Summary
A gene signature derived from EASP strongly correlated with nodal metastasis, advanced tumor stage and poor differentiation and predicted survival in three independent lung cancer data sets, including early-stage patients. It may serve as a prognostic signature to identify high-risk early-stage patients.
Abstract
In cancer cells, the process of epithelial–mesenchymal transition (EMT) confers migratory and invasive capacity, resistance to apoptosis, drug resistance, evasion of host immune surveillance and tumor stem cell traits. Cells undergoing EMT may represent tumor cells with metastatic potential. Characterizing the EMT secretome may identify biomarkers to monitor EMT in tumor progression and provide a prognostic signature to predict patient survival. Utilizing a transforming growth factor-β-induced cell culture model of EMT, we quantitatively profiled differentially secreted proteins, by GeLC-tandem mass spectrometry. Integrating with the corresponding transcriptome, we derived an EMT-associated secretory phenotype (EASP) comprising of proteins that were differentially upregulated both at protein and mRNA levels. Four independent primary tumor-derived gene expression data sets of lung cancers were used for survival analysis by the random survival forests (RSF) method. Analysis of 97-gene EASP expression in human lung adenocarcinoma tumors revealed strong positive correlations with lymph node metastasis, advanced tumor stage and histological grade. RSF analysis built on a training set (n = 442), including age, sex and stage as variables, stratified three independent lung cancer data sets into low-, medium- and high-risk groups with significant differences in overall survival. We further refined EASP to a 20 gene signature (rEASP) based on variable importance scores from RSF analysis. Similar to EASP, rEASP predicted survival of both adenocarcinoma and squamous carcinoma patients. More importantly, it predicted survival in the early-stage cancers. These results demonstrate that integrative analysis of the critical biological process of EMT provides mechanism-based and clinically relevant biomarkers with significant prognostic value.
Introduction
Lung cancers are the leading cause of cancer-related deaths worldwide. The advances made in the last decade in diagnosis and treatment have not been translated into significant improvements in overall 5 year survival rates. The tumor-node-metastasis (TNM) staging system combined with pathologic diagnosis has remained the major tool for medical decision making and predicting patient survival (1,2). However, accumulating evidence suggests that though patients with identical histology, differentiation, location and stage at diagnosis are treated by similar therapy, survival is most heterogeneous indicating that the current methods of tumor classification and staging are not sufficient for selecting the best treatment choices and defining prognosis. About 30–55% of early-stage patients who are treated primarily by surgery will have recurrence within 3 years. Recent randomized clinical trails revealed a significant survival advantage in patients receiving chemotherapy after complete resection in the stage IB–IIIA categories (3–6). This trend indicates a need to explore alternative indicators to understand the underlying prognosis of a given patient, to identify the early-stage patients at greatest risk of relapse and decide on appropriate treatment strategy to increase patient survival.
Advances in microarray analysis and proteomics have stimulated research in molecular prognostics and provided alternatives to the traditional TNM-based system for more precise classification and prognostication of human cancers. In different studies, the gene expression signatures derived from tumors at the time of diagnosis have shown promise in predicting long-term patient outcome and outperformed the standard pathologic TNM staging in stratifying breast cancer patients (7–11) and lung cancer patients (12) into high- and low-risk groups. These studies clearly established the fact that tumors contain informative signatures which can be highly valuable in prognosis and estimation of biological response of tumors to therapy over time (7–9). Based on this information, it is possible to identify the subsets of patients who are at high risk of mortality at the time of diagnosis. As a proof of this concept, recently a 70-gene signature was approved for clinical application as a prognostic classifier of breast cancers (10,11). Efforts to identify an analogous gene expression-based prognostic signature of non-small-cell lung cancer have been promising but yet to reach the stage of clinical application (12,13). The predominant approach in all the prognostic signatures to date has been identifying these signatures based on their differential expression between good and poor prognosis groups.
In this study, we attempted an alternative approach to develop a prognostic gene signature based on the cellular process of epithelial–mesenchymal transition (EMT), which plays a critical role in tumor progression. EMT is considered as an initiating event for distant dissemination of tumor cells; proteins secreted during this process may serve as potential biomarkers for patient prognosis. Utilizing a transforming growth factor (TGF)-β-induced EMT model, we quantitatively profiled differentially secreted proteins using label-free 1D gel electrophoresis followed by nanoliquid chromatography, coupled to tandem mass spectrometry (GeLC-MS/MS) analysis of the conditioned media of A549 lung adenocarcinoma cells cultured in the presence or absence of TGF-β. By integrating the secretome with the transcriptome from our earlier study (14), we identified a 97-gene EMT-associated secretory phenotype (EASP) that showed strong correlation to differentiation and stage and predicted survival of lung adenocarcinoma patients in training and independent test sets. We further refined this to a 20-gene signature (rEASP), which performed equally well in predicting survival particularly, in early-stage (stages I and II) adenocarcinomas as well as in squamous cell carcinomas of lung and stratified the lung cancer patients into low-, medium- and high-risk groups with distinct survival times.
Materials and methods
Cell culture
The A549 human lung adenocarcinoma cell line was obtained from the American Type Culture Collection (Manassas, VA) and maintained in RPMI-1640 medium with glutamine, supplemented with 10% fetal bovine serum, penicillin and streptomycin and tested for mycoplasma contamination. All tissue culture media and media supplements were purchased from Life Technologies (Gaithersburg, MD). The porcine TGF-β1 was purchased from PeproTech (Rocky Hill, NJ). In all experiments, cells at 40–50% confluency were serum starved for 24h and treated with TGF-β (5ng/ml) for 72h. At the end, conditioned media collected was centrifuged at 2000g for 20 min and filtered through 0.2 µm filter to remove the intact cells and debris and stored at −80°C until further processing. Cells in the culture dishes were lysed in radioimmunoprecipitation assay buffer and processed for western immunoblotting for assessing the expression of epithelial and mesenchymal markers. Protein concentrations were determined using the BCA Protein Assay Reagent from Pierce (Rockford, IL).
Sample preparation, sodium dodecyl sulfate–polyacrylamide gel electrophoresis and in-gel digestion
Seven milliliters of conditioned media from control and TGF-β-treated cells from two independent biological replicates was buffer exchanged into 25mM ammonium bicarbonate and the volume reduced to 100 µl using a 10kDa molecular weight cutoff filter (Millipore). Half of each replicate (~20 µg protein from controls and 10 µg protein from TGF-β treatment) was solubilized in loading buffer and resolved using Novex 4–12% gradient gels (Invitrogen Life Technologies, Carlsbad, CA). Each lane was manually excised into 40 equal slices and each slice was transferred to a well of a 96-well plate. Proteins in each gel slice were robotically reduced with 10mM dithiothreitol, alkylated with 50 mM iodoacetamide and digested with 160 ng trypsin (ProGest, Genomic Solutions, Ann Arbor, MI). Tryptic peptides were analyzed following acidification with 0.5% formic acid to a final pH 3.8. The volume of peptide mixture for each band was 40 µl.
