Summary
NSCLC is often a lethal disease but potentially curative at early stages. In this study, we showed that genetic variants in the developmental and stem cell-related Wnt signaling pathway may influence disease recurrence and survival in early-stage NSCLC patients.
Abstract
Early-stage non-small cell lung cancer (NSCLC) is potentially curative. Nevertheless, many patients will show disease recurrence after curative treatment. The Wnt signaling pathway is a developmental and stem cell pathway that plays an important role in tumorigenesis and may affect cancer progression. We hypothesize that genetic variants of the Wnt pathway may influence clinical outcome in early-stage NSCLC patients. We genotyped 441 functional and tagging single nucleotide polymorphisms (SNPs) from 54 genes of the Wnt pathway in 535 early-stage NSCLC patients treated with curative intent therapy including surgery and chemotherapy. For validation, 4 top SNPs were genotyped in 301 early-stage NSCLC patients from the Mayo Clinic. Cox proportional hazard model and combined SNP analyses were performed to identify significant SNPs correlated with recurrence-free and overall survival. Results from discovery group showed a total of 40 SNPs in 20 genes correlated with disease recurrence (P < 0.05). After correction for multiple comparisons, rs2536182 near Wnt16 remained significant (q < 0.1), which was validated in the replication population. Thirty-nine SNPs in 16 genes correlated with overall survival (P < 0.05) in the discovery group, and seven remained significant after multiple comparisons were considered (q < 0.1). In patients receiving surgery-only treatment, rs10898563 of FZD4 gene was associated with both recurrence-free and overall survival. Joint SNP analyses identified predictive markers for recurrence stratified by treatment. Our findings suggest inherited genetic variation in the Wnt signaling pathway may contribute to variable clinical outcomes for patients with early-stage NSCLC.
Introduction
Lung cancer is now the leading cause of cancer mortality for both men and women. More than 200 000 cases of lung cancer are diagnosed each year in the USA with an estimated 160 340 lung cancer deaths in 2012 (1). Only 15% of lung cancer patients are diagnosed with early-stage disease (stage I and II)—the majority of these have localized stage I cancer at presentation, potentially curable disease. Despite appropriate therapy, 30–40% of lung cancer patients with stage I disease and ~50% of patients with stage II disease ultimately relapse and die of their disease (2). Adjuvant cisplatin-based chemotherapy has been shown to improve survival in patients with stage II disease following surgery; however, this results in only a 5.4% cumulative survival benefit (3). No benefit has been shown for adjuvant chemotherapy in stage IA, and the use of adjuvant chemotherapy in stage IB is controversial. Therefore, many patients are subjected to the toxicity of adjuvant chemotherapy despite only a select few benefiting from therapy. Identifying prognostic and/or predictive markers, both tumor and host related, are important to select patients most likely to benefit from chemotherapy and to develop more targeted and effective management strategies for the highest risk patients.
Tumor-related markers for recurrence have been studied extensively; however, there is little data relating to host genetic variation and relapse in early-stage non-small cell lung cancer (NSCLC). Due to their unique properties, stem cell pathways may contribute to tumor biology within the tumor as well as within the host. Stem cells have been shown to have multiple effects in carcinogenesis, progression, invasion and metastasis (4,5). In solid tumors, a malignant stem cell population has been described, as well as a population of non-malignant host mesenchymal stem cells integral in producing tumor stroma and influencing the behavior of the malignant cells (6,7). The Wnt pathway is a stem cell pathway important in both embryogenesis and tumorigenesis (8,9). Wnt activation results in the stabilization of β-catenin and regulation of the transcription of the downstream members of the pathway. Several of these downstream molecules are important in tumorigenesis, including c-Myc, VEGF-A, cyclin D1 and MMP7 (10–12).
The Wnt pathway has been shown to be predictive of aggressive biology and progression in NSCLC (13–15). Lung adenocarcinomas have recently been shown to have a significant frequency of somatic mutations and copy number alterations within the Wnt pathway suggesting that this pathway plays a role in the pathogenesis of the disease. Specific genomic expression patterns of stem cells with an activated Wnt pathway have been described and tumors with signatures more similar to the stem cell expression pattern have a more aggressive phenotype. In lung cancer as well as several other solid tumor types, this signature has been associated with death from cancer as opposed to long-term remission after treatment for early-stage disease (16). In squamous cell cancers of the head and neck, genomic expression patterns associated with stem cell pathways have been implicated in locoregional failure after chemotherapy and radiation (17).
In this study, we determined whether germline genetic variations within the Wnt pathway correlate with relapse-free survival in patients treated for early-stage NSCLC. Tagging and functional single nucleotide polymorphisms (SNPs) were selected for each gene and genotyped in patients with early-stage NSCLC, and these variants were then correlated with the clinical outcomes of recurrence and survival.
Materials and methods
Patient population
Subjects in this study were histologically confirmed NSCLC patients, who were enrolled into an ongoing epidemiologic lung cancer study at The University of Texas MD Anderson Cancer Center between November 1995 and February 2008. Patients were included if they had clinical stage I or stage II disease and were treated with curative intent. Treatment with curative intent was defined as either surgical resection with negative margins, surgical resection followed by adjuvant chemotherapy and/or adjuvant radiation therapy, neoadjuvant chemotherapy followed by surgical resection or definitive radiation therapy for stage I patients. Patients were excluded if their surgical pathology upstaged them to stage III or IV. Patients were also excluded if they had any malignancy other than superficial skin cancer (except melanoma) within 5 years of enrollment or had any prior chemotherapy, radiation therapy or had been enrolled in a chemoprevention study. In addition, two clinical stage I patients who had pathological stage 0 diagnosis (carcinoma in situ) were excluded from further analysis. Although neoadjuvant chemotherapy is not routinely used in stage I and II NSCLC, there were patients included in this study who received neoadjuvant chemotherapy. The majority of these patients were enrolled in a clinical trial investigating neoadjuvant chemotherapy. Those who received neoadjuvant chemotherapy outside of this clinical trial were given this therapy due to a high likelihood of a positive surgical margin as assessed by the surgeon prior to surgery, or to allow for a lobectomy rather than a pneumonectomy. Although there was no restriction in terms of race/ethnicity in our patient accrual, we restricted our study to the non-Hispanic Caucasian subjects due to the small sample sizes of other groups and also to minimize population stratification.
