Summary
Overexpression of the homeodomain transcription factor Six1 augments the p38-TβRII pathway, and thus promotes EMT, increases CSC-like properties and results in malignant conversion in HPV16-immortalized HKc.
Abstract
Six1, a member of the Six family of homeodomain transcription factors, is overexpressed in various human cancers, and SIX1 overexpression is associated with tumor progression and metastasis. Six1 messenger RNA levels increase during in vitro progression of human papillomavirus type 16 (HPV16)-immortalized human keratinocytes (HKc/HPV16) toward a differentiation-resistant (HKc/DR) phenotype. In this study, we show that HKc/DR-overexpressing Six1 exhibited a more mesenchymal phenotype, as characterized by a fibroblastic appearance and increased invasion. We utilized Whole Human Genome Microarrays to explore the gene expression changes associated with Six1 overexpression in HKc/DR. We found that overexpression of Six1 downregulated epithelial-related genes and upregulated mesenchymal-related genes, which suggests that Six1 overexpression induces epithelial–mesenchymal transition (EMT). Pathway analysis of the microarray data showed alterations in the transforming growth factor-beta (TGF-β) pathway, including enhanced expression of the TGF-β receptor type II (TβRII), and activation of the mitogen-activated protein kinase (MAPK) pathway in HKc/DR-overexpressing Six1, suggesting that Smad-independent pathways of TGF-β signaling may be involved in Six1-mediated EMT. p38 MAPK activation was required for sustained Six1-induced EMT and TβRII overexpression. Finally, we determined that Six1 overexpression in HKc/DR resulted in malignant conversion and increased the cancer stem cell (CSC)-like population. Thus, Six1 overexpression promotes EMT, CSCs properties and malignant conversion in HKc/DR through MAPK activation, which supports the possible use of p38-TβRII inhibitors for the treatment of cancers overexpressing Six1.
Introduction
Cervical cancer is the second most common malignancy among women worldwide, with ~500 000 new cases and 280 000 deaths annually (1). Cervical cancer begins with persistent infection by a high-risk human papillomavirus (HPV), which may progressively lead to cervical intraepithelial neoplasia I, II and III, carcinoma in situ and ultimately invasive cervical cancer (2).
In our in vitro model for HPV-mediated carcinogensis, normal human keratinocytes (HKc) immortalized by transfection with HPV16 DNA (HKc/HPV16) progress toward malignancy through growth factor-independent (HKc/GFI) and differentiation-resistant (HKc/DR) stages (3,4). Although immortalized and differentiation resistant, HKc/DR are not tumorigenic (3,4). Gene expression profiling studies determined that Six1 is overexpressed in HKc/DR. Furthermore, Six1 was overexpressed in 23% of cervical cancer samples in a tissue microarray (5). These findings prompted us to further investigate the role of Six1 overexpression in HPV16-mediated transformation.
The Six1 homeoprotein, a member of the Six family of homeodomain transcription factors, is essential for the development of numerous organs (6–8). Six1 overexpression has been found in various human cancers including tumors of the breast (9), ovary (10), cervix (5,11) and liver (12). Six1 overexpression is associated with increased tumor progression and metastasis, and decreased survival (13). For example, Six1 overexpression in immortalized and non-tumorigenic mammary epithelial cells induced malignant transformation, leading to highly aggressive and invasive tumors in nude mice (9). In addition, Six1 overexpression in human breast cancer cell lines induced epithelial–mesenchymal transition (EMT) and metastasis, which was associated with Smad-dependent transforming growth factor-beta (TGF-β) signaling and increased TGF-β receptor type I (TβRI) expression (14,15). Six1 messenger RNA (mRNA) expression increased during in vitro progression of HPV16-immortalized cells and Six1 protein is overexpressed in cervical cancer tissues, which strongly suggests that Six1 might be involved in the progression of cervical premalignant lesions to cervical cancer (5). Six1 is also overexpressed in cervical cancer cell lines, and the extent of Six1 overexpression correlates to increased tumor malignancy and lymph node metastasis (5,11,16). However, the mechanisms by which Six1 promotes cervical carcinogenesis and progression remain to be determined.
In this study, we explored the functional consequences of Six1 overexpression in HKc/DR. HKc/DR-overexpressing Six1 exhibited a more mesenchymal phenotype compared with vector controls, including a fibroblastic appearance and increased motility and invasion. Induction of EMT by overexpression of Six1 was associated with decreased activity of Smad-dependent signaling and activation of the mitogen-activated protein kinase (MAPK) pathway. In particular, we observed increased activity of p38 MAPK and increased expression of the TβRII upon Six1 overexpression. Moreover, we determined that overexpression of Six1 in HKc/DR resulted in malignant conversion, which was associated with an increase in the population of cancer stem cell (CSC)-like cells. We conclude that Six1 overexpression in HPV16-immortalized cells induces EMT and promotes tumorigenesis by activation of p38-TβRII signaling.
Materials and methods
Cell culture and treatment
The cell lines utilized in this work have been described previously (3,4). Briefly, normal HKc immortalized by transfection with HPV16 DNA (HKc/HPV16) were cultured in keratinocyte serum-free medium supplemented with epidermal growth factor (EGF) and bovine pituitary extract (BPE) (Invitrogen, Carlsbad, CA) (4). This medium will be referred to as complete medium. Growth factor-independent HKc (HKc/GFI) lines were selected by culturing HKc/HPV16 in complete medium lacking epidermal growth factor and bovine pituitary extract. Differentiation-resistant cells (HKc/DR) were further selected from HKc/GFI in complete medium containing 1mM calcium chloride and 5% fetal bovine serum (referred to as DR medium) (3). All cells were maintained in a humidified atmosphere of 5% CO2 at 37°C.
MAPK inhibitors were dissolved in dimethyl sulfoxide and then diluted in medium to a final concentration of 10 μM. Control cultures were exposed to equivalent concentrations of dimethyl sulfoxide (0.1%) with no inhibitors. Cells were treated for 48h.
Plasmid constructs and stable transfection
Total RNA was isolated from HKc/DR using the Total RNA Isolation Mini Kit (Agilent, Wilmington, DE) according to the manufacturer’s protocol. Six1 complementary DNA (cDNA) was amplified using the Titan One Tube RT-PCR System (Roche, Indianapolis, IN). The forward primer includes a NheI restriction site: 5′-GAC GCT AGC ATG TCG ATG CTG CCG TCG TTT G-3′; the reverse primer contains a BamHI site: 5′-TAC GGA TCC TGC TGC TCC AGG AAT CCC TTC G-3′. The restriction sites are shown in bold. Six1 cDNA was then cloned into the NheI and BamHI site of the mammalian expression vector pcDNA 3.1 (Invitrogen). The Six1 cDNA insert sequence was verified by direct sequencing of both DNA strands. This construct is referred to as pcDNA3.1-Six1.
HKc/DR were transfected with pcDNA3.1 or pcDNA3.1-Six1 using TransFast (Promega, Madison, WI) following the manufacturer’s instructions. Stable transfectants, named HKc/DR-Ctrl and HKc/DR-Six1, were selected in the presence of 50 μg/ml Zeocin (Invitrogen).
Real-time PCR
Total RNA was isolated from cells using the Total RNA Isolation Mini Kit (Agilent) according to the manufacturer’s protocol. Reverse transcription was carried out with 1 μg of total RNA using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) following the manufacturer’s instructions. The sequence of the primers used for real-time PCR is shown in Supplementary Table 1, available at Carcinogenesis Online. β-Actin was used as an internal control. All samples were assayed in triplicate.
