Abstract
Regulation of mRNA decapping is a critical determinant for gene expression. We demonstrate that the poly(A) tail-mediated regulation of mRNA decapping observed in humans can be recapitulated in vitro by the cytoplasmic poly(A)-binding protein PABP through a direct and specific binding to the 5′ end of capped mRNA. The specific association of PABP with the cap occurred only within the context of the RNA whereby a cap attached to an RNA moiety served as the high-affinity substrate but not the cap structure or RNA alone. Binding of PABP to the RNA 5′ end required the presence of the cap and was accentuated by the N7 methyl moiety of the cap. Interestingly, conditions that enhanced hDcp2 decapping activity reduced the affinity of PABP for cap association and consequently its ability to inhibit decapping, suggestive of a regulated association of PABP with the cap. These observations reveal a novel direct involvement of human PABP in the stabilization of mRNA by protecting the 5′ end from decapping.
Keywords: hDcp2, mRNA decapping, poly(A)-binding protein, cap binding
Introduction
The regulation of mRNA decay is an important checkpoint in determining the expression and fate of cellular transcripts. The initial step in eucaryotic mRNA decay involves removal of polyadenosine (poly(A)) residues followed by either continual decay of the mRNA at the 3′ end or a decapping step preceding 5′ to 3′ exonucleolytic decay (Decker and Parker, 1994; Muhlrad et al, 1995; Beelman et al, 1996; Mitchell et al, 1997; Jacobs et al, 1998; Wang and Kiledjian, 2001). Following the deadenylation step in yeast, the mRNA can be decapped by the Dcp2p/Dcp1p decapping enzyme complex (Beelman et al, 1996; Dunckley and Parker, 1999; Steiger et al, 2003) and subsequently degraded progressively by the 5′ to 3′ exoribonuclease Xrn1p enzyme (Hsu and Stevens, 1993). Alternatively, the mRNA can be degraded from the 3′ end by the exosome complex of exoribonucleases (Mitchell et al, 1997; Anderson and Parker, 1998) following the initial deadenylation step and ultimately the resulting cap structure hydrolyzed by Dsc1p (Liu et al, 2002).
The pathways of mRNA decay, as well as many of the nucleases involved in the decay, are conserved between yeast and mammals where decay can occur from both the 5′ end (Wang and Kiledjian, 2001; Mukherjee et al, 2002; van Dijk et al, 2003) and the 3′ end (Ross and Kobs, 1986; Brewer, 1998; Chen et al, 2001; Wang and Kiledjian, 2001; Mukherjee et al, 2002; Rodgers et al, 2002) of an mRNA. In vitro decay assay using mammalian extract indicates that decay from the 3′ end is the predominant pathway whereby, following deadenylation, the mRNA is degraded primarily by the exosome complex and the subsequent residual cap structure hydrolyzed by the DcpS scavenger decapping enzyme (Wang and Kiledjian, 2001; Liu et al, 2002; Rodgers et al, 2002). Preferential utilization of the 3′ decay pathway has also been demonstrated for an endogenous mRNA in mammalian cells (Wang and Kiledjian, 2001), indicating that this pathway might also be prevalent in cells. The alternative and most likely regulated pathway for mRNA decay in mammals involves decapping of the mRNA following the initial deadenylation step. Decapping is carried out by the yeast homolog of Dcp2p, human Dcp2 (hDcp2) (Lykke-Andersen, 2002; Van Dijk et al, 2002; Wang et al, 2002).
A highly regulated and critical step in the 5′ decay pathway involves removal of the cap structure from the 5′ end. The intimate association of mRNA translation with mRNA turnover and decapping implies that common factors will be involved in the regulation of both processes (Jacobson and Peltz, 1996; Mangus et al, 2003). In yeast, mRNA decapping is inhibited by the eIF4E cap-binding protein as well as the poly(A) tail (Caponigro and Parker, 1995; Schwartz and Parker, 1999, 2000; Wilusz et al, 2001a; Ramirez et al, 2002). A network of interactions between the yeast eIF4E, eIF4G and poly(A)-binding protein (PABP) is thought to circularize the message to augment translation as well as prevent the access of decapping enzymes to protect the cap from hydrolysis (Wells et al, 1998; Schwartz and Parker, 2000; Wilusz et al, 2001b). In addition to the inhibitory effect of the poly(A) tail on decapping in yeast, the poly(A) tail can also negatively impact decapping in humans (Wang et al, 2002) indicating a mechanistic conservation for the utilization of the poly(A) tail to regulate decapping. However, the mechanism involved in the regulation is not clear.
PABP is generally considered the main mediator of poly(A) tail functions and is critical for protecting the poly(A) tail from degradation (Bernstein and Ross, 1989; Ford and Wilusz, 1999; Wang et al, 1999) Using in vitro decay assay systems, both yeast and human PABP were also implicated to exert the poly(A) tail-mediated inhibition of decapping (Wilusz et al, 2001a; Wang et al, 2002). In humans, PABP is a 636-amino-acid protein containing four RNA-recognition motifs (RRM; also referred to as RNP motif), RNA-binding domains at the amino-terminal half of the protein and a more evolutionarily divergent carboxyl terminus (Grange et al, 1987). Although all four RRMs are competent to bind RNA individually or in combination, the first two RRMs contain the highest affinity for poly(A) sequences and are the major contributors of the poly(A)-binding activity (Nietfeld et al, 1990; Burd et al, 1991; Deo et al, 1999).
To begin addressing the mechanism by which the poly(A) tail can negatively influence decapping, we utilized a mammalian in vitro decapping assay to reconstitute the regulation. We demonstrate that human cytoplasmic PABP can inhibit decapping independent of additional factors by a direct and specific association with the 5′ cap structure of an RNA.
Results
PABP inhibits hDcp2 decapping activity
To determine whether the poly(A) tail-dependent inhibition of hDcp2 decapping can be mediated by PABP, we carried out decapping assays in the presence of this protein. For this purpose, RNA substrate consisting of the polylinker sequence of pcDNA3 with 60 adenosines (pcP-A60) exclusively 32P-labeled at the 5′ cap was used along with bacterially expressed recombinant PABP protein containing a histidine tag at the amino terminus. As shown in Figure 1A, hDcp2 decapping activity as detected by the appearance of the m7GDP product (lane 3) was reduced to undetectable levels with the addition of PABP (lanes 4–7) but was unaffected by an unrelated RRM containing RNA-binding protein, Deleted in Azoospermia (DAZL, lane 10) (Jiao et al, 2002). Interestingly, the PABP-mediated inhibition was insensitive to cap analog competition (lanes 8 and 9). The inhibition by PABP was comparable to inhibition observed with eIF4E cap-binding protein, which as expected was sensitive to cap analog competition (Figure 1B). These results indicated that the regulation of hDcp2 decapping by the poly(A) tail can be a direct consequence of PABP, independent of any other cap-binding or adapter proteins, and unlike the inhibition by eIF4E the PABP-mediated inhibition is insensitive to cap analog competition.