Data-dependent LC/MS/MS
Thirty microliters of each digested gel slice was analyzed using nano-LC/MS/MS on a LTQ Orbitrap XL tandem mass spectrometer (ThermoFisher, San Jose, CA). Sample was loaded onto an IntegraFrit (New Objective, Woburn, MA) 75 µm × 3cm vented column packed with 0.5mm Jupiter C12 material (Phenomenex, Torrance, CA) at 10 µl/min. Peptides were eluted with a 50min gradient (0.1–30% B in 35min, 30–50% B in 10min and 50–80% B in 5min where A = 99.9% H2O, 0.1% acetonitrile in 0.1% formic acid and B = 80% acetonitrile, 20% H2O in 0.1% formic acid) at 300 nl/min using a NanoAcquity HPLC pump (Waters, Beverley, MA) over a 75 µm × 15cm IntegraFrit analytical column packed also with Jupiter C12 material. The column was coupled to a 30 µm ID × 3cm stainless steel emitter (ThermoFisher). Mass spectrometry (MS) was performed in the Orbitrap at 60 000 full width at half maximum resolution and MS/MS was performed in the LTQ on the top six ions in each MS scan using the data-dependent acquisition mode. Normalized collision energy was set at 35% and 1 microscan was used with automatic gain control implementation. Automatic gain control enables the trap to fill with ions to the set ion target values. Target values for MS and MS/MS were 5 × 104 and 1.5 × 103 counts, respectively. Dynamic exclusion and repeat settings ensured each ion was selected only once and excluded for 30 s thereafter.
Data processing
Data were processed using the MaxQuant v1.0.13.8 software (15), which provides protein identifications at a target false discovery rate. This version of MaxQuant utilizes a locally stored copy of the Mascot search engine (version 2.2; Matrix Science, London, UK) and data were searched against the International Protein Index (IPI) Human v3.53 protein database. Search parameters were: product ion mass tolerance 0.5Da, two missed cleavages allowed, fully tryptic peptides only, fixed modification of carbamidomethyl cysteine, variable modifications of oxidized methionine, N-terminal acetylation and pyroglutamic acid on N-terminal glutamine. Selected MaxQuant parameters were: ‘singlets’ mode, peptide, protein and site false discovery rate 1%, minimum peptide length of five amino acids, minimum of one unique peptide per protein. Proteins identified by this analysis are summarized in Supplementary Table S1, available at Carcinogenesis Online. In MaxQuant, the quantitative measure of each protein is based on the sum of the chromatographic peak area of each peptide matched, termed ‘intensity’. For each protein, a log2 ratio of expression is determined by comparing the average intensity for that protein between the replicates of TGF-β-treated and controls. A protein is determined as differentially expressed if it has >2-fold change in either direction. Log2 ratio >1 is considered as upregulation and <1 is considered as downregulation.
Annotation of secreted proteins and mapping to gene expression
Proteins were annotated as secreted using multiple different bioinformatic tools including SecretomeP (16) for non-classical and leaderless secreted proteins, TMHMM, an HMM-based method for prediction of transmembrane domains (17), SignalP package that detects signal peptides and predicts classical secreted proteins (18), PSORT II that predicts the protein subcellular localization (19) and Secreted Protein Database (SPD) (20). All these predictions are incorporated into Supplementary Table S2, available at Carcinogenesis Online. Others were annotated as secreted proteins based on reported empirical evidence and Gene Ontology (GO) analysis.
Entrez gene identifiers corresponding to the IPI accession numbers of identified proteins were obtained using human IPI cross reference data (‘IPI.genes.HUMAN’ for IPI human release 3.65 from ftp://ftp.ebi.ac.uk/pub/databases/IPI/current). Entrez gene identifiers were used to obtain the corresponding probe set identifiers for the associated arrays from the Affymetrix annotation. Following the above protocol, all the annotated secreted proteins were mapped to our previously published TGF-β-induced EMT time course gene expression data set (GSE 17708) from the same cell line at identical conditions (14). To match the secretome, differentially expressed genes only at 72h time point (5057 probes corresponding to 3397 genes) were used for mapping. Some probes that are identified as differentially expressed but with no assigned gene symbol were excluded.
Gene set enrichment and hierarchical clustering analysis
ConceptGen (http://conceptgen.ncibi.org) is a concept and gene set enrichment analysis tool (14). It will test a given list of genes for overlap and its significance with a specified concept or gene set, which includes GO, direct protein interactions, transcriptional regulation, miRNA targets and gene expression data sets. Using this tool, we performed GO cellular component, cellular process and KEGG pathways enrichment analysis for the 97-gene EASP. Statistically significant (P < 0.001) concepts are presented as network graphs with nodes representing concepts or gene sets and edges representing statistical significance of enrichment.
For clustering, the lists of oncogenic pathways included in the analysis were compiled from the KEGG database, except for embryonic stem cell (ESC) list, which was based on Ben-Porath et al. and Hassan et al. studies (21,22). The expression value for each pathway, including EASP, is the arithmetic mean of all genes in that pathway, giving a single value for each pathway in a given sample. Hierarchical clustering of the Shedden et al. 442 lung adenocarcinoma tumors (23) was performed for indicated oncogenic pathways along with EASP using TreeView (http://www.eisenlab.org/eisen/?page_id=42), and correlations are presented as a heat map with columns representing individual tumors and rows representing the arithmetic mean of a pathway.
Primary tumor-derived gene expression data sets and patient characteristics
Four published Affymetrix microarray data sets representing 908 lung tumors were used in the EASP survival analysis. The CEL files of microarray data were normalized using Robust Multi-array Average method (24). Shedden et al. 442 lung adenocarcinomas (Shedden) were used as training set (23). The other three data sets were used as test sets which included Bild et al. 111 adenocarcinomas and squamous cell carcinoma data set (Bild) (25), Okayama et al. 226 early-stage (stages 1 and 2) adenocarcinoma data set (Okayama) (26) and Raponi’s 129 squamous cell carcinoma data set (Raponi) (27). The patient characteristics and clinical information for these four data sets are provided in Table II. The primary end point was 5 year survival.
Statistical analysis method
The random survival forests (RSF) developed in R package by Ishwaran et al. (28,29) was employed for the EASP survival analysis of the four microarray data sets of lung cancer, as described before (12). Briefly, The RSF is an ensemble tree method for analysis of right-censored survival data. Each decision tree of forests was grown by splitting patients by comparing survival differences via log-rank test based on a randomly selected subset of variables at each node. The 1000 trees were grown for each RSF. Once trees were built, test sets were dropped down to the trees for prediction. The cumulative hazard function was derived from each tree, and an ensemble cumulative hazard function, an average over 1000 survival trees, was determined. Mortality was obtained as a weighted sum over ensemble cumulative hazard function, weighted by the number of individuals at risk at the different time points. Higher mortality values imply the higher risk. We used mortality as risk index to separate patients into three risk groups (high, medium and low risk, one-third each group) and presented Kaplan–Meier survival curves for each group. Each tree provides a measure of its predictive error as described by Ishwaran et al. (28,29), with smaller number indicating a better tree. The prediction error is calculated by C-index (i.e. the Harrell’s concordance index) in the out-of-bag data which were not used for building a tree each time.
Variable importance scores (VIMPS) for all the variables used to grow trees were also generated. Large VIMPS indicate variables as good predictors for outcome, whereas zero or negative values identify non-predictive. These scores were used to refine the 97-gene EASP to the 20-gene rEASP.
