Abstract
Objectives: The aim of this study was to develop a small interfering RNA (siRNA) against the expression of KIR3DL1 receptor on natural killer (NK) cells, in order to promote the ability of NK cells to destroy human immunodeficiency virus (HIV)-infected cells and thus prevent failure of siRNA therapy targeting human immunodeficiency virus type 1 (HIV-1) virus among HIV-1 infected patients in vitro. Methods: A siRNA targeting KIR3DL1 was synthesized and then modified with cholesterol, methylene, and sulfate. The inhibitory action of the siRNAs on primary cultured NK cells was detected. The amount of IFN-γ and TNF-α secretions in NK cells was measured. The intended functions of NK cells in vitro were analyzed by CFSE and PI methods. Results: There were no significant differences in inhibiting the expression of KIR3DL1 on NK cells between the modified and unmodified siRNAs, while inhibition by each of them differed significantly from controls. The amount of IFN-γ and TNF-α secretions in the NK cells was abundant due to unsuccessful expression of KIR3DL1 on NK cells, which further promoted function of the NK cells. Conclusion: The siRNA against KIR3DL1 could enhance the ability of the NK cells to kill the HIV-1 infected cells in vitro and successfully prevented the failure of siRNA therapy targeting the HIV-1 virus. Therefore, it can act as a potential gene therapeutic agent among HIV-1 infected people.
Introduction
During 2011 in China, the number of new human immunodeficiency virus (HIV) infections was estimated to be 48,000, with 28,000 people dying of AIDS and approximately 780,000 people living with HIV/acquired immune deficiency syndrome (AIDS), giving a national HIV prevalence of 0.58% (13). The worsening of the HIV/AIDS epidemic, as indicated by these increasing numbers of HIV infection and AIDS-related deaths, has thus culminated in a major challenge while achieving the target specified in the United Nation General Assembly Political Declaration on HIV/AIDS in China.
RNA interference (RNAi) is a safe and effective way to study genetic and therapeutic function in the desired target tissue with therapeutic doses (1). Small interfering RNA (siRNA)-based therapeutic strategies have had limited application in combatting HIV/AIDS because HIV could escape from the RNAi mechanism (15). Some studies did show that RNAi-mediated suppression could downregulate the human immunodeficiency virus type 1 (HIV-1) replication in CD4+ T-cells (19). The growing body of scientific literature dealing with HIV demonstrated that numerous siRNAs targeting a number of HIV-1 or cellular transcripts could achieve viral inhibition both in vitro and in vivo. Indeed, all the HIV-1-encoded genes such as tat, rev, gag, pol, nef, vif, env, vpr, and the long terminal repeat were found to be susceptible to RNAi-induced gene silencing in cell lines (21,25,29).
KIR3DL1 is an important inhibitory receptor on natural killer (NK) cells, the expression of which results in changes of the killing function and secretion of cytokines in the NK cells. Epidemiologic studies in humans have shown that carriers of combinations of genes encoding certain KIR3DL1 alleles and their HLA-Bw4 ligands could result in slower progression to AIDS, lowering the viral load, and preventing HIV progression (4). Previous studies have revealed that the expression of KIR3DL1 receptors in NK cells could vary with the NK cell counts from the peripheral blood mononuclear cell in HIV-1 infected people. Moreover, the changes were related to the HIV-1 viral load and CD4+ T-cell counts in HIV-1 infected patients (10,11).
We hypothesized that the inhibition of KIR3DL1 expression on NK cells could result in increasing antiviral activities in HIV-1 infected people. Thus, we developed a RNA interfering inhibition assay in vitro to test this hypothesis. In this paper, we demonstrate that synthesized siRNA could inhibit the expression of KIR3DL1 on NK cells and subsequently promote the expressions of IFN-γ and TNF-α in NK cells. Thus, the killing function would significantly increase in vitro and successfully prevent the failure of siRNA therapy targeting HIV-1 virus directly among AIDS patients. Therefore, siRNA against KIR3DL1 may be considered as the potential gene therapeutic agent in HIV-1 infected people.