For validation, we included an independent group of early-stage NSCLC cases from the Mayo Clinic. Detailed description of the subject recruitment has been described previously (18). In brief, confirmed NSCLC patients without restrictions on age, gender and ethnicity were serially identified by a daily electronic pathology system and then consented for study enrollment from 1997 to 2002. Due to the small number of minorities and patients treated with chemotherapy (<10%), only non-Hispanic white cases with curative surgery-only treatment were included in the replication analysis.
Blood samples were collected for all patients for genotyping analysis. Detailed smoking history was obtained at the time of recruitment and informed consent obtained to collect follow-up clinical data. The study was approved by the institutional review boards at MD Anderson Cancer Center and Mayo Clinic.
Clinical data and outcomes collection
Patients with NSCLC were staged clinically based on the 1997 staging system, and pathologic staging was verified by the pathologic reports. Clinical data including adjuvant chemotherapy, radiation therapy, surgical pathology with margins, histology, date of relapse and site of relapse were abstracted from the medical record. For patients without evidence of disease at last follow-up and with no follow-up within 6 months of time of abstraction, social security death index was used to confirm vital status. For patients with recurrent disease and with no follow-up within 3 months at time of abstraction, social security death index was used to confirm vital status.
The primary endpoint of this study is recurrence-free survival and the secondary outcome is overall survival. Recurrence-free survival was calculated as time from date of first treatment to date of recurrence or local and distant metastasis. Patients were censored at death or last follow-up. Overall survival was calculated from date of first treatment to date of death. Recurrence and survival data was collected from the medical record, social security death index or contact with the patient, family member or primary physician.
In the Mayo population, participants’ medical records were abstracted and interviews conducted to obtain demographic and clinical information, including smoking history. Causes of death were determined by reviewing the Mayo Clinic registration database and medical records, correspondence from patients’ next of kin, death certificates, obituary documents, the Mayo Clinic Tumor Registry and the social security death index. The same condition was used to define recurrence-free survival, and censors were applied at patient death or last follow-up.
SNP selection and genotyping
We generated a cancer-related gene list for the Wnt signaling pathway using the Gene Ontology database (http://www.geneontology.org). Extensive literature review of genes from Gene Ontology database was done using HUGO names and common aliases. A priority score was assigned to each gene based on its relevance and significance to cancer. A total of 54 genes with high priority score in the Wnt pathway were selected and their gene positions annotated using the UCSC Golden Path Genome Browser. Tagging SNPs were identified within 10 kb flanking regions upstream and downstream of each gene with r 2 of 0.8 or higher and a minor allele frequency of >5% in Caucasians using the LDSelect program (http://droog.gs.washington.edu/IdSelect.html) based on the HapMap database. Additionally, potentially functional SNPs which may affect gene expression or splicing (SNPs in coding region, promoter region, 5′-untranslated region, 3′-untranslated region or splice sites) were selected. A total of 441 SNPs were selected in these 54 genes. Complete set of SNPs was sent to Illumina for iSelect Infinium BeadChip design using proprietary program developed by Illumina. Genomic DNA was extracted from peripheral blood using QIAmp DNA extraction kit (Qiagen) and genotyped by Illumina’s BeadArray platform according to the manufacturer’s instruction. Genotypes were auto-called using the Beadstudio software. In the replication phase, the SNPs significant for recurrence in the overall group and surgery-only subgroup [q (false discovery rate-adjusted P value) < 0.1] and the top three SNPs associated with survival were selected for further genotyping using Taqman genotyping assays in 7900HT Sequence Detection System (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. For polymorphisms in the same gene that showed strong linkage disequilibrium, we selected only the most significant SNP for replication.
Statistical analysis
Hazard ratios (HR) were estimated for recurrence and survival endpoints with the Cox proportional hazards model, adjusting for sex, age, race, clinical stage, pack year tobacco history and treatment regimen. We compared three genetic models to determine the significance of each SNP: dominant (common homozygote versus variant allele carrying genotypes), recessive (wild-type allele carrying genotypes versus rare homozygous) and additive (P for trend). Only dominant model was considered if the rare homozygous genotype was observed <5% percent in both patients with event and patients without event. Differences in host characteristics between treatment groups were assessed by Pearson χ2 or Fisher exact tests. Predictive marker is defined as a SNP whose association with clinical outcome is at least significant at P < 0.05 based on the best statistical model in one treatment group but has risk estimate that is in opposite direction for different treatment group using the same statistical model. Variants with beneficial effects on both groups were not included in the predictive marker analysis since they would not be helpful for treatment selection. For the cumulative analysis of predictive markers, unfavorable genotypes were selected for each significant SNP dependent on the best model, and the total number of unfavorable genotypes was counted for each subject. Specifically, SNPs with significant HR > 1 would have homozygous variant alleles (VV) and heterozygous genotype (WV) as the unfavorable genotypes under the dominant and additive models, and VV as the unfavorable genotype under the recessive model. SNPs with HR < 1 would have homozygous wild-type alleles (WW) and heterozygous genotype (WV) as the unfavorable genotypes under the recessive model, and WW as the unfavorable genotype under the dominant and additive models. Based on the percentage of cases with adverse clinical outcome, the subjects were grouped into tertiles of low, medium and high risks by the number of unfavorable genotypes. Kaplan–Meier survival function and log-rank tests were used to assess differences in progression-free and overall survival times. Meta-analysis was performed to combine the results from discovery and validation groups adjusting for the same covariates. To correct for multiple comparisons, we computed the q value (a false discovery rate-adjusted P value) for each SNP implemented in the R-package. The above analyses were done using STATA software (version 10; STATA Corp., College Station, TX). Survival tree analyses were used to identify higher order gene–gene interactions by STREE program (http://c2s2.yale.edu/software/stree/). All tests were two sided and P < 0.05 was considered statistical significant.
Results
Host characteristics
A total of 535 patients were treated for stage I or II NSCLC at MD Anderson with curative intent between November 1995 and February 2008 (Table I). The median age of patients was 66 years (range, 29–92) with 49% of patients being male; 87% were current or former smokers with a median history of 45 pack–years. Majority of patients (88%) were white and 8% were black. Adenocarcinoma represented 59% of the patient population, squamous cell represented 28% and 13% of the patients was of other histology or not otherwise specified. More than 80% of the patients had clinical stage I disease with the remainder having clinical stage II disease. Pathological staging was available for 482 patients, and of these patients 75% had stage I disease and 24% had stage II disease. There were 340 patients who had surgical resection with negative margins and no further therapy, 142 patients who received surgery plus either adjuvant/neoadjuvant chemotherapy or adjuvant radiation and 53 patients who were treated with definitive radiation therapy without surgery for stage I disease. In the stratified analysis, there were no statistically significant differences between surgery-only and surgery plus chemotherapy treated groups with regard to age, sex, race, smoking history or histology. However, there were more patients diagnosed with stage I disease in the surgery-only group than those in the surgery and chemotherapy group (86 versus 51%, respectively; P < 0.0001). Median follow-up time for the entire cohort of cases was 58.4 months. Twenty-six percent of the patients showed disease recurrence and 40% died at the end of the follow-up period with a median survival time of 77.8 months.