Figure 1.
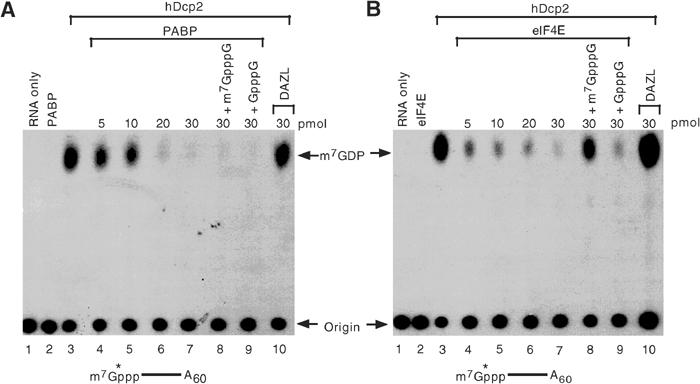
PABP can inhibit hDcp2 decapping activity. (A) In vitro decapping assays were carried out with 10 pmol recombinant histidine-tagged hDcp2 and 32P-cap-labeled pcP-A60 RNA in the presence of the indicated amounts of recombinant histidine-tagged PABP (lanes 4–9) or the control GST-tagged DAZL RNA-binding protein (lane 10). Lanes 8 and 9 also contain 100 μM of the indicated competitor cap analog. Reaction products were resolved by polyethyleneimine-TLC developed in 0.75 M LiCl. Migration of the input RNA designated as ‘origin' and m7GDP standard is shown with an arrow. A schematic denoting the cap-labeled RNA substrate containing a 60-nucleotide poly(A) tail is indicated below the figure. The asterisk denotes the labeled phosphate. (B) Labeling is as in (A) except that the indicated amount of eIF4E instead of PABP was used.
PABP specifically associates with the 5′ end of capped RNA
The inhibition of decapping by PABP could result from a direct protein–protein interaction or an association with the cap structure. We have been unable to detect an interaction of hDcp2 and PABP by either co-immunopurification or co-purification assays (data not shown). However, as shown in Figure 2A, PABP was ultraviolet light (UV) crosslinked to 32P-cap-labeled adenylated RNA (lanes 1–6) while multiple other unrelated RNA-binding proteins were not (lane 7; also see Figure 3). Surprisingly, PABP also crosslinked to the cap structure of unadenylated RNA although higher concentrations were required relative to adenylated RNA substrate (lanes 8–13). Similar to the inability of cap analog to compete PABP in the decapping assays, addition of cap analog also had no influence on the crosslinking of PABP to the cap (Figure 2A, lanes 14 and 15). The crosslinking of PABP however was not restricted to recombinant protein. Lysate from cells expressing myc epitope-tagged PABP was crosslinked to cap-labeled RNA and immunoprecipitated with α-myc-tag antibody (Figure 2B, lane 1) but not with control antisera (lane 2). These data demonstrate that cellular PABP can also gain direct access to the cap of an mRNA and this is not a unique feature of the recombinant protein. Furthermore, PABP association with the cap is not restricted to the particular substrate RNA used in these assays. PABP crosslinking to generic as well as endogenous mRNA sequences including c-myc, β-globin and α-globin could be detected (Figure 2C). PABP association with different cap-labeled RNAs demonstrates that the binding detected above is not due to a fortuitous PABP-binding site at the 5′ end of the RNA substrate.
Figure 2.
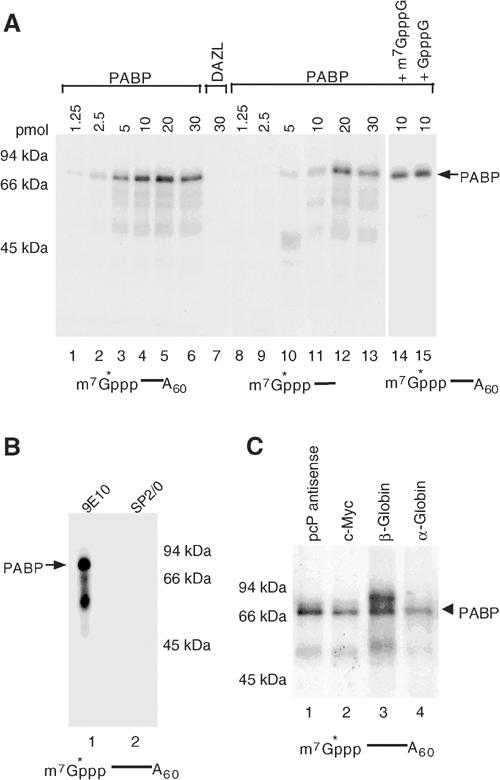
PABP can UV crosslink to the cap structure of an RNA. (A) 32P-cap-labeled RNA (5 pmol) with or without poly(A) tail as indicated was UV crosslinked to the denoted amount of PABP (lanes 1–6 and 8–13) or control DAZL protein (lane 7). Where indicated, 100 μM of the cap analog competitor was used. Crosslinked proteins were resolved on 12.5% SDS–PAGE and detected by autoradiography. Migration of 70 kDa PABP protein following RNase digestion of the RNA is shown with an arrow and molecular weight markers are indicated on the left. (B) Extract from human 293T cells (75 μg) expressing myc-tagged PABP was UV crosslinked to 10 pmol of cap-labeled pcP-A60 RNA and PABP was immunoprecipitated with the 9E10, α-myc (lane 1) or the SP2/0 control antibody (lane 2). (C) PABP crosslinks to the cap of generic as well as endogenous RNAs. PABP (10 pmol) was UV crosslinked to the following 32P-cap-labeled and adenylated RNAs: pcP anti-sense RNA (lane 1), c-myc (lane 2), β-globin (lane 3) and α-globin (lane 4) sequences.
Figure 3.