Materials and Methods
Synthesis and modification of siRNA targeting KIR3DL1
The sequence of the siRNA for KIR3DL1 (K5) was selected from GenBank accession number NM_013289 with nucleotide sequences 1042–1060. The sense oligo was 5′-ACA GAA CCA AGC UCC AAA UdT dT-3′, and the antisense oligo was 3′-dTd TUG UCU UGG UUC GAG GUU UA-5′. The siRNA was then modified with cholesterol, methylene, and sulfate mainly in the head of sense or/and antisense. The specific modifications are listed in Table 1. The siRNA targeted to Luciferase GL2 gene as the scramble siRNA control; the sense oligo was 5′-CGU ACG CGG AAU ACU UCG AdT dT-3′, and the antisense oligo was 3′-dTd TGC AUG CGC CUU AUG AAG CU-5′. The synthesis, modification, and annealing steps were conducted separately by the RiboBio Corporation, and the siRNAs were dissolved in sterilized and RNase-free water, the final concentration being 20 μM.
Table 1.
The Small Interfering RNAs (siRNAs) of K5 and the Specific Modifications with Different Chemical Groups
| Name | Sequence | Modifications |
|---|---|---|
| K5 | 5′-ACA GAA CCA AGC UCC AAA UdT dT-3′ | |
| 3′-dTd TUG UCU UGG UUC GAG GUU UA-5′ | ||
| K6 | 5′-chol- ACA GAA CCA AGC UCC AAA UdT dT-3′ | chol |
| 3′-dTd TUG UCU UGG UUC GAG GUU UA-5′ | ||
| K7 | 5′-chol- A-s-C- s-A-s- GAA CCA AGC UCC AAA-s- UdT dT-3′ | chol, s |
| 3′-dTd TUG UCU UGG UUC GAG GUU UA-5′ | ||
| K8 | 5′-chol- (mA)(mC)(mA) GAA CCA AGC UCC A(mA) (mA) (mU) dT-s -dT-3′ | chol, m, s |
| 3′-dT-s- dTU GUC (mU) UGG U(mU) CGA GGU U(mU) A-5′ | ||
| K9 | 5′-chol- A-s-C- s-AGA ACC AAG CUC CAA AUdT-s- dT-3′ | chol, s |
| 3′-dTd TUG UCU UGG UUC GAG GUU UA-5′ | ||
| K10 | 5′-chol- (mA)(mC)(mA) GAA CCA AGC UCC AAA UdT dT-3′ | chol, m |
| 3′-dTd TUG UCU UG GUU CGA GGU UUA-5′ | ||
| K11 | 5′-chol- (mA)(mC)(mA) GAA CCA AGC UCC A(mA)(mA)(mU) dT-s- dT-3′ | chol, m, s |
| 3′-dTd TUG UCU UGG UUC GAG GUU UA-5′ |
Chol, cholesterol; m, methylene; s, sulfate.
Study subjects
Ten HIV-1 infected adults from Jiangsu province, China, participated in the study, of whom five were male, aged 32–45 years (M=38.40±4.35 years). All of the subjects were married, infected with HIV-1 through the heterosexual route of transmission, and diagnosed within a period of 1 year. Epidemiologic investigations and laboratory confirmations were conducted through the China Information System for Disease Control and Prevention. According to the eligibility criteria, recruited subjects had normal liver and kidney functions, no history of cancer or other diseases that might have an effect on their immune system, had not progressed to AIDS, and were not on highly active antiretroviral therapy (HAART).
CD4+ T-cell counts of the participants were determined using EDTA-anti-coagulated whole blood samples. Cells were stained with mixed antibodies of “MultiTEST CD3-FITC/CD8-PE/CD45-PerCP/CD4-APC Reagent” (Becton Dickinson) and the labeled cells were analyzed with the BD FACSCalibur™ flow cytometer, acquiring 105 cells.
Plasma HIV-1 viral load (VL) was quantified with AMPLICOR HIV-1 MONITOR (Roche) within 6 h of blood collection and plasma separation. The detection limitation was 40 copies/μL.
Ethic statement
Signed informed consent was obtained from each participant prior to blood collection. Participants could decline or withdraw from the study at any time. The consent documents were kept confidentially in locked cupboards at the study sites, and unauthorized persons had no access to them. The study process and content were approved by the Ethics Committee at Jiangsu Provincial Center for Disease Prevention and Control (JSCDC).
