Abstract
Human papillomavirus (HPV) is the etiologic risk factor for cervical cancer. Some studies have suggested an association with a subset of lung tumors, but the etiologic link has not been firmly established. We performed an international pooled analysis of cross-sectional studies (27 datasets, n = 3249 patients) to evaluate HPV DNA prevalence in lung cancer and to investigate viral presence according to clinical and demographic characteristics. HPV16/18 were the most commonly detected, but with substantial variation in viral prevalence between geographic regions. The highest prevalence of HPV16/18 was observed in South and Central America, followed by Asia, North America and Europe (adjusted prevalence rates = 22, 5, 4 and 3%, respectively). Higher HPV16 prevalence was noted in each geographic region compared with HPV18, except in North America. HPV16/18-positive lung cancer was less likely observed among White race (adjusted odds ratio [OR] = 0.33, 95% confidence interval [CI] = 0.12–0.90), whereas no associations were observed with gender, smoking history, age, histology or stage. Comparisons between tumor and normal lung tissue show that HPV was more likely to be present in lung cancer rather than normal lung tissues (OR = 3.86, 95% CI = 2.87–5.19). Among a subset of patients with HPV16-positive tumors, integration was primarily among female patients (93%, 13/14), while the physical status in male cases (N = 14) was inconsistent. Our findings confirm that HPV DNA is present in a small fraction of lung tumors, with large geographic variations. Further comprehensive analysis is needed to assess whether this association reflects a causal relationship.
Introduction
Lung cancer is the third most common cancer in men and women of all races and the leading cause of cancer death in the United States and worldwide. Cigarette smoking is the primary risk factor and accounts for ~85% of all lung cancer cases. Other less prevalent risk factors are genetic factors, family history of lung cancer and exposures to radon, asbestos, arsenic, diesel exhaust and some forms of silica and chromium. Human papillomavirus (HPV) is a small non-enveloped DNA tumor virus of the family Papillomaviridae and is the established etiological agent of genital warts, cervical cancer and a proportion of cancers of the vulva, vagina, penis, anus and oropharynx (1,2). More than 100 different genotypes have been identified, and types 16 and 18 are the most common oncogenic types, leading to the development of ~70% of all cervical carcinomas.
In 1979, Syrjänen first hypothesized that certain types of HPV are responsible for causing cancer in the lung (3). In the last three decades, the number of reports suggesting an association between HPV and lung cancer has increased tremendously. A review of HPV in 2468 lung cancer cases was first published in 2002 and showed that, using morphological, immunohistochemical and HPV DNA detection methods (4), ~22% of the cases analyzed contained HPV. This meta-analysis was updated in 2007 and reported a prevalence of 24.5%, but considerable heterogeneity was observed between studies (5). In our meta-analysis in 2009 (6), we also identified a wide variation in the prevalence of HPV in lung cancer tissues, despite the fact that only studies utilizing PCR-based methods for HPV detection were included in order to reduce the heterogeneity between studies. Nevertheless, HPV16 and 18 were the two most common genotypes detected in lung tumors, and a higher prevalence of HPV16 and 18 was noted in Asian populations compared with European populations. In 2012, the most recent meta-analysis was published and reported that the variability of HPV prevalence may be due to differences in geographical study origin and histological types of lung cancer rather than the method of HPV detection (7). However, meta-analyses findings to date have limitations because important characteristics such as gender, tumor stage and smoking status of HPV-positive lung cancer patients have not been taken into account. To address these limitations, we have conducted an international, multi-institutional, pooled analysis of studies to further analyze the characteristics of cases observed with HPV16/18 DNA with particular emphasis on race, gender, smoking status, tumor stage as well as histology of the cancers. The most recent meta-analysis has been updated (7); prevalence rates of HPV in lung tumors by geographic region, adjusted for age, gender, smoking status and tumor stage are presented. Because the majority of studies to date have been conducted in Asian and European populations, comparisons of demographic and clinical characteristics of HPV-positive lung cancer cases were also made between North American cases and the rest of the world.
Materials and methods
Literature review and data extraction
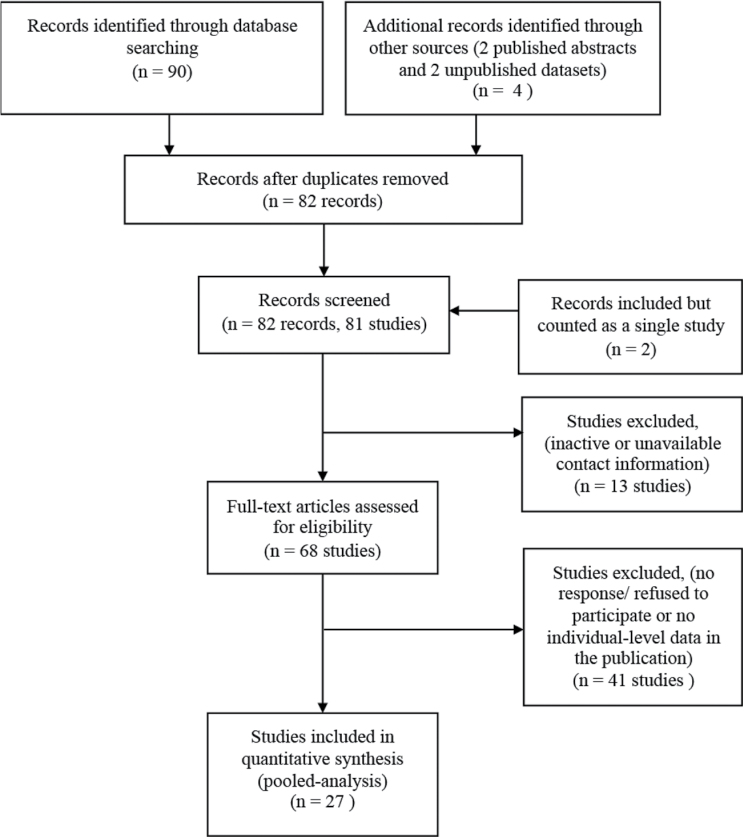
A flow diagram that summarizes the literature review process and selection of datasets for inclusion in the pooled analysis is described in Figure 1. A bibliographic search was carried out in the PubMed database to identify the studies that evaluated HPV status of primary lung tumors published up to July 2013. The search strategy used was: (HPV OR HPV) AND (lung OR bronchogenic) AND (cancer OR carcinoma). A manual review of the bibliographic references cited in the selected papers was also undertaken to retrieve papers that might have been missed in the initial search. From this search, 90 publications were identified. When the data were reported from the same cohort (8–30), only the most recent publication or one that had the larger cohort of lung cancer cases was included (10,13,14,17,20,21,25,26,30). Two publications had overlapping squamous cell carcinoma (SCC) cases (8,9) but only one reported data for adenocarcinoma cases (9). Therefore, although counted as a single study, both publications were included. To ensure that the larger cohort was included, the SCC data were extracted from one publication (8) and the adenocarcinoma data from the other (9). The majority of studies to date were conducted in Asian and European populations. Data from two published abstracts (31,32) and two unpublished datasets (E.Taioli et al. and A.Zaravinos et al., unpublished results) have also been included in this pooled analysis. These cases were recruited from three medical institutions in the United States and one from Greece. This resulted in 79 published studies and 2 unpublished studies, which include 7440 lung cancer cases. Sixteen studies also reported the prevalence of HPV in non-tumor lung tissues (i.e. in adjacent normal tissues from lung cancer cases or in lung tissues from non-cancer cases) (13,17,20,25,33–44). These data are included as a subset analysis comparing HPV prevalence between tumor and normal lung tissue. In order to describe the characteristics of all eligible studies, the following information were extracted for each study: first author, year of publication, geographic region, study size, method of HPV detection, HPV types detected and HPV status overall and when available, according to gender, smoking status, tumor stage and histology.
Fig. 1.
Flow diagram summarizing the identification and selection process of eligible studies for inclusion in the pooled analysis.