Western blot analysis
Cells lysates were prepared with RIPA buffer (Pierce, Rockford, IL). Antibodies against the following proteins were used: Six1, TβRI, TβRII, TβRIII, ALDH1 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), E-cadherin, fibronectin (1:10 000; BD Biosciences, San Jose, CA), ERK, p-ERK, JNK, p-JNK, p38, p-p38 (1:1000; Cell Signaling Technology, Danvers, MA). The blots were incubated with primary antibody overnight at 4°C, washed three times with phosphate-buffered saline (PBS) with Tween 20, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (1:10 000; Millipore, Temecula, CA). The blots were visualized with ECL Plus Western Blotting Detection System (GE Healthcare, Buckinghamshire, UK). As an internal control for equal protein loading, blots were stripped and probed with antibody against β-actin (Santa Cruz Biotechnology).
Microarray analysis
Total RNA was isolated using the Agilent Total RNA Isolation Mini Kit according to the manufacturer’s protocol. RNA quality was assessed using an Agilent 2100 Bioanalyzer and RNA Integrity Numbers ranged from 9.8 to 10.0. Microarray experiments were performed using the Agilent platform. Total RNA was amplified and labeled using Agilent’s Low Input Quick Amp Labeling Kit (Agilent) according to the manufacturer’s instructions. Labeled RNA was then purified using Qiagen’s RNeasy Mini Kit (Qiagen, Valencia, CA) and dye incorporation and cRNA yield were assessed. Labeled cRNA samples were hybridized to Agilent Whole Human Genome Microarrays 4x44K (Agilent) using Agilent’s Gene Expression Hybridization Kit (Agilent) according to the manufacturer’s instructions. Four HKc/DR-Ctrl samples were hybridized against four HKc/DR-Six1 samples in a dye swap design. After washing and drying, arrays were scanned for both the Cy3 and Cy5 channels at 5 μm resolution using a ProScanArray Express HT scanner (Perkin Elmer Life and Analytical Sciences) and the ScanArray Express SP3 software. The scanned images were saved as TIFF files and fluorescence intensities were quantitated using ImaGene 8.0.1 software (BioDiscovery). Raw intensities for backgrounds and foregrounds (spots) were uploaded into limmaGUI (17) where features were background corrected using the Normexp method with offset equal to 50 (18). Subsequently, data were normalized within arrays using the locally weighted scatterplot smoothing algorithm and between the arrays performing scale normalization. Normalized data (M and A values) were exported from limmaGUI and normalized intensities for both Cy3 and Cy5 channel were calculated for all arrays by solving the equations for M and A, using M = log2(R/G) and A = 1/2 [log2(R) + log2(G)]. R = Cy5 channel intensity (red), and G = Cy3 channel intensity (green). In the final step, normalized intensities were uploaded and analyzed using GeneSifter software (Geospiza).
Immunofluorescence
Cells were plated in lab-tek II chambers (Nalge Nunc International, Rochester, NY) for 24h. Following fixation (with 4% paraformaldehyde in PBS, pH 7.2) and permeabilization (with 0.1% Triton X-100 in PBS), samples were incubated with antibodies against E-cadherin (1:200), fibronectin (1:200), TβRI (1:100), TβRII (1:100) or TβRIII (1:100) overnight at 4°C. Samples were then washed three times with PBS with Tween 20, followed by incubation with an Alexa 568-conjugated secondary antibody (1:1000; Invitrogen, Eugene, OR). Nuclei were stained with 1:20 000 dilution of 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) before cells were mounted. Samples were viewed using either an Olympus X81 fluorescence microscope or a Zeiss LSM510 META confocal scanning laser microscope.
Cell invasion and migration assay
The invasive ability of HKc/DR-Ctrl and HKc/DR-Six1 was measured using transwell chambers (24-well plate) (Corning, Cambridge, MA) with polycarbonate membranes (8.0 μm pore size) coated with 100 μl Matrigel (BD Biosciences, Franklin Lakes, NJ) on the top side of the membrane. The upper surface of the matrix was challenged with 20 000 cells, and cells were kept in basal medium containing 0.1% bovine serum albumin. The lower chamber contained complete medium. After 24h the cells were fixed and stained. Cells and Matrigel on the upper surface of the membrane were carefully removed with a cotton swab. The cells that invaded through the matrix were fixed, stained with 1% crystal violet and counted in five randomly chosen areas or the crystal violet was extracted with ethanol and the absorbance determined in a spectrophotometer. The migration of cells was measured by direct counting cells migrated to the lower chamber. Each experiment was performed in triplicate wells and repeated three times.
Cell proliferation assay
To determine the growth rate of HKc/DR, DR-Ctrl and DR-Six1, 50 000 cells in 2ml of their respective media were plated per well in 6-well plates. Cells in triplicate wells were counted daily for up to 5 days of incubation at 37°C, using a hemocytometer.
Luciferase assays
Cells were seeded into 6-well plates. Transfections were performed using TransFast Transfection Reagent. The luciferase construct p6SBE-luc, which contains six copies of the Smad-binding element (SBE) and p6SME-luc, which contains a Smad mutated element (SME) were obtained from Dr S.Kern (19). For each well, cells were transfected with either 4 µg p6SBE-luc or 4 µg p6SME-luc, along with 4ng pRL-SV40 (Promega) Renilla luciferase as a control for transfection efficiency. Cells were treated with TGF-β1 or vehicle 24h after transfection. Both firefly and Renilla luciferase activity were measured using the Dual Luciferase Assay Kit (Promega) 48h after transfection. Firefly luciferase values were normalized to Renilla luciferase values.
Flow cytometry analysis
Cells were stained with FITC-conjugated anti-CD24 and PE-conjugated anti-CD44 (eBioscience, San Diego, CA). Flow cytomertry analysis was performed using a BD LSRII flow cytometer (BD Biosciences, San Jose, CA)
In vitro tumorsphere formation assay
HeLa cells (10 000 per well) were plated in ultra-low attachment 6-well plates (Corning) in 2ml serum-free DMEM/F12 media (Invitrogen) supplemented with 20ng/ml EGF (Sigma, St Louis, MO), 10ng/ml basic fibroblast growth factor (bFGF; Sigma), 5 μg/ml insulin (Sigma), 1×B27 supplement (Invitrogen) and 0.4% bovine serum albumin (BSA; Sigma). Cells were cultured under 5% CO2 at 37°C for 7 days.
Xenograft mouse model
Eight weeks old female athymic nude (nu/nu) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Briefly, 5 × 106 HKc/DR-Ctrl or HKc/DR-Six1 cells were suspended in 100 μl of serum-free medium containing 50% Matrigel and injected subcutaneously into the flank of nude mice. For HeLa, 3 × 106 HeLa-Ctrl or HeLa-Six1 were injected into the each side of flank of nude mice. Six weeks after implantation, mice were killed and the tumor weights were measured. All animal experiments were conducted according to the guidelines and approval of USC Institutional Animal Care and Use Committee.
Histology and immunohistochemistry
Tumors were embedded in paraffin, 5 μm sections obtained and stained with hematoxylin and eosin (H&E) (VWR, West Chester, PA) for visual examination. The stained slides were reviewed and screened for representative tumor regions by a pathologist.
Tumor sections were deparaffinized, rehydrated, then incubated in a microwave oven with 0.01M citrate buffer, pH 6.0 for 10min for antigen retrieval. Endogenous peroxidases were blocked with 3% H2O2 for 15min. Non-specific epitopes were blocked with normal horse serum (Jackson ImmunoResearch, West Grove, PA) for 1h. The sections were incubated overnight at 4°C with antibodies against one of the following proteins: proliferating cell nuclear antigen (PCNA, 1:300; Abcam, Cambridge, MA), cleaved Caspase 3 (1:100; Cell Signaling Technology), Six1 (1:100; Santa Cruz Biotechnology), Ki67 (1:100; OriGene, Rockville, MD). After incubation with the horseradish peroxidase-conjugated secondary antibody (Bio-Rad) for 1h at room temperature, antigen signals were detected using the 2-Solution Diaminobenzidine (DAB) Kit (Invitrogen, Frederick, MD) and counterstained with hematoxylin.
