Abstract
Sphingolipids are an important class of lipid molecules that play fundamental roles in our cells and body. Beyond a structural role, it is now clearly established that sphingolipids serve as bioactive signaling molecules to regulate diverse processes including inflammatory signaling, cell death, proliferation, and pain sensing. Sphingolipid metabolites have been implicated in the onset and progression of various diseases including cancer, lung disease, diabetes, and lysosomal storage disorders. Here we will review sphingolipid metabolism to introduce basic concepts as well as emerging complexities in sphingolipid function gained from modern technological advances and detailed cell and animal studies. Furthermore, we will discuss the family of neutral sphingomyelinases (N-SMases), which generate ceramide through the hydrolysis of sphingomyelin and are key enzymes in sphingolipid metabolism. Four mammalian N-SMase enzymes have now been identified. Most prominent is nSMase2 with established roles in bone mineralization, exosome formation, and cellular stress responses. Function for the other N-SMases have been more enigmatic and is an area of active investigation. The known properties and potential role(s) of each enzyme will be discussed to help guide to future studies.
Part one: Sphingolipid metabolism
Sphingolipids encompass a broad range of lipid molecules that elicit a wide range of signaling properties and cellular functions [1]. To understand the diverse functionality of sphingolipids in health and disease, it is important to be familiar with the chemical structure of these lipid molecules, their cellular metabolism, and the sphingolipid-metabolizing enzymes. Here we present a brief review of these areas in an effort to introduce the main points and emerging concepts in the field.
Introduction
Sphingolipids
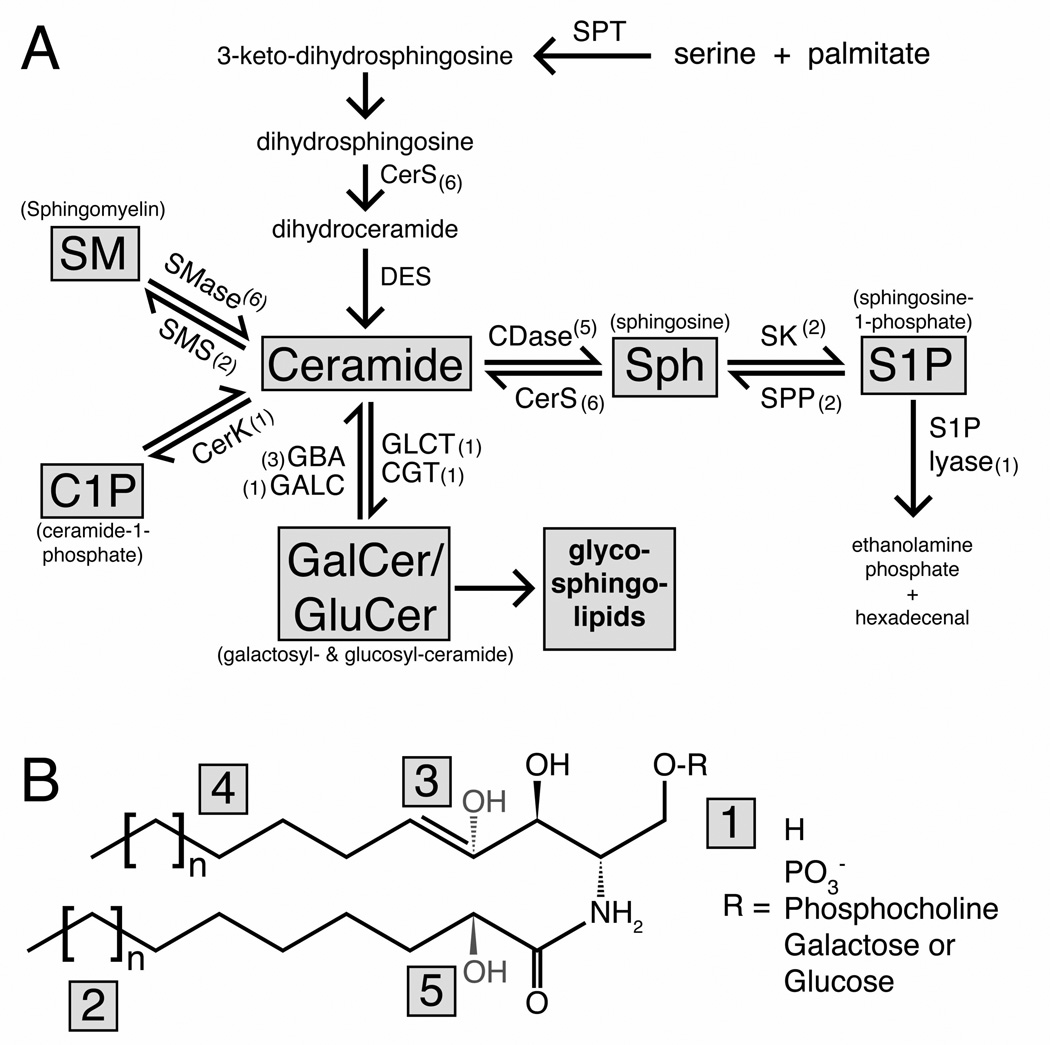
At the most basic level, sphingolipids can be defined as any lipid molecule that contains the sphingoid backbone, derived from the condensation of an amino acid (predominantly serine) and a fatty acid (predominantly palmitate) (Figure 1B) [2]. The presence or absence of an acyl chain distinguishes ceramide (Cer) from sphingosine (Sph), while phosphorylation of the 1-hydroxy group generates ceramide-1-phosphate (C1P) or sphingosine-1-phosphate (S1P). Other common sphingolipids contain different head groups at this position. Sphingomyelin (SM) contains a phosphorylcholine headgroup, and the basic glycosphingolipids, glucosylceramide (GluCer) and galactosylceramide (GalCer), contain a single sugar molecule linked to ceramide.
Figure 1. Sphingolipid metabolism and chemical structure.
(A) Diagram of sphingolipid metabolism showing the major lipid species in grey boxes and sphingolipid metabolizing enzymes. The number of mammalian genes that catalyze each conversion are denoted by brackets. (B) Generic structure for sphingolipid molecules with modification points. The presence or absence of the acyl chain (lower chain) distinguishes ceramide (shown) from sphingosine. Variation in the headgroup (1), attached to the terminal 1-oxygen, distinguishes each family of sphingolipids. The length of the sphingoid backbone (4) or acyl-chain (2) generates subspecies within each family. Further modification at the 4,5 position on the sphingoid backbone (3) can occur with different saturation levels (e.g. dihydroCer vs. Cer) and hydroxylation. In addition, the acyl chain can be hydroxylated at the 2-position (5). Abbreviations: SPT = serine phosphoryltransferase, CerS = (dihydro)ceramide synthase, DES = dihydroceramide desaturase, SMase = sphingomyelinase, SMS = sphingomyelin synthase, CerK = ceramide kinase, GBA = glucosylceramidase, GALC = galactosylceramidase, GCLT = ceramide glucosyltransferase, CGT = ceramide galactosyltransferase, CDase = ceramidase, SK= sphingosine kinase, SPP = S1P-phosphatase.