Culture of NK cells from peripheral blood mononuclear cells
Peripheral blood mononuclear cells (PBMC) were collected from the blood samples, washed twice with phosphate buffered saline (PBS), and suspended in 1 mL RPMI-1640 containing 10% fetal bovine serum (FBS). Then 200 U/mL concentrations of IL-2 (PeproTech) and 50 ng/mL PMA (Phorbol 12-Myristate 13 Acetate; Alexis Biochemicals) were added to the cells, followed by stimulation and amplification of the cells at 37°C in 5% CO2 for 24 h separately (18).
Transfection of siRNAs into NK cells in vitro
To enhance the efficiency of transfection of these siRNAs, oligofectamine (Invitrogen), a cationic liposome, was mixed with the siRNAs before the treatment. The NK cells, which were stimulated and amplified from PBMC for 24 h, were then separately transfected at a density of 5×104 cells/well in six-well plates with siRNA against KIR3DL1. According to the manufacturer's specification, 0.4 μM siRNA mixed with oligofectamine (8 μL/mL) was transfected into the former NK cells in each well for 72 h respectively. The wells of cultured NK cells were transfected with 0.4 μM scramble siRNA mixed with 8 μL/mL oligofectamine as controls.
Assay of the KIR3DL1 expression on NK cells
Fluoresence-activated cell sorting (FACS) flow cytometry analysis was performed to detect the KIR3DL1 expression on NK cells, which were interfered with siRNA as mentioned above. First, the siRNA interfered NK cells were harvested and washed with PBS. Then the cells were labeled with the mouse anti-human CD56 PE-Cy5.5/CD3 Alex Flour 488/anti-CD16-APC antibody (Invitrogen) in the dark for 15 min before anti-KIR3DL1-PE (Biolegend) was added and incubated in the dark for a further 15 min. Finally, the labeled cells were analyzed with the BD FACSCalibur flow cytometer, and 105 cells were acquired. The NK cells were CD3−/CD16+/CD56+ subpopulation cells.
Reverse transcription polymerase chain reaction (RT-PCR) was used to determine the efficacy of modified siRNAs targeting KIR3DL1 on NK cells. The primers of KIR3DL1 were: F-Primer 5′-TCA AGT AGT TGG CCT TCA C-3′, and R-Primer 5′-TAG AAG ACG TCC TCA AGG C-3′. Human GAPDH was used as the reference gene, the primers of which were as follows: F-Primer 5′-ACC ACA GTC CAT GCC ATC AC-3′, and R-Primer 5′-TCC ACC ACC CTG TTG CTG TA-3′. The RNA interfered NK cells were harvested and washed with pre-cold PBS. Then the cells were dissolved in 500 μL Trizol (Invitrogen). The RNA extraction and RT-PCR were performed according to the manufacturer's instructions.
Quantification of cytokines secreted by NK cells with FACS flow cytometry analysis
The RNA interfered NK cells were harvested and labeled with the mouse anti-human CD56 PE-Cy5.5/CD3 Alex Flour 488/anti-CD16-APC antibody. Then the labeled cells were fixed with 4% paraformaldehyde for 20 min. After washing twice with PBS, 0.1% saponin was added to destroy the cell membranes. Finally, the cells were incubated with anti-human IFN-γ-PE (eBioscience) and anti-human TNF-α-PE (eBioscience) for 30 min respectively, and 105 cells were acquired.
Assessment of the function of NK cells
The RNA interfered NK cells were harvested and suspended with 1 mL of the former NK cell culture medium, and the cells were cultured at 37°C in 5% CO2 for 24 h. Next, K562 cells were collected as the targets and were labeled with a final concentration of 2 μM CFSE (Sigma-Aldrich) for 10 min at 37°C. Then, the labeled cells were washed twice with PBS. The effector/target ratio was set at 10:1; the target cells of each sample were 105 cells according to our previous study (10). All the samples were then mixed to form a final volume of 200 μL incomplete medium without FBS, IL-2, and PMA in a 96-well flat-bottom plate, and incubated at 37°C in 5% CO2 for 4 h. The samples were then put into an ice water bath, and a final concentration of 100 μg/mL PI (Sigma-Aldrich) was added and incubated for 5 min in order to label the DNA of the dead cells. The dead target cells were analyzed within the next 60 min.