Statistical analysis
Data were expressed as the mean ± SD. Statistical analysis was performed using the Student’s t-test when only two value sets were compared, and by one-way analysis of variance followed by the Dunnett’s test when the data involved three or more groups. P ≤ 0.05, P ≤ 0.01 or P ≤ 0.001 were considered statistically significant and indicated in the figures by *, ** or ***, respectively.
Results
Overexpression of Six1 induces alterations in cell morphology and behavior
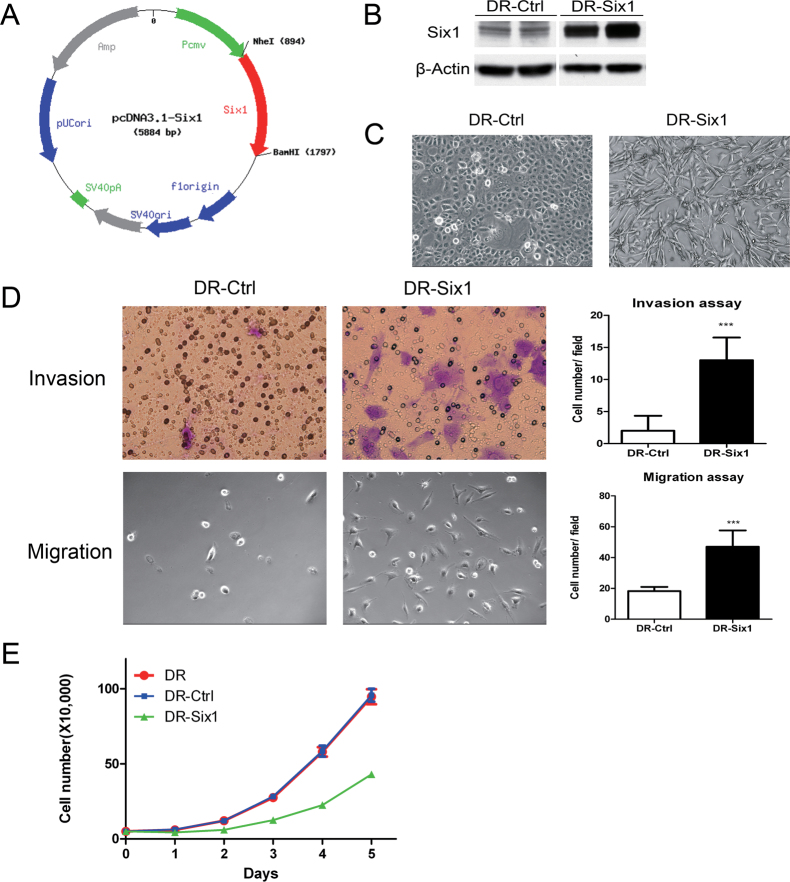
To explore the function of Six1 in HPV16-mediated transformation, we cloned a human Six1 cDNA into the expression plasmid pcDNA3.1 (pcDNA3.1-Six1, Figure 1A). HKc/DR stably overexpressing Six1 were established by transfection of HKc/DR (referred to as DR-Six1) with pcDNA3.1-Six1. Controls (referred to as DR-Ctrl) were HKc/DR transfected with pcDNA3.1. Western blots confirmed that DR-Six1 overexpressed Six1 protein (Figure 1B). DR-Six1 exhibited an elongated and fibroblastic appearance rather than the typical cobblestone epithelial-like morphology of DR-Ctrl (Figure 1C). The overexpression of Six1 in HKc/DR increased cell migration ~2.6-fold and cell invasion through Matrigel by 6.5-fold (Figure 1D). Six1 overexpression inhibited cell proliferation (Figure 1E). These data demonstrate that HKc/DR-overexpressing Six1 (DR-Six1) exhibited a much more mesenchymal phenotype with increased migration and cell invasion properties compared with the vector-transfected controls (DR-Ctrl).
Fig. 1.
Six1 overexpression in HKc/DR induces EMT-like characteristics. (A) Structure of the pcDNA3.1-Six1 expression plasmid. (B) HKc/DR were stably transfected with either pcDNA3.1 (DR-Ctrl) or pcDNA3.1-Six1 (DR-Six1). Six1 protein levels in cell extracts were determined by western blotting. β-Actin was used as a loading control. (C) Cell morphology of HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). Images are shown at ×100 magnification. (D) Invasion (upper panels) and migration (lower panels) assays for HKc/DR-Control (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). Cell invasion was determined by a Matrigel invasion assay. The number of invading cells was quantified after crystal violet staining. Cell migration was determined by a transwell migration assay. The number of migrating cells was directly counted. Images for the invasion assay are shown at ×400 and those for migration at ×100 magnification. Quantification of the invasion assay and the migration assay are shown in the upper right and lower right panels, respectively. Each column represents the mean of five different fields. Bars indicate standard deviation, and ***indicates statistically significant P values ≤ 0.001. (E) Proliferation of HKc/DR overexpressing Six1 (HKc/DR-Six1): cell proliferation was determined by directly counting cells at the days indicated in HKc/DR (DR), HKc/DR-Six1 (DR-Six1) and HKc/DR-Ctrl (DR-Ctrl).
Six1 overexpression in HKc/DR promotes EMT
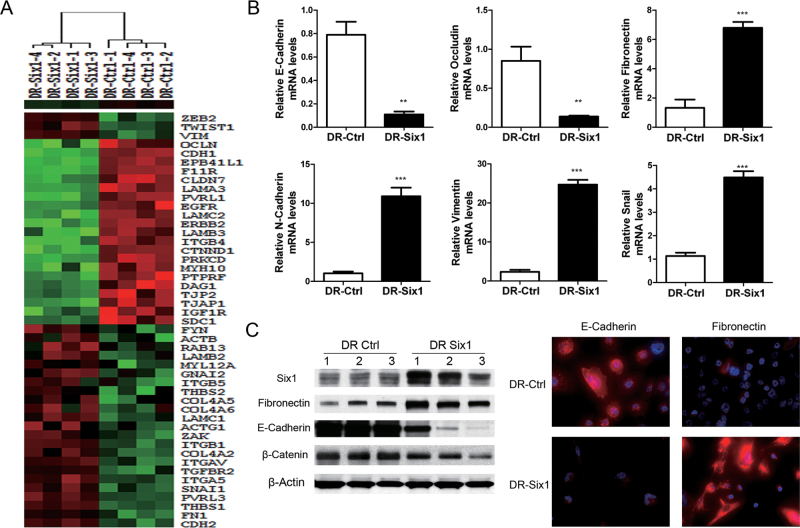
We next explored the effects of Six1 overexpression on global gene expression using Agilent Whole Human Genome Microarrays (4x44K). Paired comparisons of gene expression were made between four individual clones of DR-Ctrl and DR-Six1. We used Gene Sifter software to analyze the microarray results. To determine the differentially expressed genes in the pairwise comparisons, we used a fold change >1.5 and a P value < 0.05 as cutoff values. In addition, KEGG pathways analysis was performed on the differentially expressed genes. We noted that a number of EMT-related genes were changed in SIX1-overexpressing HKc/DR. We used Cluster and TreeView software to analyze genes involved in cell–cell junctions and a series of markers associated with EMT in DR-Ctrl and DR-Six1 (Figure 2A). We found that, in response to Six1 overexpression, cell–cell junctions and EMT markers were altered, including decreased expression of epithelial-related genes, such as E-cadherin (CDH1) and occludin (OCLN), and increased expression of mesenchymal-related genes, such as fibronectin (FN1), N-cadherin (CDH2) and vimentin (VIM), as well as increased expression of transcription factors that promote EMT, such as snail (SNAI1), twist1 and ZEB2 (Figure 2A). These microarray data suggest that Six1 overexpression induces EMT in HKc/DR.
Fig. 2.