De novo biosynthesis
Sphingolipids can be synthesized de novo by a single biosynthetic pathway, with the end product being Cer [3] (Figure 1A). The first step is catalyzed by the serine palmitoyl transferase (SPT) complex and condenses the amino acid serine and the fatty acid palmitate to form 3-keto-dihydrosphingosine. The carbonyl group of 3-keto-dihydrosphingosine is then reduced to form dihydrosphingosine, and the enzyme (dihydro)ceramide synthase (CerS) then adds a fatty acid chain (the acyl chain) by N-acylation to form dihydroceramide. Finally, desaturation of the 4,5 carbon-carbon bond on the sphingoid backbone generates Cer [4].
Sphingolipid metabolism
From Cer as a central point, sphingolipid metabolism branches out in four main directions (Figure 1A). Three of these reactions alter the head group of Cer. These include the phosphorylation of Cer by ceramide kinase (CerK) to produce C1P [5], the addition of phosphocholine by sphingomyelin synthase (SMS) to produce SM (via transfer of the phosphocholine headgroup from phosphatidylcholine) [6], and the addition of a sugar molecule by glucosyl- and galactosyl-ceramidesynthases to create GluCer or GalCer respectively [2].
Alternatively, Cer can be broken down by ceramidase (CDase), which removes the acyl chain, to produce the lyso-sphingolipid Sph [7]. Sph can be reconverted to Cer by CerS or phosphorylated by sphingosine kinase (SK) to produce S1P [8]. S1P can be either dephosphorylated back to Sph or broken down by S1P lyase to ethanolamine phosphate and hexadecenal [9]. Action by S1P lyase is notably the sole exit point for sphingolipid breakdown and is not reversible [1].
New bioactive sphingolipids
The advances in lipodomics and metabolomics provide a basis for the continuing discovery of new bioactive sphingolipids. For example, a recent study identified N,N-dimethylsphingosine (DMS) as a new bioactive molecule, inducing chronic pain in mice [10]. The pathway and enzymes responsible for DMS synthesis have not been established but may simply involve the di-methylation of Sph.
Complexity in sphingolipid metabolism
Variations in chemical structure
Each lipid species described above comprises a family of molecules that share the same basic framework (i.e. same head group) but can differ in their chemical structure in specific ways (Figure 1B). A common variation is the length of the acyl chain [2]. This occurs through utilization of different length fatty acyl-CoAs by CerS enzymes [11]. In addition, the length of the sphingoid base can vary if the SPT complex uses myristate or stearate as a substrate instead of palmitate [12]. Besides chain length, both the acyl chain and sphingoid base can be hydroxylated [13] and the sphingoid base can be saturated or desaturated [14]. Overall, these variations create a family of related molecules that have the potential for distinct molecular and cellular functions.
Compartmentalization and specificity
In mammalian sphingolipid metabolism, there is often more than one enzyme in the cell to catalyze each chemical reaction (Figure 1A) [3]. From a cellular perspective, the functional reasons for enzyme multiplicity are two-fold. First, isozymes can be localized in different cellular compartments. This allows organelle-specific activity and separate regulatory mechanisms. For example, there have been six identified SMase isoforms that localize to various compartments including the lysosome, inner or outer leaflet of the plasma membrane, ER, mitochondria, and nucleus [15,16]. Secondly, isozymes can have different substrate specificities to allow the formation or breakdown of particular molecular species. Representatively, the CerS isozymes appear to all localize to the ER but exhibit different preferences in fatty acid chain length [17].
Interconnectivity of metabolites
The metabolic network of sphingolipids is connected through a series of chemical reactions that transform one bioactive metabolite to another [3] (Figure 1A). Expanding beyond two-state equilibriums, we see a connectivity map that allows the production of distant metabolites from another; e.g. S1P from SM by the sequential action of SMases (SM to Cer), CDases (Cer to Sph) and SKs (Sph to S1P). Given the large excess of SM to S1P, it is a logical progression that a relatively small SM depletion has the ability to profoundly affect S1P production. Overall, this means the bioactive lipid generated by the activation of SMases or CDases, and the associated cellular response, does not have to directly correlate with the immediate reaction product [1].
Emerging concepts
Many ceramides
A new paradigm for sphingolipid metabolism and function has emerged that integrates the significant advances described above into a working model. In the many ceramides hypothesis, each Cer molecular species, and related bioactive sphingolipid, has the potential to illicit a unique cellular response [18]. Unique functions may arise from differences in the bioactive molecule’s structure, protein targets, and/or subcellular localization.
Molecular specificity in cellular function
An elegant example of highly selective recognition of a lipid molecule was recently reported that embodies the essence of the many ceramides hypothesis [10]. The authors found that the COPI machinery protein p24 specifically recognized and bound SM with 18 carbons. This recognition was required for efficient COPI-dependent transport. SM-18 was previously reported to be enriched in the Golgi, which suggests that this molecular specificity drives a compartment specific cellular function [10].
Many of the other current examples include the chain-length dependent correlation of Cer molecules in various processes including autophagy, apoptosis, inflammation, and cancer [19]. In general, these processes have been associated with the action of CerS enzymes, which have different chain-length specificities. Based on their chain-length specificities, the involvement of individual CerS enzymes in these pathologies has been either inferred or tested experimentally [20–23].
As another example, the three-member family of alkaline ceramidases (AlkCDase1–3) has recently been characterized to have different specificities in the hydrolysis of Cer. AlkCDase1 hydrolyzes very long chain Cers [7], AlkCDase2 has broad substrate specificity [24], and AlkCDase3 hydrolyzes long chain Cers [25] to regulate the levels of distinct Cer subclasses in the Golgi.
Mechanistic implications
It is interesting to consider how this molecular specificity is functionally translated. Consider that all the distinguishing structural characteristics of different Cer molecules are embedded in the membrane and should only be recognizable by protein transmembrane domains. Proteins working only at the membrane interface should not be able to distinguish these molecular differences. This should limit the recognition of sphingolipid specificities to integral membrane proteins or proteins with significant hydrophobic pockets or membrane insertions.
Conclusions
Although new sphingolipids and their functions are still being identified, the basic outline for sphingolipid metabolism is well established. Involvement of sphingolipids and sphingolipid metabolizing enzymes in new pathologies and signaling pathways continues to increase. Moving forward, the challenge is to understand the molecular mechanisms underlying sphingolipid functions using the advancing technology and our increased appreciation of sphingolipid complexities. Investigations of this nature will significantly advance our knowledge to benefit in the identification of novel targets and strategies for therapeutic intervention in health and disease.
Part two: Neutral Sphingomyelinases
Neutral Sphingomyelinases (N-SMases) are a family of related enzymes that catalyze the hydrolysis of SM to generate Cer and phosphorylcholine. Cer and sphingolipid metabolites are well-established regulators of many important cellular signaling pathways and are implicated in human health and disease [26]. In this section we will discuss the mammalian N-SMase enzymes highlighting biochemical properties, localization, and roles in lipid metabolism, cellular signaling, and physiology.
Introduction
The mammalian N-SMases
Since the original identification of N-SMase activity in 1967 [27] four mammalian N-SMase genes have been cloned or purified (Figure 2A). These include nSMase1 (gene name = SMPD2), nSMase2 (SMPD3), nSMase3 (SMPD4), and MA-nSMase (mitochondrial-associated nSMase) (SMPD5). NSMase2 is currently the best-studied isoform with established roles in bone mineralization, cell growth arrest, exosome formation, and the inflammatory response. The roles of the other N-SMases in mammalian physiology and biology are still ambiguous due to either an enigmatic function or an only recent identification.