Cox proportional hazards regression model, Kaplan–Meier survival curve and log-rank test were used for survival analysis of individual genes or mortality index derived from RSF. The t-test was used to assess the difference of mean expression of EASP signature in clinical and pathological groups including stage, differentiation and nodal status.
Data reporting guidelines
In preparing this manuscript, we followed Molecular & Cellular Proteomics J ournal-recommended guidelines for reporting proteomics data (http://www.mcponline.org/site/misc/PhialdelphiaGuidelinesFINALDRAFT.pdf), the Journal of Clinical Oncology-recommended REMARK guidelines for reporting tumor marker prognostic studies (30,31) and lock-down of the fully specified classifier before application to the first of the four tests of specimens, as mandated by the Institute of Medicine (IOM) (32) and by Hayes et al. (33).
Results
Quantitative identification of differentially secreted proteins during EMT
A549 lung adenocarcinoma cells were cultured in the serum-free media, stimulated with TGF-β for 72h to induce EMT and the conditioned media were collected from control and TGF-β-treated cells for the analysis of differentially secreted proteins. Induction of EMT was confirmed by assessing E-cadherin, N-cadherin and vimentin expression in the cells by western immunoblotting as described before (34,35) (data not shown). Proteins in the conditioned media from two different biological replicates were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Each lane on the gel was cut into 40 slices. Proteins in each gel slice were subjected to trypsin digestion and analyzed by LC-MS/MS on a LTQ Orbitrap mass spectrometer. The resulting MS/MS spectra were analyzed for protein identification and quantitation using MAXQUANT as described under Materials and methods. We identified a total of 2410 proteins, of which 1647 (70%) proteins were annotated as secreted using the multiple data bases and strategies described in Materials and methods (Supplementary Table S2, available at Carcinogenesis Online). With the criteria of at least 2-fold change, we identified 136 proteins as increased in secretion (log2 ratio >1) and 94 proteins as decreased in secretion (log2 ratio < −1) during EMT.
Among the differentially secreted proteins, we observed various categories of proteins including increased secretion of proteases (MMP2, MMP9, BMP1), extracellular matrix components (collagens, fibronectin, versican and SPARC), cytokines (CTGF) and cell surface receptors (mucins, CD59) that are consistent with the migratory, invasive and immune evasive abilities conferred by EMT and their regulation by TGF-β.
EMT-associated secretory phenotype
To identify a gene signature that is representative of EMT and may serve as a reliable biomarker for patient prognosis, we integrated the differentially secreted protein profile with the corresponding gene expression profile we published earlier (14), from the same cell line and under identical conditions. To match with the secretome, differentially expressed genes only at 72h time point were used for integration from the time course data set. Because the goal is to derive a measurable signature, only proteins whose secretion is induced during EMT were considered. By integrating gene and protein expression, we identified 97 genes that are upregulated at mRNA level by at least 2-fold (P > 0.01) and increased in secretion at the protein level by at least 2-fold, irrespective of P value, and defined them as EASP (Table I).
Table I.
List of genes that constitute EASP with corresponding fold change for gene and protein expression
| Gene symbol | Entrez ID | Protein accession ID | Gene title | Fold change (TGF-β/control) | |
|---|---|---|---|---|---|
| Microarray | Secretome | ||||
| ADAM19 | 8728 | IPI00011901 | A disintegrin and metalloproteinase domain 19 (meltrin beta) | 17.47 | 26.78 |
| ANGPTL4 | 51129 | IPI00153060 | Angiopoietin-like 4 | 19.67 | 2.33 |
| AP1S2 | 8905 | IPI00909244 | Adaptor-related protein complex 1, sigma 2 subunit | 2.15 | 3.23 |
| ARPC4 | 10093 | IPI00925052 | Actin-related protein 2/3 complex, subunit 4, 20 kda | 2.03 | 1.41 |
| BMP1 | 649 | IPI00014021 | Bone morphogenetic protein 1 | 4.68 | 17.90 |
| BPGM | 669 | IPI00215979 | 2,3-bisphosphoglycerate mutase | 3.85 | 4.23 |
| CD151 | 977 | IPI00298851 | Cd151 antigen | 1.81 | 2.09 |
| CD59 | 966 | IPI00011302 | Cd59 antigen | 3.99 | 2.53 |
| CHST11 | 50515 | IPI00099831 | Carbohydrate (chondroitin 4) sulfotransferase 11 | 4.62 | 4.00 |
| CHST3 | 9469 | IPI00306853 | Carbohydrate (chondroitin 6) sulfotransferase 3 | 4.30 | 3.61 |
| COL1A1 | 1277 | IPI00297646 | Collagen, type i, alpha 1 | 10.11 | 3.44 |
| COL4A1 | 1282 | IPI00743696 | Collagen, type iv, alpha 1 | 62.47 | 5.41 |
| COL4A2 | 1284 | IPI00306322 | Collagen, type iv, alpha 2 | 15.99 | 5.24 |
| COL4A3 | 1285 | IPI00010360 | Collagen, type iv, alpha 3 (good pasture antigen) | 3.18 | 4.57 |
| COL7A1 | 1294 | IPI00025418 | Collagen, type vii, alpha 1 | 3.42 | 1.35 |
| CRIP2 | 1397 | IPI00921911 | Cysteine-rich protein 2 | 2.86 | 19.22 |
| VCAN | 1462 | IPI00215628 | Chondroitin sulfate proteoglycan 2 (versican) | 2.97 | 1.89 |
| CTGF | 1490 | IPI00020977 | Connective tissue growth factor | 4.96 | 4.37 |
| CXCL12 | 6387 | IPI00719836 | Chemokine (c-x-c motif) ligand 12 (stromal cell- derived factor 1) | 5.56 | 17.71 |
| CYR61 | 3491 | IPI00299219 | Cysteine-rich, angiogenic inducer, 61 | 5.66 | 2.16 |
| DSC2 | 1824 | IPI00025846 | Desmocollin 2 | 4.12 | 3.15 |
| ECM1 | 1893 | IPI00645849 | Extracellular matrix protein 1 | 2.09 | 3.64 |
| EFNA1 | 1942 | IPI00025840 | Ephrin-a1 | 1.72 | 1.14 |
| EIF4EBP1 | 1978 | IPI00002569 | Eukaryotic translation initiation factor 4e-binding protein 1 | 1.94 | 20.54 |
| EPHB2 | 2048 | IPI00021275 | Eph receptor b2 | 8.07 | 1.76 |
| FHL2 | 2274 | IPI00396967 | Four and a half lim domains 2 | 3.15 | 3.31 |
| FN1 | 2335 | IPI00845263 | Fibronectin 1 | 5.14 | 3.15 |
| FST | 10468 | IPI00021081 | Follistatin | 6.45 | 1.30 |
| FSTL1 | 11167 | IPI00029723 | Follistatin-like 1 | 5.14 | 2.18 |