Pooled analysis data collection
Following Institutional Review Board approval, invitations to participate in the pooled analysis were prepared for the principal investigators of the 81 eligible studies. Invitations were not successfully sent to investigators of 13 studies due to inactive or unavailable contact information. Although no response or a refusal to participate was received from 47 studies, only 41 were excluded because 6 of these studies reported individual-level data in the publications. Datasets with age, gender, tumor stage, smoking status, histological type and HPV results were created for each of these studies and were included in the pooled analysis (8,9,26,38,45–47). In total, 27 studies were included in the pooled analysis (8,9,11,13,14,26,31,32,34,38,45–60) including the studies of E.Taioli et al. and A.Zaravinos et al. (unpublished results).
Verification of cancer diagnosis was reported in the corresponding publications for 21 of the 27 studies (8,9,11,13,14,26,34,38,45–50,53–60). If not explicitly stated in the published article, authors were contacted to clarify what, if any, efforts were made to avoid contamination. Efforts to avoid contamination were reported for 22 of the 27 studies (8,9,11,13,26,31,32,34,38,45–48,50–52,54–56,58,59) and for E.Taioli et al. and A.Zaravinos et al. (unpublished results), and all studies reported the inclusion of internal quality controls and/or positive and negative HPV controls. From the pooled dataset, 65 cases were excluded: 51 because of inadequate DNA based on quality control checks performed by the submitting investigator (54), 2 because they were duplicate samples (A.Zaravinos et al., unpublished results) and 12 because they were metastasis to the lung (13,57,59,60). The pooled analytic dataset included 3249 cases.
Published abstracts and unpublished studies
For E.Taioli et al. (unpublished results; n = 69), all cases were histologically confirmed lung cancer patients diagnosed at New York University (NYU)-Bellevue Hospital from 1993 to 1999. The central pathology registry at NYU-Bellevue Hospital was used to identify all consecutive African American cases; clinical charts were then retrieved using the pathology report number, and personal/behavioral information was collected. The pathologist reviewed all the tissue blocks to make sure that a block containing material from the primary lung tumor was used for testing. DNA from formalin-fixed paraffin-embedded (FFPE) tissues was extracted and tested for HPV, and physical status was determined for all HPV16-positive samples. For A.Zaravinos et al. (unpublished results; n = 17), all cases were histologically confirmed lung cancer patients diagnosed at the Department of Surgical Pathology of the University of Crete from 2007 to 2011. About 5–10 serial tissue sections of 10 µm were cut from each FFPE block and stained with hematoxylin and eosin (H&E) before microscopic examination. When the proportion of tumor cells was >70%, the FFPE block was subjected to DNA extraction. Data from Mehra et al. (32) (n = 62) were published in the 2013 Proceedings of the American Association for Cancer Research. All cases were a convenience sample of randomly selected histologically confirmed lung cancer patients (enriched for non-smoking status and adenocarcinoma) diagnosed at Fox Chase Cancer Center (FCCC) from 1993 to 2010. FFPE tissues and corresponding demographic and clinical information were obtained from the FCCC Biosample Repository. DNA from tumor tissue was extracted and tested for HPV, and physical status was determined for all HPV16-positive tumor tissues. Data from Pillai et al. (31) (n = 208), were published in the 2013 Proceedings of the American Society of Clinical Oncology. All cases were histologically confirmed surgically resected lung cancer patients diagnosed at Wellstar Health Systems, Atlanta, GA, from 2002 to 2008. The samples were consecutively acquired paraffin-embedded tissues, archived in a temperature controlled storage area until ready for analysis. Details of the tissue acquisition have been described previously (61). DNA from FFPE tissues was extracted and tested for HPV, and physical status was evaluated for all HPV16-positive tissues.
HPV detection and genotyping
The HPV testing for the published abstracts and unpublished dataset (E.Taioli et al., unpublished results) was performed in a single laboratory. DNA was extracted from FFPE lung cancer tissues using ArchivePure DNA purification kit (5 Prime). DNA concentration and quality checks were performed by evaluating 260:280 ratios as well as PCR amplification of a β-globin amplicon using the RS 42 and KM 29 primers. For the E.Taioli et al. (unpublished results) samples, HPV status was determined by nested HPV PCR reactions using PGMY09/11, followed by GP5+/GP6+ primers. Briefly, 20 µl PCR amplifications were performed using a GeneAmp PCR System® 9700 (Applied Biosystems, Life Technologies) at 95°C for 9min, 40 cycles of 95°C for 30 s, 55°C for 1min and 72°C for 10min with a final extension of 72°C for 5min. The nested PCR reaction was performed by prediluting 1:100 PCR products from the initial reaction in a final reaction volume of 25 µl. The cycling conditions were 95°C for 10min, 40 cycles of 95°C for 1min, 55°C for 1min and 72°C for 1min, followed by a final extension at 72°C for 5min. Negative and positive controls were included in each PCR reaction. The PCR products were separated in 2% agarose gel by electrophoresis and HPV genotyping of positive samples were performed using the Linear Array HPV Genotyping kit (Roche Diagnostics) according to the manufacturer’s instructions. For the Mehra et al. and Pillai et al. samples, HPV testing was performed using the INNO-LiPA genotyping Extra Amplification and Genotyping Extra kits (Innogenetics, Belgium). The SPF10 PCR primers are capable of identifying a broad spectrum of HPV genotypes by amplifying a 65bp target in the HPV L1 sequence. Genotyping of positive HPV samples was performed using the LiPA genotyping protocol that involves a reverse hybridization line probe assay to detect 18 high-risk types (16,18,26,31,33,35,39,45,51–53,56,58,59,66,68,73,82), 7 low-risk types (6,11,40,43,44,54,70) and additional types (69,71,74). Negative and positive controls (HPV6), as well as an internal control (HLA-DPB1 gene), to confirm DNA quality and the absence PCR inhibitors, were included in the assay.
HPV testing for the second unpublished study was performed by the submitting investigator (A.Zaravinos et al., unpublished results). Lung cancer tissue sections (FFPE) were deparaffinized with xylene and ethanol washes, treated with protease and then DNA from tumor tissue was extracted using the QIAmp DNA minikit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Briefly, tissue sections were digested with 0.1mg/ml proteinase K (Promega, Madison, WI) and 400 µl of digestion buffer containing 150mM NaCl, 400 mM Tris–HCl, 60 mM ethylenediaminetetraacetic acid and 15% sodium dodecyl sulfate, pH 8.0, in a 1.5 ml eppendorf tube. Samples were then incubated at 60°C for 2 days. Fresh proteinase K was added three times daily. The samples were extracted once with phenol/chloroform and once with chloroform. DNA was precipitated by the addition of 20 µl of 5 M NaCl and 1 ml of ethanol, recovered by centrifugation for 15min, washed once with cold 70% ethanol and resuspended in 50 µl of double distilled water. DNA concentration was calculated using the NanoDrop™ 1000 Spectrophotometer. Specimens were examined for the presence of amplifiable DNA using a set of primers for the β2-microglobulin gene. Amplification of HPV DNA in the specimens was performed by PCR using specific primer pairs for each E6 gene region of the HPV6, -11, -16, -18 and -33 subtypes. The samples were initially examined for the presence of non-type-specific HPV DNA using the general HPV primers, GP5+/GP6+. The following PCR amplification cycling conditions were used: 94°C for 4min; 40 cycles, 94°C for 1min, 40°C for 2min and 72°C for 1–5min. The last cycle was extended by a 4min elongation at 72°C. Appropriate negative and positive controls were included in each PCR reaction in order to exclude contamination events and to establish the specificity of primer-directed amplification. Recombinant plasmids carrying HPV type-specific sequences served as positive controls for HPV6, -11, -16, -18 and -33 genomes detection. For the general screening of HPV DNA, HeLa cells transfected with conserved L1 sequences among HPV strains were used as the positive control. PCR was carried out in a total volume of 25 µl containing 5mM of 5× Green GoTaq reaction buffer, 1.5mM MgCl2, 0.2mM of each deoxynucleotide triphosphate (dNTPs), 0.6U of GoTaq Flexi DNA polymerase (Promega) and 200ng of genomic DNA. The PCR products were examined by electrophoresis on a 2% agarose gel and photographed on an ultraviolet light transilluminator. The sensitivity of the PCR assay was determined by applying a serial-dilution amplification assay of viral-positive control DNA.
