Abstract
There are eleven zinc dependent histone deacetylases (HDAC) in humans which have histones and many non-histone substrates. The substrates of these enzymes include proteins that have a role in regulation of gene expression, cell proliferation, cell migration, cell death, immune pathways and angiogenesis. Inhibitors of HDACs (HDACi) have been developed which alter the structure and function of these proteins, causing molecular and cellular changes that induce transformed cell death. The HDACi are being developed as anti-cancer drugs and have therapeutic potential for many non-oncologic diseases.
1. Introduction
The base sequence of DNA provides the genetic code for proteins. The regulation of expression of genes is largely determined by the structure of the chromatin proteins associated with the DNA. This is referred to as epigenetic gene regulation- which is regulation of gene expression without changes in DNA sequence (1). DNA is packaged in chromatin which is structurally complex and dynamic, consisting of DNA, histones, and non-histones proteins. Nucleosomes are repeating units in chromatin composed of approximately 146 base pairs of two loops of DNA wrapped around an octamer core of pairs of histone H4, H3, H2A, and H2B. Histone amino acid tails are subject to post-translational modification by acetylation of lysines, methylation of lysines and arginines, phosphorylation of serines, ubiquitination of lysines, sumoylation, proline isomerization, and ADP-ribosylation. Among enzymes involved in chromatin protein structural modification are histone deacetylases (HDACs) and histone acetyltransferases (HATs), which determine the acetylation of histones and other proteins. Lysine acetylation catalyzed by HATs neutralizes the positive charge on histone tails and allows the negatively charged DNA to assume a transcription competent confirmation. Acetyl groups are removed by HDACs, allowing interaction between negatively charged DNA and positively charged histone proteins, which can result in heterochromatin and transcriptional silencing of genes. Specific patterns of acetylation and methylation of lysines of the histones have been identified which appear to serve as a pattern for recruitment of protein complexes regulating gene expression, DNA replication and DNA stability (2, 3). In addition, to histones, HDACs and HATs have many non-histone protein substrates that have a role in regulating gene expression, cell proliferation, cell migration, and cell death (3-7).
This review focuses on the molecular and cellular effects of HDACs and HDACi of zinc dependent HDACs and the implication of these effects for therapeutic use, primarily as anti- cancer agents. A number of recent papers have reviewed the current status of clinical trials with HDAC inhibitors as monotherapy and combination for cancers (4, 8-12).
2. Histone Deacetylases (HDACs)
Eighteen HDACs have been identified in mammals which are classified based on their homology to yeast proteins: eleven of these HDACs are zinc dependent enzymes (Table 1). Class I, HDAC 1, 2, 3, and 8 have homology to yeast RPD3; class IIa, HDACs 4, 5, 7, and 9 have homology to yeast HDA1; class IIb, HDACs 6 and 10 have two catalytic sites, and class IV, HDAC11 has conserved residues shared with both class I and class II deacetylases (Table 1) (3, 7). Class III HDACs include sirtuins 1 through 7, which have homology to yeast Sir2 and have an absolute requirement for NAD+. Phylogenetic analysis indicates that the evolution of HDACs preceded that of histones (13), consistent with the fact that HDACs have many non-histone protein substrates (Table 2). Indeed, class IIa and IIb HDACs do not have histones as primary substrates in vivo and are more properly designated lysine deacetylases (4, 14). Analyses of lysine acetylation targets found 3600 acetylated lysines in 1750 proteins (15). Inhibition of HDACs with vorinostat (suberoylanilide hydroxamic acid, SAHA, see section 4 below) altered only about 10% of these acetylation sites. These sites were found on proteins that have a role in gene expression, RNA signaling, DNA damage repair, cell cycle progression, nuclear transport, cytoskeleton function, protein chaperone, and ribosome formation and function (14).
Table 1. Zinc Dependent Histone Deacetylases.
| HDAC | Localization | Size (AA) | Chromosomal Site | Tissue Distribution | Biologic Functions* |
|---|---|---|---|---|---|
| Class I | |||||
| HDAC1 | Nucleus | 483 | 1p34.1 | Ubiquitous | Cell survival and proliferation |
| HDAC2 | Nucleus | 488 | 6p21 | Ubiquitous | Redundant function with HDACi |
| HDAC3 | Nucleus | 423 | 5q31 | Ubiquitous | Cell survival and proliferation |
| HDAC8 | Nucleus | 377 | Xq13 | Ubiquitous | Unknown |
| Class IIa | |||||
| HDAC4 | Nuc/Cyt | 1084 | 2q372 | H, SM, B | Chondrocyte and osteocyte development differentiation |
| HDAC5 | Nuc/Cyt | 1122 | 17q21 | H, SM, B | Myocardium, Endothelial |
| HDAC7 | Nuc/Cyt | 855 | 12q13 | H, PL, PA, SM | Thymocyte differentiation |
| HDAC9 | Nuc/Cyt | 1011 | 7p21-p15 | SM, B | Myocardium and skeletal muscle |
| Class IIb | |||||
| HDAC6 | Mainly Cyt | 1215 | Xp11.22-33 | H, L, K, PA | Targets tubulin, chaperones. etc. |
| HDAC10 | Mainly Cyt | 669 | 22q13.31-33 | L,S,K | Unknown |
| Class IV | |||||
| HDAC11 | Nuc/Cyt | 347 | 3p25.2 | B, H, SM, K | Immunomodulators |
SM-Skeletal Muscle; B-Brain; PL-Platelet; L-Liver; K- Kidney; S-Spleen; H- Heart; PA- Pancreas
The biological functions of the different HDACs is not completely understood. See text for details and references.
Table 2. Non-Histone Protein HDAC Substrates (Partial List).
| Pathway | Protein |
|---|---|
| HDACs | HDAC1 and ? other HDACs |
| Cell Motility | α-tubulin, cortactin |
| Chaperones | HSP90, HSP70 |
| Gene transcription factors PLAG-1 and 2, c-myc, BCL-2, Rb, And co-regulators | P53, p73, GATA-1, 2 and 3, MyoD, E2F1, 2, and 3, PGC-α |
| Chromatin structure | HMG-A1, B1, B2, N1 and 2, SRY, Histones |
| Nuclear receptors (DNA) estrogen receptor α | Androgen receptor, glucocorticoid receptor, |
| Signaling mediators | SAT3, Smad7, β-Catenin |
| DNA repair | Ku70, Ku86, WRN |
| Redox | Periredoxins |
| Nuclear import | Importin-α7 |
| Inflammation mediator | HMGB1 |
| Metabolic | GLUT1 mediated glucose transporter, HXK1 |
Class I HDACs are primarily localized in the nucleus. HDAC3 can shuttle between the nucleus and the cytoplasm. Class IIa HDACs are primarily localized in the cytoplasm but shuttle between the nucleus and the cytoplasm. Class IIb HDACs, HDAC6, is a cytoplasmic protein. HDACs do not bind directly to DNA and are recruited to target genes via their association with transcriptional activators and repressors, incorporated into large multiprotein transcription complexes (3, 4, 14, 16).
Class I HDACs are expressed ubiquitously. They have a simpler structure than class II HDACs, including a conserved deacetylase domain and relatively short amino and carboxy terminal extensions (Table 1). Class I HDACs are found in protein complexes with transcription factors and co-repressors and play a role in regulation of gene expression (16). Knock-out studies indicate that the Class I HDACs have a role in cell survival and proliferation. HDAC1 and HDAC2 have redundant and essential roles in tumor cell survival and neural precursor development. Deletion of both HDACs, but not either alone, causes cancer cell death and neural precursor maturation defects (17). HDAC1 can increase resistance of cancer cells to oxidative stress by blocking thioredoxin binding protein-2 (TBP-2) expression (18). HDAC2 is a regulator of chromatin compaction status and its down-regulation or inhibition causes chromatin decondensation and sensitization to DNA targeted anti-cancer drugs (19). Class II HDACs have more tissue specific regulatory functions (Table 1) (4, 8, 14). HDAC1 knock-out is an embryonic lethal by day 9.5 and results in proliferation defects of embryonic stem cells (20). Mice lacking HDAC2 survive until their perinatal period when they die of multiple cardiac defects. HDAC3 deletion causes early embryonic lethality. Inactivation of HDAC3 was associated with cessation in cell cycle progression, DNA damage, and impaired repair and apoptosis (21).