| FSTL3 | 10272 | IPI00025155 | Follistatin-like 3 (secreted glycoprotein) | 3.98 | 2.21 |
| C11orf41 | 25758 | IPI00852979 | G2 protein | 3.98 | 20.18 |
| GALNT2 | 2590 | IPI00004669 | Udp-n-acetyl-alpha-d-galactosamine:polypeptide n-acetylgalactosaminyltransferase 2 (galnac-t2) | 2.78 | 1.69 |
| GSN | 2934 | IPI00646773 | Gelsolin (amyloidosis, finnish type) | 3.99 | 2.70 |
| HMGA2 | 8091 | IPI00005996 | High mobility group at-hook 2 /// high mobility group at-hook 2 | 4.20 | 3.13 |
| HMOX1 | 3162 | IPI00215893 | Heme oxygenase (decycling) 1 | 3.41 | 17.36 |
| HSPB1 | 3315 | IPI00025512 | Heat shock 27 kda protein 1 | 2.28 | 2.42 |
| IGF1 | 3479 | IPI00433029 | Insulin-like growth factor 1 (somatomedin c) | 4.65 | 1.11 |
| IGF2 | 3481 | IPI00215977 | Insulin-like growth factor 2 (somatomedin a) | 1.03 | 1.87 |
| IGFBP5 | 3488 | IPI00029236 | Insulin-like growth factor-binding protein 5 | 21.60 | 7.17 |
| IGFBP7 | 3490 | IPI00016915 | Insulin-like growth factor-binding protein 7 | 63.94 | 5.33 |
| IL11 | 3589 | IPI00025820 | Interleukin 11 | 39.43 | 5.57 |
| INHBA | 3624 | IPI00028670 | Inhibin, beta a (activin a, activin ab alpha polypeptide) | 24.32 | 25.47 |
| ITGA2 | 3673 | IPI00013744 | Integrin, alpha 2 (cd49b, alpha 2 subunit of vla-2 receptor) | 5.13 | 1.15 |
| ITGA3 | 3675 | IPI00290043 | Integrin, alpha 3 (antigen cd49c, alpha 3 subunit of vla-3 receptor) | 1.90 | 1.80 |
| JAG1 | 182 | IPI00099650 | Jagged 1 (Alagille syndrome) | 3.10 | 1.87 |
| MGC17330 | 113791 | IPI00298388 | Phosphoinositide-3-kinase interacting protein 1 | 2.53 | 20.35 |
| KIAA1797 | 54914 | IPI00748360 | Kiaa1797 | 1.70 | 19.36 |
| LAMC2 | 3918 | IPI00015117 | Laminin, gamma 2 | 20.02 | 7.45 |
| LEFTY2 | 7044 | IPI00010893 | Left-right determination factor 2 | 2.23 | 23.89 |
| LIF | 3976 | IPI00009720 | Leukemia inhibitory factor (cholinergic differentiation factor) | 2.71 | 2.22 |
| XYLT1 | 64131 | IPI00183487 | Hypothetical protein loc283824 | 12.13 | 23.61 |
| LTBP1 | 4052 | IPI00784258 | Latent transforming growth factor beta-binding protein 1 | 2.86 | 2.25 |
| LTBP2 | 4053 | IPI00292150 | Latent transforming growth factor beta-binding protein 2 | 9.42 | 4.66 |
| LTBP3 | 4054 | IPI00073196 | Latent transforming growth factor beta-binding protein 3 | 3.87 | 1.55 |
| LTBP4 | 8425 | IPI00873371 | Latent transforming growth factor beta-binding protein 4 | 3.01 | 2.14 |
| PIK3IP1 | 113791 | IPI00298388 | Hgfl gene /// hgfl gene | 2.53 | 20.35 |
| MMP1 | 4312 | IPI00008561 | Matrix metalloproteinase 1 (interstitial collagenase) | 5.81 | 6.27 |
| MMP10 | 4319 | IPI00013405 | Matrix metalloproteinase 10 | 20.03 | 25.23 |
| MMP2 | 4313 | IPI00027780 | Matrix metalloproteinase 2 | 10.86 | 8.36 |
| MMP9 | 4318 | IPI00027509 | Matrix metalloproteinase 9 | 1.34 | 22.00 |
| MRC2 | 9902 | IPI00005707 | Mannose receptor, c type 2 | 4.97 | 1.47 |
| NPC2 | 10577 | IPI00301579 | Niemann-pick disease, type c2 | 2.88 | 3.33 |
| NPTX1 | 4884 | IPI00220562 | Neuronal pentraxin i | 7.29 | 7.41 |
| NRG1 | 3084 | IPI00221375 | Neuregulin 1 | 3.27 | 2.19 |
| PAWR | 5074 | IPI00001871 | Prkc, apoptosis, wt1, regulator | 2.34 | 20.17 |
| PCDH1 | 5097 | IPI00872579 | Protocadherin 1 (cadherin-like 1) | 4.48 | 22.09 |
| PDGFB | 5155 | IPI00000044 | Platelet-derived growth factor beta polypeptide | 4.86 | 18.85 |
| PDLIM2 | 64236 | IPI00007983 | Pdz and lim domain 2 (mystique) | 2.74 | 16.79 |
| PGRMC2 | 10424 | IPI00005202 | Progesterone receptor membrane component 2 | 1.85 | 2.20 |
| PLAT | 5327 | IPI00019590 | Plasminogen activator, tissue | 4.41 | 19.85 |
| PLAUR | 5329 | IPI00010676 | Plasminogen activator, urokinase receptor | 3.07 | 2.22 |
| PLOD2 | 5352 | IPI00337495 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 2.81 | 2.03 |
| PLSCR3 | 57048 | IPI00216127 | Phospholipid scramblase 3 | 2.25 | 3.22 |
| PPP1R14B | 26472 | IPI00398922 | Protein phosphatase 1, regulatory (inhibitor) subunit 14b | 2.11 | 1.55 |
| HTRA1 | 5654 | IPI00003176 | Protease, serine, 11 (igf binding) | 2.63 | 3.00 |
| PTPRK | 5796 | IPI00470937 | Protein tyrosine phosphatase, receptor type, k | 6.19 | 1.64 |
| RSU1 | 6251 | IPI00847168 | Ras suppressor protein 1 | 2.00 | 1.41 |
| SCG2 | 7857 | IPI00009362 | Secretogranin ii (chromogranin c) | 31.96 | 7.63 |
| SEMA3C | 10512 | IPI00019209 | Sema domain, immunoglobulin domain (ig), short basic domain, secreted, (semaphorin) 3c | 4.87 | 3.70 |
| SERPINE1 | 5054 | IPI00007118 | Serine (or cysteine) proteinase inhibitor, clade e (nexin, plasminogen activator inhibitor type 1), member 1 | 41.73 | 4.13 |
| SERPINE2 | 5270 | IPI00009890 | Serine (or cysteine) proteinase inhibitor, clade e (nexin, plasminogen activator inhibitor type 1), member 2 | 17.90 | 4.36 |
| SPOCK1 | 6695 | IPI00005292 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) | 70.53 | 5.84 |
| STC1 | 6781 | IPI00005564 | Stanniocalcin 1 | 10.09 | 3.99 |
| TAGLN | 6876 | IPI00216138 | Transgelin | 13.35 | 6.44 |
| TAGLN2 | 8407 | IPI00647915 | Transgelin 2 | 2.24 | 1.17 |
| TAX1BP3 | 30851 | IPI00005585 | Tax1 (human t-cell leukemia virus type i)-binding protein 3 | 1.83 | 3.54 |
| TGFB1 | 7040 | IPI00000075 | Transforming growth factor, beta 1 | 2.57 | 2.22 |
| TGFBR1 | 7046 | IPI00005733 | Transforming growth factor, beta receptor i | 5.72 | 2.72 |
| THBS1 | 7057 | IPI00296099 | Thrombospondin 1 | 16.73 | 4.71 |
| TIMP2 | 7077 | IPI00027166 | Tissue inhibitor of metalloproteinase 2 | 4.72 | 2.54 |
| TLL2 | 7093 | IPI00465231 | Tolloid-like 2 | 2.59 | 2.15 |
| TNFAIP6 | 7130 | IPI00303341 | Tumor necrosis factor, alpha-induced protein 6 | 5.03 | 7.48 |
| TNFRSF12A | 51330 | IPI00010277 | Tumor necrosis factor receptor superfamily, member 12a | 5.10 | 1.83 |
| TP53I3 | 9540 | IPI00384643 | Tumor protein p53 inducible protein 3 | 3.35 | 2.47 |
| TUBA4B | 80086 | IPI00017454 | Tubulin, alpha 4 | 2.24 | 2.20 |
| ULBP2 | 80328 | IPI00018860 | Ul16-binding protein 2 | 5.18 | 3.51 |
| VEGFA | 7422 | IPI00012567 | Vascular endothelial growth factor | 4.66 | 2.86 |
Twenty genes that are part of rEASP are in bold.