Six1 overexpression induces markers of EMT in HKc/DR. (A) Hierarchical clustering of differentially expressed genes involved in cell–cell junctions and EMT between HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1) was conducted using cluster and TreeView software. The color represents the expression level of a gene above (red), below (green) or at (black) the mean expression level of that gene across all samples. (B) Levels of mRNA expression of E-cadherin, occludin, fibronectin, N-cadherin, vimentin and snail were determined by real-time PCR in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). Data were normalized to β-actin expression. Bars indicate standard deviation, and ** and ***indicate statistically significant P values ≤ 0.01 and 0.001, respectively. (C) (Left panel) Western blot for Six1, fibronectin, E-cadherin and β-catenin in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). β-Actin was used as loading control. Right panels: immunofluorescent staining of E-cadherin (red) and fibronectin (red) merged with nuclei (blue) in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). Images are shown at ×400 magnification.
Real-time PCR was used to confirm selected gene expression changes identified by microarrays. As shown in Figure 2B, the mRNA level of E-cadherin and occludin decreased by 86 and 84%, respectively, whereas mRNA levels of fibronectin, N-cadherin, vimentin and snail increased by 5.1-, 10.5-, 10.6- and 4.0-fold, respectively, in DR-Six1 compared with DR-Ctrl. Furthermore, we examined the protein levels of E-cadherin (an epithelial maker) and fibronectin (a mesenchymal marker) by western blot analysis and immunofluorescence. The results showed changes in protein levels that corresponded with what we observed at the mRNA level (Figure 2C). We also investigated β-catenin, because it is a key component of adherens junctions in epithelial cells (20), and we found that the expression of β-catenin decreased in cells overexpressing Six1 (Figure 2C). Since the distribution of E-cadherin and β-catenin is critical for functional organization of adherens junctions, we used confocal microscopy to compare cellular localization and overall levels of E-cadherin, β-catenin and fibronectin in DR-Six1 to that found in DR-Ctrl. The results show a dramatic downregulation of E-cadherin and β-catenin, and increased levels of fibronectin in DR-Six1, which is a hallmark of EMT (Supplementary Figure 1, available at Carcinogenesis Online). Taken together, our data show that Six1 overexpression promotes EMT at the HKc/DR stage of HPV16-mediated transformation.
Increased levels of TGF-β receptors are associated with Six1 overexpression
Since Six1 overexpression induced EMT in HKc/DR, we attempted to identify the signaling pathways regulated by Six1 that might be involved in promoting EMT. The TGF-β pathway has been found to be dysregulated at early stages of cervical cancer (21), and TGF-β is a potent inducer of EMT in HKc/DR (22). Numerous EMT-associated genes regulated by TGF-β have been identified (summarized in ref. 23). Therefore, we further analyzed our microarray data for those genes involved in TGF-β signaling. In response to Six1 overexpression, we observed altered expression of EMT-associated genes targeted by TGF-β such as increased Dab2 (1.8-fold), Hic5 (1.6-fold), vimentin (11.7-fold), N-cadherin (5.3-fold), urokinase plasminogen activator (4.3-fold), α-smooth muscle actin (α-SMA) (1.7-fold), β1 integrin (2.3-fold), β3 integrin (3.3-fold) and decreased E-cadherin (6.0-fold), indicating that Six1-induced EMT may be associated with activation of TGF-β signaling.
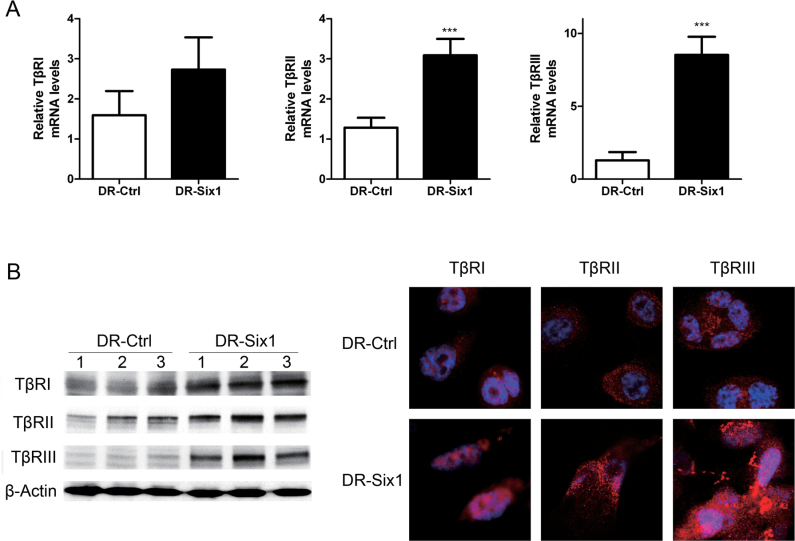
Three receptors, TβRI, TβRII and TβRIII, are involved in TGF-β signaling (24). Two of the TβR were upregulated in HKc/DR-overexpressing Six1. Real-time PCR showed upregulation of TβRII mRNA by about 2.4-fold and TβRIII expression by 6.6-fold in DR-Six1 compared with DR-Ctrl (Figure 3A). TβRI mRNA increased by ~1.7-fold in DR-Six1, but that change was not statistically significant (Figure 3A). Western blotting and immunofluorescence analysis determined increased protein levels of TβR, consistent with the changes in mRNA levels (Figure 3B). Taken together, our results show that Six1 overexpression leads to enhanced expression of the TβR, especially TβRII and TβRIII, which might contribute to activation of TGF-β signaling.
Fig. 3.
Six1 overexpression in HKc/DR increases the expression of TGF-β receptors. (A) Levels of mRNA expression of TGF-β receptors TβRI, TβRII and TβRIII were determined by real-time PCR in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). Data were normalized to β-actin expression. Bars indicate standard deviation, and ***indicates statistically significant P values ≤ 0.001. (B) (Left panel) Western blot analysis of TβRI, TβRII and TβRIII in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). β-Actin was used as control for equal protein loading. Right panels: immunofluorescent staining of TβRI, TβRII and TβRIII (red) in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). Nuclei were stained with 4′,6-diamidino-2-phenylindole (blue). Images are shown at ×630 magnification.
Smad-dependent TGF-β signaling is not activated in association with Six1 overexpression
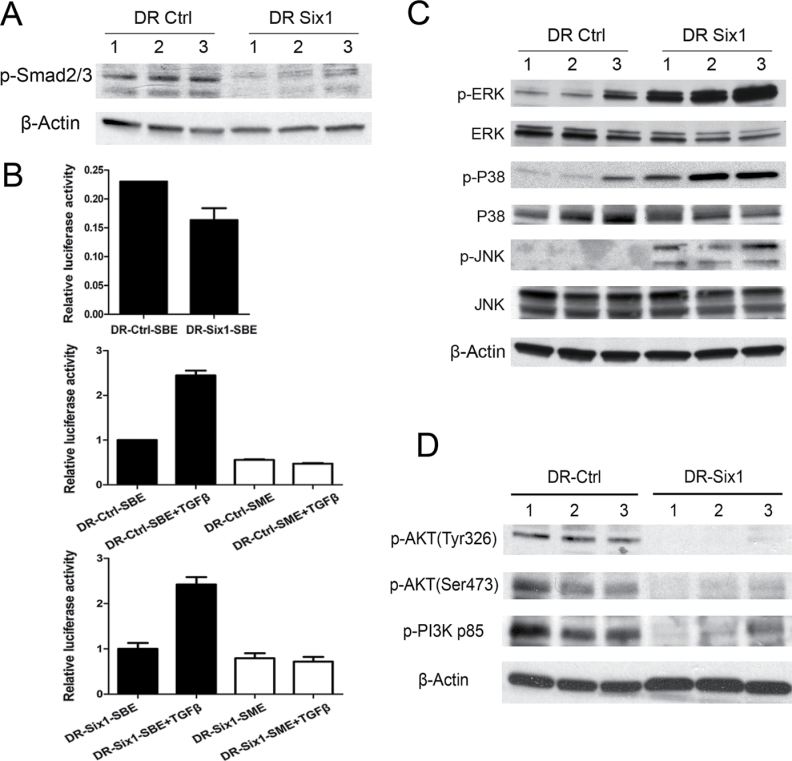
We next investigated whether increased TGF-β receptors are linked to enhanced canonical TGF-β signaling in DR-Six1. We analyzed basal phosphorylation of Smad2/3 in DR-Ctrl and DR-Six1 by western blotting. Smad2/3 phosphorylation was decreased in Six1-overexpressing cells, suggesting decreased basal canonical TGF-β signaling (Figure 4A). To further explore whether Six1 overexpression altered Smad-dependent signaling, we also compared the activity of a Smad-responsive luciferase reporter construct (p6SBE-Luc) in DR-Ctrl to DR-Six1. Overexpression of Six1 decreased somewhat the basal activity of the Smad-responsive luciferase reporter (Figure 4B, top panel). There was also no difference in fold induction of the Smad-dependent reporter construct between DR-Ctrl (Figure 4B, middle panel) and DR-Six1 (Figure 4B, bottom panel) in response to TGF-β1 treatment. These data suggest that Six1 neither increases endogenous Smad-dependent signaling nor enhances the activation of Smad-dependent signaling in response to TGF-β1 treatment.