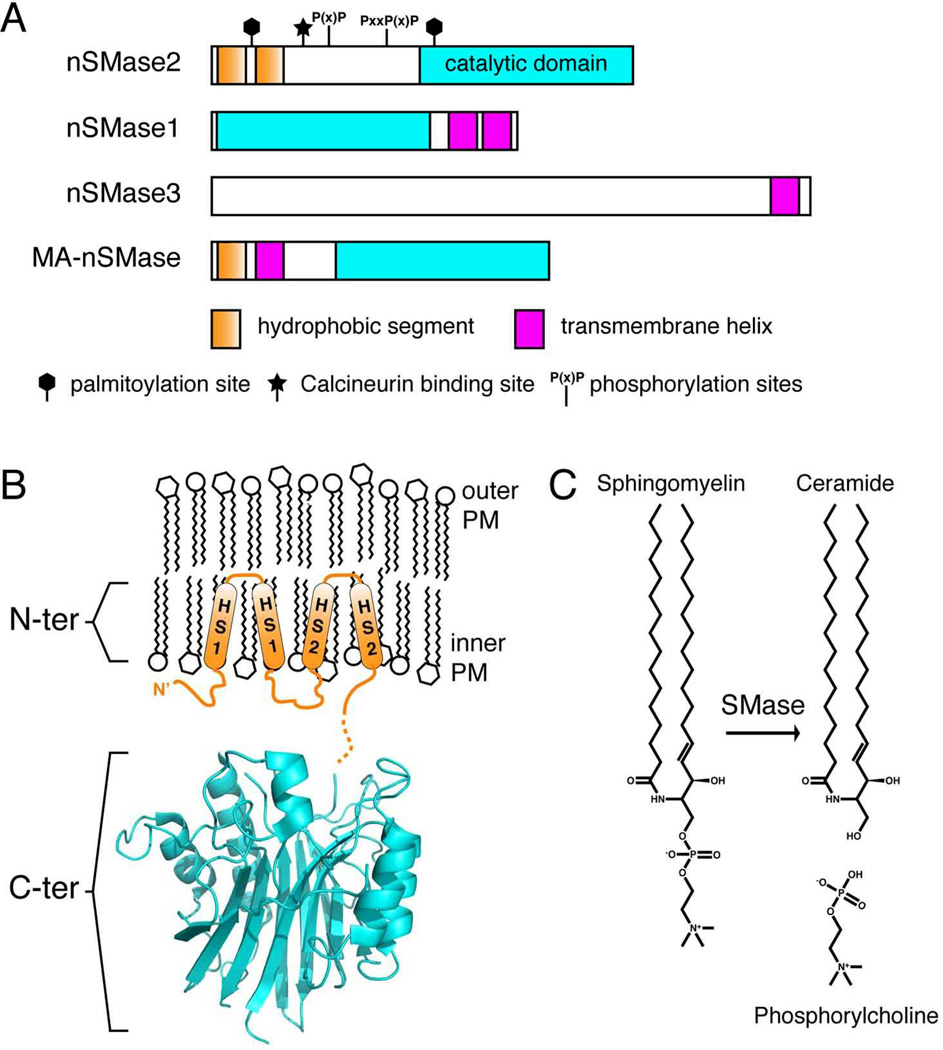
Figure 2. Domain architecture and topology of N-SMase isoforms.
(A) Domain architecture of N-SMase isoforms highlighting the catalytic domain, membrane-associated or transmembrane regions, and sites of protein binding or post-translational modifications. (B) nSMase2 contains two domains: an N-terminal domain with two hydrophobic segments (HS1 & HS2) that associate with but do not span the membrane and a C-terminal catalytic domain (blue). For the catalytic domain, the structure of a bacterial homologue (bSMase, PDB: 2DDR) is shown with the active site towards the membrane. (C) Sphingomyelinases catalyze the hydrolysis of SM to generate Cer and phosphocholine.
Biochemical and structural features
Most N-SMases, from bacteria to mammals, share a DNase I type catalytic core suggesting a common catalytic mechanism for SM hydrolysis [28]. NSMase3 is an exception and will be discussed separately. N-SMases catalyze the hydrolysis of SM in a PLC-type manner to generate the reaction products: Cer and phosphorylcholine (Figure 2C). The active site is defined by 8 metal binding residues, which together bind two Mg2+ ions [29]. Other highly conserved residues are found near the active site and may be important for catalytic activity. Two examples are Asp and Lys residues found in a P-loop like motif, which are both required for catalysis in the yeast N-SMase homolog Isc1 [30]. The crystal structures of two N-SMase bacterial homologs have defined the general protein fold and position of these conserved residues [29,31].
All mammalian N-SMases contain an extra hydrophobic domain that tethers the catalytic domain to the membrane (Figure 2A). The hydrophobic domain can also play other roles in phospholipid binding, subcellular localization, and enzyme activation to contribute to the activity and regulation of N-SMase enzymes in vivo.
Subcellular localization
In general, sphingolipids and sphingolipid metabolism are segregated into membrane compartments [1]. The localization of each N-SMase enzyme differs, suggesting each enzyme is responsible for SM hydrolysis and ceramide formation in specific organelles. In addition, enzyme localization constrains the biologically relevant activation mechanisms, as the concentration of different lipid molecules is also organelle dependent.
Neutral SMase2
NSMase2 is the best-studied mammalian N-SMase and has emerged as a key mediator of cellular stress-induced generation of Cer, as well as a somewhat surprising role in bone mineralization. Biochemical and physical characterization has identified a number of mechanisms for activation and regulation of nSMase2. These studies provide a template to investigate other N-SMase isoforms, as well as potential areas and modes for therapeutic intervention.
Biochemical properties
Two domains have been identified in nSMase2: an N-terminal domain that is hydrophobic and tethers nSMase2 to the membrane and a C-terminal domain encompassing the catalytic domain [32] (Figure 2B). Interestingly, the region separating these domains contains recently identified serine phosphorylation sites [33] and a Calcineurin binding site [34] suggesting this may be a major area for activation and regulation of nSMase2.
NSMase2 exhibits low basal SMase activity in vitro and requires activation by anionic phospholipids (APLs) [35]. The requirement of APLs is consistent with the location of nSMase2 at the plasma membrane (PM) [36,6], rich in the APL phosphatidylserine (PS) [37]. Another potential biologically relevant APL is the minor lipid phosphatidic acid (PA) transiently produced at the PM in low concentrations [38]. Given the different spatiotemporal dynamics of PS and PA at the PM, we speculate that constitutive function of nSMase2 may depend on PS, while PA generation may be one mechanism for acute activation.
The interaction of nSMase2 with the membrane has been studied in detail. The N-terminal domain contains two hydrophobic segments (Figure 2), predicted to be helical, that associate with but do not span the membrane [39] (Figure 2B). Additionally, nSMase2 harbors two palmitoylation sites that contribute to nSMase2 membrane association [40]. Recently, the APL binding domain was found to localize exclusively to the N-terminal domain [41]. A binding motif consisting of three conserved Arg residues are necessary for APL binding, APL-mediated activation, and correct trafficking of nSMase2.
Localization
NSMase2 localizes to the plasma membrane (PM), Golgi, and recycling compartments. TNF-α, PMA, H2O2, and cell confluence all induce nSMase2 translocation from the Golgi to the PM, and correlate with increased N-SMase activity. This suggests the major site of nSMase2 action is the inner leaflet of the PM. Translocation may regulate activity by controlling access to substrate SM, activating APLs, or involve other mechanisms.