Class IIa HDACs are larger molecules than class I HDACs and have conserved binding sites for transcription factors and the chaperon proteins, 14-3-3, which are involved in regulation of the shuttling of these enzymes between the nucleus and cytoplasm. Mice lacking HDAC4 expression have premature ossification of developing bones while overexpression HDAC4 inhibits chondrocyte and osteocyte differentiation (14). HDAC4 has a role in regulation in skeletalgenesis and survival of retinal neurons (22). HDAC4 inhibits nerve reinnervation by blocking expression of fibroblast growth factor protein 1 (23). The expression of HDAC4 is repressed by micro-RNA, mi-R-206, facilitating reinnervation. HDAC1 and 2 redundantly regulates cardiac morphogenesis, growth and contractility (14). HDAC5 knock-down mice have large hearts. HDAC5 and HDAC9 are involved in the development of myocardium and skeletal muscle. HDAC7 is involved in the regulation of vascular endothelial development and vascular integrity (4, 8, 24).
HDAC6 is unique among the eleven Zn dependent HDACs in having two catalytic sites and an ubiquitin binding site. Specific substrates of HDAC6 have been identified, including α-tubulin, cortacin, transmembrane proteins, such as, IFNαR, HSP90 and other chaperone proteins and peroxiredoxins (25-28). The ubiquitin site toward the c-terminal end of HDAC6 plays a critical role in aggresome formation in the pathway of proteolysis of misfolded proteins.
Little is known about the function of HDAC10. HDAC11 negatively regulates expression of the gene encoding interleukin-10 in antigen presenting cells which induce T-cell activation as well as T-cell tolerance (29). These findings suggest that HDAC11 has molecular targets that influence immune function.
Despite the ubiquitous distribution of HDACs in chromatin, inhibition of these HDACs appears to alter the transcription of a relatively small proportion of expressed genes (2-10%) in transformed cells (30-32).
The regulation of HDAC activity can occur at multiple levels including protein-protein interactions, post translational modification (sumoylation, phosphorylation, proteolysis, subcellular localization) and by metabolic cofactors (33, 34). For example, HDAC1 promoter is inducible by interleukin-2. HDAC4 promoter is regulated in part by P1/SP3 transcription factor. Phosphorylation and subsequent association with 14-3-3 regulates subcellular localization of HDAC4, HDAC5, HDAC7, and HDAC9.
The crystalline structure of the catalytic site of a histone deacetylase-like protein was solved with its binding of inhibitors, trichostatin A (TSA) and SAHA (35) (Figure 1). More recently, the crystal structure analysis of human HDACs4, 7, and 8 have been solved (36-39). Studies of the catalytic domain of HDAC4 and 7 suggest a molecular basis for the relatively low enzymatic activity of these HDACs.
Figure 1.
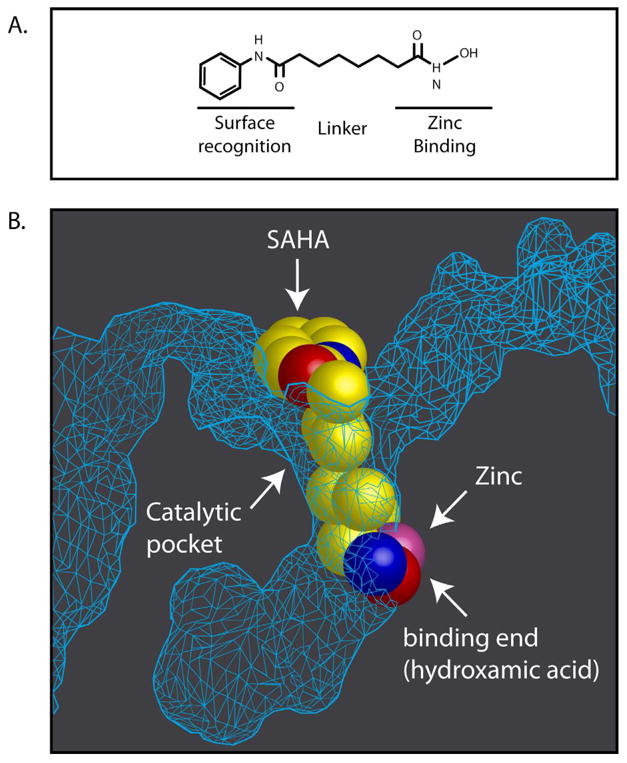
A: Structural features of hydroxamic acid based inhibitors of HDACs (see text for details). B: Schematic representation of the crystal structure of the histone deacetylase-like protein with SAHA (vorinostat) that inserts into the pocket-like catalytic site of the enzyme. At the base of the catalytic pocket is a zinc molecule with which the hydroxamic moiety of SAHA binds (after 35).
3. HDACs and Cancers
Structural mutations in HDACs associated with cancers appear to be rare, while altered expression and aberrant recruitment of different HDACs have been reported in many human cancers (4-8, 40-49). By comparison, structural alterations in HAT genes in cancers include point mutations, deletions and translocations as well as altered expression. Systematic analysis of the expression levels of the Zn dependent HDACs in cultured cancer cell lines, as well as, primary cultures of human cancer cells and various human tumor biopsy samples frequently found higher levels of expression than in corresponding normal tissue (40, 46). Over-expression of class I HDACs has been reported in esophageal and prostate cancers, as well as, other cancers. High levels of HDAC1 has been reported in non-small cell lung cancer, gastrointestinal cancers, and oral cancers and HDAC2 and HDAC3 in colon cancers. Analysis of the expression of histone deacetylases in lymphomas found that the most frequently altered HDAC was HDAC6, which was weakly expressed or undetected in 9 out of 14 lymphoid cell lines and in 83 of 89 primary lymphoma tissue specimens (47). HDAC5 and HDAC10 have been reported decreased in expression in lung cancer associated with poor prognosis. HDAC6 has been reported to be over-expressed in cutaneous T-cell lymphoma (CTCL) and peripheral T-cell lymphoma (PTCL). HDAC8 expression correlates with poor outcome in neuroblastoma and inhibition of HDAC8 induce differentiation of the transformed cells (48).
Fusion proteins involving chromosomal translocation have been identified in acute myelocytic leukemia (AML) and acute lymphoblastic leukemia (ALL) (42,43). The chromosomal translocation results in a fusion protein (MLL-CBP) consisting of the CBP and mixed lineage protein MLL. Aberrant recruitment of HDACs to the transcription factors that involve oncogenetic DNA binding fusion proteins resulting from chromosomal translocations or expression of repressive transcription factors include oncogenetic PMA-RARα or PLZF-RARα and AML1-ETO fusion protein which are found in acute promyelocytic (APL) and AML respectively.
A mutation of HDAC2 has been reported in colon cancer and endometrial cancer cell lines (50). HDAC4 mutations have been identified in breast cancer in a large scale sequencing study of breast and rectal colon cancers (51).
4. Histone Deacetylase Inhibitors
HDACi, particularly of the hydroxamic class, generally have common structural characteristics: zinc-binding moiety (ZBM) in the catalytic pocket, opposite capping group and a straight chain alkyl, vinyl, or aryl linker connecting the two (Figure 1). These functional groups interact with three relatively conserved regions of the active sites of HDACs (35-39, 52). The Zn facilitates hydrolysis of the acetyl-lysine bond and is at the bottom of a narrow catalytic pocket. A hydrophobic chain with six carbons is generally the optimal link of the ZBM and the opposite capping group. The capping group about the catalytic site opening is generally a hydrophobic structure which interacts with the rim amino acids. The amino acid sequence of the rim surrounding the catalytic site of the different HDACs has greater sequence diversity compared to the other domains and may have the most potential to be manipulated to develop selective HDACi.