Fig. 4.
Six1 overexpression in HKc/DR activates Smad-independent pathways. (A) Phospho-Smad2/3 was detected by western blot analysis in cell extracts prepared from HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). β-Actin was used as loading control. (B) Upper panel: HKc/DR-Ctrl and HKc/DR-Six1 were cotransfected with a TGF-β-inducible luciferase reporter (p6SBE-luc), which contains six tandem copies of the SBE, and pRL-SV40 Renilla luciferase, as control for transfection efficiency. Luciferase activity was determined 48h after transfection. Firefly luciferase values are normalized to Renilla luciferase. Bars indicate standard deviation. Middle/lower panels: HKc/DR-Ctrl or HKc/DR-Six1 were transfected with either p6SBE-luc or p6SME-luc, which contains mutated SBEs (SME). Twenty-four hours after transfection, cells were treated with 40 pM TGF-β1 for 24h and then luciferase activity was determined. Values are normalized to Renilla luciferase. Bars indicate standard deviation. (C) Protein levels of p-ERK, ERK, p-P38, P38, p-JNK and JNK were determined by western blotting of cell lysates prepared from HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). β-Actin was used as a loading control. (D) Western blot of p-AKT (Tyr326), p-AKT (Ser473) and p-PI3-kinase p85 (Tyr467/199) in HKc/DR-Ctrl (DR-Ctrl) and HKc/DR-Six1 (DR-Six1). β-Actin was used as a loading control.
P38 MAPK signaling contributes to Six1-induced EMT
Besides canonical signaling, TGF-β acts through a variety of non-canonical (Smad-independent) signaling pathways, including: (i) MAPK pathways, such as ERK1/ERK2, p38 MAPK and JNK, (ii) phosphatidylinositol-3-kinase (PI3K)/AKT pathways, and (iii) Rho-like GTPase signaling pathways (25). Thus, we investigated non-canonical TGF-β signaling in DR-Ctrl and DR-Six1 by western blotting using phospho-specific antibodies for p-ERK, p-p38, p-JNK, p-AKT and p-PI3K p85. As shown in Figure 4C, Six1 overexpression results in a dramatic increase in phosphorylation of ERK, p38 and JNK in DR-Six1. In contrast, overexpression of Six1 resulted in decreased phosphorylation of AKT and PI3K p85 and thus did not activate, but rather repressed the PI3K/AKT pathway (Figure 4D). Collectively, these data suggest activation of MAPKs but not PI3K/AKT upon Six1 overexpression in HKc/DR.
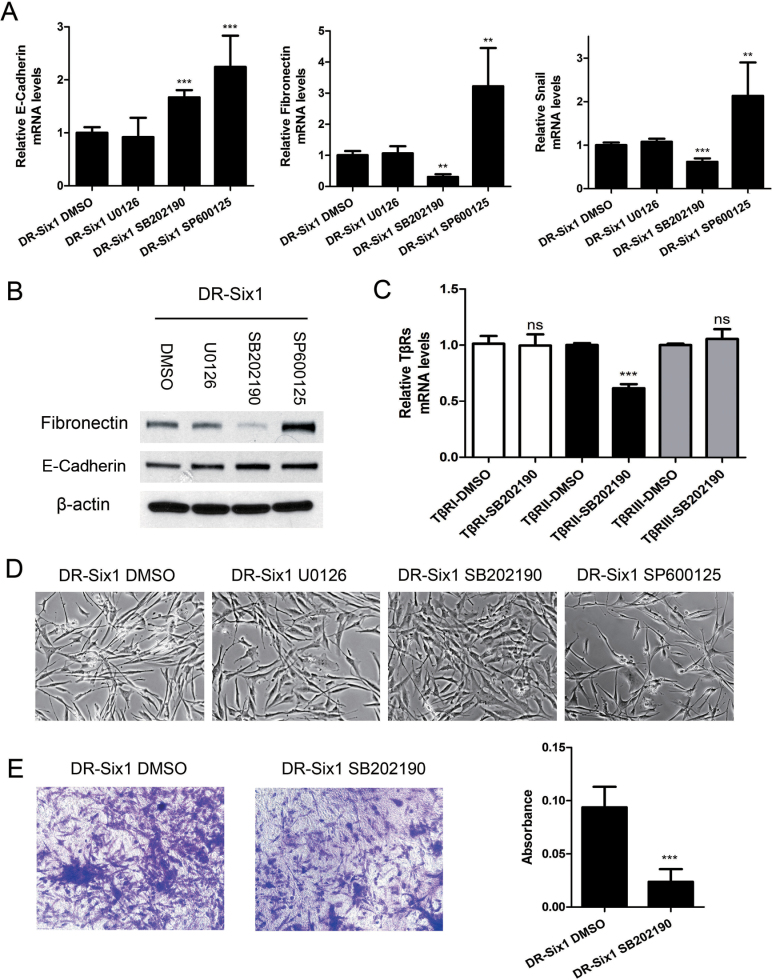
ERK1/2, JNK and p38 activation has been reported to induce EMT in various cancers (25). Therefore, we treated DR-Six1 with MAPK inhibitors to explore the role of MAPK activation in Six1 induction of EMT. Treatment of HKc/DR-Six1 with the p38 inhibitor SB202190 blocked several markers of Six1-induced EMT, increasing E-cadherin mRNA and reducing mRNA levels of fibronectin and Snail (Figure 5A). In contrast, treatment of DR-Six1 with the ERK inhibitor U0126 did not block Six1 modulation of mRNA markers of EMT, and the JNK inhibitor SP600125 increased mRNA levels of E-cadherin, but did not decrease fibronectin or snail mRNA (Figure 5A). Similar to what was observed at the mRNA level, SB202190 decreased fibronectin and increased E-cadherin protein expression in DR-Six1 (Figure 5B). Moreover, inhibition of p38 by SB202190 in DR-Six1 decreased the expression of TβRII mRNA by ~38% but had no effect on the expression of TβRI or TβRIII mRNA (Figure 5C). DR-Six1 treated with SB202190, but not U0126 or SP600125, lost some of their fibroblastic appearance (Figure 5D). Lastly, DR-Six1 treated with SB202190 exhibited a decreased ability to invade through Matrigel (Figure 5E). These results support the conclusion that p38 MAPK signaling plays an important role in promoting EMT in Six1-overexpressing HKc/DR.
Fig. 5.