GW4869: an nSMase2 specific inhibitor
NSMase2 is the only N-SMase with a specific inhibitor, GW4869 [42,43]. GW4869 has been widely used as a tool to identify and confirm nSMase2-specific functions [26]. Mechanistically, GW4869 is thought to inhibit nSMase2 by interfering with APL activation [42]. We note that any inhibitory effect of GW4869 on the recently identified MA-nSMase has not been assessed and should be tested.
Cellular signaling
NSMase2 has been implicated in the response to various cellular stresses and cytokines to affect a diverse set of signaling pathways including cancer pathogenesis [44,45], growth and development [46,47], and inflammatory responses [48,49]. It was originally cloned as a confluence arrest gene [50] and has a demonstrated role linking confluence to cell cycle arrest [51]. Most recently, it has been linked as an upstream regulator of the mTOR/S6 Kinase pathway [52] and shown that this pathway regulates the biosynthesis of the extracellular matrix component hyaluronan, which is a glycosaminoglycan produced at the PM [53].
Activation by TNF-α is currently the best-studied activation pathway and occurs through PKC-δ [54] and p38 MAPK dependent mechanisms [55]. Direct protein interaction with embryonic ectodermal development (EED) couples nSMase2 to TNF receptor 1 through FAN (Factor Associated with N-SMase activation) [56] and the scaffolding protein RACK1 [57].
Phosphorylation has emerged as one mechanism regulating nSMase2 activity. Phosphorylation has been shown to modulate the activity of nSMase2, to be regulated by the phosphatase Calcineurin [34], and to occur through p38 MAPK and PKC dependent pathways [54,55]. Recently, two clusters of specific serine residues were identified as phosphorylation sites and found to effect protein stability [33]. The actual kinase responsible for directly phosphorylating nSMase2 is unknown.
A role for nSMase2 in Cer-mediated lung injury has emerged [58]. Oxidants in cigarette smoke and hydrogen peroxide increase nSMase2 activity and Cer levels leading to increased apoptosis in bronchial epithelial cells and the lung tissues of rodents [59–61]. The effects of cigarette smoke can be blocked by the antioxidants glutathione and N-acetyl cysteine. In addition, the expression of nSMase2 was increased in lung tissues of smokers with emphysema [61].
NSMase2 may play a role in tumorigenesis with mutations identified in the SMPD3 gene in several human leukemias [44]. Two of these mutations affect function by either reducing protein stability or altering localization. Other studies suggest nSMase2 may be important for cell death in transformed cells [26]. For example, NSMase2 mRNA and protein levels were induced by the anti-cancer drug daunorubicin, leading to increase N-SMase activity, Cer levels, and cell death [45].
Several studies have now demonstrated that ceramide and nSMase2 are key regulators of exosome formation and microRNA (miRNA) secretion [62–64]. miRNAs are gene regulatory elements found both intra- and extra-cellularly [62]. It is thought they are encapsulated in exosomes, leading to their secretion outside the cell. Interestingly, overexpression of nSMase2 increased miRNA secretion, while inhibition by GW4869 or knockdown by siRNA of nSMase2 decreased miRNA secretion. This process was not dependent on the endosomal sorting complex required for transport (ESCRT) implicating a novel ceramide-dependent, ESCRT-independent secretion mechanism. In addition, these studies suggest the Golgi may be a second site of action for nSMase2 SM hydrolysis.
Physiological role in skeletal development
Animal studies of nSMase2-deficient mice have identified a role for nSMase2 in bone homeostasis. The nSMase2 knockout mice suffer from short stature [63], while the nSMase2-inactivating fro/fro (for fragilitas ossium) mutation results in bone fragility [47]. The observed skeletal abnormalities include short and bent limbs, as well as deformations in rib cages, long bones, and growth plate cartilage [64].
Recently, it was shown that nSMase2 plays cell-specific roles in skeletal development [64]. Fro/fro mice are defective in both bone and cartilage mineralization and these two events are regulated by the different cell types: osteoblasts and chondrocytes, respectively. By selectively expressing nSMase2 in the osteoblasts of fro/fro mice, the authors were able to correct the osteoblast-specific bone defects but not affect the cartilage defects [64].
The role of nSMase2 appears to involve a novel mechanism that does not involve the typical factors such as calcium, phosphate, and alkaline phosphates. This suggests a novel, nSMase2-dependent mechanism gating proper bone mineralization and may help in understanding cases of osteogenesis imperfecta in human patients that also share similar phenotypes and undetectable differences in typical mineralization parameters [64,65]. Overall, these discoveries are exciting and future work deciphering the molecular mechanisms of nSMase2 in bone mineralization is of great interest.
Neutral SMase1
Biochemical properties
NSMase1 was the first identified and cloned mammalian N-SMase based on sequence homology to bacterial SMases [66]. The domain architecture of nSMase1 is identical to Isc1, the yeast homologue to N-SMases [67], with a catalytic domain followed by two C-terminal transmembrane helices (Figure 2A). Unlike other N-SMases, nSMase1 is not activated by phospholipids [66].
Localization
The localization of nSMase1 appears to vary when comparing endogenous to overexpressed protein. Overexpressed nSMase1 mainly colocalizes with endoplasmic reticulum (ER) markers [68]. However, endogenous nSMase1, in contrast to overexpressed nSMase1, was reported to localize to the nuclear matrix [69].
A role for nSMase1 in lipid metabolism?
Despite the activity of nSMase1 on SM in vitro, overexpression in cells does not affect SM metabolism [66,70]. This ambiguity casts doubt on the role of nSMase1 in sphingolipid metabolism. Lyso-platelet activating factor (lyso-PAF) may be a biologically relevant substrate, with nSMase1 displaying both PLC-lyso-PAF activity in vitro and in cells [70]. However, nSMase1 KO mice had no detectable changes in sphingolipid metabolism by high performance thin layer chromatography (HPTLC), a relatively insensitive method compared to mass spectrometry, including no alterations in SM or lyso-PAF metabolism.
Although a function for nSMase1 is not apparent, this does not preclude a role for nSMase1 in lipid metabolism. Given the significant advancements in lipid quantification by mass spectrometry and the emerging many ceramides model [18], it would be interesting to measure lipid levels in nSMase1 KO mice to determine if discrete SM, Cer, or lyso-PAF species, undetectable by HPTLC, are altered. Quantification of ceramide levels in nSMase1 transiently transfected MCF7 cells did result in slight increases in some ceramide levels [71]. Correct substrate identification may require in vivo measurements from KO animal tissues, as recently demonstrated for a different lipid metabolizing enzyme [72]. With regards to nSMase1, this may be particularly relevant given the different subcellular localizations of endogenous and overexpressed proteins.
MA-nSMase (Mitochondrial-Associated Neutral SMase)
Biochemical properties and localization
MA-nSMase is the most recently identified mammalian N-SMase being discovered in 2010 [16] by sequence homology with nSMase2 and a zebrafish mitochondrial N-SMase [73]. The subcellular localization of overexpressed MA-nSMase protein varied with cell type, showing strong or partial co-localization with mitochondrial markers, in addition to co-localization with ER markers [16]. The mouse MA-nSMase protein conserves a mitochondrial signal peptide with the zebrafish N-SMase, which may be responsible for its mitochondrial localization.