During the past several years, numerous novel HDACi have been synthesized and are in various stages of development as potential drugs (53-57). These HDACi have been generated by modifying the ZBM, linker and/or capping group.
HDACi include hydroxamic acids, cyclic peptides, electrophilic ketones, short chain fatty acids, benzamides, boronic acid based compounds, benzofuranone and sulfonamide containing molecules and α/β peptide structures. Various chemical modifications have incorporated solubilizing functional groups which may improve pharmokinetic properties. There is a considerable effort to develop isoform selective deacetylase inhibitors, such as tubacin, which selectively inhibits HDAC6 (25), and PC-34051, a selective HDAC8 inhibitor. A new concept in HDACi structures is combining inhibition of protein kinases and HDACs in one molecule (58). The findings that structurally diverse compounds are effective inhibitors of HDACs suggest that the mechanism of action of these compounds may involve not only interaction with the catalytic site but also the surface of the enzyme which interact with other proteins independent of the deacetylase activity.
5. Biologic Effects of Histone Deacetylase Inhibitors
HDACi can induce transformed cell death by one or more pathways. HDACi can induce transformed cell growth arrest, cell death and inhibition of angiogenesis by altering the structure of many proteins targets of HDACs (Table 2). Normal cells are relatively resistant to HDACi- induced cell death (59, 60). The pathway leading to cell death of transformed cells induced by HDACi depends, in part, on the HDACi, concentration and time of exposure, and the transformed cell context.
A. DNA Damage
HDACi have not been demonstrated to directly cause mutations in DNA. Histone acetylation, which can be induced by HDACi, results in structural alterations in chromatin, which in turn, may expose portions of the DNA that may be normally protected from mutation by tightly packed heterochromatin (4-8, 61). HDACi can induce the accumulation of reactive oxygen species (ROS) resulting in DNA damage (59-62). Vorinostat induces thioredoxin binding protein (TBP-2) in certain transformed but not normal cells resulting in inactivating the reducing protein, thioredoxin (60).
HDACi induce the accumulation of the phosphorylated form of H2XA, a marker of DNA double strand breaks (DBS) in transformed cells (63). HDACi can down-regulate the expression of genes for DNA repair proteins involved in homologous recombination, including, RAD51, BRCA1, and BRCA2 (64), and in non-homologous DSB repair including Ku70, Ku86 and DNA- PKCs (64-67). The accumulation of DNA DSB is associated with altered gene expression and apoptotic cell death. Transformed cells may have many defects in pathways of repair of DNA damage and unlike normal cells, do not have the capacity to repair DNA damage (68, 69). The synergy of HDACi and DNA damaging agents such as cytotoxic drugs or radiation could result from the combined effects of HDACi in inhibiting DNA repair processes as well as activating intrinsic and extrinsic cell apoptotic death pathways.
B. Gene Expression
HDACi can alter gene transcription by inducing acetylation of histones, transcription factors and other proteins regulating gene expression (Table 2). Early differential display experiments with lymphoid cell lines cultured with TSA showed that the expression of only about 2% of expressed were altered either increased or decreased compared to untreated cells (70). More recent studies using cDNA arrays showed as many as 10%-20% of genes were altered in their expression in cell lines of leukemia, multiple myeloma, and carcinomas of colon, bladder, kidney, prostate and breast, cultured for up to 48 hrs with the HDACi, butyrate, entinostat, TSA, vorinostat or romidepsin (using 2 fold change)(30-32). The time in culture, concentration and particular HDACi used and the type of transformed cell appear to be variables that determine the number of genes altered in transcription. The number of genes whose expression is altered increased with time of culture and concentration of HDACi. Some changes in gene expression are probably direct effects of the HDACi, while many may be downstream effects. The patterns of alterations of gene expression are largely similar for different HDAC inhibitor, but there are differences induced by different agents in various transformed cells (32).
The cyclin dependent kinase (CDK) inhibitor p21 (WAF1/CIP1) is one of the most commonly induced genes by HDACi (71). HDACi induced expression of p21 is independent of p53 and correlates with an increase in the acetylation of histones associated with the p21 promoter region. In ARP-1 cells, vorinostat caused specific modification in the pattern of acetylation and methylation of lysines in histones H3 and H4 associated with the proximal promoter region of the p21 gene (72). Histone acetylation and methylation in the promoter region of the expressed p27 (KIP1) or the silent epsilon globin gene in HDACi cultured ARP-1 cells were not altered nor was the expression of these genes. Vorinostat caused a marked decrease in HDAC1 and Myc, and recruitment of RNA polymerase II, in the protein complex associated with the proximal promoter region of the p21 gene, with little detectable change in HDAC2, Brg1, GCN5, P300 or Sp1 proteins in the complex. These findings suggest that the selective alteration of transcription of a gene by HDACi may be determined by the specific composition and configuration of proteins in the transcription factor complex including the HDACs.
HDACi can inhibit gene expression mediated by STAT5 (73). HDACi can induce transcriptional repression of androgen receptor (AR) (74). HDACi can block AR mediated transcriptional activation of many genes. HDACi inhibits the assembly of coactivator/RNA polymerase II complex after AR binds to the enhancers of target genes. HDACi, such as SAHA, can alter the miRNA expression profile in transformed cells (75). These miRNAs have target genes related to angiogenesis, apoptosis, chromatin modification cell proliferation and differentiation.
C. Cell Cycle
HDACi can induce cell growth arrest in both normal and transformed cells. Vorinostat causes predominantly G1 arrest at low concentration and both G1 and G2/M arrests at higher concentration (71). G1 arrest is associated with induction of p21 which inhibits CDKs regulating G1 progression (CDK4/6) and G1/S transition (CDK2). Vorinostat induced p27 in leukemia cells K562 and LAMA-84 and breast cancer cells, MCF-7, and MDA-MB-231. In cells cultured with HDACi, the increase of CDK inhibitors and the decrease of cyclins may account for reduced CDK activity, causing dephosphorylation of Rb, blocking E2F activities in the transcription of genes for G1 progression and G1/S transition (76). HDACi can kill both proliferating and non-proliferating transformed cells (77). This is in contrast to the action of many chemotherapy drugs which are effective only on proliferating transformed cells.
D. Apoptosis
HDACi can cause death of transformed cells by inducing the extrinsic and/or intrinsic apoptotic pathways (4, 8, 10, 78-81). A number of downstream components, such as activation of caspase 3, are shared in the extrinsic and intrinsic pathways (82). The extrinsic apoptotic pathway is initiated by binding of death receptor, including Fas, tumor necrosis factor (TNF) receptor-1 (TNFR-1), TNK-related apoptosis-inducing ligand (TRAIL) receptor (DR-4 and -5), DR-3 (Apo3) and/or DR-6, to their ligands, causing activation of caspase-8 and -10. HDACi can up- regulate expression of both the death receptors and their ligands in vitro and in vivo in transformed cells, but not in normal cells. M-carboxycinnamic acid bishydroxamide (CBHA) induced Fas and Fas ligand in neuroblastoma cells (83). TNF was up-regulated by romidepsin in HL-60 and K562 cells, and C-FLIP, an inhibitor of the death receptor pathway, was down- regulated. Sequential treatment with vorinostat followed by TRAIL was shown to target multiple pathways in tumor progression, angiogenesis and metastasis. HDACi can enhance apoptosis through proteasome inhibition of TRAIL degradation, an affect which may be the basis of the synergistic apoptosis resulting from a combination of an HDACi with a proteasome inhibitor (84). Taken together, these studies indicate that the extrinsic apoptotic pathway can account for HDACi- induced cell death in many transformed cells. Combination therapy with factors inducing the extrinsic apoptotic pathway have a potential for effective therapeutic application.