Table I.
Host characteristics
| Variables | Numbera | Percent |
|---|---|---|
| Median survival time (months) | 77.80 | |
| Median follow-up time (months) | 58.42 | |
| Age, median (range) | 66 (29–92) | |
| Pack year, median (range) | 45 (0.1–256)b | |
| Total patients | 535 | |
| Sex | ||
| Male | 262 | 49 |
| Female | 273 | 51 |
| Pathological stage | ||
| Stage 0 | 2 | 0 |
| Stage 1A | 192 | 40 |
| Stage 1B | 169 | 35 |
| Stage 2A | 30 | 6 |
| Stage 2B | 89 | 18 |
| Clinical stage | ||
| Stage 1A | 245 | 46 |
| Stage 1B | 188 | 35 |
| Stage 2A | 26 | 5 |
| Stage 2B | 76 | 14 |
| Smoking status | ||
| Never | 68 | 13 |
| Former | 270 | 50 |
| Current and recently quit | 197 | 37 |
| Race | ||
| White | 469 | 88 |
| Black | 42 | 8 |
| Others | 24 | 4 |
| Histology | ||
| Adenocarcinoma | 315 | 59 |
| Squamous | 149 | 28 |
| Not otherwise specified | 27 | 5 |
| Other | 44 | 8 |
| Vital status | ||
| Alive | 322 | 60 |
| Dead | 213 | 40 |
| Recurrencec | ||
| No | 317 | 75 |
| Yes | 103 | 25 |
| Surgical procedure | ||
| Wedge resection | 16 | 4 |
| Lobectomy | 49 | 11 |
| Lobectomy + MLDd | 330 | 76 |
| Bilobectomy | 15 | 3 |
| Pneumonectomy | 26 | 6 |
| Surgery result | ||
| Complete | 470 | 98 |
| Residual | 12 | 2 |
| Treatment | ||
| Surgery-only | 340 | 64 |
| Surgery + chemotherapy | 127 | 24 |
| Surgery + adjuvant radiation | 15 | 3 |
| Treatment without surgery | 53 | 10 |
aNumbers may not add up to total number of patients due to missing clinical information.
bAmong ever smokers.
cPatients with recurrent disease at the time of first treatment were not counted in the analysis.
dMediastinal lymph node dissection.
For the Mayo Clinic group, 301 non-Hispanic white patients were treated with curative surgery for stage I or II NSCLC (Supplementary Table S1, available at Carcinogenesis Online). The median age was 69 years (range, 42–91) with 51% being male and 91% former or current smokers. Similar to the MD Anderson group, >80% of the patients were diagnosed with stage I NSCLC; however, the median survival time was longer at 133 months with a median follow-up time of 85.4 months, more likely due to the composition of surgery-only patients who usually have more favorable prognosis prior to treatment.
Associations of individual SNPs with recurrence-free survival
Analysis of SNPs in the Wnt pathway associated with recurrence-free survival in patients treated with curative therapy found 40 SNPs with P < 0.05 (Supplementary Table S2, available at Carcinogenesis Online). The top three SNPs are listed in Table II with the most significant being rs2536182 near the WNT16 gene, whose variant allele was associated with 51% decreased recurrence risk compared with wild-type [HR = 0.49, 95% confidence interval (CI): 0.33–0.73, P = 3.87 × 10− 4, q = 0.067]. When stratified by treatment regimen, rs10898563 of FZD4, was the most significant SNP associated with recurrence risk in surgery-only treated patients with borderline significance after adjusting for multiple testing (HR = 2.98, 95% CI: 1.62–5.48, P = 5.1 × 10− 4, q = 0.11; Table II). In patients treated with surgery and chemotherapy, there were no significant SNPs associated with recurrence after multiple testing adjustment, although the top SNP, rs11725638 of LEF1, was associated with a decreased risk of recurrence (HR = 0.31, 95% CI: 0.14–0.67, P = 3.05 × 10− 3, q = 0.24; Table II).
Table II.
Individual SNPs associated with recurrence risk in NSCLC patients treated with curative therapy
| SNP | Gene | Chr | MAF | SNP location | Allelic change | Overall | |||
|---|---|---|---|---|---|---|---|---|---|
| Best model | HR (95% CI)a | P value | q value | ||||||
| rs2536182 | WNT16 | 7 | 0.48 | 3′ flanking | G>C | Dominant | 0.49 (0.33–0.73) | 3.87×10−4 | 0.067 |
| rs7296283 | LRP6 | 12 | 0.18 | 5′ flanking | G>A | Dominant | 1.84 (1.24–2.72) | 2.30×10−3 | 0.188 |
| rs10898563 | FZD4 | 11 | 0.37 | 3′ UTR | A>G | Recessive | 2.00 (1.26–3.17) | 3.30×10−3 | 0.188 |
| SNP | Surgery-only | Surgery and chemotherapy | |||||||
| Best model | HR (95% CI)b | P value | q value | Best model | HR (95% CI)b | P value | q value | ||
| rs2536182 | Dominant | 0.47 (0.28–0.80) | 5.24×10−3 | 0.154 | Dominant | 0.64 (0.27–1.53) | 0.3554 | 0.259 | |
| rs10898563c | Recessive | 2.98 (1.60–5.35) | 4.31×10−4 | 0.105 | Dominant | 0.60 (0.29–1.22) | 0.1554 | 0.259 | |
| rs11725638d | Dominant | 1.29 (0.75–2.22) | 0.3486 | 0.368 | Dominant | 0.31 (0.14–0.67) | 3.05×10−3 | 0.242 | |
Underlined bold numbers denote significant association at q < 0.1. MAF, minor allele frequency; UTR, untranslated region.
aAdjusted for sex, age, race, clinical stage, pack year tobacco history and treatment regimen.
bAdjusted for sex, age, race, clinical stage and pack year tobacco history.
cTop SNP in surgery-only group.
dTop SNP in surgery and chemotherapy group.