HPV physical status
The physical status of HPV16 was assessed in HPV16-positive lung cancer tissues for three published studies that were included in the pooled analysis (32,48,49) using the same quantitative PCR assay as described previously (62). Each HPV16 DNA-positive sample was amplified for 76bp of the E2 gene using the following primers: forward 5′-AACGAAGTATCC TCTCCTGAAATTATTAG-3′ (3361–3389 nt); reverse 5′-CCAAGGCG ACGGCTTTG-3′ (3427–3443 nt), as well as 81bp of the E6 gene, primers forward 5′-GAGAACTGCAA TGTTTCAGGACC-3′ (94–116 nt); reverse 5′-TGTATAAGTTGTTTGCAGC TCTGTGC-3′ (150–169 nt), in the presence of specific hybridization probes for E2-(FAM-CACCC CGCCGCGACCCATA-TAMRA) (3406–3424 nt) and E6-(FAM-C AGGAGCGAC CCAGAAAGTTACCACAGTT-TAMRA) (119–147 nt). The cycling conditions were 2min at 50°C, 10min at 95°C, and a two-step cycle of 95°C for 15 s and 60°C for 60 s for a total of 40 cycles. A standard curve was generated from 8-fold serial dilution of p1203 PML2d HPV16 (Addgene plasmid 10869, deposited by Peter Howley, MD; Addgene, Cambridge, MA). Both HPV16-positive cell lines, SiHa and Caski DNA were included as controls. The assay was performed in duplicate for each sample. Physical status was determined by calculating the ratios of E2 to E6 where a ratio <1.0 indicates a predominance of integrated HPV genomes, >1.0 indicates a predominance of episomal genomes. The results were recorded as copy numbers per 20ng of DNA.
Statistical analysis
All statistical analyses were carried out using SAS 9.3 (SAS Institute, Cary, NC) with a significant level of 0.05 and evaluation of publication bias was performed using the Comprehensive Meta Analysis Version 2 software (Biostat, Englewood, NJ). The adjusted HPV prevalence of each region was calculated after adjusting for age, gender, smoking history, tumor stage and study and reported as an estimated probability of HPV-positive lung cancer patients with 95% confidence intervals (CIs). Random effects logistic regression models were used to determine the association between HPV16/18 status and clinical and demographic variables. Study was used as a random effect to allow deviation caused by different study conditions. In the international pooled dataset, one out of three of the subjects was missing either smoking history or tumor stage. Both variables, we believe, are important in understanding the nature of HPV16/18 prevalence. However, exclusion of subjects with missing values is inefficient and can lead to biased results if those dropped are atypical in some respect. Therefore, multiple imputations were performed prior to regression analyses using IVEware. Ten imputation datasets were generated and final model estimates were combined using SAS 9.3. For each imputed dataset, the missing values were drawn from other observed data (63). The uncertainty about the correct model for non-response was captured by the variance across multiple imputed datasets. Subjects with one or part of the covariates missing were included in the analysis and hence, no information was lost due to exclusion.
Results
Twenty-seven of the 81 eligible studies elected to participate in this pooled analysis and the resulting pooled dataset included 3249 cases, whereas the remaining 54 studies had 4199 cases. Detailed characteristics of each study in the pooled dataset are summarized in Table I. The majority of cases in the pooled dataset were from Asia (40%, 1312 cases) and Europe (34%, 1100 cases). North/South American studies represented 26% of all cases in the pooled dataset (n = 837). For the non-included studies, the majority of cases were also from Asia (55%, 2332 cases) and Europe (33%, 1404 cases), whereas 7% of cases (n = 299) were from North/South America and 4% (n = 164 cases) from other geographic regions (Australia and the Middle East). The size of studies in the pooled dataset varied from 17 to 399 cases, whereas the non-included study sizes ranged from 5 to 319 cases. All studies included in the pooled analysis performed HPV DNA testing on surgical tissue (FFPE and/or frozen) or biopsy specimens. Similarly the non-included studies performed HPV DNA testing on surgical or biopsy tissue specimens except for one study which utilized bronchial aspirates from lung cancer patients (64). Most of the studies in the pooled analysis as well as the non-included studies used PCR-based methods for HPV DNA detection, 93 and 80%, respectively. Although adenocarcinoma and SCC were the most common histological types included in the pooled dataset (56%, 1808 cases and 38%, 1220 cases, respectively) and in the non-included studies (33%, 1404 cases and 57%, 2379 cases, respectively), the pooled analysis consisted of a larger proportion of adenocarcinoma cases compared with the non-included studies. Similar proportions of other histological types were represented in both the pooled dataset and non-included studies (7%, 214 cases and 8%, 350 cases, respectively). Histology was not specified for 7 (0.2%) cases in the pooled dataset and for 68 (1.6%) cases in the non-included studies.
Table I.
Characteristics of studies included in the pooled analysis and adjusted HPV prevalence
| Study | Tissue source | Method of HPV detection | Histological type | HPV types detected |
|---|---|---|---|---|
| Asia (N = 1312) | ||||
| Baba et al. (N = 77) (49) | FFPE | PCR (SPF10, Inno-LIPA) | AC, SCC | HPV 6, 16, 18, 33 |
| Kinoshita et al. (N = 34) (57) | FF | PCR (TS: HPV 16, 18, 33) | AC, SCC, ASqa, LCCa, SmCCa | HPV 18 |
| Iwakawa et al. (N = 275) (53) | FF | PCR (TS: HPV 16, 18, 33) | AC | HPV negative |
| Hiroshima et al. (N = 49) (52) | FFPE | PCR (TS: HPV 16, 18, 33) | AC, SCC | HPV 16 |
| Goto et al. (N = 296) (51) | FFPE | PCR GP5+/GP6+ | AC, SCC, CSrca | HPV 6, 11, 16, 18 |
| Lim et al, 2009 (N = 99) (56) | FFPE | ISH (HPV6, 11, 16, 18, 31, 33, 35, 45, 51, 52, 56, 58, 66) | AC | HPV negative |
| Miyagi et al. (N = 176) (8,9) | FFPE | PCR (TS: HPV 6, 11, 16, 18) | AC, SCC | HPV 6, 11, 16, 18 |
| Hirayasu et al. (N = 73) (26) | FFPE | PCR (TS: HPV 6, 11, 16, 18) | SCC | HPV 6, 16, 18 |
| Wang et al. (N = 210) (58) | FFPE, Bxb, PEb | PCR (MY09/MY11 and TS: HPV 16, 18) | AC | HPV16, HPV18 |
| Tsuhako et al. (N = 23) (47) | FFPE | PCR (TS: HPV 6, 11, 16, 18) | ASq | HPV 6, 11, 16, 18 |
| Adjusted HPV 16/18 prevalence (95% CI)c = 4.60% (3.48, 5.73) | ||||
| Adjusted HPV 16 prevalence (95% CI)c = 1.49% (0.86, 2.11) | ||||
| Adjusted HPV 18 prevalence (95% CI)c = 1.09% (0.66, 1.52) | ||||
| Europe (N = 1100) | ||||
| Syrjanen et al. (N = 131) (14) | FFPE | ISH (HPV6, 11, 16, 18, 30) | SCC | HPV 16, Xd |
| Syrjanen et al. (N = 77) (55) | FFPE | PCR (MY09/MY11/GP5+/GP6+) | AC, SCC, other | HPV 6, 16 |
| Giuliani et al. (N = 77) (13) | FFPE and FF | PCR (MY09/MY11/GP5+) | AC, SCC, ASqa, LCCa, SmCCa, NSCLCa, othera | HPV 6, 16, 18, 31, 53, |
| Gorgoulis et al. (N = 68) (11) | FFPE and FF | PCR (MY09/MY11/GP5+/GP6+) | AC, SCC, LCCa | HPV negative |
| Zaravinos et al. (unpublished results, N = 17) | FFPE | PCR (GP5+/GP6+) | ACe, SCCe, LCCa, NSCLCa | HPV negative |
| Koshiol et al. (N = 399) (54) | FFPE | PCR (TS: HPV 16, 18; SPF10 primers (n = 92) | AC, SCC, LCC, SmCC, other | HPV 16, 18 |
| Carpagnano et al. (N = 89) (34) | FFPE and BBb | PCR (consensus HPV primers) | AC, SCC, SmCC | HPV 16, 30, 31, 39, |
| Nuorva et al. (N = 22) (46) | FFPE | PCR (MY09/MY11) | AC | HPV 6, 11, 16, 18, 31, 33 |