Intrinsic apoptosis pathway is mediated by disruption of mitochondria with the release of mitochondrial intermembrane proteins, including cytochrome c, AIF and Smac, leading to activation of caspases (76, 78, 79, 81, 84-86). HDACi can induce the intrinsic apoptotic pathway by inactivation or suppression of anti-apoptotic proteins and activation of pro-apoptotic proteins. High levels of expression of Bcl-2 or Bcl-XL, which protect mitochondria, have been found in some transformed cells resistant to HDACi induced cell death (81). Inhibition of Bcl-2 by a chemical inhibitor, such as, HA14-1, can increase HDACi- induced cell death. HDACi can up- regulate pro-apoptotic proteins of Bcl-2 family, such as Bim, Bmf, Bax, Bak and Bik, decrease anti-apoptotic proteins of Bcl-2 family, such as Bcl-2, Bcl-XL, Bcl-w and Mcl-1 and suppress the expression of the pro-survival gene inhibitor of apoptosis, XIAP, and induce degradation of survivin.
The level of the pro- and anti-apoptotic proteins and the effects of HDACi vary dramatically in different tumor cells, even of the same clinical diagnosis, e.g. prostate (81).
E. Mitotic Disruptions
HDACi can cause aberrant acetylation of histones in heterochromatin and centromere domains with death of transformed cells (87-91). In culture with TSA, histones in newly synthesized chromatin remain acetylated and disrupt the structure and function of the centromere and the pericentric heterochromatin with loss of binding to heterochromatin binding proteins. Histone acetylation also blocks histone phosphorylation disrupting the function of mitotic spindle checkpoint proteins, such as BubR1, hBUB1, CENP-F and CENP-E, causing transient arrest at prometaphase and aberrant mitosis with chromosomal disruption resulting in cell death by either apoptosis or mitotic cell death.
F. Autophagic
The HDACi, vorinostat or butyrate, induced autophagic cell death with vacuoles in the cytoplasm of HeLa cells with Apaf-1 knockout or overexpression of Bcl-XL (92). A senescence phenotype with polyploidy was induced by vorinostat in HCT1165 colon cancer cells (87).
G. Reactive Oxidative Species and Redox Pathways
HDACi cause an accumulation of ROS in transformed cells but not in normal cells (59, 60, 93- 95). Increased cellular ROS can occur within 2 hrs of culture with HDACi, before disruption of mitochondria. Free radical scavengers such as N-acetylcysteine decrease HDACi- induced apoptosis and support the conclusion that the generation of ROS facilitates apoptotic transformed cell death. Inhibition of HDAC6 leads to acetylation of its target redox proteins, peroxiredoxin I and II, which neutralize H2O2 (26). The acetylated form of the redox proteins have increased activity in reducing H2O2.
Thioredoxin (Trx) is a hydrogen donor that is required for activation of several proteins, including, ribonucleotide reductases, essential for DNA synthesis, and transcription factors, e.g. NF-kB. Reduced Trx is an antioxidant scavenger of ROS (94). Vorinostat up-regulates the expression of TBP-2 (60, 95), which binds and inhibits reduced Trx activity (96), causing a down-regulation of Trx in transformed but not in normal cells. Trx is an inhibitor of apoptosis signal- regulating kinase 1 (ASK1). Inhibition of Trx by binding to TBP2 activates ASK1 which, in turn, promotes apoptosis by inducing SET1-JNK and MKK3/MKK6-p38 signaling cascades, and enhancing the expression of pro-apoptotic protein, Bim (97).
H. Inhibition of HDAC6 and Target Proteins
HDAC6 is primarily a cytoplasmic protein where it associates with non-histone substrates, such HSP90 and α-tubulin (16, 26-28, 98, 99). Overexpression of HDAC6 leads to deacetylation of α-tubulin and an increase of cell motility. HDAC6 can bind both mono and poly-ubiquitinated proteins and promotes its own ubiquitination. Specific inhibition of HDAC6 activity with tubacin or downregulation by siRNA, causes accumulation of acetylated α-tubulin, HSP90, peroxiredoxins and other client proteins. HSP90 acetylation causes loss of chaperone function and exposes its client proteins, such as pro-survival and pro-proliferation proteins, Akt, Bcr-Abl, c-Raf and ErbB2, to poly-ubiquitination and degradation via proteasome pathway (100-103). HSP90 chaperon function is also essential for the stability and function of proteins, such as, steroid hormone receptors and protein kinases involved in cell signaling pathways and cellular homeostasis. Recent studies have demonstrated both a direct physical interaction between HDAC6 and HSP90 and HDAC6 as a regulator of HSP90 activity through its deacetylation. Considering the number of client proteins of HSP90, many molecular alterations can be a result of HSP90 inactivation. HDAC6 can also bind directly to protein phosphatase (PP1) and cause simultaneous changes in cellular protein phosphorylation and acetylation, which contribute to the anti-tumor activity of HDACi. Therapeutic strategies combing HDAC6 inhibition with an HSP90 inhibitor have potential to be effective anti-cancer regimens.
HDAC6 is a component of the aggresome, a cellular structure that constitutes the major site of degradation for misfolded protein aggregates, both non-ubiquitinated and ubiquitinated misfolded proteins (102). Misfolded proteins are susceptible to forming cytotoxic aggregates that can interfere with normal cell function. Aggresome formation requires that microtubule network and microtubule- associated motor, dynein. HDAC6 acts as a bridge between the dynein and the ubiquitination process, directing the poly-ubiquitinated proteins to the aggresome. The BUZ domain of HDAC6 has high affinity for ubiquitin molecule and is involved in the transport of poly-ubiquitinated proteins (102, 103). Loss of HDAC6 function increases the sensitivity of transformed cells to misfolded protein stress induced by proteasome inhibitor. These findings have therapeutic implications for developing HDACi and proteasome inhibitors in combination treatment of certain malignancies.
I. Anti-Angiogenesis
HDACi can exert anti-cancer activity by inhibiting tumor angiogenesis (104). Solid tumors including breast, lung, and prostate cancers are frequently angiogenesis dependent. Tumor angiogenesis can be mediated by hypoxia secondary to tumor growth or by increased oncogenic signaling and, as a consequence, hypoxia inducible factor- 1α (HIF-1α) and its transcriptional target, vascular endothelial growth factor (VEGF), can be elevated. HDACi have been shown to inhibit angiogenesis via the suppression of HIF-1α and its target, VEGF, in animal models (104-107). Under normoxic conditions, HIF-1α binds to von Hippel-Lindau protein (pVHL) and is inactivated by ubiquitination-proteasome degradation. Hypoxic conditions can increase transcription of HDAC1, 2, and 3 in transformed cells causing decreased expression of pVHL, and, as a consequence, increased expression of HIF-1α which promotes angiogenesis, a sequence of events that may be blocked by HDACi. HDACi can also induce HIF-1α degradation in a VHL-independent mechanism. Class II HDACs, HDAC4 and HDAC6, physically associate with HIF-1α and their selective inhibition by siRNA induces HIF-1α degradation. HIF-1α binds to HSP90, disrupting the HSP90 chaperone function by acetylation, which exposes HIF-1α to proteasomal degradation. The anti-angiogenic effects of HDACi can contribute to anti-tumor activity and support the development of combination therapies of HDACi with VEGF inhibitors.
J. Metastasis
HDACi upregulates metastasis suppressor genes, e.g., kangai (KAII), Ras homologue genes, RhoB, reversion inducing cysteine rich protein with KAZAL motifs (RECK) and tissue inhibitor of metalloproteinases (TIMP-1) (108). Metastasis promoting genes can be down-regulated by HDACi- including genes for matrix metalloproteinases (MMPs), integrin-α5 and collagen proteins (109).
K. Metabolic
In addition to targeting gene expression and altering the structure and activity of proteins in the pro and anti-apoptotic pathways, in aggresome and protesome complexes, redox pathway and angiogenesis, HDACi target glucose transporter 1 (GLUT1 mediated glucose transport) and hexakinase I (HXK1) proteins, blocking enzymatic activity and, thereby inhibiting glucose utilization in transformed cells (110).
The present evidence, primarily based on studies with cells in culture, indicates that HDACs have multiple targets which are involved in almost every cellular pathway critical to cell survival, differentiation, proliferation, migration, and death (Table 2).