Six1 overexpression in HKc/DR activates p38-TβRII signaling. (A) HKc/DR-Six1 were treated with the ERK inhibitor U0126 (10 μM), the p38 inhibitor SB202190 (10 μM), the JNK inhibitor SP600125 (10 μM) or dimethyl sulfoxide (DMSO) for 48h. mRNA expression of E-cadherin, fibronectin and snail was then determined by real-time PCR. Data were normalized to β-actin expression. (B) Western blot analysis for fibronectin, E-cadherin and β-actin (loading control) after treatment of DR-Six1 for 48h with the ERK inhibitor U0126 (10 μM), the p38 inhibitor SB202190 (10 μM), the JNK inhibitor SP600125 (10 μM) or DMSO. (C) mRNA levels of TβRI, TβRII and TβRIII were determined by real-time PCR after treatment of HKc/DR-Six1 with 10 μM SB202190 for 48h. Data were normalized to β-actin expression. Bars indicate standard deviation, and ** and ***indicate P values < 0.01 and < 0.001, respectively. (D) Phase-contrast photographs of DR-Six1 treated with the ERK inhibitor U0126 (10 μM), the p38 inhibitor SB202190 (10 μM), the JNK inhibitor SP600125 (10 μM) or DMSO for 48h. Images are shown at ×200 magnification. (E) Invasion assays for DR-Six1 treated with DMSO or SB202190 (10 μM for 24h). Cell invasion was determined by a Matrigel invasion assay. The number of invading cells was quantified after crystal violet staining. Images are shown at ×100 magnification. Quantification of the invasion assay is presented in the bar graph next to the photographs. Each column represents the mean of three repeats.
Overexpression of Six1 results in malignant conversion in HKc/DR and larger tumors in HeLa
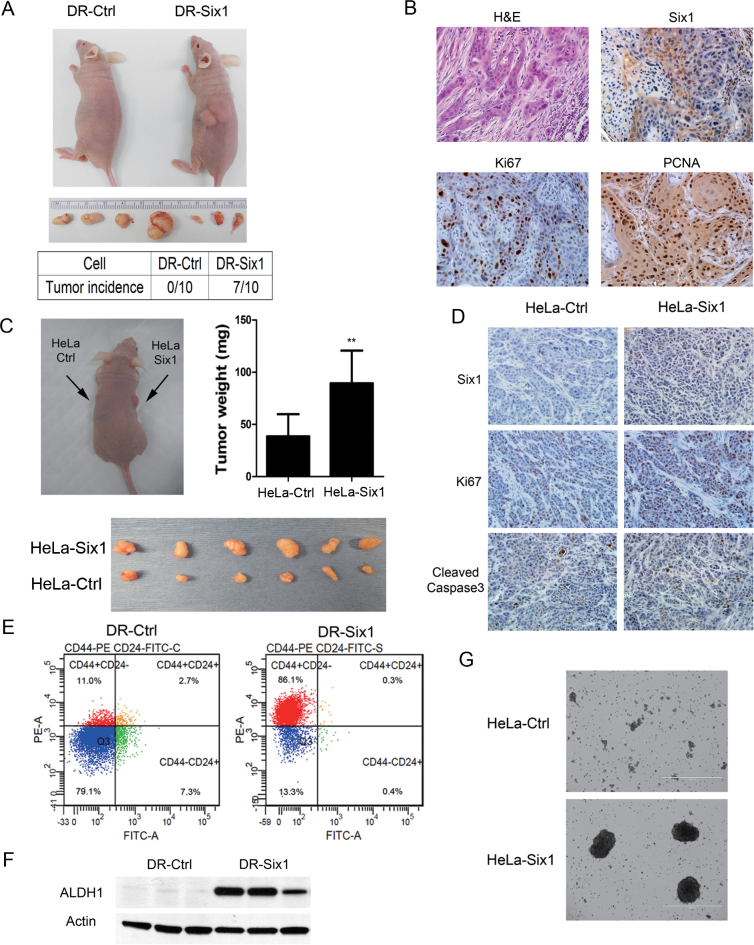
Next, we tested whether overexpression of Six1 in the non-tumorigenic HKc/DR resulted in malignant conversion. We found that HKc/DR-Six1 formed tumors in nude mice (7 out of 10 mice), whereas HKc/DR-Ctrl failed to form tumors (0 out of 10 mice) (Figure 6A). Histologic examination indicated high-grade squamous cell carcinoma, characterized by poor differentiation and high mitotic activity (Figure 6B). We detected by immunohistochemistry the expression of Six1 in the tumors as well as Ki67 and proliferating cell nuclear antigen (markers of cell proliferation) (Figure 6B). We dissected the tumor tissue, isolated tumor cells and then cultured the tumor cells in vitro: these cells exhibited similar fibroblastic-like morphology as HKc/DR-Six1 (Supplementary Figure 2, available at Carcinogenesis Online) and showed even higher levels of Six1 mRNA than HKc/DR-Six1 (data not shown).
Fig. 6.
(A–D) Six1 overexpression promotes tumorigenesis. (A) Six1-overexpressing HKc/DR form aggressive tumors in vivo. Upper panel: Six1-overexpressing HKc/DR (DR-Six1) or controls (DR-Ctrl) were injected into the flank of nude mice for 6 weeks. Tumors arose in DR-Six1, but not in DR-Ctrl. Middle panel: Six1-induced tumors. Lower panel: tumor incidence rates in DR-Six1 and DR-Ctrl groups (n = 10). (B) Histology of tumors formed by Six1-overexpressing HKc/DR. Hematoxylin and eosin (H&E)-stained sections of DR-Six1 tumors (upper left), the expression of Six1 (upper right), Ki67 (lower left) and proliferating cell nuclear antigen (PCNA) (lower right) in DR-Six1 tumors by immunohistochemistry (×400). (C) Upper panel: Six1-overexpressing HeLa (HeLa-Six1) were injected into the right flank of nude mice, and vector controls (HeLa-Ctrl) were injected into the left flank for 6 weeks. The weight of tumors in HeLa-Six1 group increases as compared with that in HeLa-Ctrl group. Lower panel: tumors formed from HeLa-Six1 (upper) and HeLa-Ctrl (lower). (D) The expression of Six1, Ki67 and cleaved caspase-3 in tumors from HeLa-Ctrl and HeLa-Six1 (×400). (E–G) Six1 overexpression in HKc/DR and HeLa increases CSC properties. (E) Analysis of CD24 and CD44 expression in Six1-overexpressing HKc/DR (DR-Six1) or HKc/DR controls (DR-Ctrl) by flow cytometry. (F) Western blot of ALDH1 in DR-Six1 and DR-Ctrl. β-Actin was used as a loading control. (G) Tumorsphere formation ability in HeLa-Ctrl and HeLa-Six1 (×40).
We also used the human cervical cancer cell line HeLa to explore the enhanced tumorigenic properties associated with Six1 overexpression. We found that HeLa stably overexpressing Six1 (HeLa-Six1) formed larger tumors in nude mice as compared with vector controls (HeLa-Ctrl) (Figure 6C). Since the increased tumor size may be caused by increased proliferation or decreased apoptosis, we compared the cell proliferation marker Ki67 and the apoptosis marker cleaved caspase-3 in HeLa-Ctrl and HeLa-Six1. Tumors-overexpressing Six1 (HeLa-Six1) showed a greater percentage of Ki67-positive cells compared with controls (HeLa-Ctrl) (Figure 6D), indicating Six1 enhanced tumor cell proliferation in vivo. There was no marked difference in the expression of cleaved caspase-3 between tumors from HeLa-Six1 and HeLa-Ctrl (Figure 6D), suggesting the effect of Six1 on tumor growth is associated with increased cell proliferation rather than alterations in apoptosis.
Six1 overexpression induces features of CSCs
CSCs are proposed to drive tumor onset and maintain tumor progression. TGF-β signaling has been known to play a vital role in regulating CSCs by induction of EMT (26). Therefore, we analyzed CSC markers in HKc/DR-Ctrl and HKc/DR-Six1. Cell surface antigens CD44+/CD24− have been widely used as breast and ovarian CSCs markers (27,28) and have recently been identified as cervical CSCs markers (29). Another important stem cell marker is aldehyde dehydrogenase 1 (ALDH1) (30,31). We thus measured CD44+/CD24− populations and the expression level of ALDH1 in DR-Ctrl and DR-Six1 by flow cytometry. HKc/DR-Six1 had a markedly increased CD44+/CD24− population (86% in DR-Six1 versus 11% in DR-Ctrl) (Figure 6E) and increased ALDH1 expression as compared with DR-Ctrl (Figure 6F). To determine whether Six1 also enhances characteristics of CSCs in HeLa, we examined tumorsphere formation ability of HeLa-Ctrl and HeLa-Six1. As shown in Figure 6G, increased tumorsphere formation ability was observed in HeLa-Six1 as compared with HeLa-Ctrl. Taken together, our studies demonstrate that Six1 overexpression increases the CSC-like cell population in HKc/DR and HeLa.