| van Boerdonk et al. (N = 220) (59) | FFPE | PCR (GP5+/GP6+) | AC, SCC, LCC, NSCLCe | HPV negative |
| Adjusted HPV 16/18 prevalence (95% CI)f = 3.03% (2.76, 3.30) | ||||
| Adjusted HPV 16 prevalence (95% CI)f = 2.94% (2.68, 3.21) | ||||
| Adjusted HPV 18 prevalence (95% CI)f = 0.82%(0.73, 0.92) | ||||
| South and Central America (N = 105) | ||||
| Castillo et al. (N = 36) (50) | FFPE | PCR (GP5+6+) and SB | AC, SCC, SmCCe | HPV 16, 18, 33 |
| Aguayo et al. (N = 69) (48) | FFPE | PCR (GP5+/GP6+) and SB | AC, SCC | HPV 6, 16, 18, 31, 45 |
| Adjusted HPV 16/18 prevalence (95% CI)f= 21.90% (19.61, 24.20) | ||||
| Adjusted HPV 16 prevalence (95% CI)f = 19.18% (16.88, 21.49) | ||||
| Adjusted HPV 18 prevalence (95% CI)f = 7.78% (6.61, 8.95) | ||||
| North America (N = 732) | ||||
| Taioli et al. (unpublished results, N = 69) | FFPE | PCR (PGMY09/PGMY11/ GP5+/6+) | AC, SCC, LCCa, NSCLCa, othere | HPV 51, X |
| Pillai et al. (N = 208) (31) | FFPE | PCR (SPF10, Inno-LIPA) | AC, SCC, ASqe, LCC, othere | HPV 6, 16, 18, 39, 53, X |
| Mehra et al. (N = 62) (32) | FFPE | PCR (SPF10, Inno-LIPA) | AC, SCCe, ASqa, LCCa, | HPV 6, 16, 18, 52, 53, 44, 68, 39, 74, 82, X |
| Joh et al. (N = 29) (38) | FF | PCR (GP5+/GP6+) | AC, SCCe, LCCa, NSCLCa | HPV 11, 16 |
| Yanagawa et al. (N = 330) (60) | FFPE | PCR (GP5+/GP6+) and ISH | AC, SCC | HPV negative |
| Bohlmeyer et al. (N = 34) (45) | FFPE | PCR (MY09/MY11) | SCC | HPV 18 |
| Adjusted HPV 16/18 prevalence (95% CI)f = 3.78% (3.35, 4.22) | ||||
| Adjusted HPV 16 prevalence (95% CI)f = 2.03% (1.68, 2.39) | ||||
| Adjusted HPV 18 prevalence (95% CI)f = 2.49% (2.23, 2.75) | ||||
Other: histological type, not otherwise specified. AC, adenocarcinoma; ASq, adenosquamous carcinoma; BB, bronchial brushing; Bx, biopsy tissue; CSrc, carcinosarcoma; FF, fresh frozen; ISH, in situ hybridization; LCC, large cell carcinoma; NSCLC, non-small-cell lung carcinoma, not otherwise specified; PE, pleural effusion; SB, Southern blot; SmCC, small cell carcinoma; TS, type-specific primers.
aNumber of cases ≤6.
bCancer cells present and confirmed by pathologist.
cStudy is included in the model as a random effect.
dType not defined; HPV+ (for the HPV6/11/16/18/30 panel).
eNumber of cases 7–10.
fStudy is included as a covariate in the model.
The predominant high-risk HPV types found in lung cancer patients were HPV 16 and HPV18 (Table I). Other high-risk HPV types (HPV31, 33, 35, 45, 51, 52, 68 and 82) were also detected. For all geographic regions (Asia, Europe as well as North America) and with the exception of South and Central America, large-to-moderate heterogeneity was observed between the studies with no evidence of publication bias (data not shown). In order to address the heterogeneity, HPV prevalence rates were calculated after adjusting for age, gender, smoking history and tumor stage; study was included as a random effect where appropriate (Table I). Significant differences in HPV16/18 prevalence were noted between geographic regions. HPV 16/18 was most prevalent in South and Central America (adjusted prevalence [AdjPr] = 21.90%, 95% CI = 19.61–24.20), followed by Asia (AdjPr = 4.60%, 95% CI = 3.48–5.73), North America (AdjPr = 3.78%, 95% CI = 3.35–4.22) and Europe (AdjPr = 3.03%, 95% CI = 2.76–3.30). For each geographic region, the prevalence of HPV16 was significantly higher than HPV18, with the exception of North America and Asia. South and Central America had the highest prevalence of HPV16 followed by Europe, North America and Asia, whereas HPV 18 was also highest in South and Central America, followed by North America and Asia. There was virtually no HPV 18 detected in European patients (AdjPr = 0.82%, 95% CI = 0.73–0.92).
Clinical and demographic characteristics of HPV16/18-positive lung cancer cases
The mean age of all lung cancer patients in the pooled dataset was 65.0±10.1 (SD) years. The demographic and clinical characteristics for all cases are presented in Table II; comparisons were made between North American cases and the rest of the world. For the studies conducted in North America as well as the rest of the world, a higher proportion of the patients were male (53 and 72%, respectively), ever smokers (84 and 76%, respectively) and most of the lung cancers were not SCC (64 and 62%, respectively). North American patients were mostly White (89%) in contrast to the rest of the world where the majority were non-White (57%). Early-stage lung cancers were more represented in the North American studies (63%) in contrast to only 50% in studies from the rest of the world. For each geographic subgroup, there were no noted differences in the proportion of HPV16/18 patients according clinical and demographic characteristics with the exception of race. The majority of patients in North America were White and a higher proportion of HPV 16/18-positive lung cancers were also noted among White patients (72%). However, for the rest of the world, although a near equal proportion of White and non-White patients were included, the majority of HPV 16/18-positive lung cancers were observed in non-White patients (88%).
Table II.
Comparisons of demographic characteristics of patients according to HPV16/18 status (published datasets only)
| Characteristic | Asia, Europe and South/Central America | North America | ||||||
|---|---|---|---|---|---|---|---|---|
| Total (N = 2500) | HPV16/18 positive (N = 233, 9.3%) | HPV16/18 negative (N = 2267, 90.7%) | P valuea | Total (N = 663) | HPV16/18 positive (N = 23, 3.5%) | HPV16/18 negative (N = 640, 96.5%) | P valuea | |
| Gender | 0.157 | 0.632 | ||||||
| Male | 1749 (71.92) | 154 (66.96) | 1595 (72.43) | 349 (52.64) | 10 (43.48) | 339 (52.97) | ||
| Female | 683 (28.08) | 76 (33.04) | 607 (27.57) | 314 (47.36) | 13 (56.52) | 301 (47.03) | ||
| Age (years) | 0.529 | 0.557 | ||||||
| ≤66 | 1212 (51.33) | 92 (42.4) | 1120 (52.24) | 315 (47.51) | 10 (43.48) | 305 (47.66) | ||
| >66 | 1149 (48.67) | 125 (57.6) | 1024 (47.76) | 348 (52.49) | 13 (56.52) | 335 (52.34) | ||
| Race | 0.059 | 0.102 | ||||||
| White | 1083 (43.32) | 27 (11.59) | 1056 (46.58) | 239 (88.52) | 13 (72.22) | 226 (89.68) | ||
| Othersb | 1417 (56.68) | 206 (88.41) | 1211 (53.42) | 31 (11.48) | 5 (27.78) | 26 (10.32) | ||
| Smoking history | 0.762 | 0.256 | ||||||
| Never smoked | 602 (24.06) | 67 (28.93) | 534 (23.56) | 104 (15.69) | 3 (13.04) | 101 (15.78) | ||
| Ever smoked | 1898 (75.94) | 166 (71.07) | 1733 (76.44) | 559 (84.31) | 20 (86.96) | 539 (84.22) | ||
| Histology | 0.536 | 0.345 | ||||||
| SCC | 953 (38.12) | 107 (45.92) | 846 (37.32) | 239 (36.43) | 5 (22.73) | 234 (36.91) | ||
| Othersc | 1547 (61.88) | 126 (54.08) | 1421 (62.68) | 417 (63.57) | 17 (77.27) | 400 (63.09) | ||
| Stage | 0.614 | 0.099 | ||||||
| 0/Id | 1240 (49.60) | 108 (46.52) | 1132 (49.92) | 422 (63.63) | 15 (65.65) | 407 (63.59) | ||
| II | 460 (18.41) | 36 (15.41) | 424 (18.72) | 106 (16.01) | 1 (5.59) | 105 (16.39) | ||
| III | 479 (19.14) | 43 (18.37) | 436 (19.22) | 109 (16.43) | 3 (13.91) | 106 (16.53) | ||
| IV | 321 (12.84) | 46 (19.70) | 275 (12.14) | 26 (3.93) | 4 (16.52) | 22 (3.48) | ||
Data are presented as number of patients (%).
a P value was calculated by a logistic regression model with study as a random effect.
bOthers include Black, Hispanic, Asian and race not specified.
cOthers include adenocarcinoma and histology not specified.
dIncludes four cases with stage 0 (in situ).