All samples were analyzed on a BD FACSCalibur using the CellQuest software, and 5,000 target events were collected. CFSE+/PI+ cells were considered as dead target cells. The percentage of specific target cell death (killing ratio) was determined with the help of the CellQuest software.
Statistical analysis
Results were expressed as mean±standard deviation (SD). Statistical significance was assessed by nonparametric tests performed using SPSS v11.5 (SPSS, Inc.). Results were considered to be statistically significant at p<0.05.
Results
CD4+ T-cell counts and HIV-1 viral load among HIV-1 infected adults
The CD4+ T-cell counts among study subjects ranged from 323 to 452 (365.40±37.52)/μL while the CD4+ T-cell counts of healthy people in China ranged from 410 to 1041/μL. Among these participants, HIV-1 viral loads could not be detected because the numbers of copies of HIV-1 viral load were below the lower threshold for the detection limit.
Screening of the effective siRNAs from modified siRNAs
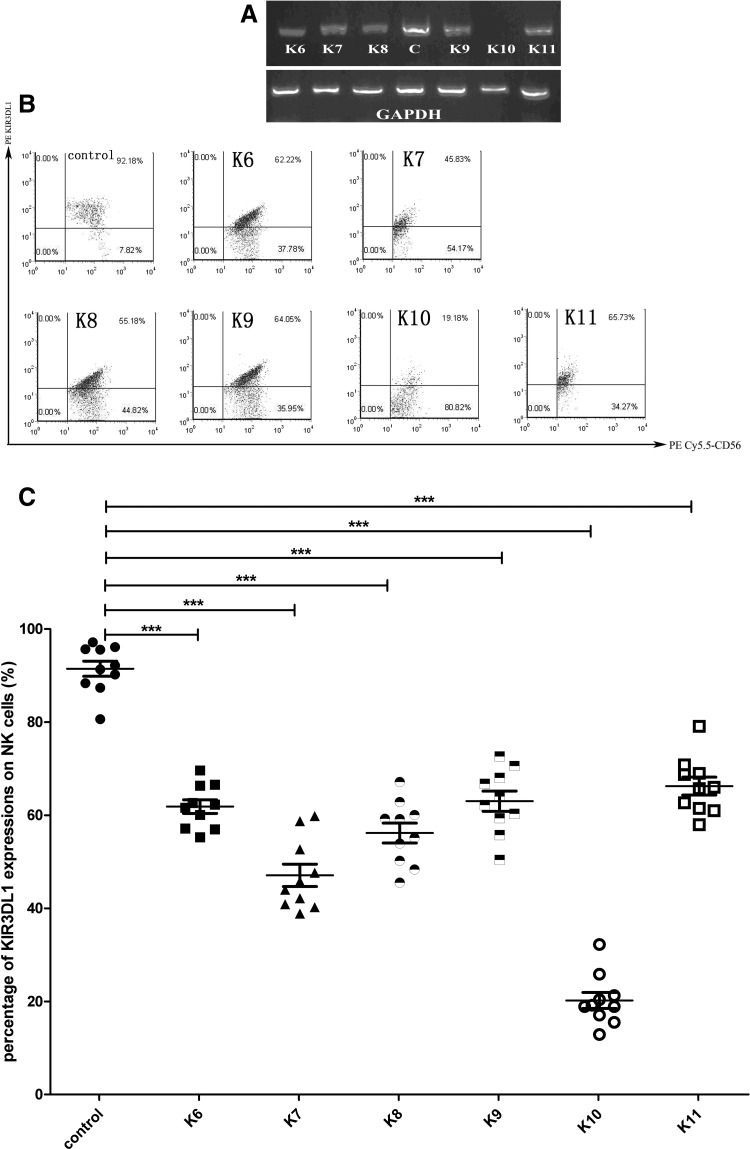
Previous studies have shown that the sequence of the siRNA K5 was the effective siRNA targeting the KIR3DL1 gene (27). Here, we modified the K5 using different chemical groups in order to promote the transfection into the target cells and to slow down the degradation in the target cells. Our results indicate that different modification could result in different inhibition of KIR3DL1. The most effective siRNA was K10, which was modified with cholesterol and methylene. The RT-PCR results were consistent with the FACS flow cytometry analysis results. There were significant differences between the groups of K6-K11 and the control group (p<0.001; Fig. 1A–C).