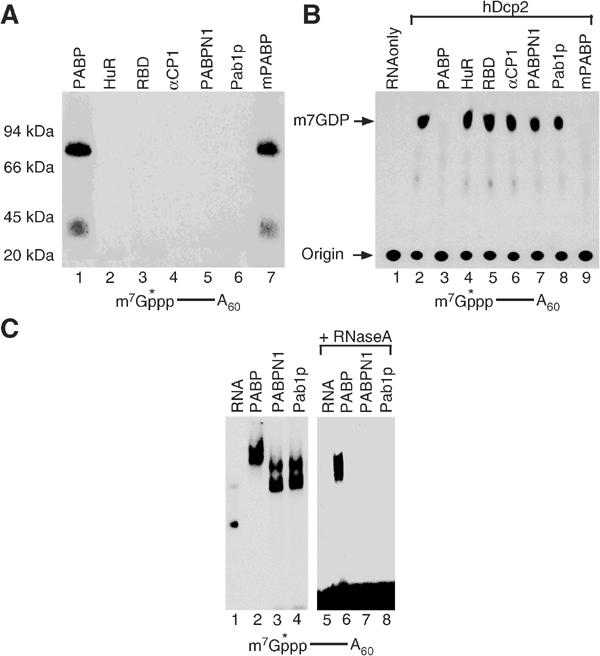
Cap association is not a general feature of all RNA- and poly(A)-binding proteins. (A) UV crosslinking of 10 pmol of the indicated proteins with 32P-cap-labeled adenylated RNA is shown. The RRM protein, His-HuR (36 kDa), is shown in lane 2 and the RGG box RNA-binding domain of hnRNP U (GST-RBD; 39 kDa) is in lane 3 and the KH domain protein, His-αCP1 (40 kDa), is in lane 4. Human nuclear PABP His-PABPN1 (35 kDa) and yeast His-Pab1p (64 kDa) are shown in lanes 5 and 6, respectively. The histidine-tagged mouse PABP (70 kDa) is shown in lane 7. Only human and mouse PABP molecules can crosslink to the 5′ end of cap-labeled RNA. (B) In vitro decapping assays were carried out as above with 10 pmol of the indicated proteins incubated with hDcp2 and 32P-cap-labeled pcP-A60 RNA. Whereas the human and mouse PABP efficiently inhibited hDcp2 decapping (lanes 3 and 9), other RNA-binding proteins could not (lanes 4–8). (C) Not all PABPs are capable of binding the 5′ end of capped RNA. A 20 pmol portion of the indicated proteins was incubated with 20 pmol of 32P-cap-labeled pcP-A60 RNA followed by digestion with RNase A as described above. Lane 1 contains the RNA only, lanes 2–4 contain RNA bound with his-PABP, his-Pab1p and his-PABPN1 respectively, lane 5 contains RNA digested with RNase A and lanes 6–8 contain the digested RNA bound to the proteins. All three PABPs can bind polyadenylated RNA.
To address whether the 5′ cap association of PABP was a general property of other RNA-binding proteins, several different classes of RNA-binding proteins were tested. Since PABP is a multiple RRM-containing protein, we tested another multiple RRM-containing protein, HuR. HuR is an ELAV family member RNA-binding protein in humans with three RRMs (Ma et al, 1997). Although PABP efficiently crosslinked to the 32P-cap-labeled RNA, crosslinking of HuR could not be detected (Figure 3A, lane 2). Similarly, the RGG-box RNA-binding domain of hnRNP U (RBD; Kiledjian and Dreyfuss, 1992) or the KH domain containing αCP1 protein (Kiledjian et al, 1995) also did not crosslink to the cap-labeled RNA (lanes 3 and 4, respectively). The inability of these proteins to associate with the cap is not a unique feature of the crosslink assay as they also failed to associate with the cap and inhibit decapping in a functional decapping assay (Figure 3B, lanes 4–6).
The association of PABP to the cap is not a general feature of all PABPs. The human nuclear PABP (PABPN1) and the yeast PABP (Pab1p) were neither able to crosslink to cap-labeled RNA (Figure 3A, lanes 5 and 6) nor inhibit decapping (Figure 3B, lanes 7 and 8). The inability of these two PABPs to associate with the 5′ cap was not due to their inability to bind poly(A) RNA but rather their inability to associate with the 5′ cap. All three PABPs can bind the 32P-cap-labeled polyadenylated RNA as shown by an electrophoretic mobility shift assay (Figure 3C, lanes 2–4). However, consistent with the crosslinking data, only PABP can remain stably associated with the labeled 5′ cap following RNase digestion (lane 6) while PABPN1, which has no obvious homology with PABP, did not (lane 7) and neither did Pab1p (lane 8), which shares 44% identity with the human PABP. However, the highly conserved mouse cytoplasmic PABP (mPABP) was able to bind to the 5′ cap of an RNA (Figure 3A, lane 7) and consequently inhibit decapping (Figure 3B, lane 9). These data demonstrate that the 5′ cap association and inhibition of decapping is not a general feature of all mRNA-binding proteins or PABPs but is also not a unique property of human PABP and is conserved at least in mouse, suggesting that the inhibition could be conserved in mammals.
Specific cap association of PABP requires a cap linked to an RNA moiety
To determine the specificity of PABP association with the cap, crosslinking of PABP to capped (32P-5′-cap-labeled) or uncapped (32P-5′-triphosphate-labeled) RNAs containing a poly(A) tail was carried out. As shown in Figure 4A, PABP crosslinking required the cap structure since it did not crosslink to the uncapped RNA (Figure 4A, lane 2). A similar result was also obtained with an electrophoretic mobility shift assay using the same two RNAs (Figure 4B). PABP was incubated with excess labeled RNA to enable binding and the RNA body subsequently digested with RNase A. The RNase-resistant RNA–protein complex at the 5′ end was detected by native gel electrophoresis (Figure 4B). A PABP cap complex was observed only with capped RNA (lanes 3 and 4) and not with uncapped RNA (lanes 7 and 8). This is consistent with the UV crosslinking data where PABP association with the 5′ end required the presence of the cap. Interestingly, unlike the ability of eIF4E to associate with the cap structure lacking an RNA moiety, binding of PABP was not detected (Figure 4C), indicating that the cap structure in conjunction with the RNA is the substrate recognized by PABP and neither is recognized efficiently alone. PABP association with the cap was also dependent on the N7 methyl moiety. Crosslinking of PABP was carried out with capped RNA substrates either containing or lacking the methyl group. The presence of the N7 methyl moiety on the cap structure significantly enhanced the association of PABP to the 5′ cap (Figure 4D). Collectively, the results in Figures 2, 3 and 4 demonstrate that human cytoplasmic PABP is capable of associating directly with or adjacent to the cap structure in the absence of translation initiation factors and that the extent of its cap binding is significantly enhanced in the presence of the poly(A) tail and methylated cap.
Figure 4.