6. HDACi in Combination Therapy for Cancers
Numerous studies have examined the use of HDACi in combination with radiation; antimetabolites; such as, 5-flurodeoxyuridine and gemcitabine; antitubule agents such as, docetaxel, paclitaxel and epothilone B; topoisomerase, (Topo) II inhibitors doxorubicin, epirubicin, VP-16 (etopside) and ellipticine; DNA cross-linking agent, cisplatin; HSP90 antagonist, 17-ally-amino-demethoxy geldanamycin; and targeted agents, such as rituximab, trastuzumab, and EFGR inhibitor, erlotinib (4, 8-10, 12, 111-115). The synergistic effects may depend on the sequence of drug administration. HDACi also have been reported to have synergy with the transcription modulator, all-trans retinoic acid; DNA demethylating agent, 5- aza-2′deoxycytidine, and the Bcr-Abl kinase inhibitor, imatinib. Up-regulation of death receptors and/or reducing the inhibitory regulators of death receptor pathway by HDACi, sensitize tumor cells to TRAIL. HDACi achieve synergy with TRAIL by simultaneous activation of the intrinsic and the extrinsic apoptotic pathways without changing the expression of TRAIL receptors or the inhibitory protein c-FLIP. Many kinase inhibitors, including CDK inhibitor flavopyridol, phosphatidyl-inositol 3 kinase inhibitor, LY294002, FLT3 inhibitor, PKC412 and MEK1/2 inhibitor PD184352, potentiate the cell killing effect of HDACi. Blocking NF-KB activation by I Kappa B Alpha (IKBα) phosphorylation inhibitor, bay 11-7082, markedly increased HDACi-induced apoptosis.
Taken together, these findings indicate that combination therapies with HDACi is likely to be the best therapeutic strategy using these agents.
7. HDACi Development for Non-Cancer Therapies
HDACi have the potential for the treatment of various non-oncologic disease including auto- immune disorders, inflammation, and malaria (116- 119). HDACi may be useful in the treatment of central nervous system diseases (120-124). Vorinostat was shown to slow the progression of Huntington like syndrome in mice (123) and to increase expression of SNM protein in spinal muscular atrophy fibroblasts (122). HDAC2 modulates synaptic plasticity and long lasting changes of neural circuits and, in turn, plays a role in regulating learning and memory. These observations suggest that HDAC2 selective inhibition may be useful in memory impairment (124). HDACi have been reported to be potent inducers of γ-globin gene expression with therapeutic value in the treatment of sickle cell anemia (125). HDACi have anti-rheumatic activity in rodent models (126). HDACi can modulate stem cell survival and mobilization in in vitro studies (127). HDACi have been shown to decrease multilineage differentiation potential of human mesenchymal stem cells (128). HDACi can improve animal survival after hemorrhagic shock (129).
8. Clinical Development of HDACi as Anti-Cancer Drugs
A number of structurally different HDACi are in clinical trials either as monotherapy or in combination therapy for many different hematologic and solid tumors (Table 3). Four major classes of HDACi are currently in clinical trials, including aliphatic acids (butyrates, and valproic acid), hydroxmates (vorinostat, panobinostat, givinostat, belinostat, PCI24781), benzamides (entinostat and MGCD-103) and cyclic peptide (romidepsin). There are on-going clinical trials with HDACi in combination therapy with radiation, cytotoxic agents, and different targeted anti-cancer agents ([NCI website: CTEP clinical trials.gov], 4, 8, 11, 111-115, 130-139). These clinical trials include patients with cancer of lung, breast, pancreas, renal and bladder, melanoma, glioblastoma, leukemias, lymphomas, and multiple myeloma.
Table 3. Histone Deacetylase Inhibitors in Clinical Trials (Partial list*).
| Hydroxamates |
| Vorinostat (SAHA, Zolinza ™) |
| Panobinostat (LBH589) |
| Belinostat (PXD101) |
| Givinostat (ITF-2357) |
| PCI-24781 (CRA-024781) |
| JNJ-26481585 |
| SB-639 |
| Cyclic Peptide |
| Romidepsin (Depsipeptide, FK228) |
| Aliphatic Acids |
| Valproic Acid (Baceca, Savicol) |
| Phenylbutyrate (VP-101, El-532) |
| Pivanex (AN-9) |
| Benzamides |
| Entinostat (MS 275, SNDX 275) |
| MGCD0103 |
Review References cited in text
Vorinostat was the first of the HDACi approved by the US Food and Drug Administration (FDA) in 2006 for the clinical use in cancer patients, for the treatment of cutaneous T-cell lymphoma (CTCL) (52, 134). Vorinostat is being evaluated in phase II and phase III clinical trials as monotherapy and in combination with various anti-cancer agents for both hematologic and solid tumors (12), Ongoing clinical trials in combination therapy for vorinostat include azacitidine, decitabine, the proteasome inhibitor, bortezomib, and taxanes.
Panobinostat (LBH589) is more potent than vorinostat in pre-clinical models (111). It is in phase I and II clinical trials for hematologic and solid tumors as monotherapy and various combination therapy protocols, including with proteasome inhibitors, as well as, DNA methylase inhibitor, azacitidine.
Other hydroxamic acid-based HDACi in clinical trials include belinostat, givinostat, JNJ26481585 and 5B639. Each of these HDACi has shown anti-tumor activity, including stable disease, partial response and in a few cases, complete responses of transient duration at doses generally well tolerated by the patients. Adverse effects observed with hydroxamate HDACi include fatigue, nausea, dehydration, diarrhea, and thrombocytopenia. With certain hydroxamic acid-based HDACi, electrocardiogram changes occurred. These side effects have been reversible upon cessation of the administration of the drug.
Two benzamide HDACi in clinical trials are entinostat (entinostat, Sndx-275) and MGCD103. These agents are in trials as monotherapy and in combination with other anti-cancer drugs.
Romidepsin, a cyclic peptide HDACi, is in clinical trials as monotherapy, as well as, in combination in phase I and phase II in patients with high risk myelodysplastic syndrome and acute myelogenous leukemia and certain solid tumors. Romidepsin has been approved by the FDA in 2008 for the therapy of CTCL (113, 139).
9. Biomarkers for Predicting Clinical Response to HDACi
The identification and development of assays for biomarkers that can predict resistance or sensitivity to HDACi clearly have significant potential for clinical utility. The accumulation of acetylated histones in peripheral mononuclear cells is a marker for the biological activity of HDACi (140) but does not correlate with therapeutic response.
A biomarker that may inform on the therapeutic response to the HDACi is HR23B, a protein which shuttles ubiquitinated cargo proteins to the protostome. HR23B is found expressed at high levels in CTCL lymphomas which respond favorably to the HDACi, vorinostat (141).
10. Summary and Future Directions
Histone deacetylase inhibitors (HDACi) are a promising new group of targeted ant-cancer agents with potential application in the therapy of hematologic and solid neoplasms, as well as, several non-oncologic diseases. There are considerable on-going efforts to develop new and more effective HDACi.
HDAC enzymes have histones and many non-histone proteins as targets. Current evidence indicates that HDACi act not only to block the catalytic activity of the enzyme but may also affect the protein-protein interaction of specific HDACs with various critical protein partners. These target proteins are involved in many cell pathways including gene expression, cell proliferation, cell migration and cell death and have a role in angiogenesis and immune response. While not completely understood, it is clear that the mechanism of HDACi induced transformed cell death involves more than one pathway. Normal cells are relatively resistant to HDACi induced cell death. A possible explantation for this therapeutic window is that the majority of cancer cells have multiple genetic and molecular defects. Normal cells compared to transformed cells have the capacity to relatively rapidly reverse the adverse effects of HDACi given an intermittent regimen of exposure to the drugs.
Understanding the biological activities of the eleven Zn dependent HDACs is incomplete. An important question is whether a pan HDACi, such as, vorinostat, which inhibits class I HDACs and class IIb HDAC6, are potentially more effective therapeutic agents than an HDAC selective inhibitors. The development of HDAC isoform selective inhibitors will be useful in further dissecting their biological function and addressing this therapeutic issue.