FIG. 1.
(A) Reverse transcription polymerase chain reaction (RT-PCR) analysis of the different modified small interfering RNAs (siRNAs) interfering to KIR3DL1, GAPDH being the reference gene. (B) Fluoresence-activated cell sorting (FACS) flow cytometry analysis of the different modified siRNAs. The average of KIR3DL1 expression on NK cells that were interfered with scramble siRNA as the control and the KIR3DL1 expressions on NK cells that were interfered with K6-K11 were 91.47±5.10, 61.86±4.70, 47.09±7.58, 56.18±6.79, 63.03±6.86, 20.22±5.45, and 66.25±6.08 respectively. (C) The distributions of expressing percentage of KIR3DL1 on NK cells that were interfered with control and K6-K11 siRNAs. Groups of the KIR3DL1 expressions on NK cells that were interfered with K6-K11 were significantly different compared to the control group (***p<0.001).
Comparative effects of K5 and K10 in inhibiting the expression of KIR3DL1 on NK cells
The amplified NK cells of study subjects were used to compare the differences in inhibiting the expression of KIR3DL1 on NK cells by K5 and K10. The results of RT-PCR and the FACS flow cytometry analysis showed that there was no statistically significant difference in inhibiting the expression of KIR3DL1 on NK cells by K5 and K10 (p>0.05). On the other hand, significant differences in inhibition were observed while separately comparing K5 (p<0.001) and K10 (p<0.001) with the control group (Fig. 2A and B).
FIG. 2.
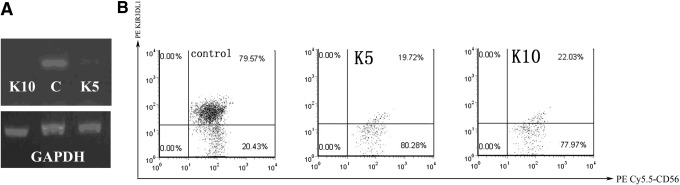
(A) RT-PCR analysis of K5 and K10 interfering to KIR3DL1, GAPDH being the reference gene. (B) FACS flow cytometry analysis of KIR3DL1 expression on NK cells that were interfered with K5 and K10. The average of KIR3DL1 expression on NK cells that were interfered with scramble siRNA as the control and the KIR3DL1 