MA-nSMase has comparable domain architecture and biochemical properties with nSMase2. Catalytic activity requires Mg2+ or Mn2+ ions and is strongly increased by the presence of the phospholipids cardiolipin (CL), PS, or phosphatidylglycerol (PG) (Figure 2). Both CL and PG are present in mitochondria providing a putative mechanism for activation and/or regulation of MA-nSMase activity in vivo [16]. The N-terminal hydrophobic domain of MA-nSMase is similar to nSMase2 by sequence homology, but the membrane topology and APL binding motif has yet to be characterized.
Human vs. murine MA-nSMase
The human homolog to murine MA-nSMase has yet to be cloned and characterized. However, an open reading frame for the human SMPD5 gene (XP_001714084) is present in the NCBI database. Surprisingly, the human and mouse proteins contain key differences in the N-terminal APL activation domain. Importantly, the putative mitochondrial signal peptide is conserved suggesting the human protein will also localize to mitochondria. In the future it is important to clone the human gene and compare the amino acid sequence, biochemical properties, localization, and cellular functions.
Future directions
At present, little is known about MA-nSMase beyond basic properties. However, the identification of a mammalian mitochondrial N-SMase presents another potential endogenous mechanism, in addition to action of ceramide synthases, for mitochondrial ceramide generation. This is exciting considering the numerous studies linking ceramide to mitochondrial activation of apoptosis [74].
Another potential role for MA-nSMase is in fertilization. Activation of an unidentified SMase during fertilization was inferred by a corresponding decrease in SM and increase in Cer levels [75]. In support of this hypothesis, MA-nSMase gene expression in organ tissues is the highest in testis [16] and the MA-nSMase gene appears to be bicistronic with a spermatogenesis associated factor (NCBI database, unpublished observation). It is our hope that future studies taking advantage of gene silencing, antibodies to endogenous proteins, and knockout animals will better illuminate the role of MA-nSMase in mammalian physiology and biology.
Neutral SMase3
Biochemical properties
Although human nSMase3 was identified in 2006 [76], relatively little work has been reported since regarding further biochemical and functional characterization. Identification was accomplished by sequence comparison using a peptide (KGLPYLEQLFR) from a previously purified N-SMase from bovine brain [77]. The peptide sequence only matches 7 of the 11 residues in the identified human protein and bovine homolog. Given the short peptide sequence and low identity, this raises the question if the original purified bovine and identified human proteins correspond to the same protein.
NSMase3 shares no sequence homology with any N-SMases or any other characterized type of enzyme catalytic domain. The region comprising the catalytic domain is yet to be identified. A C-terminal transmembrane helix is predicted to embed nSMase3 in the membrane [76].
Two conflicting reports have characterized nSMase3 activity. In the original identification, nSMase3 activity is reported to occur at neutral pH and require Mg2+ or Mn2+ [76]. The observed activity was slightly enhanced, approximately two-fold, by the phospholipid PS. In a later study, MCF-7 cells transiently and stably overexpressing nSMase3 did not have significant N-SMase activity over vector controls [71]. We hope that future studies will determine the underlying reasons behind this major discrepancy.
Localization
Subcellular localization studies found nSMase3 to display an ER distribution pattern [76]. Later work confirmed this and found localization was not dependent on a putative ER localization motif or the C-terminal transmembrane helix [78].
Cellular signaling
It has been suggested nSMase3 may play a role in TNF-α mediated signaling [76,78]. However, another report found that nSMase2 was the primary N-SMase activated by TNF-α in MCF-7 cells [71]. In support of this, a comparison of mouse fibroblasts from normal and nSMase2 deficient fro-/fro- mice found that nSMase2 deficiency abrogated TNF-α induced increases in N-SMase activity [49] suggesting nSMase3 is not required in these cells.
The other identified putative function of nSMase3 involves a role in tumorigenesis [78]. In this study, nSMase3 expression was found to be both upregulated and downregulated to varying degrees (−70 to +70%) in different tumor samples. In addition, nSMase3 mRNA levels were down regulated by the tumor suppressor p53.
Overall, a clear functional role for nSMase3 has yet to emerge. Further studies investigating the biological function and molecular mechanisms of nSMase3 are required to validate the current findings.
Conclusions
Four N-SMase isoforms have now been cloned and purified. However, beyond nSMase2 there is relatively little known about N-SMase physiology and cellular function. In addition, at a biochemical and mechanistic level, there is still much to learn about N-SMase activation and regulation. The roles of nSMase2 in bone homeostasis, exosome secretion of miRNAs, and cigarette-induced lung injury are essential and provide the potential for nSMase2-targeted therapies.
Acknowledgements
This work was supported by the NIH grant R37GM43825 (Y.A.H.). We thank Chris J. Clarke, Bill X. Wu, and David M. Perry for helpful review of this manuscript.
References
- 1.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9(2):139–150. doi: 10.1038/nrm2329. doi:nrm2329 [pii] 10.1038/nrm2329 [doi] [DOI] [PubMed] [Google Scholar]
- 2.Merrill AH., Jr Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chemical Reviews. 2011;111(10):6387. doi: 10.1021/cr2002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res. 2009;50(Suppl):S91–S96. doi: 10.1194/jlr.R800080-JLR200. doi:R800080-JLR200 [pii] 10.1194/jlr.R800080-JLR200 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. Sphingolipids as Signaling and Regulatory Molecules. 2010:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arana L, Gangoiti P, Ouro A, Trueba M, Gómez-Muñoz A. Review Ceramide and ceramide 1-phosphate in health and. 2010 doi: 10.1186/1476-511X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milhas D, Clarke CJ, Hannun YA. Sphingomyelin metabolism at the plasma membrane: implications for bioactive sphingolipids. FEBS Letters. 2010;584(9):1887–1894. doi: 10.1016/j.febslet.2009.10.058. doi:S0014-5793(09)00846-1 [pii] 10.1016/j.febslet.2009.10.058 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2008;1781(9):424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitson SM. Regulation of sphingosine kinase and sphingolipid signaling. Trends in Biochemical Sciences. 2011;36(2):97–107. doi: 10.1016/j.tibs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Serra M, Saba JD. Sphingosine 1-phosphate lyase, a key regulator of sphingosine 1-phosphate signaling and function. Advances in enzyme regulation. 2010;50(1):349. doi: 10.1016/j.advenzreg.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patti GJ, Yanes O, Shriver LP, Courade JP, Tautenhahn R, Manchester M, Siuzdak G. Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nat Chem Biol. 2012;8(3):232–234. doi: 10.1038/nchembio.767. doi:nchembio.767 [pii] 10.1038/nchembio.767 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281(35):25001–25005. doi: 10.1074/jbc.R600010200. doi:R600010200 [pii] 10.1074/jbc.R600010200 [doi] [DOI] [PubMed] [Google Scholar]