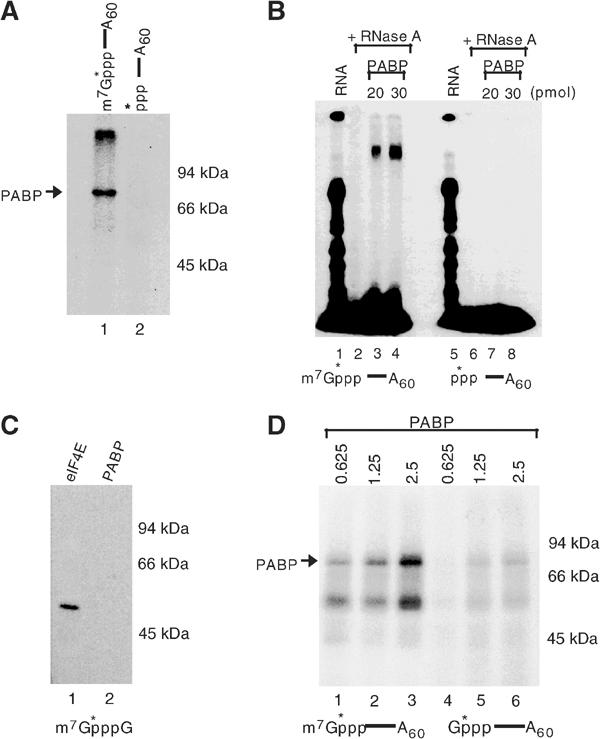
Specificity of PABP crosslinking to methylated capped RNA. (A) Crosslinking of 2.5 pmol PABP to 5′-capped or -uncapped adenylated RNA as described in the legend to Figure 2A is shown. A schematic of the RNA used is shown above each lane with lane 1 containing and lane 2 lacking a cap, and the asterisk denotes the 32P-labeled phosphate. PABP crosslinking to only the capped RNA was detected (lane 1). (B) Electrophoretic mobility shift assays were carried out by incubation of the indicated amount of PABP with capped and uncapped RNAs at room temperature. Following RNase digestion, the complex was resolved by a native polyacrylamide gel. Binding of PABP to only the 5′ end of capped RNA was detected (lanes 3 and 4). (C) UV crosslinking of 10 pmol of 47 kDa GST-eIF4E (lane 1) or His-PABP (lane 2) to 32P-labeled methylated cap analog (m7GpppG) is shown. PABP did not crosslink to the cap structure devoid of RNA (lane 2). (D) PABP UV crosslinking to 32P-cap-labeled and adenylated RNA containing or lacking a methyl group (lanes 1–3 and 4–6, respectively) is shown. The PABP cap association was enhanced by the presence of N7 methylated moiety.
A filter binding assay was next used to assess the efficiency of PABP binding to the 5′ cap. A titration binding of PABP to 5′-end-labeled polyadenylated pcP-A60 RNA with and without a cap was carried out as described above for the electrophoretic mobility shift assays. An increasing titration of recombinant PABP was used in the binding reactions until concentrations of protein were obtained to saturate the limiting RNA substrate. Following the binding reaction, the RNA–protein complex that remained after digestion of the RNA with RNase A was trapped by vacuum filtration through nitrocellulose filters. A graph of RNA bound relative to PABP concentration was plotted to derive dissociation constants. The apparent dissociation constant (Kd) was determined as the concentration of PABP at which 50% of the labeled RNA substrate was bound (Wilson and Brewer, 1999). Consistent with the data presented in the above figures, efficiency of PABP binding to the 5′ end of capped polyadenylated RNA was greater than that to uncapped polyadenylated RNA (Table I). The apparent Kd of PABP to the capped RNA was 1.5 × 10−7 M, which is 14-fold lower than that to uncapped RNA (2.1 × 10−6). No appreciable binding to the 5′ end of unadenylated RNA was detected under these assay conditions (data not shown) underscoring the requirement of the poly(A) tail to recruit PABP onto the 5′ cap. These data confirm the UV crosslinking and functional inhibition of decapping and demonstrate that PABP can specifically bind the 5′ cap of a polyadenylated RNA in vitro.
Table 1.
Binding affinity of PABP
| Binding of PABP to | Kd (μM)a |
|---|---|
| 5′ end of capped and polyadenylated RNAb | 0.15 |
| 5′ end of uncapped and polyadenylated RNAb | 2.1 |
| Poly(A) RNAc |
0.03 |
| aDissociation constant determined as the concentration of protein at which 50% of His-PABP was bound to the substrate. Average of three independent experiments. | |
| bpcP-A60 RNA containing or lacking a cap. | |
| cPoly(A) homopolymer with an average of 60 adenosines. | |
eIF4E can displace PABP from the cap
The above data demonstrate that PABP has the potential to associate with the cap of an mRNA and hinder decapping. To determine whether PABP can gain access to the cap in the presence of the eIF4E cap-binding protein, the ability of eIF4E to displace PABP was tested. Crosslinking of recombinant PABP to cap-labeled polyadenylated RNA in the presence of glutathione S-transferase (GST) eIF4E fusion protein was tested. Although direct UV crosslinking of eIF4E is not detected with these assay conditions, PABP was efficiently displaced by eIF4E from the cap (compare lane 1 to 2 and 3). Binding of PABP however was unaffected by the HuR RNA-binding protein (lanes 6 and 7). Interestingly, a GST fusion of eIF4G adaptor protein containing the N-terminal 401-amino-acid fragment of eIF4G, which retains the PABP interaction region, did not interfere with the binding of PABP to the cap (lanes 4 and 5). On the contrary, a subtle enhancement of PABP cap binding was detected in the presence of eIF4G, but a corresponding functional increase in the ability of PABP to inhibit hDcp2 decapping was not detected (data not shown). These data indicate that eIF4E has a higher affinity to the cap than PABP and the presence of eIF4G does not preclude the ability of PABP to associate with the cap. Further indications that the association of PABP with the cap must occur following removal of eIF4E was obtained with binding analysis in the presence of cellular extract. Using a concentration of recombinant PABP where binding could not be detected in the presence of extract, addition of cap analog competitor, which would sequester eIF4E, resulted in detectable PABP binding to the cap (Figure 5, lanes 8 and 9). These data suggest that PABP association with the cap can occur following removal of eIF4E from the cap.
Figure 5.
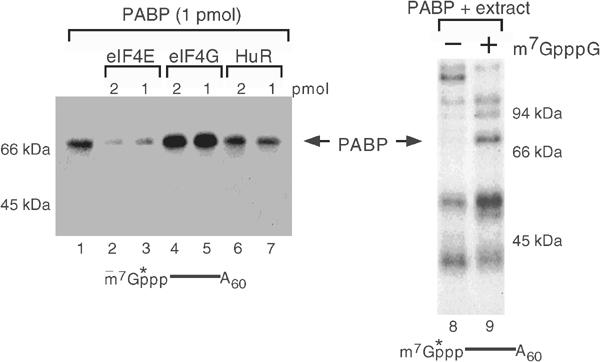
PABP association with the cap is competed by eIF4E. The ability of 1 pmol of His-PABP to bind the 5′ cap of 32P-cap-labeled pcP-A60 RNA in the presence of competitor proteins is shown in lanes 1–7. The indicated amounts of GST-eIF4E, GST-eIF4G and His-HuR proteins were used as competitors. Binding was detected by UV crosslinking, and cap-associated PABP remaining after RNase digestion was resolved by SDS–PAGE. Crosslinking of 2.5 pmol His-PABP was tested in the presence of 25 μg of human erythroleukemia K562 cell extract in the absence or presence of 100 μM m7GpppG cap analog. Migration of the recombinant PABP is denoted by the arrows, and molecular weight markers and a schematic of the RNA substrate are indicated.