No statistically significant associations were observed between HPV16/18-positive lung tumors and gender, age, smoking history, stage and histology (Table III). However, HPV 16/18-positive lung cancer cases were less likely to be observed among White patients compared with non-White patients (adjusted odds ratio [Adj OR] = 0.33, 95% CI = 0.12–0.90). For larger studies, (≥100 cases, 4 studies with a minimum of 131 cases and a maximum of 399 cases), the association of HPV 16/18 with race persisted (White: Adj OR = 0.08, 96% CI = 0.02–0.33). For smaller studies (<100 cases, 16 studies with a minimum of 22 cases and a maximum of 99 cases), the HPV 16/18-positive patients were more likely to be female (Adj OR = 1.61, 95% CI = 1.00–2.59), older than 66 years (Adj OR = 1.65, 95% CI = 1.05–2.59) and diagnosed with stage IV tumors than stage I/II (Adj OR = 2.55, 95% CI = 1.08–6.04). There were no differences in the association of HPV with stage IV tumors when we compared studies that reported verification of cancer diagnosis to those that did not report verification of cancer diagnosis in their publication. Similarly, for studies that reported efforts to avoid contamination, there was no difference in the association of HPV16/18 with stage IV tumors compared with those studies that did not include this information in their publication (data not shown). When North American studies were evaluated separately, there was no association of HPV 16/18 with gender, age, smoking history or histology and a marginal association with race was observed, which was significant for larger studies only (data not shown).
Table III.
Association of demographic and clinical variables with HPV 16/18 (published datasets only in all regions)
| Variable | HPV16/18 positive | |||||
|---|---|---|---|---|---|---|
| All studies (N = 3163) | Small studiesa (N = 918) | Large studiesb (N = 2245) | ||||
| ORc (95% CI) | P value | ORc (95% CI) | P value | ORc (95% CI) | P value | |
| Gender | 0.141 | 0.049 | 0.824 | |||
| Male | 1.0 (ref.) | 1.0 (ref.) | 1.0 (ref.) | |||
| Female | 1.27 (0.92, 1.75) | 1.61 (1.00, 2.59) | 1.05 (0.68, 1.62) | |||
| Age (years) | 0.420 | 0.029 | 0.361 | |||
| ≤66 | 1.0 (ref.) | 1.0 (ref.) | 1.0 (ref.) | |||
| >66 | 1.13 (0.84, 1.54) | 1.65 (1.05, 2.59) | 0.82 (0.54, 1.25) | |||
| Race | 0.029 | 0.708 | < 0.001 | |||
| Othersd | 1.0 (ref.) | 1.0 (ref.) | 1.0 (ref.) | |||
| White | 0.33 (0.12, 0.90) | 0.80 (0.25, 2.59) | 0.08 (0.02, 0.33) | |||
| Smoking history | 0.930 | 0.859 | 0.974 | |||
| Never smoked | 1.0 (ref.) | 1.0 (ref.) | 1.0 (ref.) | |||
| Ever smoked | 1.02 (0.66, 1.57) | 1.06 (0.58, 1.91) | 1.01 (0.59, 1.74) | |||
| Histology | 0.812 | 0.576 | 0.827 | |||
| Otherse | 1.0 (ref.) | 1.0 (ref.) | 1.0 (ref.) | |||
| SCC | 1.05 (0.71, 1.54) | 1.16 (0.69, 1.93) | 0.94 (0.53, 1.66) | |||
| Tumor stage | 0.561 | 0.044 | 0.338 | |||
| I/II | 1.0 (ref.) | 1.0 (ref.) | 1.0 (ref.) | |||
| III | 0.87 (0.56, 1.37) | 0.94 (0.53, 1.66) | 0.78 (0.36, 1.68) | |||
| IV | 1.10 (0.66, 1.85) | 2.55 (1.08, 6.04) | 0.68 (0.36, 1.27) | |||
Ref.: reference level.
aSmall studies include studies with 22 ≤ number of cases ≤ 99.
bLarge studies include studies with 131 ≤ number of cases ≤ 399.
cStudy was included as a random effect.
dOthers include Asian, Black, Hispanic and race not specified.
eOthers include adenocarcinoma and histology not specified.
HPV prevalence in normal versus lung cancer tissues
There were 16 studies that evaluated HPV prevalence in lung cancer tissues (1208 cases) in addition to normal lung tissues (732 controls). Normal lung tissues were either from lung cancer patients (i.e. adjacent normal or non-adjacent normal, n = 255) or from normal lung tissues from non-cancer patients (n = 477). Lung cancer tissues were almost four times more likely to be positive for any HPV DNA compared with normal (OR = 3.86, 95% CI = 2.87–5.19; study was included as a random effect).
Evidence of HPV16/18 DNA integration in lung cancer
Because integration of high-risk HPV DNA correlates with the overexpression of HPV oncogenes, we investigated the physical status of HPV16 in a subset of cases. In our pooled international dataset, physical status data were available for three studies conducted in Asia, South/Central America and North America (32,48,49) (n = 28, HPV16-positive patients). For the HPV16-positive lung tumors, 75% (21/28) carried integrated HPV16 genomes, 3.6% (1/28) carried episomal HPV16 genomes and 21.4% (6/28) carried both integrated and episomal forms of HPV16 DNA. There was no significant difference in the distribution of HPV physical status according to each study (P = 0.436).
An equal number of male and female patients were HPV16 positive, but the majority of female patients, i.e. 92.9% (13/14), carried integrated HPV 16 DNA compared with 57% (8/14) of male patients (Table IV). None of the female patients had episomal HPV genomes in their tumors and only 7.1% (1/14) had both integrated and episomal forms. On the other hand, 7.1% (1/14) of male patients had episomal HPV genomes, and 35.7% (5/14) had both integrated and episomal HPV genomes in their tumors.
Table IV.
HPV 16 physical status in primary lung carcinomas
| Case | Age | Sex | Histology | Physical status |
|---|---|---|---|---|
| 1 | 69 | Female | Adenocarcinoma | Mixed |
| 2 | 84 | Male | Adenocarcinoma | Integrated |
| 3 | 67 | Male | Adenocarcinoma | Integrated |
| 4 | 74 | Male | SCC | Mixed |
| 5 | 62 | Female | Adenocarcinoma | Integrated |
| 6 | 58 | Male | SCC | Integrated |
| 7 | 66 | Female | Adenocarcinoma | Integrated |
| 8 | 75 | Male | SCC | Integrated |
| 9 | 70 | Female | Adenocarcinoma | Integrated |
| 10 | 72 | Female | Adenocarcinoma | Integrated |
| 11 | 80 | Male | SCC | Mixed |
| 12 | 72 | Female | Adenocarcinoma | Integrated |
| 13 | 55 | Female | Adenocarcinoma | Integrated |
| 14 | 79 | Male | SCC | Integrated |
| 15 | 59 | Female | Adenocarcinoma | Integrated |
| 16 | 74 | Female | SCC | Integrated |
| 17 | 71 | Female | Adenocarcinoma | Integrated |
| 18 | 70 | Male | SCC | Mixed |
| 19 | 74 | Male | SCC | Integrated |
| 20 | 69 | Male | Adenocarcinoma | Integrated |
| 21 | 78 | Male | SCC | Mixed |
| 22 | 68 | Male | SCC | Episomal |
| 23 | 59 | Female | Adenocarcinoma | Integrated |
| 24 | 63 | Male | Adenocarcinoma | Integrated |
| 25 | 69 | Female | Adenocarcinoma | Integrated |
| 26 | 39 | Female | SCC | Integrated |
| 27 | 66 | Male | SCC | Mixed |
| 28 | 62 | Female | Adenocarcinoma | Integrated |
| SiHa | Integrated | |||
| Caski | Mixed |
Mixed: integrated and episomal genomes.