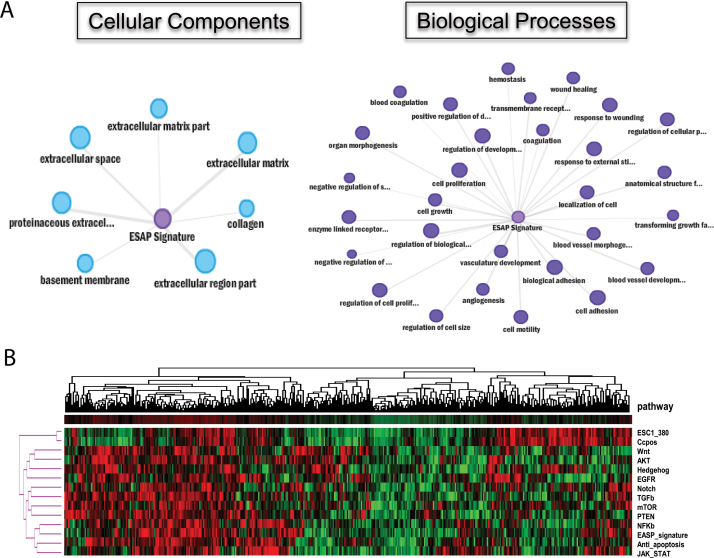
For functional interpretation, EASP was subjected to gene set enrichment analysis using ConceptGen (14). Analysis for cellular components has associated EASP with extracellular matrix, proteinaceous extracellular, collagen, basement membrane, matrix space, matrix part and matrix region part (Figure 1A), consistent with their annotation as secretory proteins. More importantly, enrichment analysis for biological processes has associated EASP with the cellular processes including cell adhesion, motility, actin cytoskeleton reorganization, coagulation, acute inflammatory response, proteolysis and response to wounding and external stimuli (Figure 1A), consistent with the biology of EMT. Moreover, this also demonstrates that EASP is a true representation of EMT and may serve as reliable biomarker to track EMT.
Fig. 1.
(A) Analysis of EMT secretome for enriched biological processes and cellular components. All the genes listed in EASP signature (Table I) were uploaded to ConceptGen (http://conceptgen.ncibi.org) tool. The enriched GO cellular components (P < 0.0001) and biological processes (P < 0.0001) are shown. Each node size is proportional to the number of genes in the enriched process or pathway and each edge represents a statistically significant enrichment with defined P value. (B) Cluster analysis of EASP signature and other cancer-related pathways in 442 lung adenocarcinomas. Column represents individual cancer sample, row represents different pathway (mean value was used for each specific pathway). Ccpos, positive cell cycle gene; ESC1_380, embryonic stem cell 380 genes. Red indicated higher expression and green is lower expression.
To assess the correlation of EASP with other known oncogenic pathways, we performed hierarchical clustering of 442 lung adenocarcinomas based on their mean gene expression of the indicated pathway. Clustering analysis yielded two distinct lung adenocarcinoma tumor groups with 50% tumors demonstrating higher expression of all pathways. Mean EASP expression pattern correlated with mean gene expression of all the oncogenic pathways tested. These include NF-κβ, antiapoptosis, JAK-STAT, Notch, AKT, WNT pathways and ESC signature (Figure 1B). All these pathways are known to be deregulated in lung adenocarcinomas and were implicated in the regulation of EMT.
Correlation of EASP with clinical variables
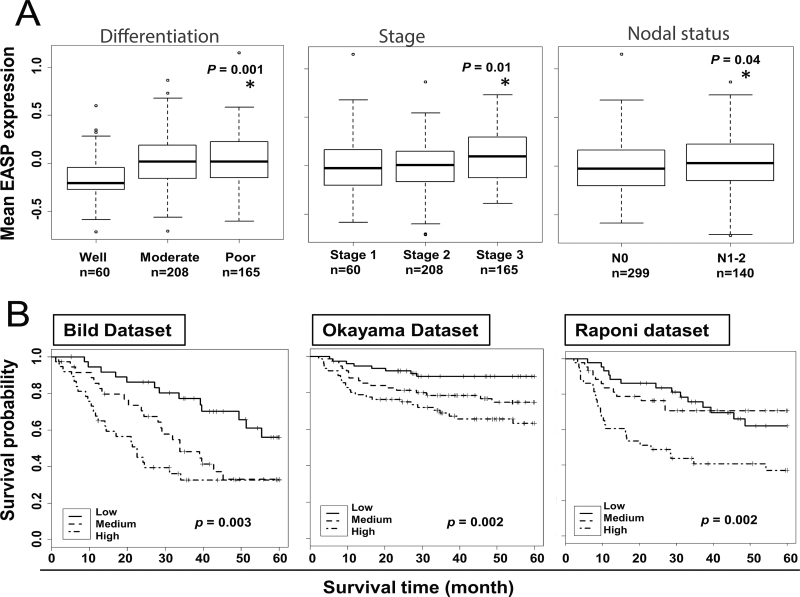
We determined the ability of the EASP signature to stratify the patients based on tumor stage, differentiation and nodal status using the gene expression data derived from the Shedden et al. 442 lung adenocarcinoma patients (23) (Figure 2A). The EASP signature was able to identify the patients with well-differentiated tumors from moderately and poorly differentiated tumors (P < 0.001). Similarly, the EASP signature was able to separate patients with stage I tumors from stage II and III (P ≤ 0.01). Furthermore, the EASP signature expression is high in patients with positive nodal status (N1–2) compared with patients with negative nodal status (N0). Together, these results indicate the potential clinical utility of EASP in predicting aggressive tumor behavior.
Fig. 2.