PABP association with the cap resides in the second RRM
In an effort to define the regions of PABP required for the cap binding, fragments of PABP were tested for their ability to crosslink to cap-labeled RNA. The first 350 amino acids of the protein containing the four RRMs (PABP 1-4) were able to crosslink the cap of both adenylated and unadenylated RNA (Figure 6A, lanes 2 and 6), while the remaining C-terminal half of the protein (PABP CT) did not (lanes 4 and 8). A smaller N-terminal fragment containing the first two RRMs and linker region between the second and third RRMs (PABP 1-2), which are the essential poly(A)-binding domains of the protein (Nietfeld et al, 1990; Burd et al, 1991), did not crosslink to the unadenylated RNA (lane 3) but crosslinked only to the adenylated RNA (lane 7). These results further support a role of the poly(A) tail to recruit PABP to the RNA and facilitate association with the 5′ cap. We hypothesize that the truncated minimal protein is compromised in its ability to detect the cap structure relative to the full-length proteins and is therefore more dependent on the poly(A) tail to recruit and position the protein to the cap. The N-terminal truncation removing only the first RRM (PABP 2-CT) was still capable of crosslinking to the cap (Figure 6A, lane 9), while removal of the first two RRMs (PABP 3-CT) was not (lane 10). Collectively, these data indicate that PABP crosslinking to the cap is mediated through the second RRM (amino acids 99–190).
Figure 6.
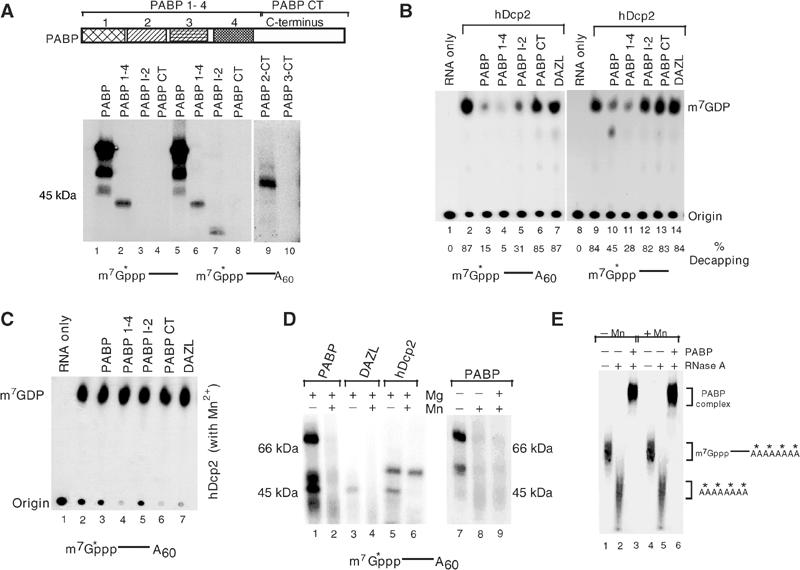
Crosslinking of PABP is confined to the second RRM and regulated by Mn2+. (A) The upper panel shows a schematic of PABP with its domain organization. The numbers denote the individual RRM, and RNA-binding domains and the carboxyl terminus are also indicated. The lower panel shows the crosslinking of 10 pmol PABP or PABP truncation proteins as indicated to unadenylated (lanes 1–4) or adenylated (lanes 5–10) RNA substrate 32P-labeled at the cap. (B) In vitro decapping assays were carried out with 10 pmol of hDcp2 and either cap-labeled adenylated (lanes 1–7) or unadenylated (lanes 8–14) RNA in the absence (lanes 2 and 9) or presence of PABP and its derived truncation proteins as indicated. The DAZL control protein is shown in lanes 7 and 14. Inhibition by PABP 1-2 was only seen with the adenylated RNA substrate (lane 4). The % decapping is shown below each respective lane. (C) PABP is unable to inhibit decapping in the presence of Mn2+. In vitro decapping assays were carried out as in B above, except 0.5 mM Mn2+ was included in the reaction buffer. (D) PABP crosslinking to the cap is inhibited by Mn2+. UV crosslinking of 5 pmol of the indicated proteins was carried out with 32P-cap-labeled pcP-A60 RNA in the presence or absence of the indicated cation. The 70 kDa His-PABP only crosslinked when Mn2+ was omitted from the buffer (lanes 1, 2 and 7–9). Crosslinking of the 48 kDa His-hDcp2 protein to the 5′ cap was unaffected by Mn2+ (lanes 5 and 6) while the 57 kDa GST-DAZL protein failed to crosslink (lanes 3 and 4). (E) Binding of PABP to the poly(A) tail is not affected by Mn2+. PABP (10 pmol) was incubated with pcP RNA containing a 32P-labeled poly(A) tail as described previously (Wang et al, 1999) in the presence or absence of Mn2+. Lanes 1 and 4 contain the RNA only while lanes 2 and 5 contain RNA treated with RNase. Binding of PABP to the poly(A) sequence is shown in lanes 3 and 6 where the complex is treated with RNase A to hydrolyze the RNA body and leave the 32P-labeled poly(A) tail intact. The binding of PABP to poly(A) tail is unaffected by the presence of Mn2+. The RNAs corresponding to the detected bands are shown on the right as is the PABP-labeled poly(A) complex. The asterisks denote the 32P labeling.
To determine whether a functional correlation existed between binding of the truncated PABP proteins and regulation of decapping, decapping assays were carried out with hDcp2 in the presence of the truncated PABP proteins. As expected, inhibition of decapping occurred when the wild type and N-terminal half of PABP proteins were used with adenylated as well as unadenylated RNA (Figure 6B, lanes 3, 4, 10 and 11), although the extent of inhibition of decapping was greater with the adenylated RNA. Consistent with the crosslinking data, the minimal PABP 1-2 protein had an inhibitory effect on the decapping of adenylated RNA (lane 5) but minimal effect on unadenylated RNA (lane 12). The C-terminal fragment of PABP or DAZL did not inhibit hDcp2 decapping activity (lanes 6, 7 and 13, 14). These results demonstrate a correlation between the crosslinking of PABP and its truncated derivatives to the cap structure and their ability to prevent hDcp2 decapping activity. Therefore, at least one function of PABP association with the cap of an mRNA could be to regulate decapping.
Association of PABP with the cap is regulated by Mn2+
We recently demonstrated that the hydrolysis activity of hDcp2 is significantly enhanced when Mn2+ was the divalent cation source (Piccirillo et al, 2003). Since all the reactions reported above were carried out exclusively with Mg2+, we determined what the consequence of Mn2+ would be on the PABP-mediated regulation of decapping. Contrary to the inhibition of decapping by PABP and its N-terminal fragments when Mn2+ was omitted from the reactions (Figure 6B), neither the wild type nor the truncated PABP proteins could inhibit decapping when Mn2+ was included in the decapping reactions (Figure 6C). The lack of competition for decapping directly correlated with the inability of PABP to bind the cap, as the crosslinking of PABP to the cap was significantly and reproducibly reduced in the presence of Mn2+ (Figure 6D, lane 2). However crosslinking of hDcp2 was detected with both divalent cations (lanes 5 and 6). The effect of Mn2+ on PABP crosslinking appears to be an active inhibition, since divalent cations were not required for the crosslinking to occur (lane 7) and addition of Mg2+ did not affect the interaction (lane 1). However, the inclusion of Mn2+ inhibited the crosslinking of PABP to the cap (lanes 2, 8 and 9).