expressions on NK cells that were interfered with K5 and K10 were 89.25±5.08, 22.10±4.75, and 20.79±4.64 respectively. Groups of the KIR3DL1 expressions on NK cells that were interfered with K5 and K10 were significantly different compared to the control group (p<0.001).
Secretion of cytokines in NK cells
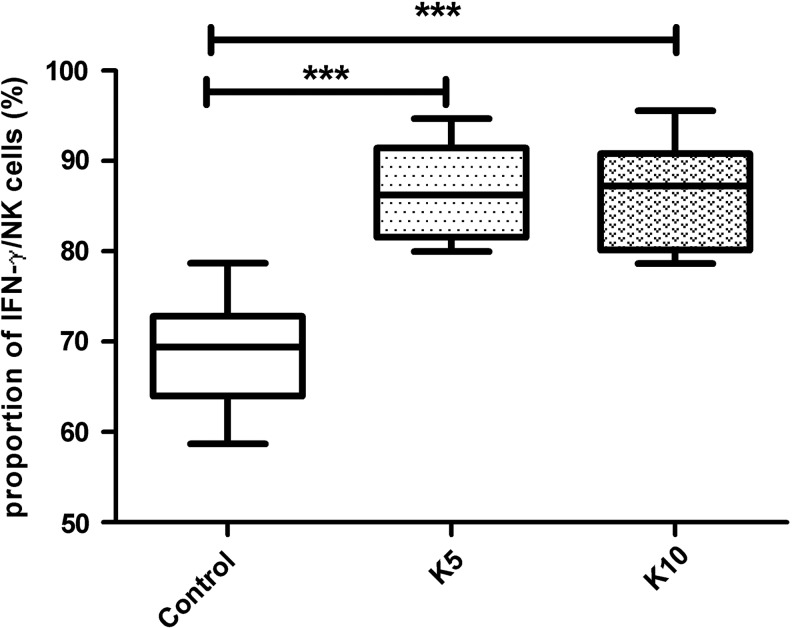
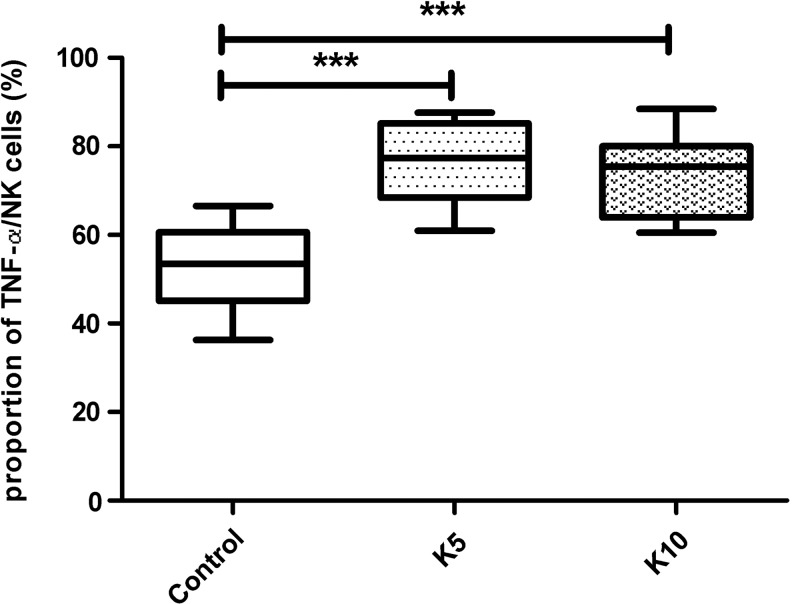
In humans, IFN-γ and TNF-α are mainly secreted by NK cells. Our results revealed that, compared to the control group, secretion of IFN-γ and TNF-α differed significantly among the subjects treated with K5 and K10 (p<0.001; Figs 3 and 4).
FIG. 3.
The proportions of IFN-γ secreted by NK cells that were interfered with K5 and K10. The proportions of IFN-γ in the control group and interfering with the K5 and K10 groups were 68.84±6.09, 86.70±4.98, and 86.52±5.51 respectively. There were significant differences between the K5 and K10 interfering groups and the control group (***p<0.001).
FIG. 4.
The proportions of TNF-α secreted by NK cells that were interfered with K5 and K10. The proportions of TNF-α in control and interfering with the K5 and K10 groups were 52.89±9.25, 76.67±8.78, and 73.77±9.13 respectively. There were significant differences between the K5 and K10 interfering groups and the control group (***p<0.001).
Killing function of NK cells
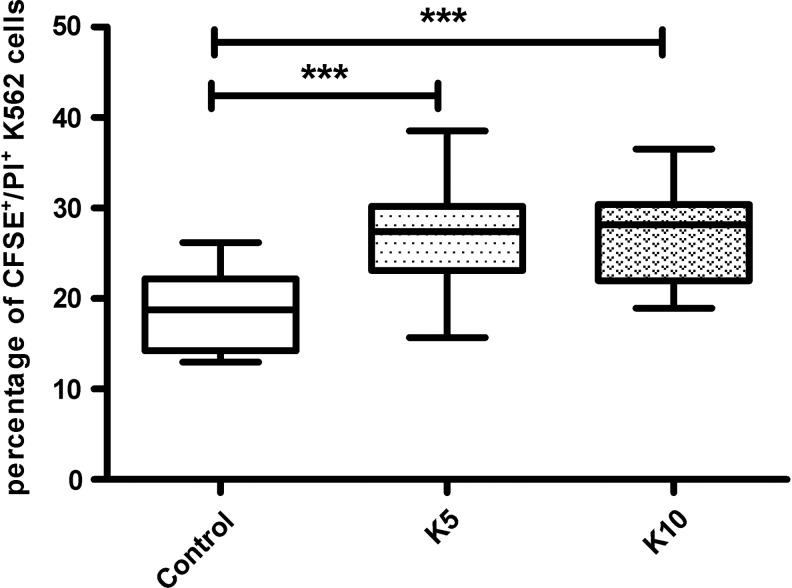
The killing function of NK cells was assessed by K562 as the target cell. The percentage of CFSE+/PI+ cells in control subjects was 18.62±4.50%. However, the percentage of CFSE+/PI+ cells in K5 and K10 treated groups was 26.98±6.13% and 27.27±5.20% respectively, and the differences were significant (p<0.001; Fig. 5), indicating that the NK cell killing function was promoted by RNA interference targeting KIR3DL1.