HDAC inhibitors developed for clinical use to date are structurally diverse and inhibit more than a single Zn dependent HDAC. HDACi have anti-tumor activity across a broad variety of hematologic and solid tumors but only a proportion of patients with a given diagnosis show a therapeutic response. There is the need to identify markers of potential response or resistance to one or another HDACi. Optimization of the pharmaceutical properties of HDACi is another challenging area for the future development of HDACi.
To date, the evidence indicates that HDACi may be most useful in combination with other anti- cancer agents including radiation and cytotoxic and targeted drugs. As we gain a better understanding of the mechanisms of action of the Zn dependent HDAC, as well as the different HDACi, we will be able to develop more effective therapeutic strategies.
The large number of defects present in most cancer cells suggests that therapeutic strategies that target multiple biological pathways are likely to be more effective than drugs targeted at a single pathway. This concept supports the need for continued development of HDACi in combination with other anti-cancer agents.
Acknowledgments
The author is grateful to Joann Perrone and Mable Miranda for their excellent assistance in the preparation of this manuscript. Studies referred to in this review from the author's laboratory were supported, in part, by the National Institute of Cancer Grant P30CA08748-44, The David Koch Foundation, The Jack and Susan Rudin Foundation, The CapCure Foundation, and Experimental Therapeutics at Memorial Sloan-Kettering Cancer Center. Memorial Sloan- Kettering Cancer Center and Columbia University hold patents on suberoylanilide hydroxamic acid (SAHA, vorinostat) and related compounds that were exclusively licensed to ATON Pharma, a biotechnology start-up that was wholly acquired by Merck Inc. in April 2004. The author was a founder of ATON and may receive royalty with the further development of vorinostat by Merck.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Struhl K. Gene regulation. A paradigm for precision. Science. 2001;293(5532):1054–5. doi: 10.1126/science.1064050. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 4.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009 doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteller M. Epigenetics in cancer. New England Journal of Medicine. 2008;358(11):1148–59. doi: 10.1056/NEJMra072067. The. [DOI] [PubMed] [Google Scholar]
- 6.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8(6):1409–20. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: current status and overview of recent clinical trials. Drugs. 2009;69(14):1911–34. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Smith KT, Workman JL. Histone deacetylase inhibitors: anticancer compounds. Int J Biochem Cell Biol. 2009;41(1):21–5. doi: 10.1016/j.biocel.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Schrump DS. Cytotoxicity Mediated by Histone Deacetylase Inhibitors in Cancer Cells: Mechanisms and Potential Clinical Implications. Clin Cancer Res. 2009;15(12):3947–3957. doi: 10.1158/1078-0432.CCR-08-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: The next step? Cancer Lett. 2009 doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280(2):201–10. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 16.Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13(2):143–53. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- 17.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21(14):1790–802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato T, et al. Characterization of the HDAC1 complex that regulates the sensitivity of cancer cells to oxidative stress. Cancer Res. 2009;69(8):3597–604. doi: 10.1158/0008-5472.CAN-08-4368. [DOI] [PubMed] [Google Scholar]
- 19.Marchion DC, et al. HDAC2 regulates chromatin plasticity and enhances DNA vulnerability. Mol Cancer Ther. 2009;8(4):794–801. doi: 10.1158/1535-7163.MCT-08-0985. [DOI] [PubMed] [Google Scholar]
- 20.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. Embo J. 2002;21(11):2672–81. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30(1):61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323(5911):256–9. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Richardson WD. Genetics meets epigenetics: HDACs and Wnt signaling in myelin development and regeneration. Nat Neurosci. 2009;12(7):815–7. doi: 10.1038/nn0709-815. [DOI] [PubMed] [Google Scholar]
- 24.Chang S, et al. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126(2):321–34. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 25.Haggarty SJ, et al. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci U S A. 2003;100(8):4389–94. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parmigiani RB, et al. HDAC6 is a specific deacetylase of peroxiredoxins and is involved in redox regulation. Proc Natl Acad Sci U S A. 2008;105(28):9633–8. doi: 10.1073/pnas.0803749105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27(2):197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18(5):601–7. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Villagra A, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol. 2009;10(1):92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsiades CS, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci U S A. 2004;101(2):540–5. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray SG, et al. Microarray profiling of the effects of histone deacetylase inhibitors on gene expression in cancer cell lines. Int J Oncol. 2004;24(4):773–95. doi: 10.3892/ijo.24.4.773. [DOI] [PubMed] [Google Scholar]
- 32.Peart MJ, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102(10):3697–702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dokmanovic M, et al. Histone deacetylase inhibitors selectively suppress expression of HDAC7. Mol Cancer Ther. 2007;6(9):2525–34. doi: 10.1158/1535-7163.MCT-07-0251. [DOI] [PubMed] [Google Scholar]
- 34.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93(1):57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 35.Finnin MS, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401(6749):188–93. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 36.Somoza JR, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12(7):1325–34. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Vannini A, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004;101(42):15064–9. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottomley MJ, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008:M803514200. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ficner R. Novel structural insights into class I and II histone deacetylases. Curr Top Med Chem. 2009;9(3):235–40. doi: 10.2174/156802609788085304. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa M, et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep. 2007;18(4):769–74. [PubMed] [Google Scholar]
- 41.Zhang Z, et al. Quantitation of HDAC1 mRNA expression in invasive carcinoma of the breast*. Breast Cancer Res Treat. 2005;94(1):11–6. doi: 10.1007/s10549-005-6001-1. [DOI] [PubMed] [Google Scholar]
- 42.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20(40):5695–707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 43.Lin RJ, et al. Transcriptional regulation in acute promyelocytic leukemia. Oncogene. 2001;20(49):7204–15. doi: 10.1038/sj.onc.1204853. [DOI] [PubMed] [Google Scholar]
- 44.Marquard L, et al. Histone deacetylase 1, 2, 6 and acetylated histone H4 in B- and T- cell lymphomas. Histopathology. 2009;54(6):688–698. doi: 10.1111/j.1365-2559.2009.03290.x. [DOI] [PubMed] [Google Scholar]
- 45.Chang HH, et al. Histone deacetylase 2 expression predicts poorer prognosis in oral cancer patients. Oral oncology. 2009;45(7):610–614. doi: 10.1016/j.oraloncology.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 46.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280(2):168–76. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 47.Gloghini A, et al. Expression of histone deacetylases in lymphoma: implication for the development of selective inhibitors. Br J Haematol. 2009;147(4):515–25. doi: 10.1111/j.1365-2141.2009.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oehme I, et al. Targeting of HDAC8 and investigational inhibitors in neuroblastoma. Expert Opin Investig Drugs. 2009;18(11):1605–17. doi: 10.1517/14728220903241658. [DOI] [PubMed] [Google Scholar]
- 49.Dokmanovic M, Clarke C, Marks PA. Histone Deacetylase Inhibitors: Overview and Perspectives. Mol Cancer Res. 2007;5(10):981–989. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 50.Ropero S, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet. 2006;38(5):566–9. doi: 10.1038/ng1773. [DOI] [PubMed] [Google Scholar]