(A) Correlation of EASP expression with differentiation, stage and nodal status. All the patients in Shedden data set (n = 442) were classified into different subgroups based on tumor differentiation, tumor stage or lymph node status recorded at the time of diagnosis. The mean expression of EASP signature (mean centered value) in tumors is significantly different in differentiation (well vs poor), tumor stage (stage 1 vs stage 3) and lymph node status (N0 vs N1–2). (B) Testing of 97-gene EASP signature as a predictor of patient survival in lung cancer: lung cancer patients from Bild, Okayama and Raponi data sets were stratified using 97-gene EASP signature into low-, medium- and high-risk groups (one-third in each group) based on survival analysis by RSF model built on Shedden 442 training set. Shown are the Kaplan–Meir survival curves based on mortality. The log-rank P values compare the groups stratified by EASP signature.
EASP stratifies lung cancer patients into low-, medium- and high-risk groups with distinct survival
In order to investigate whether the 97-gene EASP signature could predict the overall survival in non-small-cell lung cancer patients, the Shedden data set (n = 442) was used as training set. As detailed in Materials and methods, a mathematical model based on an RSF algorithm was built in the training set to predict the prognostic significance of EASP with stage, age and sex included. After locking down the model, it was tested in three independent publicly available lung cancer data sets, Bild et al. (n = 111) (25), Okayama et al. (n =226) (26) and Raponi et al. (n = 129) (27). These cohorts include lung adenocarcinoma and squamous cell carcinoma patients. The prediction error rates were 33.6, 30.0 and 36.7%, respectively, for the Bild, Okayama and Raponi data sets (Supplementary Table S3, available at Carcinogenesis Online). We tested the usefulness of RSF predictors using a univariate Cox model with the mortality index as a continuous measure. The RSF prediction was significant for the Bild test set (likelihood ratio test [LRT] P = 0.00008), Okayama test set (LRT P = 0.005) and Raponi test set (LRT P = 0.02). In all three test sets, low-, medium- and high-risk groups were clearly separated by mortality index (Figure 2B). The hazard ratios (HRs) were 1.00, 2.17 and 3.16 for the Bild data set (log-rank test, P = 0.003); 1.00, 2.60 and 5.37 for Okayama data set (log-rank test, P = 0.002) and 1.00, 0.96 and 2.61 for the Raponi data set (log-rank test, P = 0.002), for low-, medium- and high-risk groups, respectively (Supplementary Table S3, available at Carcinogenesis Online).
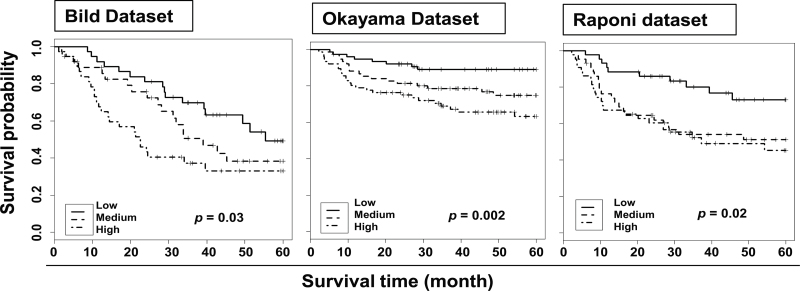
Refining the 97-gene EASP to the 20-gene rEASP
In order to refine the EASP into a smaller gene subset that remains as effective in predicting survival of lung cancer patients, the 97-gene EASP was filtered based on the VIMPS generated by RSF analysis of the Shedden et al. 442 data set. Higher VIMP values indicate variables with predictive ability, whereas zero or negative values identify non-predictive variables. This resulted in a refined EASP (rEASP) comprising of top 20 genes with higher VIMPS (Supplementary Figure S1, available at Carcinogenesis Online). To assess whether the 20-gene rEASP performs as well as the 97-gene EASP signature in predicting patient survival, another model based on RSF algorithm was built to predict the prognostic significance of rEASP with stage, age and sex included, using Shedden data set (n = 442) as training set. Next, we tested the predictive power of this 20-gene rEASP model in all three independent test sets described above (Bild, Raponi and Okayama) with clinical information in Table II. The prediction error rates were 35.6, 29.9 and 36.4%, respectively for the Bild, Okayama and Raponi data sets (Table III). We tested the usefulness of RSF predictors using a univariate Cox model with the mortality index as a continuous measure. The RSF prediction was significant for the Bild test set (LRT P = 0.002), Okayama test set (LRT P = 0.0004) and Raponi test set (LRT P = 0.007). In all three test sets, low-, medium- and high-risk groups were clearly separated by mortality index (Figure 3). The HRs were 1.00, 1.54 and 2.34 for the Bild data set (log-rank test, P = 0.03); 1.00, 2.37 and 3.75 for Okayama data set (log-rank test, P = 0.002) and 1.00, 2.38 and 2.79 for the Raponi data set (log-rank test, P = 0.02), for low-, medium- and high-risk groups, respectively (Table III).
Table II.
Clinical characteristics of samples used in this study
| Data set | Shedden set | Bild set | Raponi set | Okayama set |
|---|---|---|---|---|
| Sample number | 442 | 111 | 129 | 226 |
| Type of cancer | Ad | 58 Ad/53 SCC | SCC | Ad |
| Age average | 64.4 | 64.8 | 67.5 | 59.6 |
| Gender | ||||
| Female | 219 | 48 | 48 | 121 |
| Male | 224 | 63 | 82 | 105 |
| Stage | ||||
| Stage I | 276 | 67 | 73 | 168 |
| Stage II | 105 | 18 | 34 | 58 |
| Stage III | 59 | 21 | 23 | 0 |
| Differentiation | ||||
| Well | 60 | NA | 15 | NA |
| Moderate | 209 | NA | 76 | NA |
| Poor | 167 | NA | 39 | NA |
| Dead (5 year) | 188 | 58 | 52 | 32 |
| Alive | 255 | 53 | 78 | 194 |
| Adjuvant therapy | ||||
| Yes | 109 | 48 | ||
| No | 330 | 69 | 204 | |
| Unknown | 3 | 111 | 12 | 22 |
Ad, adenocarcinomas; NA, not available; SCC, squamous cell cancer.
Adjuvant therapy includes chemo- and/or radiotherapy.
Table III.
Prediction results of 20-gene rEASP signature on three test sets
| RSFa | Cox modelb | HR | Log rank testc | P | |
|---|---|---|---|---|---|
| Test error rate | P | 95% Confidence interval | |||
| Okayama test set (n = 226) | |||||
| 29.9% | 0.0004 | ||||
| Low risk | 1 | 0.002 | |||
| Medium risk | 2.37 | 1.03–5.46 | |||
| High risk | 3.75 | 1.70–8.28 | |||
| Raponi test set (n = 129) | |||||
| 36.4% | 0.007 | ||||
| Low risk | 1 | 0.02 | |||
| Medium risk | 2.38 | 1.12–5.10 | |||
| High risk | 2.79 | 1.32–5.90 | |||
| Bild test set (n = 111) | |||||
| 35.6% | 0.002 | ||||
| Low risk | 1 | 0.03 | |||
| Medium risk | 1.54 | 0.78–3.02 | |||
| High risk | 2.34 | 1.24–4.42 | |||
aRSF prediction model built from the 442 training set including 20 genes, age, gender and stage.
bMortality risk index (MRI) as continuous value, LRT was used in univariate Cox model.
cMRI separated test patients to three risk groups (low, medium and high-risk, one-third in each group).