Validation of the top SNPs with recurrence
We assessed the association of rs2536182 near Wnt16 and rs10898563 in FZD4 (the latter SNP reached borderline significance at q = 0.11 with recurrence in stratified analysis) in 301 early-stage, surgery-only treated NSCLC patients from the Mayo Clinic. In recurrence-free survival analysis, only rs2536182 near Wnt16 but not the FZD4 variant showed significant association with similar effect as the discovery group (HR = 0.39, 95% CI: 0.18–0.84, P = 0.01; meta-analysis HR = 0.44, 95% CI: 0.29–0.68, P = 2.3 × 10− 4) (Table III).
Table III.
Replication result of top SNPs associated with recurrence in NSCLC patients treated with surgery-only
| SNP | MD Anderson | P | MOIa | Mayo Clinic recur (yes/no) | Mayo Clinic | P | Meta | Meta P | ||
|---|---|---|---|---|---|---|---|---|---|---|
| HRb (95% CI) | WW | WV | VV | HRb (95% CI) | HRb (95% CI) | |||||
| rs2536182 | 0.47 (0.28–0.80) | 5.24×10 −3 | Dom | 12/67 | 9/146 | 5/57 | 0.39 (0.18–0.84) | 0.0162 | 0.44 (0.29–0.68) | 2.30×10 −4 |
| rs10898563c | 2.98 (1.60–5.35) | 0.0004 | Rec | 12/11 | 100/129 | 3/38 | 0.81 (0.24–2.75) | 0.7409 | 2.31 (1.33–3.96) | 0.0246 |
Significant P values (P < 0.05) in bold type. VV, homozygous variant genotype;
WW, homozygous wild-type genotype; WV, heterozygous variant genotype.
aMOI: model of inheritance, the model with the smallest P value for both discovery and replication groups; Dom, dominant; Rec, recessive.
bAdjusted by age, sex, smoking status, cancer stage and treatment regimen.
cInformation for three samples from the Mayo Clinic was missing due to failed genotyping assay.
Predictive markers for recurrence-free survival
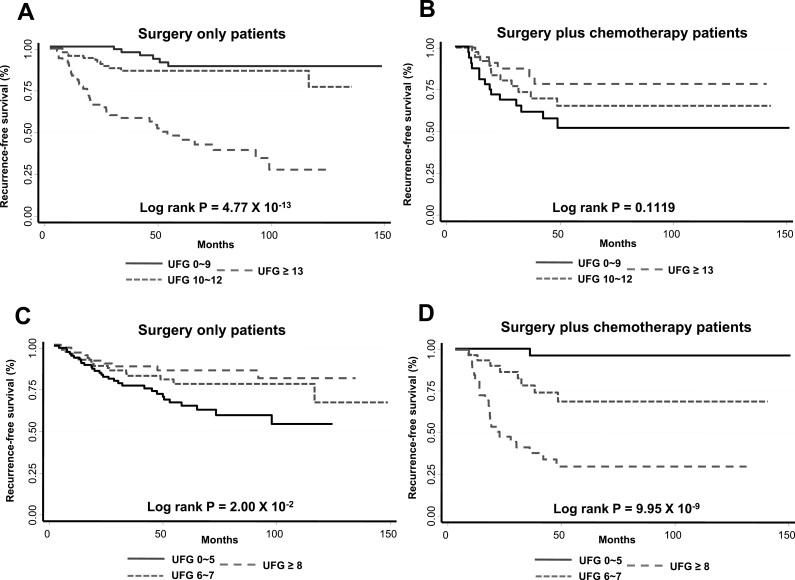
Since recurrence was the primary endpoint of this study, we sought to identify a group of SNPs that are predictive of recurrence-free survival in patients treated with each curative regimen. As seen in Supplementary Tables S3 and S4, available at Carcinogenesis Online, 26 SNPs were associated with recurrence risk in surgery-only group and 21 SNPs, including rs11725638 of LEF1, were associated with recurrence in surgery- and chemotherapy-treated patients (P < 0.05), respectively. Table IV shows the cumulative effects of these predictive markers on the risk for recurrence in early-stage NSCLC patients stratified by treatment. In this analysis, we counted the number of unfavorable genotypes for all predictive markers in each patient. Using patients carrying 0–9 unfavorable genotypes for markers based on surgery-only patients as reference, those with 10–12 unfavorable genotypes and 13 or more favorable genotypes had 2.2-fold (HR = 2.24, 95% CI: 0.81–6.19) and 11-fold (HR = 11.09, 95% CI: 4.59–26.80) increased risk of recurrence, respectively (P trend < 0.0001) in the surgery-only group. Consistent with the single SNP analysis, cumulative analysis of the same predictive markers identified for surgery-only group in the surgery and chemotherapy-treated subjects demonstrated less significant association and opposite risk effect. Similarly for markers based on surgery- and chemotherapy-treated group, individuals with 6–7 unfavorable genotypes and 8 or more unfavorable genotypes had 11-fold (HR = 11.1, 95% CI: 1.36–89.8) and >54-fold (HR = 54.6, 95% CI: 7.2–413.2), respectively (P trend < 0.0001) increased recurrence risk compared with the reference group of 0–5 unfavorable genotypes in patients treated with surgery and chemotherapy. Again, these markers had less significant association and opposite effect on recurrence risk in surgery-only treated patients. The cumulative effects of the predictive markers on recurrence-free survival are graphically shown by the Kaplan–Meier estimates for the three groups of patients in each treatment regimen (Figure 1). There was a dose-dependent decrease in recurrence-free survival for patients with increasing number of unfavorable genotypes for each set of predictive markers. This dose dependence was specific for the markers that were predictive of each therapy. In surgery-only patients, subjects from the high-risk group with >12 unfavorable genotypes from markers based on surgery-only group had recurrence-free median survival time of 51 months compared with >150 months for those with 12 or fewer unfavorable genotypes. Similarly, in patients treated with surgery and chemotherapy, those with >7 unfavorable genotypes from markers based on the same treatment regiment had median survival time of <19.5 months compared with >150 months for patients in the middle- and low-risk groups (Figure 1).
Table IV.