Survival analysis
A subset of patients had available follow-up data for four published studies (11,31,55,56) (n = 451). For all patients combined, the 2 year and 5 year overall survival rates for HPV16/18-negative patients were 60.9 and 38.5%, respectively, whereas the 2 year and 5 year overall survival rates for HPV16/18-positive patients were 71.4 and 64.3%, respectively. The Kaplan–Meier curves according to HPV16/18 status (data not shown) were not statistically significantly different (log-rank P value = 0.263).
Discussion
To date, >100 studies (including case reports) have analyzed HPV DNA in lung cancers (7). Although the majority of these studies have reported the prevalence of HPV16 and/or 18 DNA and a few publications have shown evidence of HPV oncogene (E6 and E7) expression (12,57), definite evidence of a causal relationship is still missing. Previously published meta-analyses of HPV in lung cancer show that HPV16 and HPV18 are the two most common genotypes detected in lung tumors worldwide (5–7); however, the characteristics of patients with these tumors were not determined. This study is the first international pooled analysis of individual data received from various research groups and was focused to define the demographic, behavioral and clinical characteristics of lung cancer cases with HPV16 and/or 18 DNA. We have also for the first time reported and compared published data from five North American studies with other parts of the world. Similar to previous reports, as expected, in our study (5–7), HPV16 and/or 18 were the most prevalent high-risk genotypes detected in lung cancers, but still counting for a low fraction of lung cancers and with heterogeneity among studies.
It is plausible that differences in the geographic location, study size, demographic and clinical makeup of each study could contribute to heterogeneity in HPV prevalence between studies. Furthermore, the majority of studies included in this pooled analysis were small (<100 cases) and may be less representative samples, thus contributing to heterogeneity in HPV prevalence. Therefore, for each geographic region, the prevalence of HPV 16/18 was adjusted for each study as well as potential confounders yet variations in HPV16/18 prevalence remained. The highest prevalence of HPV16/18-positive lung cancers was observed in South and Central American, whereas North American and Asian studies had similar prevalence and European studies had significantly lower prevalence of HPV 16/18. Similar observations were made for HPV 16 and HPV 18 independently. However, within each geographic region, distinct differences in the prevalence of HPV 16 and HPV18 genotypes were noted with the exception of Asia and North America. In North America, although the prevalence of each genotype was low, the prevalence of HPV 18 was slightly higher than that of HPV 16. In contrast, for Asia, Europe and South and Central America, if any HPV DNA detected in lung cancer, HPV 16 appeared to be more prominent than HPV18. In Europe, the prevalence of HPV18 was near zero. Factors that may be responsible for variability in HPV prevalence could be sexual behaviors of participants, genetics and possible environmental contamination as argued in the study by Koshiol et al. (54).
In addition to geographic location, the recent meta-analysis by Syrjanen et al. (7) reported that the heterogeneity between studies might be related to differences in histologic type. In this study, there were no differences in the proportion of histologic types in North America compared with the rest of the world and we observed no association of HPV 16/18 status with histology. Although South and Central American studies had the highest HPV prevalence, there were only two studies from this region; therefore, this finding will need to be confirmed by additional studies. Second to South and Central America, we observed that Asia had the highest prevalence of HPV 16 and 18. A significant association of HPV 16/18 DNA was also observed with race, where non-White lung cancer patients were more likely to have HPV-positive tumors compared with White lung cancer patients. There may be a number of reasons for this observed difference in HPV prevalence some of which might include differences in sexual practices or differences in susceptibility. Further investigation of the potential reasons for the racial difference in the association of HPV16/18 is needed.
Park et al. (65) evaluated HPV status in 112 non-small-cell lung cancer patients from Korea and reported that HPV16 was more common in younger lung cancer patients and HPV18 was more common in patients diagnosed with advanced stage. Although we categorized HPV16 and HPV18-positive tumors into a single group, we found no associations with age or stage, neither for gender, smoking history or histology. However, for small studies only, HPV16/18 was associated with old age (>66 years), female gender and advanced stage. It is possible that patients with metastatic disease (by definition, stage IV) could include metastases from other or mixed tumors, such as head and neck or cervical cancers (59), and the resulting misclassification could account for some of these findings. Therefore, the association with older females with stage IV tumors warrants further investigation in a single, larger and more representative study.
Although HPV-positive oropharyngeal cancers have been found to be associated with non-smokers, these tumors are also associated with patients having lower number of pack-years of cigarettes compared with heavy smokers (66). In vitro studies also show that tobacco smoke carcinogens have been shown to increase HPV16 and 18 viral synthesis as well as interact with HPV16 E6/E7 oncoproteins to increase lung cell proliferation (67). Furthermore, smoking suppresses the host innate immunity including functional and structural changes in the respiratory ciliary epithelium, lung surfactant protein and immune cells in the lung (68), thus may facilitate HPV infection and persistence in the lung in a subset of tumors. Mechanistically, it is not yet clear how HPV might promote lung carcinogenesis; however, the cooperation between HPV and tobacco smoke carcinogens for lung carcinogenesis is plausible. In this study, the association of HPV with smoking status was not conclusive. Therefore, further studies are needed to investigate the relationship between HPV and smoking status among patients with HPV-positive lung tumors.
Although our study cannot conclusively confirm the carcinogenetic role of HPV in lung cancer, we have shown that lung cancer tissues were almost 4-fold more likely to be HPV-positive compared with normal lung tissues. Secondly, our preliminary investigation of HPV16/18 physical status among a subset of tumors shows that the majority of the female tumors carried integrated HPV DNA while the physical status of HPV16/18 in male tumors was inconsistent. Given the predominance of integrated HPV genomes in female lung cancer patients, it is possible that HPV may play a role in lung cancer development but is unlikely to contribute to a large proportion of lung cancer cases. Although the presence of integrated DNA would suggest that the DNA detected was not simply due to contamination, it is important to note that there are no published studies comparing E2/E6-based integration with direct integration detection methods, which are more reliable. Thus, the results must be interpreted with caution. Our findings suggest an association of HPV DNA with a small fraction of lung tumors, with large geographic variations, but further comprehensive analysis is needed to assess whether this association reflects a causal relationship. Such detailed analysis should include not only HPV DNA testing but evaluation of all criteria that were postulated to prove a causal involvement of HPV in carcinogenesis, such as p16 expression and HPV E6/E7 oncogene expression (2,69), and measures to exclude pulmonary metastases (59).
The survival data in this pooled analysis only included 14 HPV16/18-positive cases and 5 events, and there were no significant differences between the Kaplan–Meier curves. However, based on the 2 and 5 year survival rates between HPV16/18-positive and HPV16/18-negative patients, there was some suggestion that HPV16/18-positive lung cancer patients had improved survival. This trend toward improved survival is consistent with HPV-related carcinogenesis in non-cervical sites, such as the head and neck and penile cancer, where HPV-positive cancer patients have an improved overall and disease-free survival compared with HPV-negative patients (70–72). This observation needs to be verified in a much larger number of cases/events and adjustment for known prognostic factors including a history of HPV-related disease as well as treatment for previous HPV-related disease.
The availability of individual data from each study included in the analysis reduces the potential errors associated with aggregate data. However, a number of limitations are inherent in this study design as well. One of these limitations is the lack of a uniform gold standard in the HPV DNA detection methods leading to variability in HPV prevalence. Only 27 of the 81 eligible studies participate in this analysis. We have compared the characteristics of the cases in the pooled dataset and with that of the non-included studies and remain confident that both subsets are similar in characteristics. Nevertheless, possible selection bias cannot completely be ruled out. Although the overall sample size was large, some of the subgroup analyses included small numbers. Although this study has good representation of populations from the Americas, Europe as well as Asia, China, Korea and Taiwan were not as well represented. In addition, the North American dataset was limited, in that, the majority of cases were of European descent; a larger US study with broad ethnic diversity is needed to allow for further investigation related to HPV and race. Although the present analysis provides evidence in favor of an association of HPV DNA with a small subset of lung cancer, these findings need to be validated and further comprehensive analysis is needed to examine a potential causal relationship. A detailed analysis of a larger and more representative case series, including the testing of all criteria that were postulated to prove a causal involvement of HPV in carcinogenesis and excluding pulmonary metastases, needs to be performed. Most importantly, the preliminary evidence on the potential prognostic role of HPV in lung cancer needs confirmation because of its potential clinical implications, e.g. in therapy of lung cancer.