FIG. 5.
NK cells killing ratio analyzed by K562 cells. The killing ratio of NK cells that were interfered with scramble siRNA as the control and the killing ratios of NK cells that were interfered with K5 and K10 were 18.62±4.50, 26.98±6.13, and 27.27±5.20 respectively. There was a statistically significant difference between the control group and K5 and K10 interfering groups (***p<0.001).
Discussion
NK cells constitute an integral part of innate immunity, and these cells play an important role in controlling viral infections (6). They are regulated by the integration of signals transmitted internally by activating and inhibiting surface receptors such as killer immunoglobulin-like receptors (KIRs), which include alleles encoded by the KIR3DL1 framework locus (17). NK cell function and KIR have not only been linked to the prevention of HIV disease progression, but also to the development of resistance against HIV infection in certain individuals who remain seronegative despite repeated exposure to HIV (3). Costa et al. observed low expression of inhibitory NK receptors in CD8+ cytotoxic T-lymphocytes in long-term nonprogressor HIV-1 infected patients (7). KIR3DL1 was found to act in concert with HLA-B locus for controlling the progression of disease among HIV-1 infected patients in a specific sample of Zambian patients (16). Several studies have also revealed that NK cell activity might well influence the progression of many other infectious diseases, including hepatitis, tuberculosis, and cancers (12,20,22).
Since the discovery of RNAi in 1998 (9), it has become a popular method for gene function analysis (28), and it has also been explored rapidly for therapeutic applications (5,8,14). Although current HAART treatment for HIV/AIDS has been therapeutically very effective in the majority of patients, drug resistance and toxicity issues remain serious concerns (23). RNAi is considered to have some unique therapeutic attributes for the treatment of HIV-1 infection in the 21st century (2). Despite several years of devoted effort in search of the best therapeutic siRNA targeting HIV, researchers have so far failed because of the increased rate of mutation in HIV. Moreover, many promising therapeutic siRNAs effective in vitro were not found to be functional in vivo (24,26).
We have developed a modified therapeutic siRNA targeting KIR3DL1 that has been successfully transferred to cells and remained unaffected by mutations of the virus. No obvious differences between modified siRNA and nonmodified siRNA were observed while inhibiting the KIR3DL1 expression on NK cells in vitro. Both modified and nonmodified siRNAs significantly lowered KIR3DL1 expression on NK cells.
It was observed that lower expression of KIR3DL1 could improve the ability of the secretion of some cytokines, such as IFN-γ and TNF-α in NK cells, which also in turn could enhance the killing function of NK cells. These findings suggested that KIR3DL1 could play a crucial role in CD8+ CTL and NK cell responses to HIV-1 virus. Therefore, lower expression of KIR3DL1 could also slow down the progression of HIV infection into AIDS, and could also protect the individuals to remain seronegative despite repeated exposure to HIV. Thus, it can be concluded that siRNA against KIR3DL1 may be considered as a potential gene therapeutic agent for HIV-1 infected people.