- 51.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 52.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 53.Wang H, Dymock BW. New patented histone deacetylase inhibitors. Expert Opin Ther Pat. 2009;19(12):1727–57. doi: 10.1517/13543770903393789. [DOI] [PubMed] [Google Scholar]
- 54.Price S, Dyke HJ. Histone deacetylase inhibitors: An analysis of recent patenting activtiy. Expert Opinion on Therapeutic Patents. 2007;17(7):745–765. [Google Scholar]
- 55.Weinmann E, Ottow E. Histone deacetylase inhibitors: A survey of recent patents. Expert Opinion on Therapeutic Patents. 2005;15:1677–1690. [Google Scholar]
- 56.Marson C. Histone deacetylase inhibitors: design, structure-activity relationships and therapeutic implications for cancer. Anti-cancer agents in medicinal chemistry. 2009;9(6):661–692. doi: 10.2174/187152009788679976. [DOI] [PubMed] [Google Scholar]
- 57.Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des. 2008;14(6):505–28. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 58.Mahboobi S, et al. Design of chimeric histone deacetylase- and tyrosine kinase- inhibitors: a series of imatinib hybrides as potent inhibitors of wild-type and mutant BCR-ABL, PDGF-Rbeta, and histone deacetylases. J Med Chem. 2009;52(8):2265–79. doi: 10.1021/jm800988r. [DOI] [PubMed] [Google Scholar]
- 59.Ruefli AA, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci U S A. 2001;98(19):10833–8. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ungerstedt JS, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005;102(3):673–8. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eot-Houllier G, et al. Histone deacetylase inhibitors and genomic instability. Cancer Lett. 2009;274(2):169–76. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Gaymes TJ, et al. Histone deacetylase inhibitors (HDI) cause DNA damage in leukemia cells: a mechanism for leukemia-specific HDI-dependent apoptosis? Mol Cancer Res. 2006;4(8):563–73. doi: 10.1158/1541-7786.MCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 63.Pilch DR, et al. Characteristics of gamma-H2AX foci at DNA double-strand breaks sites. Biochem Cell Biol. 2003;81(3):123–9. doi: 10.1139/o03-042. [DOI] [PubMed] [Google Scholar]
- 64.Adimoolam S, et al. HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination. Proc Natl Acad Sci U S A. 2007;104(49):19482–7. doi: 10.1073/pnas.0707828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munshi A, et al. Histone deacetylase inhibitors radiosensitize human melanoma cells by suppressing DNA repair activity. Clin Cancer Res. 2005;11(13):4912–22. doi: 10.1158/1078-0432.CCR-04-2088. [DOI] [PubMed] [Google Scholar]
- 66.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280(2):125–33. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 67.Chen CS, et al. Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res. 2007;67(11):5318–27. doi: 10.1158/0008-5472.CAN-06-3996. [DOI] [PubMed] [Google Scholar]
- 68.Fernandez-Capetillo O, Nussenzweig A. Linking histone deacetylation with the repair of DNA breaks. Proc Natl Acad Sci U S A. 2004;101(6):1427–8. doi: 10.1073/pnas.0307342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munshi A, et al. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol Cancer Ther. 2006;5(8):1967–74. doi: 10.1158/1535-7163.MCT-06-0022. [DOI] [PubMed] [Google Scholar]
- 70.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5(4-5):245–53. [PMC free article] [PubMed] [Google Scholar]
- 71.Richon VM, et al. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A. 2000;97(18):10014–9. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gui CY, et al. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci U S A. 2004;101(5):1241–6. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rascle A, Johnston JA, Amati B. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol Cell Biol. 2003;23(12):4162–73. doi: 10.1128/MCB.23.12.4162-4173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang LG, Ossowski L, Ferrari AC. Androgen receptor level controlled by a suppressor complex lost in an androgen-independent prostate cancer cell line. Oncogene. 2004;23(30):5175–84. doi: 10.1038/sj.onc.1207654. [DOI] [PubMed] [Google Scholar]
- 75.Lee EM, et al. Suberoylanilide hydroxamic acid (SAHA) changes microRNA expression profiles in A549 human non-small cell lung cancer cells. International journal of molecular medicine. 2009;24(1):45–50. doi: 10.3892/ijmm_00000204. [DOI] [PubMed] [Google Scholar]
- 76.Rosato RR, Grant S. Histone deacetylase inhibitors: insights into mechanisms of lethality. Expert Opin Ther Targets. 2005;9(4):809–24. doi: 10.1517/14728222.9.4.809. [DOI] [PubMed] [Google Scholar]
- 77.Burgess A, et al. Histone deacetylase inhibitors specifically kill nonproliferating tumour cells. Oncogene. 2004;23(40):6693–701. doi: 10.1038/sj.onc.1207893. [DOI] [PubMed] [Google Scholar]
- 78.Insinga A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;11(1):71–6. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 79.Nakata S, et al. Histone deacetylase inhibitors upregulate death receptor 5/TRAIL-R2 and sensitize apoptosis induced by TRAIL/APO2-L in human malignant tumor cells. Oncogene. 2004;23(37):6261–71. doi: 10.1038/sj.onc.1207830. [DOI] [PubMed] [Google Scholar]
- 80.Zhao Y, et al. Inhibitors of histone deacetylases target the Rb-E2F1 pathway for apoptosis induction through activation of proapoptotic protein Bim. Proc Natl Acad Sci U S A. 2005;102(44):16090–5. doi: 10.1073/pnas.0505585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu W, et al. Intrinsic apoptotic and thioredoxin pathways in human prostate cancer cell response to histone deacetylase inhibitor. Proc Natl Acad Sci U S A. 2006;103(42):15540–5. doi: 10.1073/pnas.0607518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 83.Coffey DC, et al. The histone deacetylase inhibitor, CBHA, inhibits growth of human neuroblastoma xenografts in vivo, alone and synergistically with all-trans retinoic acid. Cancer Res. 2001;61(9):3591–4. [PubMed] [Google Scholar]
- 84.Borbone E, et al. Histone deacetylase inhibitors induce thyroid cancer-specific apoptosis through proteasome-dependent inhibition of TRAIL degradation. Oncogene. 2009 doi: 10.1038/onc.2009.306. [DOI] [PubMed] [Google Scholar]
- 85.Rosato RR, et al. The histone deacetylase inhibitor LAQ824 induces human leukemia cell death through a process involving XIAP down-regulation, oxidative injury, and the acid sphingomyelinase-dependent generation of ceramide. Mol Pharmacol. 2006;69(1):216–25. doi: 10.1124/mol.105.017145. [DOI] [PubMed] [Google Scholar]
- 86.Zhang XD, et al. The histone deacetylase inhibitor suberic bishydroxamate regulates the expression of multiple apoptotic mediators and induces mitochondria-dependent apoptosis of melanoma cells. Mol Cancer Ther. 2004;3(4):425–35. [PubMed] [Google Scholar]
- 87.Xu WS, et al. Induction of polyploidy by histone deacetylase inhibitor: a pathway for antitumor effects. Cancer Res. 2005;65(17):7832–9. doi: 10.1158/0008-5472.CAN-04-4608. [DOI] [PubMed] [Google Scholar]
- 88.Cimini D, et al. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol Biol Cell. 2003;14(9):3821–33. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taddei A, et al. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3(2):114–20. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 90.Dowling M, et al. Mitotic spindle checkpoint inactivation by trichostatin a defines a mechanism for increasing cancer cell killing by microtubule-disrupting agents. Cancer Biol Ther. 2005;4(2):197–206. [PubMed] [Google Scholar]
- 91.Robbins AR, et al. Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle. 2005;4(5):717–26. doi: 10.4161/cc.4.5.1690. [DOI] [PubMed] [Google Scholar]
- 92.Shao Y, et al. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2004;101(52):18030–5. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosato RR, Almenara JA, Grant S. The histone deacetylase inhibitor MS-275 promotes differentiation or apoptosis in human leukemia cells through a process regulated by generation of reactive oxygen species and induction of p21CIP1/WAF1 1. Cancer Res. 2003;63(13):3637–45. [PubMed] [Google Scholar]
- 94.Lillig CH, Holmgren A. Thioredoxin and related molecules--from biology to health and disease. Antioxid Redox Signal. 2007;9(1):25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 95.Butler LM, et al. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci U S A. 2002;99(18):11700–5. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nishiyama A, et al. Identification of thioredoxin-binding protein-2/vitamin D(3) up- regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem. 1999;274(31):21645–50. doi: 10.1074/jbc.274.31.21645. [DOI] [PubMed] [Google Scholar]