Funding
National Cancer Institute (R03 CA141483 to C.R. and P30 CA006927); Commonwealth of Pennsylvania.
Acknowledgements
The authors would like to thank Chaka Peters, Helina Tadesse and Justin Wong for their contributions in coordinating the collection of datasets included in this analysis. Special thanks to Dr Wen-Chi Chang for assisting with the translation of Chinese publications and to laboratory personnel Elizabeth Blackman for her contribution in this work. We thank Ms JoEllen Weaver and the Fox Chase Cancer Center Biosample Repository for their support of this study.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AdjPr
adjusted prevalence
- CI
confidence interval
- FFPE
formalin fixed paraffin embedded
- HPV
human papillomavirus
- OR
odds ratio
- SCC
squamous cell carcinoma.
References
- 1. Gillison M.L., et al. (2000). Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst., 92, 709–720 [DOI] [PubMed] [Google Scholar]
- 2. zur Hausen H. (1996). Papillomavirus infections–a major cause of human cancers. Biochim. Biophys. Acta, 1288, F55–F78 [DOI] [PubMed] [Google Scholar]
- 3. Syrjänen K.J. (1979). Condylomatous changes in neoplastic bronchial epithelium. Report of a case. Respiration., 38, 299–304 [DOI] [PubMed] [Google Scholar]
- 4. Syrjänen K.J. (2002). HPV infections and lung cancer. J. Clin. Pathol., 55, 885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klein F., et al. (2009). Incidence of human papilloma virus in lung cancer. Lung Cancer, 65, 13–18 [DOI] [PubMed] [Google Scholar]
- 6. Srinivasan M., et al. (2009). Human papillomavirus type 16 and 18 in primary lung cancers–a meta-analysis. Carcinogenesis, 30, 1722–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Syrjänen K. (2012). Detection of human papillomavirus in lung cancer: systematic review and meta-analysis. Anticancer Res., 32, 3235–3250 [PubMed] [Google Scholar]
- 8. Miyagi J., et al. (2000). Recent striking changes in histological differentiation and rate of human papillomavirus infection in squamous cell carcinoma of the lung in Okinawa, a subtropical island in southern Japan. J. Clin. Pathol., 53, 676–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miyagi J., et al. (2001). Extremely high Langerhans cell infiltration contributes to the favourable prognosis of HPV-infected squamous cell carcinoma and adenocarcinoma of the lung. Histopathology, 38, 355–367 [DOI] [PubMed] [Google Scholar]
- 10. Gorgoulis V.G., et al. (1998). Alterations of the p16-pRb pathway and the chromosome locus 9p21-22 in non-small-cell lung carcinomas: relationship with p53 and MDM2 protein expression. Am. J. Pathol., 153, 1749–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gorgoulis V.G., et al. (1999). Human papilloma virus (HPV) is possibly involved in laryngeal but not in lung carcinogenesis. Hum. Pathol., 30, 274–283 [DOI] [PubMed] [Google Scholar]
- 12. Ciotti M., et al. (2006). Detection and expression of human papillomavirus oncogenes in non-small cell lung cancer. Oncol. Rep., 16, 183–189 [PubMed] [Google Scholar]
- 13. Giuliani L., et al. (2007). Detection of oncogenic viruses SV40, BKV, JCV, HCMV, HPV and p53 codon 72 polymorphism in lung carcinoma. Lung Cancer, 57, 273–281 [DOI] [PubMed] [Google Scholar]
- 14. Syrjänen K., et al. (1989). Human papillomavirus (HPV) type 6 and 16 DNA sequences in bronchial squamous cell carcinomas demonstrated by in situ DNA hybridization. Lung, 167, 33–42 [DOI] [PubMed] [Google Scholar]
- 15. Syrjänen K.J., et al. (1987). Human papillomavirus DNA in bronchial squamous cell carcinomas. Lancet, 1, 168–169 [DOI] [PubMed] [Google Scholar]
- 16. Cheng Y.W., et al. (2001). The association of human papillomavirus 16/18 infection with lung cancer among nonsmoking Taiwanese women. Cancer Res., 61, 2799–2803 [PubMed] [Google Scholar]
- 17. Cheng Y.W., et al. (2004). Gender difference in human papillomarvirus infection for non-small cell lung cancer in Taiwan. Lung Cancer, 46, 165–170 [DOI] [PubMed] [Google Scholar]
- 18. Cheng Y.W., et al. (2007). Human papillomavirus 16/18 E6 oncoprotein is expressed in lung cancer and related with p53 inactivation. Cancer Res., 67, 10686–10693 [DOI] [PubMed] [Google Scholar]
- 19. Hu K., et al. (1997). Detection of human papillomavirus types 16, 18 DNA related sequences in bronchogenic carcinoma. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi, 11, 147–149 [PubMed] [Google Scholar]
- 20. Li Q., et al. (1995). Detection of human papillomavirus types 16, 18 DNA related sequences in bronchogenic carcinoma by polymerase chain reaction. Chin. Med. J. (Engl)., 108, 610–614 [PubMed] [Google Scholar]
- 21. Thomas P., et al. (1995). Detection of human papillomavirus DNA in primary lung carcinoma by nested polymerase chain reaction. Cell. Mol. Biol. (Noisy-le-grand)., 41, 1093–1097 [PubMed] [Google Scholar]
- 22. Thomas P., et al. (1996). Detection of human papillomavirus by polymerase chain reaction in primary lung carcinoma. Bull. Cancer, 83, 842–846 [PubMed] [Google Scholar]
- 23. Wang J., et al. (2006). Frequent FHIT gene loss of heterozygosity in human papillomavirus-infected non-smoking female lung cancer in Taiwan. Cancer Lett., 235, 18–25 [DOI] [PubMed] [Google Scholar]
- 24. Yu Y., et al. (2009). Correlation of HPV-16/18 infection of human papillomavirus with lung squamous cell carcinomas in Western China. Oncol. Rep., 21, 1627–1632 [DOI] [PubMed] [Google Scholar]
- 25. Yu Y., et al. (2013). Significance of human papillomavirus 16/18 infection in association with p53 mutation in lung carcinomas. Clin. Respir. J., 7, 27–33 [DOI] [PubMed] [Google Scholar]
- 26. Hirayasu T., et al. (1996). Human papillomavirus DNA in squamous cell carcinoma of the lung. J. Clin. Pathol., 49, 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwamasa T., et al. (2000). Prognostic implication of human papillomavirus infection in squamous cell carcinoma of the lung. Pathol. Res. Pract., 196, 209–218 [DOI] [PubMed] [Google Scholar]
- 28. Tung M.C., et al. (2013). Association of epidermal growth factor receptor mutations with human papillomavirus 16/18 E6 oncoprotein expression in non-small cell lung cancer. Cancer, 119, 3367–3376 [DOI] [PubMed] [Google Scholar]
- 29. Liu H.R., et al. (1994). A study of human papillary virus infection by in situ hybridization and histopathology in squamous cell carcinoma of the lung. Zhonghua Bing Li Xue Za Zhi, 23, 299–301 [PubMed] [Google Scholar]
- 30. Xing L.Q., et al. (1994). Analysis of the characteristics of human papilloma virus infection in 85 neoplasms of the respiratory system in adult patients. Zhonghua Zhong Liu Za Zhi, 16, 424–427 [PubMed] [Google Scholar]
- 31. Pillai R.N., et al. (2013). Human papillomavirus (HPV)-associated early stage non-small cell lung cancer (NSCLC). ASCO Meeting Abstracts, Chicago IL, 31, Abstract no. 7560. [Google Scholar]
- 32. Mehra R., et al. (2013). A pilot study of the association and prevalence of the human papillomavirus (HPV) in non-small cell lung cancer (NSCLC). AACR Meeting Abstracts, Washington, DC., Abstract no. 4785. [Google Scholar]
- 33. Béjui-Thivolet F., et al. (1990). Detection of human papillomavirus DNA in squamous bronchial metaplasia and squamous cell carcinomas of the lung by in situ hybridization using biotinylated probes in paraffin-embedded specimens. Hum. Pathol., 21, 111–116 [DOI] [PubMed] [Google Scholar]