Cumulative effect of treatment-based exploratory predictive markers on recurrence risk in NSCLC patients stratified by treatment
| Markers based on surgery-only | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of unfavorable genotypes | Surgery-only | Surgery and chemotherapy | ||||||||
| Recur, n (%) | No recur, n (%) | HR | P value | Log- rank P | Recur, n (%) | No recur, n (%) | HR | P value | Log- rank P | |
| 0–9 | 6 (10.3) | 78 (37.5) | 1 (reference) | 14 (45.2) | 20 (27.4) | 1 (reference) | ||||
| 10–12 | 12 (20.7) | 87 (41.8) | 2.24 (0.81–6.19) | 0.1186 | 11 (35.5) | 27 (37.0) | 0.56 (0.25–1.24) | 0.1523 | ||
| ≥13 | 40 (69.0) | 43 (20.7) | 11.09 (4.59–26.80) | 8.93×10−8 | 4.77×10−13 | 6 (19.4) | 26 (35.6) | 0.30 (0.11–0.82) | 0.0184 | 0.1119 |
| Markers based on surgery and chemotherapy | ||||||||||
| Number of unfavorable genotypes | Surgery-only | Surgery and chemotherapy | ||||||||
| Recur, n (%) | No recur, n (%) | HR | P value | Log- rank P | Recur, n (%) | No recur, n (%) | HR | P value | Log- rank P | |
| 0–5 | 30 (52.6) | 68 (32.2) | 1 (reference) | 1 (3.2) | 35 (47.9) | 1 (reference) | ||||
| 6–7 | 17 (29.8) | 78 (37.0) | 0.57 (0.31–1.04) | 0.0682 | 8 (25.8) | 24 (32.9) | 11.07 (4.38–79.05) | 0.0244 | ||
| ≥8 | 10 (17.5) | 65 (30.8) | 0.41 (0.20–0.85) | 0.0171 | 0.0201 | 22 (71.0) | 14 (19.2) | 54.56 (7.21–413.2) | 1.08×10−4 | 9.95×10−9 |
Unfavorable genotypes for surgery-only markers: rs10898563 (VV), rs757558 (WV + VV), rs4074947 (WV + VV),
rs3781586 (WW + WV), rs3765351 (WV + VV), rs11922919 (WW + WV), rs557077 (WW), rs4257529 (WV + VV),
rs3923087 (WV + VV), rs7591 (WV + VV), rs7526484 (VV), rs566926 (WV + VV), rs288326 (WV + VV),
rs648872 (VV), rs8192633 (WV + VV), rs10127943 (WV + VV), rs12816278 (WV + VV), rs2163910 (WV + VV),
rs8068404 (WV + VV), rs11650531 (WV + VV), rs12632968 (WV + VV), rs4968281 (WV + VV), rs314756 (WV + VV),
rs640569 (WV + VV), rs4919139 (VV) and rs390258 (WV + VV).
Unfavorable genotypes for surgery and chemotherapy markers: rs11725638 (WV + VV), rs10891310 (VV), rs4919139 (VV),
rs9864832 (WV + VV), rs9675157 (WV + VV), rs4958216 (WV + VV), rs2163910 (WV + VV), rs1722846 (VV), rs395901 (WV + VV),
rs2039826 (VV), rs7478323 (VV), rs2707761 (WV + VV), rs2277268 (WV + VV), rs2240511 (WV + VV), rs9872276, (VV),
rs4730775 (WV + VV), rs4956157 (VV), rs7301320 (WV + VV), rs10917155 (WV + VV) and rs7296858 (WV + VV). V, variant;
W, wild-type.
Fig. 1.
Kaplan–Meier estimates of recurrence-free survival stratified by treatment for early-stage NSCLC patients grouped by the number of unfavorable genotypes in the predictive markers for surgery-only (A and B) and predictive markers for surgery plus chemotherapy (C and D). UFG: number of unfavorable genotypes as depicted in Table IV.
Associations of individual SNPs with overall survival
Variant genotypes were analyzed for association with survival after treatment with curative intent for patients with stage I or II NSCLC. In all our analyses, we minimized false discovery by using a false discovery rate of 10% (q < 0.1). There were 39 SNPs in 16 genes that were associated with overall survival at P < 0.05 (Supplementary Table S5, available at Carcinogenesis Online). After adjustment for multiple testing, seven SNPs had q < 0.1 (Table V). Among these, the most significant polymorphisms were rs4135385 of CTNNB1 (also known as B-catenin, HR = 3.74, 95% CI: 2.23–6.29, P = 6.02 × 10− 7, q = 1.43 × 10− 4), rs10898563 of FZD4 (HR = 2.29, 95% CI: 1.60–3.28, P = 5.19 × 10− 6, q = 3.46 × 10− 4) and two SNPs in WNT5A, rs503022 and rs629537, which were highly linked (r 2 = 0.99; HR = 4.34, 95% CI: 2.30–8.18, P = 5.82 × 10− 5, q = 3.46 × 10− 4). Other SNPs that were significantly correlated with survival include rs11658976 of WNT3 (HR = 1.89; 95% CI: 1.31–2.73; P = 6.43 × 10− 4), rs3765351 of WNT4 (HR = 1.82, 95% CI: 1.28–2.59, P = 9.24 × 10− 4) and another SNP of FZD4, rs713065 (HR = 0.63, 95% CI: 0.47–0.85, P = 0.0022).
Table V.
Individual SNPs associated with survival in NSCLC patients treated with curative therapy (q < 0.1)
| SNP | Gene | Chr | SNP location | Allelic change | MAF | Overall | |||
|---|---|---|---|---|---|---|---|---|---|
| Best model | HR (95% CI)a | P value | q value | ||||||
| rs4135385 | CTNNB1 | 3 | Intron | A>G | 0.25 | Recessive | 3.74 (2.23–6.29) | 6.02×10−7 | 1.43× 10−4 |
| rs10898563 | FZD4 | 11 | 3′ UTR | A>G | 0.35 | Recessive | 2.29 (1.60–3.28) | 5.19×10−6 | 3.46×10−4 |
| rs503022 | WNT5A | 3 | 3′ flanking | C>A | 0.16 | Recessive | 4.34 (2.30–8.18) | 5.82×10−6 | 3.46×10−4 |
| rs629537b | WNT5A | 3 | 3′ flanking | G>A | 0.16 | Recessive | 4.34 (2.30–8.18) | 5.82×10−6 | 3.46×10−4 |
| rs11658976 | WNT3 | 17 | Intron | A>G | 0.40 | Recessive | 1.89 (1.31–2.73) | 6.43×10−4 | 0.0306 |
| rs3765351 | WNT4 | 1 | 3′ UTR | A>G | 0.44 | Recessive | 1.82 (1.28–2.59) | 9.24×10−4 | 0.0366 |
| rs713065 | FZD4 | 11 | 3′ UTR | A>G | 0.37 | Dominant | 0.63 (0.47–0.85) | 0.0022 | 0.0752 |
Underlined bold numbers denote significant association at q < 0.1. MAF, minor allele frequency; UTR, untranslated region.
aAdjusted for sex, age, race, clinical stage, pack year tobacco history and treatment regimen.
bShowed linkage with rs503022, r 2 = 0.99.