- 12.Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, Brown RH, Harmon JM, Dunn TM. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proceedings of the National Academy of Sciences. 2009;106(20):8186. doi: 10.1073/pnas.0811269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hama H. Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids. 2010;1801(4):405–414. doi: 10.1016/j.bbalip.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, Merrill AH., Jr Biodiversity of sphingoid bases ("sphingosines") and related amino alcohols. J Lipid Res. 2008;49(8):1621–1639. doi: 10.1194/jlr.R800012-JLR200. doi:R800012-JLR200 [pii] 10.1194/jlr.R800012-JLR200 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins RW, Canals D, Hannun YA. Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell Signal. 2009;21(6):836–846. doi: 10.1016/j.cellsig.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu BX, Rajagopalan V, Roddy PL, Clarke CJ, Hannun YA. Identification and characterization of murine mitochondria-associated neutral sphingomyelinase (MA-nSMase), the mammalian sphingomyelin phosphodiesterase 5. J Biol Chem. 2010;285(23):17993–18002. doi: 10.1074/jbc.M110.102988. doi:M110.102988 [pii] 10.1074/jbc.M110.102988 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J. 2012;441(3):789–802. doi: 10.1042/BJ20111626. doi:BJ20111626 [pii] 10.1042/BJ20111626 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannun YA, Obeid LM. Many ceramides. J Biol Chem. 2011;286(32):27855–27862. doi: 10.1074/jbc.R111.254359. doi:R111.254359 [pii] 10.1074/jbc.R111.254359 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosch S, Schiffmann S, Geisslinger G. Chain length-specific properties of ceramides. Prog Lipid Res. 2012;51(1):50–62. doi: 10.1016/j.plipres.2011.11.001. doi:S0163-7827(11)00042-7 [pii] 10.1016/j.plipres.2011.11.001 [doi] [DOI] [PubMed] [Google Scholar]
- 20.Mullen TD, Jenkins RW, Clarke CJ, Bielawski J, Hannun YA, Obeid LM. Ceramide Synthase-dependent Ceramide Generation and Programmed Cell Death. Journal of Biological Chemistry. 2011;286(18):15929. doi: 10.1074/jbc.M111.230870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiffmann S, Ziebell S, Sandner J, Birod K, Deckmann K, Hartmann D, Rode S, Schmidt H, Angioni C, Geisslinger G. Activation of ceramide synthase 6 by celecoxib leads to a selective induction of C16: 0-ceramide. Biochemical pharmacology. 2010;80(11):1632–1640. doi: 10.1016/j.bcp.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, Haimovitz-Friedman A, Fuks Z, Kolesnick R. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cellular signalling. 2010;22(9):1300–1307. doi: 10.1016/j.cellsig.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-David O, Futerman AH. The role of the ceramide acyl chain length in neurodegeneration: involvement of ceramide synthases. Neuromolecular medicine. 2010;12(4):341–350. doi: 10.1007/s12017-010-8114-x. [DOI] [PubMed] [Google Scholar]
- 24.Sun W, Jin J, Xu R, Hu W, Szulc ZM, Bielawski J, Obeid LM, Mao C. Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2) J Biol Chem. 2010;285(12):8995–9007. doi: 10.1074/jbc.M109.069203. doi:M109.069203 [pii] 10.1074/jbc.M109.069203 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu W, Xu R, Sun W, Szulc ZM, Bielawski J, Obeid LM, Mao C. Alkaline ceramidase 3 (ACER3) hydrolyzes unsaturated long-chain ceramides, and its down-regulation inhibits both cell proliferation and apoptosis. J Biol Chem. 2010;285(11):7964–7976. doi: 10.1074/jbc.M109.063586. doi:M109.063586 [pii] 10.1074/jbc.M109.063586 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu BX, Clarke CJ, Hannun YA. Mammalian neutral sphingomyelinases: regulation and roles in cell signaling responses. Neuromolecular Med. 2010;12(4):320–330. doi: 10.1007/s12017-010-8120-z. doi:10.1007/s12017-010-8120-z [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider PB, Kennedy EP. Sphingomyelinase in normal human spleens and in spleens from subjects with Niemann-Pick disease. J Lipid Res. 1967;8(3):202–209. [PubMed] [Google Scholar]
- 28.Clarke CJ, Snook CF, Tani M, Matmati N, Marchesini N, Hannun YA. The extended family of neutral sphingomyelinases. Biochemistry. 2006;45(38):11247–11256. doi: 10.1021/bi061307z. doi:10.1021/bi061307z [doi] [DOI] [PubMed] [Google Scholar]
- 29.Ago H, Oda M, Takahashi M, Tsuge H, Ochi S, Katunuma N, Miyano M, Sakurai J. Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus. J Biol Chem. 2006;281(23):16157–16167. doi: 10.1074/jbc.M601089200. doi:M601089200 [pii] 10.1074/jbc.M601089200 [doi] [DOI] [PubMed] [Google Scholar]
- 30.Okamoto Y, Vaena de Avalos S, Hannun YA. Functional analysis of ISC1 by site-directed mutagenesis. Biochemistry. 2003;42(25):7855–7862. doi: 10.1021/bi0341354. doi:10.1021/bi0341354 [doi] [DOI] [PubMed] [Google Scholar]
- 31.Openshaw AE, Race PR, Monzo HJ, Vazquez-Boland JA, Banfield MJ. Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria. J Biol Chem. 2005;280(41):35011–35017. doi: 10.1074/jbc.M506800200. doi:M506800200 [pii] 10.1074/jbc.M506800200 [doi] [DOI] [PubMed] [Google Scholar]
- 32.Clarke CJ, Wu BX, Hannun YA. The neutral sphingomyelinase family: identifying biochemical connections. Adv Enzyme Regul. 2011;51(1):51–58. doi: 10.1016/j.advenzreg.2010.09.016. doi:S0065-2571(10)00075-0 [pii] 10.1016/j.advenzreg.2010.09.016 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filosto S, Ashfaq M, Chung S, Fry W, Goldkorn T. Neutral sphingomyelinase 2 activity and protein stability are modulated by phosphorylation of five conserved serines. J Biol Chem. 2011 doi: 10.1074/jbc.M111.315481. doi:M111.315481 [pii] 10.1074/jbc.M111.315481 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filosto S, Fry W, Knowlton AA, Goldkorn T. Neutral sphingomyelinase 2 (nSMase2) is a phosphoprotein regulated by calcineurin (PP2B) J Biol Chem. 2010;285(14):10213–10222. doi: 10.1074/jbc.M109.069963. doi:M109.069963 [pii] 10.1074/jbc.M109.069963 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann K, Tomiuk S, Wolff G, Stoffel W. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc Natl Acad Sci U S A. 2000;97(11):5895–5900. doi: 10.1073/pnas.97.11.5895. doi:97/11/5895 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milhas D, Clarke CJ, Idkowiak-Baldys J, Canals D, Hannun YA. Anterograde and retrograde transport of neutral sphingomyelinase-2 between the Golgi and the plasma membrane. Biochim Biophys Acta. 2010;1801(12):1361–1374. doi: 10.1016/j.bbalip.2010.08.001. doi:S1388-1981(10)00179-4 [pii] 10.1016/j.bbalip.2010.08.001 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. doi:10.1146/annurev.biophys.093008.131234 [doi] [DOI] [PubMed] [Google Scholar]