Acknowledgments
The project was supported by National Natural Science Foundation of China (Grant No. 81373125), Jiangsu Provincial Natural Science Program (BK2009435), and Jiangsu Province's Outstanding Medical Academic Leader Program (RC2011086, 2011087).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Akinc A, Zumbuehl A, Goldberg M, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol 2008;26:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout B, and Das AT. HIV-1 escape from RNAi antivirals: yet another Houdini action? Mol Ther Nucleic Acids 2012;1:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulet S, Kleyman M, Kim JY, et al. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 2008;22:1487–1491 [DOI] [PubMed] [Google Scholar]
- 4.Boulet S, Song R, Kamya P, et al. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol 2010;184:2057–2064 [DOI] [PubMed] [Google Scholar]
- 5.Castanotto D, and Rossi JJ. The promises and pitfalls of RNA-interference based therapeutics. Nature 2009;457:426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerwenka A, and Lewis LL. Natural killer cells, viruses and cancer. Nat Rev Immunol 2001;1:41–49 [DOI] [PubMed] [Google Scholar]
- 7.Costa P, Rusconi S, Fogli M, et al. Low expression of inhibitory natural killer receptors in CD8 cytotoxic T lymphocytes in long-term non-progressor HIV-1–infected patients. AIDS 2003;17:257–260 [DOI] [PubMed] [Google Scholar]
- 8.De Fougerolles A, Vornlocher HP, Maraganore J, et al. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov 2007;6:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998;391:806–811 [DOI] [PubMed] [Google Scholar]
- 10.Fu GF, Chen X, Hao S, et al. Differences in natural killer cell quantification and receptor profile expression in HIV-1 infected Chinese children. Cell Immunol 2010;265:37–43 [DOI] [PubMed] [Google Scholar]
- 11.Fu GF, Hao S, Zhao JL, et al. Changes in NK cell counts and receptor expressions and emergence of CD3dim/CD56+ cells in HIV-1 infected patients in China. Viral Immunol 2009;22:105–116 [DOI] [PubMed] [Google Scholar]
- 12.Gross E, Sunwoo JB, Bui JD. Cancer immunosurveillance and immunoediting by natural killer cells. Cancer J 2013;19:483–489 [DOI] [PubMed] [Google Scholar]
- 13.Hvistendahl M. China partners with gay groups on HIV screening. Science 2013;339:17–18 [DOI] [PubMed] [Google Scholar]
- 14.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed Engl 2009;48:1378–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence D. RNAi could hold promise in the treatment of HIV. Lancet 2002;359:2007. [DOI] [PubMed] [Google Scholar]
- 16.López-Vázquez A, Miña-Blanco A, Martínez-Borra J, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum Immunol 2005;66:285–289 [DOI] [PubMed] [Google Scholar]
- 17.Martin MP, Qi Y, Gao X, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 2007;39:733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montoya CJ, Velilla PA, Chougnet C. Increased IFN-γ production by NK and CD3+/CD56+ cells in sexually HIV-1-exposed but uninfected individuals. Clin Immunol 2006;120:138–146 [DOI] [PubMed] [Google Scholar]
- 19.Neff CP, Zhou J, Remling L, et al. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4+ T cell decline in humanized mice. Sci Transl Med 2011;3:66ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parasa VR, Sikhamani R, Raja A. Effect of recombinant cytokines on the expression of natural killer cell receptors from patients with TB or/and HIV infection. Plos One 2012;7:e37448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podlekareva D, Mocroft A, Dragsted UB, et al. Factors associated with the development of opportunistic infections in HIV-1-infected adults with high CD4+ cell counts. J Infect Dis 2006;194:633–641 [DOI] [PubMed] [Google Scholar]
- 22.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat Med 2013;19:859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richman DD, Margolis DM, Delaney M, et al. The challenge of finding a cure for HIV infection. Science 2009;323:1304–1307 [DOI] [PubMed] [Google Scholar]
- 24.Sanghvi VR, and Steel LF. A re-examination of global suppression of RNA interference by HIV-1. PLoS One 2011;6:e17246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsygankov AY. Current developments in anti-HIV/AIDS gene therapy. Curr Opin Investig Drugs 2009;10:137–149 [PubMed] [Google Scholar]
- 26.Unwalla HJ, Li MJ, Kim JD, et al. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat Biotechnol 2004;22:1573–1578 [DOI] [PubMed] [Google Scholar]
- 27.Yang HT, Fu GF, Huan XP, et al. The application of small interfering RNA targeting to the NK cell receptor of KIR3DL1. 2014, patent number: 201210383220.8
- 28.Zamore PD. RNA interference: big applause for silencing in Stockholm. Cell 2006;127:1083–1086 [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, and Rossi JJ. Current progress in the development of RNAi-based therapeutics for HIV-1. Gene Ther 2011;18:1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]