SNPs associated with survival stratified by treatment
Since survival of early-stage NSCLC patients might be affected by the type of treatment administered, we also performed single SNP analysis stratified by treatment regimen (Supplementary Table S6, available at Carcinogenesis Online). Four SNPs were associated with overall survival in patients treated with surgery-only and four SNPs were associated with survival in surgery plus chemotherapy-treated patients (q < 0.1). Genetic variants of the two FZD4 SNPs, rs10898563 and rs713065, were associated with 3.2-fold increased death risk (HR = 3.2, 95% CI: 2.08–4.92) and 55% decreased death risk (HR = 0.45, 95% CI: 0.31- 0.65), respectively, in surgery-only group. As shown previously, rs10898563 also correlated with recurrence and survival in the overall curative therapy group. Other significant SNPs associated with survival in surgery-only treated patients were rs4135385 of CTNNB1 (HR = 3.34, 95% CI: 1.75–6.39) and rs75264 of WNT4 (HR = 3.04, 95% CI: 1.64–5.62). Additional SNPs associated with survival in surgery- and chemotherapy-treated patients included a SNP in APC2 (rs4807928), two SNPs in WNT5A (rs590386 and rs508407), a SNP in AXIN1 (rs11645554). For validation, we also tested the top three SNPs (q < 0.01) associated with survival (Supplementary Table S5, available at Carcinogenesis Online), including rs10898563 of FZD4 and rs4135358 of CTNNB1, in the Mayo Clinic surgery-only treated patients; however, none of the SNPs were replicated in this population (data not shown).
Discussion
The Wnt signaling pathway is one of the critical signaling pathways involved in stem cell maintenance and in embryonic development which has also been significantly implicated in tumorigenesis, tumor progression and metastasis in many malignancies including lung cancer (19–22). Several recent independent studies have reported alterations of the Wnt pathway in tumors were associated with poor clinical outcomes in NSCLC (23–26). However, the influence of germline Wnt pathway genetic polymorphisms on disease recurrence and survival of early-stage NSCLC patients treated with curative therapy has not been previously reported.
In this hospital-based study, we observed significant associations between multiple SNPs within the Wnt pathway and clinical outcomes including both recurrence and survival in patients with early-stage NSCLC. There were 43 SNPs associated with recurrence risk and 49 SNPs associated with overall survival at P < 0.05 in individual SNP analysis; 11 of these were associated with both recurrence and overall survival. Due to the relatively large number of SNPs tested, we limited the following discussion to those with at least borderline significance (defined as q value < 0.1) for either survival or recurrence after multiple comparison test was performed.
For individual SNP analysis, rs2536182 near the WNT16 gene has the strongest association with recurrence. This SNP was also replicated in the Mayo Clinic group for its association with recurrence-free survival among patients treated with surgery-only curative therapy. WNT16 has been implicated in acute lymphoblastic leukemia and in chronic lymphocytic leukemia, with increased expression or activity being associated with proliferation and tumorigenesis and decreased expression associated with apoptosis (27–29). In addition, upregulation of a WNT16 isoform (WNT16B) was found in basal cell carcinoma and could enhance keratinocyte proliferation (30). However, the involvement of Wnt16 in NSCLC has not been previously reported. It is possible that this 3′ flanking SNP could affect the expression of different WNT16 isoforms in the lungs or is tagging for another causal SNP. Alternatively, this SNP which resides in the last intron of a nearby gene FAM3C could affect the gene’s expression or activity. Further functional characterization may be necessary to dissect the mechanistic basis for its association with NSCLC recurrence. Another SNP, rs10898563 of FZD4, was most significantly associated with recurrence and survival in patients treated with surgery-only but not in patients treated with surgery and chemotherapy. Patients carrying the variant allele of this SNP had 2- to 3-fold increased recurrence and death risk compared with those of the wild-type homozygous genotype. Interestingly, stratified analysis by treatment showed opposite effects on risk suggesting its potential role as predictive marker for both recurrence and survival. FZD4 encodes a member of the seven transmembrane receptors for the Wnt signaling ligands. FZD4 expression has been linked to aberrant Wnt signaling in the pathogenesis of acute myelogenous leukemia (31) and prostate cancer oncogenesis involving epithelial-to-mesenchymal transition (EMT) (32,33). It is possible that increased expression of FZD4 may predispose tumor cells toward EMT facilitating anchorage independence and tumor progression. The functional basis for the association of this gene with clinical outcome in lung cancer requires further investigation.
An intronic variant (rs4135385) located in the CTNNB1 gene showed the strongest association with overall survival in NSCLC, but less significant association with recurrence risk. This association was also highly significant in the subgroup analysis in surgery-only patients but not in patients treated with surgery and chemotherapy (Table V). CTNNB1 is the gene that encodes beta-catenin, a dual function protein involved in the coordination of cell adhesion and gene transcription. This well-studied gene has been implicated in the carcinogenesis and progression of numerous malignancies (34,35). Mutations of this gene, although rare, have been reported in lung cancers (26). The value of beta-catenin as prognostic factor for NSCLC is controversial; a recent meta-analysis showed reduced expression of beta-catenin in tumors correlated with poor prognosis (36). Nevertheless, genetic variants of CTNNB1, including rs4135385, have been associated with the prognosis of gastric and advanced NSCLC in Chinese and Caucasian populations (37,38).
Other SNPs that are associated with overall survival in NSCLC patients involved genes of WNT5A (rs503022 and rs629537), WNT3 (rs11658976) and WNT4 (rs3765351). rs503022 and rs629537 are two linked SNPs located in the 3′ far end of WNT5A. The association of these SNPs with survival was seen in patients treated with surgery and chemotherapy but not surgery-only patients (Table V). Wnt5a signals via the non-canonical pathway to promote cellular movement and has been shown to activate calcium-dependent pathway which results in stimulation of cancer invasion and EMT (39,40). Wnt5a can also either inhibit or activate the canonical B-catenin pathway and in this way can be either oncogenic or act as a tumor suppressor depending on the cellular context and cancer sites (41–44). WNT3 gene acts in the canonical WNT-beta-catenin signaling pathway and is implicated in some types of human cancers. WNT3 was overexpressed in hepatocellular carcinoma as compared with normal surrounding liver tissues (45) and also highly expressed in mantle cell lymphoma (46). WNT4 is a non-canonical Wnt mainly expressed in epithelial cells. It is involved in the alteration of cell adhesion, motility and invasion associated with EMT (47). WNT4 expression is altered in several cancers (47–49) and might act as a potential target of Ras oncogenic signaling (50). Further research is necessary to determine the involvement of the Wnt pathway genes in the clinical outcome of NSCLC patients and whether any of the significant SNPs have functional activity in lung cancer cells or are linked to other functional SNPs in the Wnt pathway.