Interestingly, the influence of Mn2+ on PABP is restricted to the cap-binding activity and not poly(A) tail-binding activity, as inclusion of Mn2+ in the binding buffer did not adversely affect the binding of PABP to the poly(A) tail (Figure 6E). These data demonstrate that Mn2+ can serve a reciprocal function on hDcp2 and PABP. The presence of Mn2+ increases the ability of hDcp2 to hydrolyze the cap (Piccirillo et al, 2003) and simultaneously specifically reduces the ability of PABP to associate with and protect the cap.
PABP can simultaneously bind the cap and poly(A) sequence in vitro
The enhanced crosslinking of PABP to the cap in the presence of the poly(A) tail raises the interesting possibility that PABP might be able to bind the poly(A) tail and cap simultaneously. To test this possibility, PABP was crosslinked to the cap of the cap-labeled pcP-A60 RNA as shown in Figure 2 to trap PABP onto the cap structure. The RNA was subsequently degraded with micrococcal nuclease and the cap-bound protein was incubated with either poly(A)- or poly(C)-agarose beads. As shown in Figure 7, the cap-crosslinked (and 32P-labeled) PABP efficiently bound to poly(A)- (lane 1) but not poly(C)-agarose beads (lane 2). Similar results were obtained with the PABP 1-2 minimal protein (lanes 3 and 4). These data demonstrate that PABP has the capacity to associate with the poly(A) tail and the 5′ cap simultaneously. It also demonstrates that the association of PABP with the labeled cap and poly(A) sequence was not a function of protein–protein interaction, which requires the C-terminus (Kuhn and Pieler, 1996).
Figure 7.
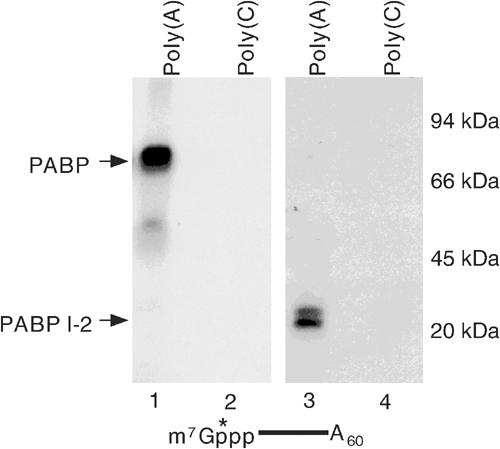
Simultaneous binding of PABP to the cap and poly(A) sequence. A 10 pmol portion of PABP and PABP 1-2 crosslinked to 32P-cap-labeled pcP-A60 RNA as described in Figure 2A, and bound after RNA degradation to either poly(A)-agarose or poly(C)-agarose matrix. Bound protein was eluted and resolved by SDS–PAGE and detected by autoradiography. Migration of PABP and PABP 1-2 is shown on the left.
Discussion
We recently demonstrated that the hDcp2 decapping activity can be negatively regulated through the poly(A) tail (Wang et al, 2002). In this report, we demonstrate that the inhibition of decapping can be directly mediated through PABP. The mechanism by which PABP can inhibit decapping was demonstrated to be through the direct and specific association of PABP with the 5′ end of capped RNA. The cap association of PABP is facilitated by the poly(A) tail (Figures 2 and 6) and mediated by the second RRM RNA-binding domain (Figure 6), which is essential for the association with the cap. The inhibition of decapping by PABP is comparable to the inhibition observed by the eIF4E cap-binding protein (Figure 1B). However, unlike the eIF4E inhibition of decapping that can be reversed upon the addition of cap analog to sequester eIF4E, PABP-mediated inhibition was resistant to cap analog competition. These findings are consistent with the endogenous hDcp2 decapping we previously reported, which was dependent on the poly(A) tail and resistant to cap analog competition (Wang et al, 2002). Collectively, our data demonstrate that PABP singularly can be a critical mediator of deadenylation-dependent decapping.
Binding of PABP to the 5′ end of capped RNA was shown by three independent assays: UV crosslinking, electrophoretic mobility shift and filter binding. Curiously, PABP can only associate with the cap within the context of RNA. Unlike eIF4E, which can bind both the cap structure and capped RNA, PABP was only detected to bind the cap when it was linked to an RNA moiety and failed to associate stably with either the cap structure alone or to the 5′ end of an RNA lacking the cap (Figure 4B and C). Similarly, the ability of PABP to inhibit decapping or bind capped RNA was not inhibited by cap analog competition (Figures 1A and 2A, respectively). The requirement of a dual cap and RNA substrate is not unique to PABP and is analogous to the cap hydrolysis property of the hDcp2 decapping enzyme. hDcp2 also does not bind the cap analog (Piccirillo et al, 2003), cannot hydrolyze the cap analog and is not competed by cap analog (Van Dijk et al, 2002; Wang et al, 2002; Piccirillo et al, 2003) even though it is the decapping enzyme. These data indicate that similar to hDcp2, PABP association with the 5′ cap is a function of a dual binding requirement for the cap and RNA.
PABP binding to the cap is accentuated by the presence of the poly(A) tail. The data presented in this report are consistent with the poly(A) tail serving to recruit PABP onto the cap as is evident by the crosslinking of the minimal PABP truncation protein (PABP 1-2) only to the adenylated and not unadenylated RNA (Figure 6A). Furthermore, the simultaneous association of the minimal PABP 1-2 with the 5′ cap and poly(A) sequences (Figure 7) implies a deposition of PABP onto the cap following poly(A) tail binding. Our data suggest that at least one function of a direct association of PABP with the cap could be to regulate decapping. Whereas the primary function of PABP is to bind the poly(A) tail and protect the 3′ end of an mRNA, an alternative function would be to bind the 5′ cap and protect it from hydrolysis. The apparent dissociation constant of PABP for poly(A) sequence in our assay conditions was found to be five-fold lower compared to binding of PABP to the 5′ cap of an RNA, (Table I) which further supports this premise.
An intriguing property of PABP is its capacity to bind simultaneously both a poly(A) sequence and the 5′ end of a capped RNA in trans (Figure 7). This in vitro demonstration implies that PABP can singularly juxtapose the two ends of an mRNA and protect the mRNA from both deadenylation and decapping. However, whether this can occur in cells remains to be determined and future studies will begin addressing the functional link between PABP cap binding and its correlation to mRNA decapping in cells.