- 38.Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta. 2006;1761(8):913–926. doi: 10.1016/j.bbalip.2006.03.006. doi:S1388-1981(06)00067-9 [pii] 10.1016/j.bbalip.2006.03.006 [doi] [DOI] [PubMed] [Google Scholar]
- 39.Tani M, Hannun YA. Analysis of membrane topology of neutral sphingomyelinase 2. FEBS Lett. 2007;581(7):1323–1328. doi: 10.1016/j.febslet.2007.02.046. doi:S0014-5793(07)00214-1 [pii] 10.1016/j.febslet.2007.02.046 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tani M, Hannun YA. Neutral sphingomyelinase 2 is palmitoylated on multiple cysteine residues. Role of palmitoylation in subcellular localization. J Biol Chem. 2007;282(13):10047–10056. doi: 10.1074/jbc.M611249200. doi:M611249200 [pii] 10.1074/jbc.M611249200 [doi] [DOI] [PubMed] [Google Scholar]
- 41.Wu BX, Clarke CJ, Matmati N, Montefusco D, Bartke N, Hannun YA. Identification of novel anionic phospholipid binding domains in neutral sphingomyelinase 2 with selective binding preference. J Biol Chem. 2011;286(25):22362–22371. doi: 10.1074/jbc.M110.156471. doi:M110.156471 [pii] 10.1074/jbc.M110.156471 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luberto C, Hassler DF, Signorelli P, Okamoto Y, Sawai H, Boros E, Hazen-Martin DJ, Obeid LM, Hannun YA, Smith GK. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J Biol Chem. 2002;277(43):41128–41139. doi: 10.1074/jbc.M206747200. doi:10.1074/jbc.M206747200 [doi] M206747200 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Canals D, Perry DM, Jenkins RW, Hannun YA. Drug targeting of sphingolipid metabolism: sphingomyelinases and ceramidases. Br J Pharmacol. 2011;163(4):694–712. doi: 10.1111/j.1476-5381.2011.01279.x. doi:10.1111/j.1476-5381.2011.01279.x [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim WJ, Okimoto RA, Purton LE, Goodwin M, Haserlat SM, Dayyani F, Sweetser DA, McClatchey AI, Bernard OA, Look AT, Bell DW, Scadden DT, Haber DA. Mutations in the neutral sphingomyelinase gene SMPD3 implicate the ceramide pathway in human leukemias. Blood. 2008;111(9):4716–4722. doi: 10.1182/blood-2007-10-113068. doi:blood-2007-10-113068 [pii] 10.1182/blood-2007-10-113068 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito H, Murakami M, Furuhata A, Gao S, Yoshida K, Sobue S, Hagiwara K, Takagi A, Kojima T, Suzuki M, Banno Y, Tanaka K, Tamiya-Koizumi K, Kyogashima M, Nozawa Y, Murate T. Transcriptional regulation of neutral sphingomyelinase 2 gene expression of a human breast cancer cell line, MCF-7, induced by the anti-cancer drug, daunorubicin. Biochim Biophys Acta. 2009;1789(11–12):681–690. doi: 10.1016/j.bbagrm.2009.08.006. doi:S1874-9399(09)00100-X [pii] 10.1016/j.bbagrm.2009.08.006 [doi] [DOI] [PubMed] [Google Scholar]
- 46.Stoffel W, Jenke B, Block B, Zumbansen M, Koebke J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc Natl Acad Sci U S A. 2005;102(12):4554–4559. doi: 10.1073/pnas.0406380102. doi:0406380102 [pii] 10.1073/pnas.0406380102 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stoffel W, Jenke B, Holz B, Binczek E, Gunter RH, Knifka J, Koebke J, Niehoff A. Neutral sphingomyelinase (SMPD3) deficiency causes a novel form of chondrodysplasia and dwarfism that is rescued by Col2A1-driven smpd3 transgene expression. Am J Pathol. 2007;171(1):153–161. doi: 10.2353/ajpath.2007.061285. doi:S0002-9440(10)61951-7 [pii] 10.2353/ajpath.2007.061285 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolova-Karakashian M, Karakashian A, Rutkute K. Role of neutral sphingomyelinases in aging and inflammation. Subcell Biochem. 2008;49:469–486. doi: 10.1007/978-1-4020-8831-5_18. doi:10.1007/978-1-4020-8831-5_18 [doi] [DOI] [PubMed] [Google Scholar]
- 49.Devillard R, Galvani S, Thiers JC, Guenet JL, Hannun Y, Bielawski J, Negre-Salvayre A, Salvayre R, Auge N. Stress-induced sphingolipid signaling: role of type-2 neutral sphingomyelinase in murine cell apoptosis and proliferation. PLoS One. 2010;5(3):e9826. doi: 10.1371/journal.pone.0009826. doi:10.1371/journal.pone.0009826 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayashi Y, Kiyono T, Fujita M, Ishibashi M. cca1 is required for formation of growth-arrested confluent monolayer of rat 3Y1 cells. J Biol Chem. 1997;272(29):18082–18086. doi: 10.1074/jbc.272.29.18082. [DOI] [PubMed] [Google Scholar]
- 51.Marchesini N, Osta W, Bielawski J, Luberto C, Obeid LM, Hannun YA. Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J Biol Chem. 2004;279(24):25101–25111. doi: 10.1074/jbc.M313662200. doi:10.1074/jbc.M313662200 [doi] M313662200 [pii] [DOI] [PubMed] [Google Scholar]
- 52.Clarke CJ, Mediwala K, Jenkins RW, Sutton CA, Tholanikunnel BG, Hannun YA. Neutral sphingomyelinase-2 mediates growth arrest by retinoic acid through modulation of ribosomal S6 kinase. J Biol Chem. 2011;286(24):21565–21576. doi: 10.1074/jbc.M110.193375. doi:M110.193375 [pii] 10.1074/jbc.M110.193375 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin J, Berdyshev E, Poirier C, Schwartz NB, Dawson G. Neutral sphingomyelinase 2 deficiency increases hyaluronan synthesis by up-regulation of hyaluronan Synthase 2 through decreased ceramide production and activation of Akt. J Biol Chem. 2012 doi: 10.1074/jbc.M111.304857. doi:M111.304857 [pii] 10.1074/jbc.M111.304857 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke CJ, Guthrie JM, Hannun YA. Regulation of neutral sphingomyelinase-2 (nSMase2) by tumor necrosis factor-alpha involves protein kinase C-delta in lung epithelial cells. Mol Pharmacol. 2008;74(4):1022–1032. doi: 10.1124/mol.108.046250. doi:mol.108.046250 [pii] 10.1124/mol.108.046250 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clarke CJ, Truong TG, Hannun YA. Role for neutral sphingomyelinase-2 in tumor necrosis factor alpha-stimulated expression of vascular cell adhesion molecule-1 (VCAM) and intercellular adhesion molecule-1 (ICAM) in lung epithelial cells: p38 MAPK is an upstream regulator of nSMase2. J Biol Chem. 2007;282(2):1384–1396. doi: 10.1074/jbc.M609216200. doi:M609216200 [pii] 10.1074/jbc.M609216200 [doi] [DOI] [PubMed] [Google Scholar]