- 34. Carpagnano G.E., et al. (2011). HPV in exhaled breath condensate of lung cancer patients. Br. J. Cancer, 105, 1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fei Y., et al. (2006). Different human papillomavirus 16/18 infection in Chinese non-small cell lung cancer patients living in Wuhan, China. Jpn. J. Clin. Oncol., 36, 274–279 [DOI] [PubMed] [Google Scholar]
- 36. Galvan A., et al. (2012). Testing of human papillomavirus in lung cancer and non-tumor lung tissue. BMC Cancer, 12, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gatta L.B., et al. (2012). Human papillomavirus DNA and p16 gene in squamous cell lung carcinoma. Anticancer Res., 32, 3085–3089 [PubMed] [Google Scholar]
- 38. Joh J., et al. (2010). Human papillomavirus (HPV) and Merkel cell polyomavirus (MCPyV) in non small cell lung cancer. Exp. Mol. Pathol., 89, 222–226 [DOI] [PubMed] [Google Scholar]
- 39. Krikelis D., et al. (2010). Frequent presence of incomplete HPV16 E7 ORFs in lung carcinomas: memories of viral infection. J. Clin. Virol., 49, 169–174 [DOI] [PubMed] [Google Scholar]
- 40. Nadji S.A., et al. (2007). Relationship between lung cancer and human papillomavirus in north of Iran, Mazandaran province. Cancer Lett., 248, 41–46 [DOI] [PubMed] [Google Scholar]
- 41. Niyaz H., et al. (2000). Detection and significance of HPV16, 18 infection, P53 overexpression and telomerase activity in patients with lung cancer. Zhonghua Jie He He Hu Xi Za Zhi, 23, 679–682 [PubMed] [Google Scholar]
- 42. Wang Y.H., et al. (2010). The relationship among human papilloma virus infection, survivin, and p53 gene in lung squamous carcinoma tissue. Saudi Med. J., 31, 1331–1336 [PubMed] [Google Scholar]
- 43. Xu Y., et al. (2009). The Relationship between the Status of Human Papillomavirus 16/18 Infection and the Expression of Bcl-2 and Bax in Squamous Cell Carcinomas of the Lung. Zhongguo Fei Ai Za Zhi, 12, 849–852 [DOI] [PubMed] [Google Scholar]
- 44. Yang Y., et al. (2008). Association of the XRCC1 and hOGG1 polymorphisms with the risk of laryngeal carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 25, 211–213 [PubMed] [Google Scholar]
- 45. Bohlmeyer T., et al. (1998). Detection of human papillomavirus in squamous cell carcinomas of the lung by polymerase chain reaction. Am. J. Respir. Cell Mol. Biol., 18, 265–269 [DOI] [PubMed] [Google Scholar]
- 46. Nuorva K., et al. (1995). p53 protein accumulation and the presence of human papillomavirus DNA in bronchiolo-alveolar carcinoma correlate with poor prognosis. Int. J. Cancer, 64, 424–429 [DOI] [PubMed] [Google Scholar]
- 47. Tsuhako K., et al. (1998). Human papillomavirus DNA in adenosquamous carcinoma of the lung. J. Clin. Pathol., 51, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aguayo F., et al. (2007). Human papillomavirus-16 is integrated in lung carcinomas: a study in Chile. Br. J. Cancer, 97, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baba M., et al. (2010). Human papillomavirus is frequently detected in gefitinib-responsive lung adenocarcinomas. Oncol. Rep., 23, 1085–1092 [DOI] [PubMed] [Google Scholar]
- 50. Castillo A., et al. (2006). Human papillomavirus in lung carcinomas among three Latin American countries. Oncol. Rep., 15, 883–888 [PubMed] [Google Scholar]
- 51. Goto A., et al. (2011). Human papillomavirus infection in lung and esophageal cancers: analysis of 485 Asian cases. J. Med. Virol., 83, 1383–1390 [DOI] [PubMed] [Google Scholar]
- 52. Hiroshima K., et al. (1999). Ultrastructural study of intranuclear inclusion bodies of pulmonary adenocarcinoma. Ultrastruct. Pathol., 23, 383–389 [DOI] [PubMed] [Google Scholar]
- 53. Iwakawa R., et al. (2010). Prevalence of human papillomavirus 16/18/33 infection and p53 mutation in lung adenocarcinoma. Cancer Sci., 101, 1891–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koshiol J., et al. (2011). Assessment of human papillomavirus in lung tumor tissue. J. Natl. Cancer Inst., 103, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Syrjänen K., et al. (2012). Detection of human papillomavirus genotypes in bronchial cancer using sensitive multimetrix assay. Anticancer Res., 32, 625–631 [PubMed] [Google Scholar]
- 56. Lim W.T., et al. (2009). Assessment of human papillomavirus and Epstein-Barr virus in lung adenocarcinoma. Oncol. Rep., 21, 971–975 [DOI] [PubMed] [Google Scholar]
- 57. Kinoshita I., et al. (1995). Human papillomavirus type 18 DNA and E6-E7 mRNA are detected in squamous cell carcinoma and adenocarcinoma of the lung. Br. J. Cancer, 71, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J.L., et al. (2014). Human papillomavirus infections as a marker to predict overall survival in lung adenocarcinoma. Int. J. Cancer, 134, 65–71 [DOI] [PubMed] [Google Scholar]
- 59. van Boerdonk R.A., et al. (2013). High-risk human papillomavirus-positive lung cancer: molecular evidence for a pattern of pulmonary metastasis. J. Thorac. Oncol., 8, 711–718 [DOI] [PubMed] [Google Scholar]
- 60. Yanagawa N., et al. (2013). Human papilloma virus genome is rare in North American non-small cell lung carcinoma patients. Lung Cancer, 79, 215–220 [DOI] [PubMed] [Google Scholar]
- 61. Behera M., et al. (2013). Survival analysis of patients with stage I non-small-cell lung cancer using clinical and DNA repair pathway expression variables. Clin. Lung Cancer, 14, 128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Peitsaro P., et al. (2002). Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol., 40, 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raghunathan T.E., et al. (2001). A multivariate technique for multiply imputing missing values using a sequence of regression models. Surv. Meth., 27, 85–95 [Google Scholar]
- 64. Branica B.V., et al. (2010). Detection of human papillomaviruses type 16, 18 and 33 in bronchial aspirates of lung carcinoma patients by polymerase chain reaction: a study of 84 cases in Croatia. Coll. Antropol., 34, 159–162 [PubMed] [Google Scholar]
- 65. Park M.S., et al. (2007). The prevalence of human papillomavirus infection in Korean non-small cell lung cancer patients. Yonsei Med. J., 48, 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ang K.K., et al. (2010). Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med., 363, 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Muñoz J.P., et al. (2012). Functional interaction between human papillomavirus type 16 E6 and E7 oncoproteins and cigarette smoke components in lung epithelial cells. PLoS One, 7, e38178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mehta H., et al. (2008). Cigarette smoking and innate immunity. Inflamm. Res., 57, 497–503 [DOI] [PubMed] [Google Scholar]
- 69. Pagano J.S., et al. (2004). Infectious agents and cancer: criteria for a causal relation. Semin. Cancer Biol., 14, 453–471 [DOI] [PubMed] [Google Scholar]
- 70. Ragin C.C., et al. (2007). Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int. J. Cancer, 121, 1813–1820 [DOI] [PubMed] [Google Scholar]
- 71. Rietbergen M.M., et al. (2013). Human papillomavirus detection and comorbidity: critical issues in selection of patients with oropharyngeal cancer for treatment De-escalation trials. Ann. Oncol., 24, 2740–2745 [DOI] [PubMed] [Google Scholar]
- 72. Lont A.P., et al. (2006). Presence of high-risk human papillomavirus DNA in penile carcinoma predicts favorable outcome in survival. Int. J. Cancer, 119, 1078–1081 [DOI] [PubMed] [Google Scholar]