Patients diagnosed with NSCLC have a high incidence of smoking-related comorbid illnesses. Therefore, SNPs associated with survival without association with recurrence may be markers for mortality risk due to comorbidities. Because the median time to recurrence and relapse was >5 years in this study, it is possible that our follow-up time was not long enough to detect the association of some SNPs with recurrence and/or survival for early-stage NSCLC patients.
We have included a second independent population from the Mayo Clinic for replication of the top SNPs associated with recurrence and survival. Although both Mayo Clinic and MD Anderson Cancer Center are considered specialized tertiary medical centers, it is likely that heterogeneity between the two patient populations exists as shown by the discrepancies in clinical stage, median survival and follow-up times and in proportion of patients with recurrent disease, which may contribute to the failure of replicating some of the significant SNPs from the discovery group. Due to limiting resource, we only aimed to replicate the top SNPs associated with recurrence and survival. Hence, other significant variants might have been missed. For obvious reason, we could not validate markers for surgery and chemotherapy since the Mayo Clinic group consisted of mostly surgery-only patients.
We did not perform stratified analysis for ethnicity, smoking, disease stage and histology, due to the limited numbers in some categories and the fact that some minor constituents consisted of only a small proportion of the total population studied (e.g. non-smokers). Moreover, it is possible that some of the significant associations might be attained by chance due to multiple testing. Nevertheless, we have limited our analysis to white subjects and performed adjustment for multiple comparisons to minimize population heterogeneity and false discovery, respectively. Since different chemotherapeutic regimens in different time periods (Supplementary Table S7, available at Carcinogenesis Online) might cause differential effects on NSCLC recurrence and survival, we also adjusted by types of chemotherapy agents and obtained mostly similar results (Supplementary Tables S8 and S9, available at Carcinogenesis Online, respectively). Further studies in stratified populations and validation in larger independent populations might reveal more specific associations and reduce potential confounder effects.
There are several strengths of our study. First, we have examined a relatively large patient population with early-stage (I or II) non-small lung cancer, whose clinical information and follow-up data were detailed and complete with replication of the top association with recurrence in an independent population. Second, we have utilized a targeted pathway-based approach which covered most of the major genes in the Wnt signaling pathway. Both tagging and potentially functional SNPs were selected from these genes for greater sensitivity and specificity of detecting association with clinical outcome. Third, we have identified sets of predictive markers that were specific and prognostic for recurrence in patients treated with two different curative therapies, which may be applied clinically to identify high-risk population for poor treatment outcome.
Overall, this is the first report showing the association of the genetic variants in the Wnt signaling pathway with clinical outcome for NSCLC. Future epidemiological and functional studies are warranted to confirm these findings for translational application and to uncover the mechanistic basis of the Wnt pathway genes in affecting cancer progression and mortality.
Supplementary material
Supplementary Tables S1–S9 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute grant (P50 CA070907 to Dr X.W.).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- CI
confidence interval
- EMT
epithelial-to-mesenchymal transition
- HR
hazard ratio
- NSCLC
non-small cell lung cancer
- SNP
single nucleotide polymorphism.
References
- 1. Siegel R., et al. (2012). Cancer statistics, 2012. Cancer J. Clin., 62, 10–29 [DOI] [PubMed] [Google Scholar]
- 2. Fry W.A., et al. (1999). Ten-year survey of lung cancer treatment and survival in hospitals in the United States: a national cancer data base report. Cancer, 86, 1867–1876 [DOI] [PubMed] [Google Scholar]
- 3. Pignon J.P., et al. ; LACE Collaborative Group. (2008). Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J. Clin. Oncol., 26, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 4. Bjerkvig R., et al. (2005). Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat. Rev. Cancer, 5, 899–904 [DOI] [PubMed] [Google Scholar]
- 5. Wicha M.S., et al. (2006). Cancer stem cells: an old idea–a paradigm shift. Cancer Res., 66, 1883–90; discussion 1895. [DOI] [PubMed] [Google Scholar]
- 6. Hall B., et al. (2007). The participation of mesenchymal stem cells in tumor stroma formation and their application as targeted-gene delivery vehicles. Handb. Exp. Pharmacol., 180, 263–283 [DOI] [PubMed] [Google Scholar]
- 7. Mishra P.J., et al. (2009). Mesenchymal stem cells: flip side of the coin. Cancer Res., 69, 1255–1258 [DOI] [PubMed] [Google Scholar]
- 8. Reya T., et al. (2001). Stem cells, cancer, and cancer stem cells. Nature, 414, 105–111 [DOI] [PubMed] [Google Scholar]
- 9. Dale T.C. (1998). Signal transduction by the Wnt family of ligands. Biochem. J., 329(Pt 2), 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. You Z., et al. (2002). Wnt signaling promotes oncogenic transformation by inhibiting c-Myc-induced apoptosis. J. Cell Biol., 157, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X., et al. (2001). Regulation of vascular endothelial growth factor by the Wnt and K-ras pathways in colonic neoplasia. Cancer Res., 61, 6050–6054 [PubMed] [Google Scholar]
- 12. Katoh M. (2008). WNT signaling in stem cell biology and regenerative medicine. Curr. Drug Targets, 9, 565–570 [DOI] [PubMed] [Google Scholar]
- 13. Huang C.L., et al. (2005). Wnt5a expression is associated with the tumor proliferation and the stromal vascular endothelial growth factor–an expression in non-small-cell lung cancer. J. Clin. Oncol., 23, 8765–8773 [DOI] [PubMed] [Google Scholar]
- 14. Wei Q., et al. (2008). Dishevelled family proteins are expressed in non-small cell lung cancer and function differentially on tumor progression. Lung Cancer, 62, 181–192 [DOI] [PubMed] [Google Scholar]