Fig. 3.
Testing of 20-gene rEASP signature as a predictor of patient survival in lung cancer: lung cancer patients from Bild’s, Okayama’s and Raponi’s data sets were stratified using 20-gene rEASP signature into low-, medium- and high-risk groups based on survival analysis by RSF model built on Shedden 442 training set. Shown are the Kaplan–Meir survival curves based on mortality. The log-rank P values compare the groups stratified by EASP signature.
Discussion
Contrary to the perception that tumor metastasis progresses in a linear and step-wise fashion, recent evidence suggests that a subset of tumors harbor molecular alterations at an early stage that are indicative of bad prognosis and poor patient survival (36). This demonstrates the importance of identifying molecular changes at an early stage that dictate clinical behavior. The current system of TNM staging cannot identify such changes. There is an urgent need to develop prognostic tests that can predict recurrence and identify high-risk patients at an early stage when they would benefit from adjuvant therapy (5,37–44). Demonstrating the utility of such a prognostic test, a 70-gene signature (10) (MammaPrint; Agendia, The Netherlands) has been approved by the Food and Drug Administration for breast cancer patients (32). A 21-gene signature (45) (Oncotype DX; Genome Health, CA) is approved for breast cancers, with analogous signatures under development for prostate and colon cancers. Even though multiple gene, protein, autoantibodies and miRNA-based profiles have been proposed for lung cancer prognosis, none to date has been approved for clinical use.
The predominant approach in deriving most of the prognostic signatures has been profiling differentially occurring molecular changes between good versus bad outcome groups, without any consideration to the underlying tumor biology. Here, we adopted a new, mechanism-based approach to identify predictive biomarkers by profiling the complex cellular process of EMT, which is implicated in the initiation of tumor metastasis. The rationale behind this approach is that identifying proteins secreted during the course of a critical biological process that promotes a metastatic phenotype would provide relevant, reliable and robust prognostic biomarkers. The other novel aspect of this approach is that one can measure EASP at the mRNA level and also at the protein level, because the biomarkers of EASP are based on the strong concordant expression of both mRNA and protein. Given that EMT is the initiating event for metastasis and may result in the dissemination of tumor cells, measuring EASP in the primary tumor, tumor cells in bone marrow compartment and circulating tumor cells may allow the ability to track disease progression.
Consistent with the functional attributes conferred by EMT and its regulation by TGF-β, we identified proteins that are implicated in tumor cell adhesion, migration, invasion, immune evasive mechanisms, extracellular matrix components and tumor-stromal interactions. Gene set enrichment analysis of the 97-gene EASP, which is the subset of all up regulated proteins and their mRNAs, also identified biological processes that are reflective of EMT and TGF-β biology. Similarly, even the proteins in rEASP are representative of the functional EMT phenotype. Furthermore, clustering analysis of EASP with key oncogenic pathways showed a similar correlation to the expression of various pathways that are deregulated in lung adenocarcinomas. These include NF-κβ, antiapoptosis, JAK-STAT, PTEN, AKT, WNT, Notch, Hedgehog and EGFR signaling pathways (46,47). Most importantly, the correlation of EASP expression with ESC signature is consistent with the recent finding that the ESC signature is associated with poor prognosis and worse overall survival in lung adenocarcinoma patients (22). This correlation is also consistent with finding that EMT may confer stem cell-like properties to breast cancer cells (48). Together, these observations demonstrate that EASP not only reflects the heterogeneity and complexity associated with oncogenesis of lung cancer, but also demonstrates the significance and relevance of EASP biomarkers to the underlying biology of tumor metastasis.
Consistent with its prognostic significance, EASP distinguished well from moderate or poorly differentiated tumors and stage 1 from stage 2 and 3 patients. Most importantly, it was strongly correlated with positive lymph node status, which is an important prognostic factor that influences the therapeutic decision making and probability of lung cancer recurrence. To test the clinical utility of EASP, the RSF analysis-based survival model was built and trained on 442 primary lung adenocarcinoma tumor-derived gene expression data set, the largest lung cancer gene expression data set available with pathological, clinical and treatment annotations (23). Using VIMPS from the training set, we refined the EASP into a subset of 20 genes (rEASP) with highest VIMPS and tested its prognostic significance in three independent lung cancer data sets. Because the EASP was derived from an adenocarcinoma cell line, we asked whether EASP is specific to adenocarcinomas or does it have any relevance to other subtypes of lung cancer. To address this, we selected the Okayama et al. data set of only adenocarcinomas (n = 226), the Bild et al. data set of adenocarcinomas (n = 58) and squamous cell carcinomas (n = 53) and the Raponi et al. data set of only squamous cell carcinomas (n = 129) for independent testing. Interestingly, in all three independent test sets, both EASP- and rEASP-based models were able to stratify patients into low-, medium- and high-risk groups with clearly separated mortality indexes and distinct HRs. This demonstrates the relevance of these models even for lung squamous cell cancer. It is important to note that both EASP and rEASP performed very well in the Okayama et al. data set, which is comprised of only early-stage patients (stages I and II). Because rEASP predicted the survival of early-stage patients, it might serve as an ideal prognostic signature to identify the high-risk early-stage patients who might benefit the most from adjuvant therapy. In earlier studies, we identified multiple inhibitors of EMT in lung cancer (49,50), and it might be beneficial to test these agents or other EMT blockers as adjuvants for patients with high EASP expression.
In conclusion, this study demonstrates the importance of a mechanism-based approach that integrates multiple omics data sets, to identify clinically relevant biomarkers for patient prognosis. Biomarkers rooted in underlying molecular and cellular biology of the tumors may provide very useful and actionable information for patient care.
Supplementary material
Supplementary Tables S1–S3 and Figures S1 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health/ National Cancer Institute (CA132571-01); American Cancer Society (RSG-CSM-116801 to V.G.K.); NIH (RM-08-029, P30U54ES017885 to G.S.O).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- EASP
EMT-associated secretory phenotype
- EMT
epithelial–mesenchymal transition
- ESC
embryonic stem cell
- GO
Gene Ontology
- HR
hazard ratio
- IPI
International Protein Index
- LC
liquid chromatography
- LRT
likelihood ratio test
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- RSF
random survival forests
- TGF
transforming growth factor
- TNM
tumor-node-metastasis
- VIMPS
variable importance scores.