Discussion
The Six1 homeoprotein is essential for the development of numerous organs, but also contributes to cancer cell proliferation and survival. Six1 overexpression has been found in various human cancers and is associated with increased tumor progression and metastasis, and poor prognosis (13). Although overexpression of Six1 had been observed in cervical cancer cell lines and tissues (5,11,16), the role of Six1 in the progression of HPV-mediated cancer was unknown. We demonstrate here that Six1 overexpression promotes EMT at a late premalignant stage of HPV16-mediated transformation (HKc/DR). We further show that increases in TβRII and TβRIII and activation of non-canonical (Smad-independent) TGF-β signaling are associated with Six1 overexpression and that p38 MAPK activation is pivotal in Six1-induced EMT in HPV16-immortalized cells. Finally, we show that Six1 overexpression increases the CSC-like cell population in HKc/DR and results in malignant conversion.
Previous studies showed that Six1 promotes cell proliferation and cell cycle progression in breast cancer by upregulating cyclin A1 (9,32). Six1 promotes proliferation of pancreatic cancer cells by upregulation of cyclin D1 expression (33). However, in our study, we found decreased cell growth in Six1-overexpressing HKc/DR as compared with controls in vitro (Figure 1E). We did not observe significant cell cycle changes in response to Six1 overexpression in HKc/DR. The in vitro growth rate of Six1-overexpressing HeLa is comparable with the vector control (data not shown) but in vivo Six1-overexpressing HeLa form larger tumors. Clinically, Six1 overexpression is associated with cervical cancer metastasis but not with tumor size (11), indicating that Six1 is not critical for the regulation of tumor cell growth in cervical cancer. Therefore, Six1-induced cell proliferation is context dependent, and cell cycle-associated growth promotion is not the main mechanism by which increased Six1 expression leads to enhanced aggressiveness and poor prognosis in cervical cancer.
Dysregulation of TGF-β signaling has been related to cervical cancer progression and metastasis (21,34). At early stages of tumorigenesis, TGF-β acts as a potent tumor suppressor, strongly inhibiting the proliferation of epithelial cells. However, at late stages of tumorigenesis, tumors become resistant to growth inhibition by TGF-β and exploit TGF-β signaling to promote EMT, thereby facilitating tumor metastasis and invasion (24). To date, several studies showed that imbalances in the activation status of canonical and non-canonical TGF-β signaling may underlie the ability of TGF-β to induce EMT in normal and malignant cells, and might be critical in the switch in TGF-β signaling from tumor suppression to tumor promotion (25). Therefore, we analyzed both signaling pathways in response to Six1 overexpression in HKc/DR and observed significant MAPK activation, including ERK1/2, JNK and p38 MAPK. By using inhibitors, we identified p38 MAPK as the primary mediator of Six1-induced EMT. The activation of p38 MAPK by TGF-β, which induces EMT in normal and malignant cells, requires the expression and activity of either β1 or β3 integrin, and the involvement of TβRII (35,36). Based on our microarray data, β1 integrin increased 2.3-fold and β3 integrin increased 3.3-fold in DR/SIX1. Our finding that the inhibition of p38 MAPK in DR-Six1 decreases the expression of TβRII is consistent with the finding that TβRII is induced by a transcriptional mechanism involving p38 activation in breast cancer (37). The activation of ERK1/2 and JNK may be due to increased expression of TβRII, or the dramatically enhanced fibronectin expression, which activates ERK1/2 and JNK (38). Taken all together, these data suggest for the first time that Six1 augments the p38-TβRII pathway, and thus promotes EMT during HPV16-mediated transformation of human epithelial cells.
TGF-β-induced EMT is also thought to drive cells toward a more ‘cancer stem cell-like’ phenotype. Although very few studies demonstrate the role of TGF-β-induced EMT on regulating CSCs in cervical cancer, it has been reported that induction of EMT by TGF-β1 or its downstream targets, Snail or Twist, promoted the expression of cell surface markers associated with CSCs (CD44+/CD24− phenotype) and tumorsphere-forming features in breast cancer (26). Interestingly, the specific activation of TGF-β signaling in CD44+/CD24− breast cancer cells is due to the dramatically increased expression of TβRII in these cells as compared with the corresponding CD44−/CD24+ population (39). Moreover, non-canonical TGF-β effectors β1 and β3 integrin can induce CSC-like properties in breast cancer cells (36,40). We found that overexpression of Six1 promotes EMT and enhances features of CSCs in HKc/DR, including promoting tumorigenesis in vivo, augmenting the population of CD44+/CD24− and increasing expression of ALDH1. Importantly, we found increased expression of TβRII, β1 and β3 integrin, which are consistent with TGF-β-induced CSCs in breast cancer. These findings strongly suggest that Six1 promotes tumor progression by inducing characteristics of CSCs, and TGF-β-associated EMT plays an important role in this process.
The precise role that the p38 MAPK pathway plays in tumor initiation and progression is somewhat controversial. For example, p38 MAPK activation is implicated in the suppression of tumorigenesis because it can inhibit cell growth by decreasing the expression of cyclin D1 (41) However, p38 is essential for the expression of vascular endothelial growth factor, which promotes tumor angiogenesis (42). p38 kinase is also important in promoting H-ras-specific cell invasion and migration in human breast epithelial cells (43). The inhibition of p38 kinase suppresses the proliferation of human estrogen receptor-negative breast cancer cells (44). In addition, it has been reported that an increased CSC population found under hypoxia conditions and serum depletion is the result of activation of p38 signaling (45). We determined that p38 MAPK plays an important role in Six1-induced EMT and activation of non-canonical TGF-β signaling. To date, a large number of small molecule p38 MAPK inhibitors have been developed, some of which are in clinical trials (46). Based on our studies, p38 inhibitors might prove effective in the treatment of Six1-overexpressing malignancies.
In summary, we demonstrate that Six1 overexpression alters the TGF-β-signaling network, induces EMT, promotes malignant progression and increases the CSC population during HPV16-mediated transformation of HKc. We further establish a link between Six1 overexpression and an activated p38-TβRII pathway, which contributes to Six1-associated EMT and CSC characteristics. Recent studies have identified genes related to organ-specific metastasis and suggested that tumor progression and metastasis is determined by the genetic characteristics of early-stage transformed cells (47–49). Therefore, the pathways of Six1-induced EMT and malignant conversion we uncovered in HKc/DR are likely to be relevant to cancer progression and metastasis in general. This work provides a rationale for future studies targeting the p38-TβRII pathway for the treatment of cancers overexpressing Six1.
Supplementary material
Supplementary Table 1 and Figures 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
National Institute on Minority Health and Health Disparities ; National Institute of General Medical Sciences (5P20MD001770 to K.E.C., P20GM103499).
Supplementary Material
Acknowledgement
We thank the University of South Carolina Microarray Core Facility for assistance with microarrays.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ALDH1
aldehyde dehydrogenase 1
- cDNA
complementary DNA
- CSC
cancer stem cell
- EMT
epithelial–mesenchymal transition
- HKc/DR
differentiation-resistant human keratinocytes
- HKc/GFI
growth factor-independent human keratinocytes
- HPV16
human papillomavirus type 16
- MAPK
mitogen-activated protein kinase
- mRNA
messenger RNA
- PBS
phosphate-buffered saline
- PI3K
phosphatidylinositol-3-kinase
- SBE
Smad-binding element
- TβR
TGF-β receptor
- TGF-β
transforming growth factor-beta.