- 56.Adam-Klages S, Adam D, Wiegmann K, Struve S, Kolanus W, Schneider-Mergener J, Kronke M. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell. 1996;86(6):937–947. doi: 10.1016/s0092-8674(00)80169-5. doi:S0092-8674(00)80169-5 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Philipp S, Puchert M, Adam-Klages S, Tchikov V, Winoto-Morbach S, Mathieu S, Deerberg A, Kolker L, Marchesini N, Kabelitz D, Hannun YA, Schutze S, Adam D. The Polycomb group protein EED couples TNF receptor 1 to neutral sphingomyelinase. Proc Natl Acad Sci U S A. 2010;107(3):1112–1117. doi: 10.1073/pnas.0908486107. doi:0908486107 [pii] 10.1073/pnas.0908486107 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldkorn T, Filosto S. Lung injury and cancer: Mechanistic insights into ceramide and EGFR signaling under cigarette smoke. American journal of respiratory cell and molecular biology. 2010;43(3):259. doi: 10.1165/rcmb.2010-0220RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy M, Castillo SS, Goldkorn T. nSMase2 activation and trafficking are modulated by oxidative stress to induce apoptosis. Biochem Biophys Res Commun. 2006;344(3):900–905. doi: 10.1016/j.bbrc.2006.04.013. doi:S0006-291X(06)00788-1 [pii] 10.1016/j.bbrc.2006.04.013 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levy M, Khan E, Careaga M, Goldkorn T. Neutral sphingomyelinase 2 is activated by cigarette smoke to augment ceramide-induced apoptosis in lung cell death. Am J Physiol Lung Cell Mol Physiol. 2009;297(1):L125–L133. doi: 10.1152/ajplung.00031.2009. doi:00031.2009 [pii] 10.1152/ajplung.00031.2009 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Filosto S, Castillo S, Danielson A, Franzi L, Khan E, Kenyon N, Last J, Pinkerton K, Tuder R, Goldkorn T. Neutral sphingomyelinase 2: a novel target in cigarette smoke-induced apoptosis and lung injury. Am J Respir Cell Mol Biol. 2011;44(3):350–360. doi: 10.1165/rcmb.2009-0422OC. doi:2009-0422OC [pii] 10.1165/rcmb.2009-0422OC [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends in Cell Biology. 2012 doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Aubin I, Adams CP, Opsahl S, Septier D, Bishop CE, Auge N, Salvayre R, Negre-Salvayre A, Goldberg M, Guenet JL, Poirier C. A deletion in the gene encoding sphingomyelin phosphodiesterase 3 (Smpd3) results in osteogenesis and dentinogenesis imperfecta in the mouse. Nat Genet. 2005;37(8):803–805. doi: 10.1038/ng1603. doi:ng1603 [pii] 10.1038/ng1603 [doi] [DOI] [PubMed] [Google Scholar]
- 64.Khavandgar Z, Poirier C, Clarke CJ, Li J, Wang N, McKee MD, Hannun YA, Murshed M. A cell-autonomous requirement for neutral sphingomyelinase 2 in bone mineralization. J Cell Biol. 2011;194(2):277–289. doi: 10.1083/jcb.201102051. doi:jcb.201102051 [pii] 10.1083/jcb.201102051 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Glorieux FH, Ward LM, Rauch F, Lalic L, Roughley PJ, Travers R. Osteogenesis imperfecta type VI: a form of brittle bone disease with a mineralization defect. J Bone Miner Res. 2002;17(1):30–38. doi: 10.1359/jbmr.2002.17.1.30. doi:10.1359/jbmr.2002.17.1.30 [doi] [DOI] [PubMed] [Google Scholar]
- 66.Tomiuk S, Hofmann K, Nix M, Zumbansen M, Stoffel W. Cloned mammalian neutral sphingomyelinase: functions in sphingolipid signaling? Proc Natl Acad Sci U S A. 1998;95(7):3638–3643. doi: 10.1073/pnas.95.7.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matmati N, Hannun YA. Thematic review series: sphingolipids. ISC1 (inositol phosphosphingolipid-phospholipase C), the yeast homologue of neutral sphingomyelinases. J Lipid Res. 2008;49(5):922–928. doi: 10.1194/jlr.R800004-JLR200. doi:R800004-JLR200 [pii] 10.1194/jlr.R800004-JLR200 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tomiuk S, Zumbansen M, Stoffel W. Characterization and subcellular localization of murine and human magnesium-dependent neutral sphingomyelinase. J Biol Chem. 2000;275(8):5710–5717. doi: 10.1074/jbc.275.8.5710. [DOI] [PubMed] [Google Scholar]
- 69.Mizutani Y, Tamiya-Koizumi K, Nakamura N, Kobayashi M, Hirabayashi Y, Yoshida S. Nuclear localization of neutral sphingomyelinase 1: biochemical and immunocytochemical analyses. J Cell Sci. 2001;114(Pt 20):3727–3736. doi: 10.1242/jcs.114.20.3727. [DOI] [PubMed] [Google Scholar]
- 70.Sawai H, Domae N, Nagan N, Hannun YA. Function of the cloned putative neutral sphingomyelinase as lyso-platelet activating factor-phospholipase C. J Biol Chem. 1999;274(53):38131–38139. doi: 10.1074/jbc.274.53.38131. [DOI] [PubMed] [Google Scholar]
- 71.Clarke CJ, Cloessner EA, Roddy PL, Hannun YA. Neutral sphingomyelinase 2 (nSMase2) is the primary neutral sphingomyelinase isoform activated by tumour necrosis factor-alpha in MCF-7 cells. Biochem J. 2011;435(2):381–390. doi: 10.1042/BJ20101752. doi:BJ20101752 [pii] 10.1042/BJ20101752 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Long JZ, Cisar JS, Milliken D, Niessen S, Wang C, Trauger SA, Siuzdak G, Cravatt BF. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat Chem Biol. 2011;7(11):763–765. doi: 10.1038/nchembio.659. doi:nchembio.659 [pii] 10.1038/nchembio.659 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yabu T, Shimuzu A, Yamashita M. A novel mitochondrial sphingomyelinase in zebrafish cells. J Biol Chem. 2009;284(30):20349–20363. doi: 10.1074/jbc.M109.004580. doi:M109.004580 [pii] 10.1074/jbc.M109.004580 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mullen TD, Obeid LM. Ceramide and Apoptosis: Exploring the Enigmatic Connections Between Sphingolipid Metabolism and Programmed Cell Death. Anticancer Agents Med Chem. 2011 doi: 10.2174/187152012800228661. doi:BSP/ACAMC/E-Pub/ 00187 [pii] [DOI] [PubMed] [Google Scholar]
- 75.Petcoff DW, Holland WL, Stith BJ. Lipid levels in sperm, eggs, and during fertilization in Xenopus laevis. J Lipid Res. 2008;49(11):2365–2378. doi: 10.1194/jlr.M800159-JLR200. doi:M800159-JLR200 [pii] 10.1194/jlr.M800159-JLR200 [doi] [DOI] [PubMed] [Google Scholar]
- 76.Krut O, Wiegmann K, Kashkar H, Yazdanpanah B, Kronke M. Novel tumor necrosis factor-responsive mammalian neutral sphingomyelinase-3 is a C-tail-anchored protein. J Biol Chem. 2006;281(19):13784–13793. doi: 10.1074/jbc.M511306200. doi:M511306200 [pii] 10.1074/jbc.M511306200 [doi] [DOI] [PubMed] [Google Scholar]
- 77.Bernardo K, Krut O, Wiegmann K, Kreder D, Micheli M, Schafer R, Sickman A, Schmidt WE, Schroder JM, Meyer HE, Sandhoff K, Kronke M. Purification and characterization of a magnesium-dependent neutral sphingomyelinase from bovine brain. J Biol Chem. 2000;275(11):7641–7647. doi: 10.1074/jbc.275.11.7641. [DOI] [PubMed] [Google Scholar]
- 78.Corcoran CA, He Q, Ponnusamy S, Ogretmen B, Huang Y, Sheikh MS. Neutral sphingomyelinase-3 is a DNA damage and nongenotoxic stress-regulated gene that is deregulated in human malignancies. Mol Cancer Res. 2008;6(5):795–807. doi: 10.1158/1541-7786.MCR-07-2097. doi:6/5/795 [pii] 10.1158/1541-7786.MCR-07-2097 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]