References
- 1. Detterbeck F.C., et al. (2009). The new lung cancer staging system. Chest, 136, 260–271 [DOI] [PubMed] [Google Scholar]
- 2. Arribalzaga E.B. (2009). New tumor, node, metastasis staging system for lung cancer. J. Thorac. Oncol., 4, 1301; author reply 1301–1301; author reply 1302 [DOI] [PubMed] [Google Scholar]
- 3. Visbal A.L., et al. (2005). Adjuvant chemotherapy for early-stage non-small cell lung cancer. Chest, 128, 2933–2943 [DOI] [PubMed] [Google Scholar]
- 4. Waller D., et al. (2004). Chemotherapy for patients with non-small cell lung cancer: the surgical setting of the Big Lung Trial. Eur. J. Cardiothorac. Surg., 26, 173–182 [DOI] [PubMed] [Google Scholar]
- 5. Dômont J., et al. (2005). Adjuvant chemotherapy in early-stage non-small cell lung cancer. Semin. Oncol., 32, 279–283 [DOI] [PubMed] [Google Scholar]
- 6. Azzoli C.G. (2005). Can adjuvant chemotherapy improve survival in patients with early-stage, resected non-small-cell lung cancer? Nat. Clin. Pract. Oncol., 2, 552–553 [DOI] [PubMed] [Google Scholar]
- 7. Bhattacharjee A., et al. (2001). Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl Acad. Sci. USA, 98, 13790–13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lacroix L., et al. (2008). Gene expression profiling of non-small-cell lung cancer. Expert Rev. Mol. Diagn., 8, 167–178 [DOI] [PubMed] [Google Scholar]
- 9. Garber M.E., et al. (2001). Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl Acad. Sci. USA, 98, 13784–13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buyse M., et al. ; TRANSBIG Consortium. (2006). Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J. Natl. Cancer Inst., 98, 1183–1192 [DOI] [PubMed] [Google Scholar]
- 11. Slodkowska E.A., et al. (2009). MammaPrint 70-gene signature: another milestone in personalized medical care for breast cancer patients. Expert Rev. Mol. Diagn., 9, 417–422 [DOI] [PubMed] [Google Scholar]
- 12. Chen G., et al. (2011). Development and validation of a quantitative real-time polymerase chain reaction classifier for lung cancer prognosis. J. Thorac. Oncol., 6, 1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Subramanian J., et al. (2010). Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J. Natl. Cancer Inst., 102, 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sartor M.A., et al. (2010). ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics, 26, 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox J., et al. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol., 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 16. Bendtsen J.D., et al. (2004). Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel., 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 17. Möller S., et al. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics, 17, 646–653 [DOI] [PubMed] [Google Scholar]
- 18. Bendtsen J.D., et al. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol., 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 19. Nakai K., et al. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 20. Chen Y., et al. (2005). SPD–a web-based secreted protein database. Nucleic Acids Res., 33(Database issue), D169–D173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ben-Porath I., et al. (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet., 40, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hassan K.A., et al. (2009). An embryonic stem cell-like signature identifies poorly differentiated lung adenocarcinoma but not squamous cell carcinoma. Clin. Cancer Res., 15, 6386–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shedden K., et al. (2008). Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med., 14, 822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Irizarry R.A., et al. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 25. Bild A.H., et al. (2006). Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature, 439, 353–357 [DOI] [PubMed] [Google Scholar]
- 26. Okayama H., et al. (2012). Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res., 72, 100–111 [DOI] [PubMed] [Google Scholar]
- 27. Raponi M., et al. (2006). Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res., 66, 7466–7472 [DOI] [PubMed] [Google Scholar]
- 28. Ishwaran H., et al. (2007). Random survival forests for R. R News 7, 7 [Google Scholar]
- 29. Ishwaran H., et al. (2008). Random survival forests. Ann. Appl. Statist., 2, 20 [Google Scholar]
- 30. McShane L.M., et al. ; Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics. (2005). Reporting recommendations for tumor marker prognostic studies. J. Clin. Oncol., 23, 9067–9072 [DOI] [PubMed] [Google Scholar]
- 31. Alonzo T.A. (2005). Standards for reporting prognostic tumor marker studies. J. Clin. Oncol., 23, 9053–9054 [DOI] [PubMed] [Google Scholar]
- 32. Omenn G.S., et al. (2012). Evolution of Translational Omics: Lessons Learned and the Path Forward. Institute of Medicine of National Academies Press, Washington, DC; [PubMed] [Google Scholar]
- 33. Hayes D.F., et al. (2013). Breaking a vicious cycle. Sci. Transl. Med., 5, 196cm6. [DOI] [PubMed] [Google Scholar]
- 34. Keshamouni V.G., et al. (2009). Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-Induced epithelial-mesenchymal transition. J. Proteome Res., 8, 35–47 [DOI] [PubMed] [Google Scholar]
- 35. Keshamouni V.G., et al. (2006). Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. J. Proteome Res., 5, 1143–1154 [DOI] [PubMed] [Google Scholar]
- 36. Ramaswamy S., et al. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet., 33, 49–54 [DOI] [PubMed] [Google Scholar]
- 37. Felip E., et al. ; ESMO Guidelines Task Force. (2005). ESMO Minimum Clinical Recommendations for diagnosis, treatment and follow-up of non-small-cell lung cancer (NSCLC). Ann. Oncol., 16 (suppl. 1), i28–i29 [DOI] [PubMed] [Google Scholar]
- 38. Scagliotti G.V., et al. ; Adjuvant Lung Project Italy/European Organisation for Research Treatment of Cancer-Lung Cancer Cooperative Group Investigators. (2003). Randomized study of adjuvant chemotherapy for completely resected stage I, II, or IIIA non-small-cell Lung cancer. J. Natl. Cancer Inst., 95, 1453–1461 [DOI] [PubMed] [Google Scholar]
- 39. Johnson B.E., et al. (2005). Patient subsets benefiting from adjuvant therapy following surgical resection of non-small cell lung cancer. Clin. Cancer Res., 11(13 Pt 2), 5022s–5026s [DOI] [PubMed] [Google Scholar]
- 40. Keller S.M., et al. (2000). A randomized trial of postoperative adjuvant therapy in patients with completely resected stage II or IIIA non-small-cell lung cancer. Eastern Cooperative Oncology Group. N. Engl. J. Med., 343, 1217–1222 [DOI] [PubMed] [Google Scholar]
- 41. Douillard J.Y., et al. (2006). Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol., 7, 719–727 [DOI] [PubMed] [Google Scholar]
- 42. Arriagada R., et al. ; International Adjuvant Lung Cancer Trial Collaborative Group. (2004). Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N. Engl. J. Med., 350, 351–360 [DOI] [PubMed] [Google Scholar]
- 43. Jang R.W., et al. (2009). Quality-adjusted time without symptoms or toxicity analysis of adjuvant chemotherapy in non-small-cell lung cancer: an analysis of the National Cancer Institute of Canada Clinical Trials Group JBR.10 trial. J. Clin. Oncol., 27, 4268–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arriagada R., et al. (2010). Long-term results of the international adjuvant lung cancer trial evaluating adjuvant Cisplatin-based chemotherapy in resected lung cancer. J. Clin. Oncol., 28, 35–42 [DOI] [PubMed] [Google Scholar]
- 45. Paik S. (2007). Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. Oncologist, 12, 631–635 [DOI] [PubMed] [Google Scholar]
- 46. Faivre S., et al. (2006). New paradigms in anticancer therapy: targeting multiple signaling pathways with kinase inhibitors. Semin. Oncol., 33, 407–420 [DOI] [PubMed] [Google Scholar]
- 47. Sun S., et al. (2007). New molecularly targeted therapies for lung cancer. J. Clin. Invest., 117, 2740–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mani S.A., et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell, 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reka A.K., et al. (2010). Peroxisome proliferator-activated receptor-gamma activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol. Cancer Ther., 9, 3221–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reka A.K., et al. (2011). Identifying inhibitors of epithelial-mesenchymal transition by connectivity map-based systems approach. J. Thorac. Oncol., 6, 1784–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.