References
- 1. Peralta-Zaragoza O., et al. (2012). Targeted treatments for cervical cancer: a review. Onco. Targets. Ther., 5, 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saslow D., et al. ; ACS-ASCCP-ASCP Cervical Cancer Guideline Committee. (2012). American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Cancer J. Clin., 62, 147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pirisi L., et al. (1988). Continuous cell lines with altered growth and differentiation properties originate after transfection of human keratinocytes with human papillomavirus type 16 DNA. Carcinogenesis, 9, 1573–1579 [DOI] [PubMed] [Google Scholar]
- 4. Pirisi L., et al. (1987). Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA. J. Virol., 61, 1061–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wan F., et al. (2008). Gene expression changes during HPV-mediated carcinogenesis: a comparison between an in vitro cell model and cervical cancer. Int. J. Cancer, 123, 32–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zheng W., et al. (2003). The role of Six1 in mammalian auditory system development. Development, 130, 3989–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ikeda K., et al. (2010). Six1 is indispensable for production of functional progenitor cells during olfactory epithelial development. Int. J. Dev. Biol., 54, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 8. Xu P.X., et al. (2003). Six1 is required for the early organogenesis of mammalian kidney. Development, 130, 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coletta R.D., et al. (2008). Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res., 68, 2204–2213 [DOI] [PubMed] [Google Scholar]
- 10. Behbakht K., et al. (2007). Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res., 67, 3036–3042 [DOI] [PubMed] [Google Scholar]
- 11. Zheng X.H., et al. (2010). Expression and clinical implications of homeobox gene Six1 in cervical cancer cell lines and cervical epithelial tissues. Int. J. Gynecol. Cancer, 20, 1587–1592 [PubMed] [Google Scholar]
- 12. Ng K.T., et al. (2010). Suppression of tumorigenesis and metastasis of hepatocellular carcinoma by shRNA interference targeting on homeoprotein Six1. Int. J. Cancer, 127, 859–872 [DOI] [PubMed] [Google Scholar]
- 13. Christensen K.L., et al. (2008). The six family of homeobox genes in development and cancer. Adv. Cancer Res., 101, 93–126 [DOI] [PubMed] [Google Scholar]
- 14. Micalizzi D.S., et al. (2010). Homeoprotein Six1 increases TGF-beta type I receptor and converts TGF-beta signaling from suppressive to supportive for tumor growth. Cancer Res., 70, 10371–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Micalizzi D.S., et al. (2009). The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J. Clin. Invest., 119, 2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan J., et al. (2011). Expression and significance of Six1 and Ezrin in cervical cancer tissue. Tumour Biol., 32, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 17. Wettenhall J.M., et al. (2004). limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics, 20, 3705–3706 [DOI] [PubMed] [Google Scholar]
- 18. Ritchie M.E., et al. (2007). A comparison of background correction methods for two-colour microarrays. Bioinformatics, 23, 2700–2707 [DOI] [PubMed] [Google Scholar]
- 19. Dai J.L., et al. (1998). Dpc4 transcriptional activation and dysfunction in cancer cells. Cancer Res., 58, 4592–4597 [PubMed] [Google Scholar]
- 20. Schock F., et al. (2002). Molecular mechanisms of epithelial morphogenesis. Annu. Rev. Cell Dev. Biol., 18, 463–493 [DOI] [PubMed] [Google Scholar]
- 21. Noordhuis M.G., et al. (2011). Involvement of the TGF-beta and beta-catenin pathways in pelvic lymph node metastasis in early-stage cervical cancer. Clin. Cancer Res., 17, 1317–1330 [DOI] [PubMed] [Google Scholar]
- 22. Kowli S., et al. (2013). TGF-β regulation of gene expression at early and late stages of HPV16-mediated transformation of human keratinocytes. Virology, 447, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wendt M.K., et al. (2009). Mechanisms of the epithelial-mesenchymal transition by TGF-beta. Future Oncol., 5, 1145–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meulmeester E., et al. (2011). The dynamic roles of TGF-β in cancer. J. Pathol., 223, 205–218 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y.E. (2009). Non-Smad pathways in TGF-beta signaling. Cell Res., 19, 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mani S.A., et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell, 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meng E., et al. (2012). CD44+/CD24- ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival. Clin. Exp. Metastasis, 29, 939–948 [DOI] [PubMed] [Google Scholar]
- 28. Sheridan C., et al. (2006). CD44+/CD24- breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res., 8, R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gu W., et al. (2011). Silencing oncogene expression in cervical cancer stem-like cells inhibits their cell growth and self-renewal ability. Cancer Gene Ther., 18, 897–905 [DOI] [PubMed] [Google Scholar]
- 30. Rao Q.X., et al. (2012). Expression and functional role of ALDH1 in cervical carcinoma cells. Asian Pac. J. Cancer Prev., 13, 1325–1331 [DOI] [PubMed] [Google Scholar]
- 31. Yao T., et al. (2011). The expression of ALDH1 in cervical carcinoma. Med. Sci. Monit., 17, HY21–HY26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coletta R.D., et al. (2004). The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl Acad. Sci. USA, 101, 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Z., et al. (2013). Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS One, 8, e59203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baritaki S., et al. (2007). Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: implication of YY1 in cervical tumorigenesis and HPV infection. Int. J. Oncol., 31, 69–79 [PubMed] [Google Scholar]
- 35. Bhowmick N.A., et al. (2001). Integrin beta 1 signaling is necessary for transforming growth factor-beta activation of p38MAPK and epithelial plasticity. J. Biol. Chem., 276, 46707–46713 [DOI] [PubMed] [Google Scholar]
- 36. Galliher A.J., et al. (2007). Src phosphorylates Tyr284 in TGF-beta type II receptor and regulates TGF-beta stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res., 67, 3752–3758 [DOI] [PubMed] [Google Scholar]
- 37. Buck M.B., et al. (2006). TGF-beta signaling in breast cancer. Ann. N. Y. Acad. Sci., 1089, 119–126 [DOI] [PubMed] [Google Scholar]
- 38. Jiang F., et al. (2002). Fibronectin- and protein kinase C-mediated activation of ERK/MAPK are essential for proplateletlike formation. Blood, 99, 3579–3584 [DOI] [PubMed] [Google Scholar]
- 39. Shipitsin M., et al. (2007). Molecular definition of breast tumor heterogeneity. Cancer Cell, 11, 259–273 [DOI] [PubMed] [Google Scholar]
- 40. Galliher A.J., et al. (2006). Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res., 8, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lavoie J.N., et al. (1996). Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem., 271, 20608–20616 [DOI] [PubMed] [Google Scholar]
- 42. Yoshino Y., et al. (2006). Activation of p38 MAPK and/or JNK contributes to increased levels of VEGF secretion in human malignant glioma cells. Int. J. Oncol., 29, 981–987 [PubMed] [Google Scholar]
- 43. Kim M.S., et al. (2003). p38 kinase is a key signaling molecule for H-Ras-induced cell motility and invasive phenotype in human breast epithelial cells. Cancer Res., 63, 5454–5461 [PubMed] [Google Scholar]
- 44. Chen L., et al. (2009). Inhibition of the p38 kinase suppresses the proliferation of human ER-negative breast cancer cells. Cancer Res., 69, 8853–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin S.P., et al. (2012). Survival of cancer stem cells under hypoxia and serum depletion via decrease in PP2A activity and activation of p38-MAPKAPK2-Hsp27. PLoS One, 7, e49605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wagner E.F., et al. (2009). Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer, 9, 537–549 [DOI] [PubMed] [Google Scholar]
- 47. Minn A.J., et al. (2005). Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Invest., 115, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Minn A.J., et al. (2007). Lung metastasis genes couple breast tumor size and metastatic spread. Proc. Natl Acad. Sci. USA, 104, 6740–6745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bos P.D., et al. (2009). Genes that mediate breast cancer metastasis to the brain. Nature, 459, 1005–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.