev Microbiol 47: 565–596 [DOI] [PubMed] [Google Scholar]
- Taha MK, Morand PC, Pereira Y, Eugene E, Giorgini D, Larribe M, Nassif X (1998) Pilus-mediated adhesion of Neisseria meningitidis: the essential role of cell contact-dependent transcriptional upregulation of the PilC1 protein. Mol Microbiol 28: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Vale RD (2000) AAA proteins. Lords of the ring. J Cell Biol 150: F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M (1998a) PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol 29: 321–330 [DOI] [PubMed] [Google Scholar]
- Wolfgang M, Park HS, Hayes SF, van Putten JP, Koomey M (1998b) Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA 95: 14973–14978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, Dorward D, Koomey M (2000) Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J 19: 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]