The inability of recombinant PABP to displace recombinant eIF4E from the cap (Figure 5 and data not shown) indicates that association of PABP with the cap occurs at some point following the removal of eIF4E. This is further substantiated by the ability of recombinant PABP to associate with the cap in the presence of cellular extract upon the inclusion of cap analog competitor to sequester eIF4E. The recent demonstration that decapping of an mRNA can occur in distinct foci within the cytoplasm of yeast (Sheth and Parker, 2003) and the presence of similar foci in mammalian cells (Ingelfinger et al, 2002; Van Dijk et al, 2002) suggests that the mammalian foci might also be sites of decapping. This hypothesis implies that the mRNA would be transported to the foci subsequent to translational cessation and the cap remains intact during the transit. Following translation and deadenylation, when the eIF4E–eIF4G–PABP association is disrupted, PABP could gain assess to the cap. Although speculative, deposition of PABP onto the cap following deadenylation and exit from the polysomes might serve to protect the 5′ end of the mRNA during its transit to these foci. However, it should be noted that it is currently not clear how and when eIF4E is displaced from the cap post-translation, but removal of eIF4E from the cap of an RNA would ensure that a translation initiation complex does not reform on the RNA. Future experiments will focus on determining whether PABP could be involved in the transition between translationally active state of an mRNA, when it is indirectly associated with the cap through eIF4E and eIF4G, and the translationally inactive state, when it is directly associated with the cap following deadenylation.
Materials and methods
Construction of recombinant plasmids
Plasmids expressing an amino-terminal histidine-tagged recombinant protein of PABP (pET28-PABP), the N-terminus of PABP containing RRMs 1–4 (pET28-PABP NT) and the C-terminus of PABP (pET28-PABP CT) have previously been described (Wang et al, 1999). pET28-PABP 1-2 was constructed by digesting the pET28-PABP plasmid with EcoRI, which cleaves within the PABP cDNA and polylinker to remove amino acids 185–636. The pET28-PABP 2-CT plasmid expressing amino acids 99–636 was constructed by PCR with primers that introduce BamHI and XhoI sites (5′ GCCGTGGATCCATGGATCCATCACTTCGCAAA 3′ and 5′ CCCTGACTCGAGTTACATATGAAGAAGTTCTGA 3′) and inserted into the same sites of pET 28a (Invitrogen). The pET28-PABP 3-CT plasmid encoding amino acids 191–636 was similarly constructed with primers 5′ GCCGTGGATCCATGACCAATGTTTACATC 3′ and 5′ CCCTGACTCGAGTTACATATGAAGAAGTTCTGA 3′. The expression and purification of histidine-tagged or GST-tagged proteins were carried out according to the manufacturer (Novagen and Pharmacia, respectively). The pCMV-PABP plasmid expressing myc tag at the N-terminus of PABP was generated by PCR using 5′ GCCGTGGATCCATGAACCCCAGTGCCCCC 3′ and 5′ CCCTGACTCGAGTTACATATGAAGAAGTTCTGA 3′, which introduce BamHI and XhoI sites respectively, and inserted into the same sites of pCMV vector (Stratagene). The yeast Pab1p was PCR-amplified from reverse-transcribed yeast mRNA and was cloned into the BamHI and XhoI sites of pET 28a (5′ GCCGTGGATCCATGGCTGATATTACTGATAAG 3′ and 5′ CCCTGACTCGAGTTAAGCTTGCTCAGTTTGTTG 3′) to express histidine-tagged Pab1p. Likewise, the mouse PABP was PCR-amplified from murine erythroleukemia MEL cell reverse-transcribed RNA using the primers 5′ GCCGTGCCATGAACCCCAGCGCCCCCC3′ and 5′ CCCTGACTCGAGTTACATGTGAAGTAATTC 3′, and inserted into the BamHI and XhoI sites of pET 28a.
RNA production
Template to transcribe the unadenylated RNA substrate corresponding to the pcDNA3 polylinker containing a G16 track at the 3′ end (pcP) was generated by PCR using the SP6 promoter 5′ primer and C16T7 3′ primer (CCCCCCCCCCCCCCCCGT AATACGACTCACTATAGGG). The template for the adenylated RNA substrate containing 60 adenosine residues at the 3′ end (pcP-A60) was PCR-amplified from the pcP template using SP6 promoter 5′ primer and C16T60 3′ primer (16 cytosine followed by 60 thymine nucleotides). Both templates were transcribed with SP6 RNA polymerase. RNAs exclusively 32P-labeled at the 5′ cap were generated with the vaccinia virus capping enzyme and gel-purified prior to use as described previously (Wang et al, 1999) where the label is on the first phosphate following the methylated guanosine (m7G*pppN). 32P-labeled cap structure lacking an RNA was generated by hydrolysis of 32P-cap-labeled RNA with 1 U of Nuclease P1 (Roche) for 1 h at 37°C as described (Wang and Kiledjian, 2001). Nuclease P1 cleaves the phosphodiester linkages within the RNA and leaves the triphosphate cap structure intact. The samples were extracted once with an equal mixture of phenol:chloroform (1:1) and twice with chloroform, and the resulting supernatant containing the labeled cap analog was used as substrate for UV crosslinking assays.
UV crosslinking
UV crosslinking was carried out in a 20 μl reaction volume with 32P-cap-labeled RNA (10 pmol was used in each reaction unless otherwise stated) and incubated with PABP for 15 min at room temperature in Eppendorf caps in IVDA buffer (10 mM Tris (7.5), 100 mM KAc, 2 mM MgAc, 1 mM DTT, 10 mM creatine phosphate, 1 mM ATP and 0.1 mM spermine). The samples were subsequently transferred onto ice and covalently crosslinked by UV irradiation for 10 min with a 15 W germicidal lamp. Following UV crosslinking, the RNA body was cleaved with RNase buffer (10 mM Tris (pH 7.5), 400 U/ml micrococcal nuclease, 1 mM CaCl2, 1% aprotinin, 2 μg/ml leupeptin/pepstatin, 100 mM PMSF, 0.1 mg/ml RNase A and RNase U2) for 30 min at 37°C. The proteins were resolved by a 12.5% SDS–PAGE and visualized by autoradiography.
Immunoprecipitation
293T cells were transfected with 10 μg of pCMV-PABP plasmid using Lipofectamine (Gibco BRL). The cells were harvested after 24 h, extract was prepared and immunoprecipitated with the α-myc (9E10) and SP2/0 antibodies as described (Ishigaki et al, 2001). The incubation was extended for additional 10 min in 1 mg/ml heparin at 4°C and washed five times with PBS containing 0.1% NP-40. The bound protein was eluted in SDS sample buffer, resolved on 12.5% SDS–PAGE and visualized by autoradiography.
In vitro RNA decapping assay
In vitro RNA decapping assays were carried out by incubating cap-labeled pcP and pcP-A60 RNA with recombinant proteins at room temperature for 10 min in IVDA buffer, followed by the addition of 10 pmol of hDcp2 and incubation at 37°C for 20 min. Following decapping, 5 μl aliquots were spotted onto polyethyleneimine-TLC plates (Sigma) and developed in 0.75 M LiCl. Cold standards and 32Pi were visualized by UV shadowing or autoradiography, respectively.