- 15. Campioni M., et al. (2008). Identification of genes down-regulated during lung cancer progression: a cDNA array study. J. Exp. Clin. Cancer Res., 27, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glinsky G.V., et al. (2005). Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest., 115, 1503–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pramana J., et al. (2007). Gene expression profiling to predict outcome after chemoradiation in head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys., 69, 1544–1552 [DOI] [PubMed] [Google Scholar]
- 18. Yang P., et al. (2005). Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest, 128, 452–462 [DOI] [PubMed] [Google Scholar]
- 19. Katoh M., et al. (2007). WNT signaling pathway and stem cell signaling network. Clin. Cancer Res., 13, 4042–4045 [DOI] [PubMed] [Google Scholar]
- 20. Reya T., et al. (2005). Wnt signalling in stem cells and cancer. Nature, 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 21. Van Scoyk M., et al. (2008). Wnt signaling pathway and lung disease. Transl. Res., 151, 175–180 [DOI] [PubMed] [Google Scholar]
- 22. Nguyen D.X., et al. (2009). WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell, 138, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang C.L., et al. (2008). Wnt1 overexpression promotes tumour progression in non-small cell lung cancer. Eur. J. Cancer, 44, 2680–2688 [DOI] [PubMed] [Google Scholar]
- 24. Nakashima T., et al. (2008). Wnt1 overexpression associated with tumor proliferation and a poor prognosis in non-small cell lung cancer patients. Oncol. Rep., 19, 203–209 [PubMed] [Google Scholar]
- 25. Brock M.V., et al. (2008). DNA methylation markers and early recurrence in stage I lung cancer. N. Engl. J. Med., 358, 1118–1128 [DOI] [PubMed] [Google Scholar]
- 26. Sunaga N., et al. (2001). Constitutive activation of the Wnt signaling pathway by CTNNB1 (beta-catenin) mutations in a subset of human lung adenocarcinoma. Genes. Chromosomes Cancer, 30, 316–321 [DOI] [PubMed] [Google Scholar]
- 27. Román-Gómez J., et al. (2007). Epigenetic regulation of Wnt-signaling pathway in acute lymphoblastic leukemia. Blood, 109, 3462–3469 [DOI] [PubMed] [Google Scholar]
- 28. Lu D., et al. (2004). Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA, 101, 3118–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mazieres J., et al. (2005). Inhibition of Wnt16 in human acute lymphoblastoid leukemia cells containing the t(1;19) translocation induces apoptosis. Oncogene, 24, 5396–5400 [DOI] [PubMed] [Google Scholar]
- 30. Teh M.T., et al. (2007). Role for WNT16B in human epidermal keratinocyte proliferation and differentiation. J. Cell Sci., 120(Pt 2), 330–339 [DOI] [PubMed] [Google Scholar]
- 31. Tickenbrock L., et al. (2008). Activation of Wnt signalling in acute myeloid leukemia by induction of Frizzled-4. Int. J. Oncol., 33, 1215–1221 [PubMed] [Google Scholar]
- 32. Gupta S., et al. (2010). FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res., 70, 6735–6745 [DOI] [PubMed] [Google Scholar]
- 33. Acevedo V.D., et al. (2007). Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell, 12, 559–571 [DOI] [PubMed] [Google Scholar]
- 34. Rubinfeld B., et al. (1993). Association of the APC gene product with beta-catenin. Science, 262, 1731–1734 [DOI] [PubMed] [Google Scholar]
- 35. Reed K.R., et al. (2008). B-catenin deficiency, but not Myc deletion, suppresses the immediate phenotypes of APC loss in the liver. Proc. Natl Acad. Sci. USA, 105, 18919–18923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mei X.D., et al. (2013). Prognostic significance of β-catenin expression in patients with non-small cell lung cancer: a meta-analysis. Biosci. Trends, 7, 42–49 [PubMed] [Google Scholar]
- 37. Wang S., et al. (2012). Genetic variation of CTNNB1 gene is associated with susceptibility and prognosis of gastric cancer in a Chinese population. Mutagenesis, 27, 623–630 [DOI] [PubMed] [Google Scholar]
- 38. Hu L., et al. (2012). Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Clin. Cancer Res., 18, 5507–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weeraratna A.T., et al. (2002). Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell, 1, 279–288 [DOI] [PubMed] [Google Scholar]
- 40. Dissanayake S.K., et al. (2007). The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J. Biol. Chem., 282, 17259–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamamoto H., et al. (2009). Laminin gamma2 mediates Wnt5a-induced invasion of gastric cancer cells. Gastroenterology, 137, 242–52, 252.e1 [DOI] [PubMed] [Google Scholar]
- 42. Da Forno P.D., et al. (2008). WNT5A expression increases during melanoma progression and correlates with outcome. Clin. Cancer Res., 14, 5825–5832 [DOI] [PubMed] [Google Scholar]
- 43. Jönsson M., et al. (2002). Loss of Wnt-5a protein is associated with early relapse in invasive ductal breast carcinomas. Cancer Res., 62, 409–416 [PubMed] [Google Scholar]
- 44. Dejmek J., et al. (2005). Expression and signaling activity of Wnt-5a/discoidin domain receptor-1 and Syk plays distinct but decisive roles in breast cancer patient survival. Clin. Cancer Res., 11(2 Pt 1), 520–528 [PubMed] [Google Scholar]
- 45. Kim M., et al. (2008). Functional interaction between Wnt3 and Frizzled-7 leads to activation of the Wnt/beta-catenin signaling pathway in hepatocellular carcinoma cells. J. Hepatol., 48, 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gelebart P., et al. (2008). Constitutive activation of the Wnt canonical pathway in mantle cell lymphoma. Blood, 112, 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taki M., et al. (2003). Down-regulation of Wnt-4 and up-regulation of Wnt-5a expression by epithelial-mesenchymal transition in human squamous carcinoma cells. Cancer Sci., 94, 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bui T.D., et al. (1997). Expression and hormone regulation of Wnt2, 3, 4, 5a, 7a, 7b and 10b in normal human endometrium and endometrial carcinoma. Br. J. Cancer, 75, 1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huguet E.L., et al. (1994). Differential expression of human Wnt genes 2, 3, 4, and 7B in human breast cell lines and normal and disease states of human breast tissue. Cancer Res., 54, 2615–2621 [PubMed] [Google Scholar]
- 50. De Menna M., et al. (2013). Wnt4 inhibits cell motility induced by oncogenic Ras. Oncogene, 32, 4110–4119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.