- 97.Tan J, et al. Apoptosis signal-regulating kinase 1 is a direct target of E2F1 and contributes to histone deacetylase inhibitor-induced apoptosis through positive feedback regulation of E2F1 apoptotic activity. J Biol Chem. 2006;281(15):10508–15. doi: 10.1074/jbc.M512719200. [DOI] [PubMed] [Google Scholar]
- 98.Bali P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–34. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 99.Westendorf JJ, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22(22):7982–92. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen CS, et al. Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J Biol Chem. 2005;280(46):38879–87. doi: 10.1074/jbc.M505733200. [DOI] [PubMed] [Google Scholar]
- 101.Solit DB, Rosen N. Hsp90: a novel target for cancer therapy. Curr Top Med Chem. 2006;6(11):1205–14. doi: 10.2174/156802606777812068. [DOI] [PubMed] [Google Scholar]
- 102.Kawaguchi Y, et al. The Deacetylase HDAC6 Regulates Aggresome Formation and Cell Viability in Response to Misfolded Protein Stress. Cell. 2003;115(6):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 103.Boyault C, et al. HDAC6-p97/VCP controlled polyubiquitin chain turnover. Embo J. 2006;25(14):3357–66. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ellis L, Hammers H, Pili R. Targeting tumor angiogenesis with histone deacetylase inhibitors. Cancer Lett. 2009;280(2):145–53. doi: 10.1016/j.canlet.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liang D, Kong X, Sang N. Effects of Histone Deacetylase Inhibitors on HIF-1. Cell Cycle. 2006;5(21) doi: 10.4161/cc.5.21.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kong X, et al. Histone deacetylase inhibitors induce VHL and ubiquitin-independent proteasomal degradation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2006;26(6):2019–28. doi: 10.1128/MCB.26.6.2019-2028.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qian DZ, et al. Class II histone deacetylases are associated with VHL-independent regulation of hypoxia-inducible factor 1 alpha. Cancer Res. 2006;66(17):8814–21. doi: 10.1158/0008-5472.CAN-05-4598. [DOI] [PubMed] [Google Scholar]
- 108.Joseph J, et al. Expression profiling of sodium butyrate (NaB)-treated cells: identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene. 2004;23(37):6304–15. doi: 10.1038/sj.onc.1207852. [DOI] [PubMed] [Google Scholar]
- 109.Liu LT, et al. Histone deacetylase inhibitor up-regulates RECK to inhibit MMP-2 activation and cancer cell invasion. Cancer Res. 2003;63(12):3069–72. [PubMed] [Google Scholar]
- 110.Wardell SE, et al. Glucose metabolism as a target of histone deacetylase inhibitors. Mol Endocrinol. 2009;23(3):388–401. doi: 10.1210/me.2008-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Atadja P. Development of the pan-DAC inhibitor panobinostat (LBH589): successes and challenges. Cancer Lett. 2009;280(2):233–41. doi: 10.1016/j.canlet.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 112.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15(12):3970–7. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 113.Piekarz RL, Bates SE. Epigenetic modifiers: basic understanding and clinical development. Clin Cancer Res. 2009;15(12):3918–26. doi: 10.1158/1078-0432.CCR-08-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prince HM, Bishton MJ, Harrison SJ. Clinical Studies of Histone Deacetylase Inhibitors. Clin Cancer Res. 2009;15(12):3958–3969. doi: 10.1158/1078-0432.CCR-08-2785. [DOI] [PubMed] [Google Scholar]
- 115.Nolan L, et al. Will histone deacetylase inhibitors require combination with other agents to fulfil their therapeutic potential? Br J Cancer. 2008;99(5):689–94. doi: 10.1038/sj.bjc.6604557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jennifer LB, et al. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood. 2007;109(3):1123–1130. doi: 10.1182/blood-2006-04-019711. [DOI] [PubMed] [Google Scholar]
- 117.Adcock IM. HDAC inhibitors as anti-inflammatory agents. Br J Pharmacol. 2007;150(7):829–831. doi: 10.1038/sj.bjp.0707166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Villagra A, Sotomayor EM, Seto E. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 2009 doi: 10.1038/onc.2009.334. [DOI] [PubMed] [Google Scholar]
- 119.Andrews KT, et al. Targeting histone deacetylase inhibitors for anti-malarial therapy. Curr Top Med Chem. 2009;9(3):292–308. doi: 10.2174/156802609788085313. [DOI] [PubMed] [Google Scholar]
- 120.Papait R, Monti E, Bonapace IM. Novel approaches on epigenetics. Curr Opin Drug Discov Devel. 2009;12(2):264–75. [PubMed] [Google Scholar]
- 121.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7(10):854–68. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 122.Antonello M, et al. Histone Deacetylase Inhibitors and Neurodegenerative Disorders: Holding the Promise. Curr Pharm Des. 2009 doi: 10.2174/138161209789649349. [DOI] [PubMed] [Google Scholar]
- 123.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci U S A. 2003;100(4):2041–6. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kilgore M, et al. Inhibitors of Class 1 Histone Deacetylases Reverse Contextual Memory Deficits in a Mouse Model of Alzheimer's Disease. Neuropsychopharmacology. 2009 doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cao H, Jung M, Stamatoyannopoulos G. Hydroxamide derivatives of short-chain fatty acid have erythropoietic activity and induce [gamma] gene expression in vivo. Experimental Hematology. 2005;33(12):1443–1449. doi: 10.1016/j.exphem.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 126.Lin HS, et al. Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br J Pharmacol. 2007;150(7):862–72. doi: 10.1038/sj.bjp.0707165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Romagnani P, et al. Pharmacological modulation of stem cell function. Curr Med Chem. 2007;14(10):1129–39. doi: 10.2174/092986707780362880. [DOI] [PubMed] [Google Scholar]
- 128.Lee S, et al. Histone deacetylase inhibitors decrease proliferation potential and multilineage differentiation capability of human mesenchymal stem cells. Cell Prolif. 2009;42(6):711–20. doi: 10.1111/j.1365-2184.2009.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Butt MU, et al. Pharmacologic resuscitation: cell protective mechanisms of histone deacetylase inhibition in lethal hemorrhagic shock. J Surg Res. 2009;156(2):290–6. doi: 10.1016/j.jss.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 130.Schrump DS, et al. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin Cancer Res. 2008;14(1):188–98. doi: 10.1158/1078-0432.CCR-07-0135. [DOI] [PubMed] [Google Scholar]
- 131.Lee MJ, et al. Histone deacetylase inhibitors in cancer therapy. Current opinion in oncology. 2008;20(6):639–49. doi: 10.1097/CCO.0b013e3283127095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramalingam SS, et al. Phase II study of belinostat (PXD101), a histone deacetylase inhibitor, for second line therapy of advanced malignant pleural mesothelioma. J Thorac Oncol. 2009;4(1):97–101. doi: 10.1097/JTO.0b013e318191520c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kelly WK, Marks PA. Drug insight: Histone deacetylase inhibitors--development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat Clin Pract Oncol. 2005;2(3):150–7. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 134.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16(7):1111–20. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 135.Botrugno OA, Santoro F, Minucci S. Histone deacetylase inhibitors as a new weapon in the arsenal of differentiation therapies of cancer. Cancer Lett. 2009;280(2):134–44. doi: 10.1016/j.canlet.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 136.Cang S, Ma Y, Liu D. New clinical developments in histone deacetylase inhibitors for epigenetic therapy of cancer. J Hematol Oncol. 2009;2:22. doi: 10.1186/1756-8722-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fujiwara Y, et al. Phase I and pharmacokinetic study of vorinostat (suberoylanilide hydroxamic acid) in Japanese patients with solid tumors. Cancer Sci. 2009;100(9):1728–34. doi: 10.1111/j.1349-7006.2009.01237.x. [DOI] [PubMed] [Google Scholar]
- 138.Marks P, et al. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1(3):194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 139.Piekarz RL, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27(32):5410–7. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kelly WK, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23(17):3923–31. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stimson L, La Thangue NB. Biomarkers for predicting clinical responses to HDAC inhibitors. Cancer Lett. 2009;280(2):177–83. doi: 10.1016/j.canlet.2009.03.016. [DOI] [PubMed] [Google Scholar]