Poly(A)-agarose bead binding
In all, 10 pmol of PABP or PABP 1-2 was crosslinked to cap-labeled pcP A60 RNA and RNase treated as described above. The samples were subsequently bound to poly(A)- or poly(C)-agarose beads in RNA binding buffer (RBB; 10 mM Tris–HCl (7.5), 1.5 mM MgCl2, 0.5 mM DTT and 0.5% Triton X-100) containing 500 mM KCl and incubated at 4°C for 1 h with end-over-end rotation. The beads were subsequently washed once in RBB containing 750 mM KCl and 1 mg/ml heparin for 10 min at 4°C, followed by two washes in RBB with 500 mM KCl followed by a PBS wash. Bound proteins were eluted by boiling in SDS sample buffer and resolved and detected as above.
Electrophoretic mobility shift assays
Gel shift assays were carried out in a 20 μl reaction volume at room temperature for 30 min in RBB (150 mM KCl, 100 mM Tris–HCl (pH 7.5), 1.5 mM MgCl2 and 5 mM DTT) containing capped (32P-5′-cap-labeled) or uncapped (32P-5′-triophosphate-labeled) adenylated RNA and the indicated amounts of PABP. The RNA was subsequently cleaved with 50 ng of RNase A for 10 min followed by an additional 10 min incubation with 1 mg/ml heparin. The resulting protein–cap complexes were resolved on a 5% native polyacrylamide gel and visualized by autoradiography.
Filter binding assays
Filter binding assays were carried out as described for the electrophoretic mobility shift analysis with a limiting concentration of 32P-labeled polyadenylated pcP-A60 RNA (0.1 pmol; ∼10 000 cpm) and an increasing concentration of recombinant histidine-tagged PABP. The polyadenylated RNA either contained a 32P-labeled 5′ cap or lacked a cap (32P-5′-triphosphate-labeled). Following a 30 min incubation at room temperature, the reaction was treated with RNase A to separate proteins that were bound to the labeled 5′ end from protein bound to other regions of the RNA, and RNA–protein complexes were captured by retention on 0.2 μm Millipore nitrocellulose filters prewashed with RBB. The unbound RNA was washed twice with 2 ml ice-cold RBB and the filters dried. The amount of isotope bound to the filters, which corresponds to the retained PABP–cap complex, was determined by a liquid scintillation counter and the values were corrected by subtraction of background counts obtained with RNA filtered in the absence of protein. The values for the bound RNA–protein complex were plotted relative to protein concentration, and apparent dissociation constants were determined as the concentration of protein at which 50% of RNA was bound (Wilson and Brewer, 1999). An average of three independent experiments is reported. The dissociation constant of recombinant PABP for poly(A) sequences was similarly determined except that the poly(A) substrate was generated with the bovine poly(A) polymerase using 32P-labeled αATP to polyadenylate the unlabeled pcP RNA as described previously (Wang et al, 1999). Following adenylation, the RNA body was digested with RNase A as above and the resulting uniformly labeled poly(A) sequence (60 nucleotides average length) was used as the substrate.
Acknowledgments
We thank Z Wang and AC Schmid for valuable input and expertise at the onset of this study. We are grateful to AC Gingras and N Sonenberg for the GST-eIF4E plasmid, J-Y Lu and R Schneider for the eIF4G plasmid and AB Shyu for the HuR plasmid. We also thank A Shatkin, R Parker and members of the Kiledjian lab for helpful discussions and critical reading of the manuscript. This work was supported by funds from the NIH, DK51611 to MK.
References
- Anderson JSJ, Parker RP (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 [DOI] [PubMed] [Google Scholar]
- Bernstein P, Ross J (1989) Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci 14: 373–377 [DOI] [PubMed] [Google Scholar]
- Brewer G (1998) Characterization of c-myc 3′ to 5′ mRNA decay activities in an in vitro system. J Biol Chem 273: 34770–34774 [DOI] [PubMed] [Google Scholar]
- Burd CG, Matunis EL, Dreyfuss G (1991) The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol 11: 3419–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Parker R (1995) Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev 9: 2421–2432 [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R (1994) Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci 19: 336–340 [DOI] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845 [DOI] [PubMed] [Google Scholar]
- Dunckley T, Parker R (1999) The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Wilusz J (1999) 3′-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′ → 5′ exonuclease in vitro. Nucleic Acids Res 27: 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange T, de Sa CM, Oddos J, Pictet R (1987) Human mRNA polyadenylate binding protein: evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res 15: 4771–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Stevens A (1993) Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T (2002) The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y, Li X, Serin G, Maquat LE (2001) Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617 [DOI] [PubMed] [Google Scholar]
- Jacobs JS, Anderson AR, Parker RP (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW (1996) Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem 65: 693–739 [DOI] [PubMed] [Google Scholar]
- Jiao X, Trifillis P, Kiledjian M (2002) Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod 66: 475–485 [DOI] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G (1992) Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J 11: 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Wang X, Liebhaber SA (1995) Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J 14: 4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Pieler T (1996) Xenopus poly(A) binding protein: functional domains in RNA binding and protein–protein interaction. J Mol Biol 256: 20–30 [DOI] [PubMed] [Google Scholar]
- Liu H, Rodgers ND, Jiao X, Kiledjian M (2002) The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 21: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J (2002) Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WJ, Chung S, Furneaux H (1997) The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res 25: 3564–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol 15: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J (2002) The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J 21: 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietfeld W, Mentzel H, Pieler T (1990) The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J 9: 3699–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo C, Khanna R, Kiledjian M (2003) Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9: 1138–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CV, Vilela C, Berthelot K, McCarthy JE (2002) Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J Mol Biol 318: 951–962 [DOI] [PubMed] [Google Scholar]
- Rodgers ND, Wang Z, Kiledjian M (2002) Regulated alpha-globin mRNA decay is a cytoplasmic event proceeding through 3′-to-5′ exosome-dependent decapping. RNA 8: 1526–1537 [PMC free article] [PubMed] [Google Scholar]
- Ross J, Kobs G (1986) H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3′ terminus and proceeds 3′ to 5′. J Mol Biol 188: 579–593 [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Parker R (1999) Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 19: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R (2000) mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R (2003) Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Le Hir H, Seraphin B (2003) DcpS can act in the 5′–3′ mRNA decay pathway in addition to the 3′–5′ pathway. Proc Natl Acad Sci USA 100: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M (1999) An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19: 4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M (2002) The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M (2001) Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762 [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140 [DOI] [PubMed] [Google Scholar]
- Wilson GM, Brewer G (1999) Identification and characterization of proteins binding A+U-rich elements. Methods 17: 74–83 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Gao M, Jones CL, Wilusz J, Peltz SW (2001a) Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA 7: 1416–1424 [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW (2001b) The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2: 237–246 [DOI] [PubMed] [Google Scholar]


