Abstract
Kinase signaling is under tight spatiotemporal control, with signaling hubs within the cell often coordinated by protein scaffolds. Genetically encoded kinase activity reporters afford a unique tool to interrogate the rate, amplitude, and duration of kinase signaling at specific locations throughout the cell. This protocol describes how to assay kinase activity at a protein scaffold in live cells using a fluorescence resonance energy transfer (FRET)-based kinase activity sensor for protein kinase D (PKD) as an example.
Keywords: FRET, kinase activity reporter, DKAR, protein kinase D, scaffold protein, NHERF
1. Introduction
Phosphorylation of substrate proteins by protein kinases affords one of nature’s most effective mechanisms to reversibly regulate protein function. The simple addition of phosphate alters the chemical properties of the targeted surface, thus altering protein function by many mechanisms. For example, phosphorylation can modulate the intrinsic catalytic activity of the phosphorylated substrate; this includes other kinases and even the kinase, itself, via autophosphorylation. In addition, protein phosphorylation can regulate the subcellular localization of the substrate protein by affecting its association with other proteins or with lipids, either by altering the protein conformation or by altering the electrostatic properties of the interacting interface. Control of localization is particularly critical in cell signaling, where activation of kinases occurs at precise locations to effect localized signaling. Protein phosphatases oppose protein kinases, allowing acute regulation of the time period during which a protein is modified by phosphate. Thus, phosphorylation events are usually transient. Signaling by protein kinase D (PKD) family members affords one example of tight regulation of the spatial and temporal dynamics of kinase activity.
The PKD family plays a role in numerous processes, including cell proliferation and survival, immune cell signaling, gene expression, vesicle trafficking, and neuronal development [1]. The role this family plays thus depends on cell type (e.g. immune versus cancer cells) and subcellular localization (e.g. regulation of vesicle transport at the Golgi). The family comprises three members, PKD1, PKD2, and PKD3, each consisting of a conserved catalytic core, an amino-terminal regulatory domain containing tandem C1 domains, and, for PKD1 and PKD2, a PDZ-binding motif at the C-terminus [2]. The C1 domains bind diacylglycerol (DAG), a lipid second messenger that recruits PKD isozymes to membranes, a first step in PKD activation. Binding of the regulatory domain to membrane-embedded DAG results in a conformational change that poises PKD for subsequent phosphorylation by novel protein kinase C (PKC) family members at two sites within its catalytic core; this event is followed by PKD autophosphorylation at a site within its C-terminal tail [3,4]. Because phosphorylation is a hallmark of PKD activation, as it is for many other kinases, activity is traditionally demonstrated via Western blotting using phospho-specific antibodies to these activating sites. However, both the temporal and spatial resolution of this method are poor, limiting the approach for assessing kinase signaling in cells. Furthermore, while the sites probed are indicative of kinase activation, there may be other means of activating the kinase or opposing inactivating phosphorylations elsewhere on the kinase, neither of which will be taken into account when probing a specific phosphorylated site. These problems are all circumvented by use of genetically encoded, fluorescence resonance energy transfer (FRET)-based kinase activity reporters.
Genetically encoded, FRET-based kinase activity reporters enable real-time monitoring of localized kinase activity within cells. Such reporters often utilize a modular design whereby a FRET pair flanking a phospho-peptide binding domain and a substrate sequence undergoes a conformational change following phosphorylation of a consensus substrate sequence (Figure 1). Considerations in reporter design involve selection of a suitable FRET pair, identification of a kinase-specific substrate sequence, and selection of a compatible phosphoamino-binding module that binds efficiently to the phosphorylated substrate sequence, yet not with such high affinity that the phosphorylation cannot be reversed by phosphatases (detailed in [5]). For some kinases, additional modules that facilitate recognition by the kinase may be necessary; for example, the reporter of ERK activity includes a docking domain for ERK on its C-terminus [6]. The prototypical kinase activity reporters were designed in 2001 to read out activity from the tyrosine kinases Src, Abl, and EGFR [7] and PKA [8]. Since then, many new reporters have been developed based on this modular design; those reporters designed for protein kinases A through D (PKA through PKD) as well as their variants (usually improvements made to increase their sensitivity) are depicted in Table 1 [8-16].
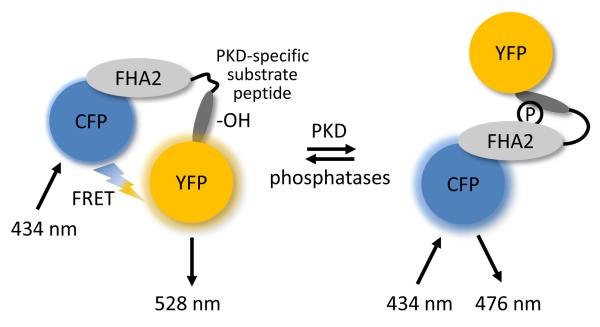
Figure 1.
Schematic diagram showing the modular structure of DKAR. Kinase activity reporters consist of a FRET donor (CFP) and acceptor (YFP) flanking a phosphoamino-acid binding domain (FHA2) and a consensus phosphorylation sequence (substrate peptide). In DKAR, the unphosphorylated reporter exists in a conformation wherein the FRET pair is undergoing FRET. When PKD is active, it phosphorylates the threonine within the consensus sequence (-OH), inducing a conformational change as the FHA2 domain binds the newly phosphorylated (circle with P) substrate peptide sequence. This intramolecular association alters the distance and/or relative orientation between the FRET pair, resulting in a decrease in FRET.
Table 1.
Evolution of kinase activity reporters for the serine/threonine protein kinases A through D.
| Kinase | Reporter | Notes | References |
|---|---|---|---|
| PKA | AKAR AKAR2 AKAR3 |
first serine/threonine kinase reporter, irreversible reversible response increased response range |
[8] [9] [10] |
| PKB/Akt | Aktus BKAR AktAR |
first PKB/Akt reporter increased sensitivity increased response range |
[11] [12] [13] |
| PKC | CKAR δCKAR |
first PKC reporter isozyme-specific (PKCδ) reporter |
[14] [15] |
| PKD | DKAR | first PKD reporter | [16] |
Genetically encoded reporters are introduced into cells by simple transfection, where they read out the rate, amplitude, and duration of endogenous (or exogenous) kinase activity in response to specific stimuli. Because they are genetically encoded, they can be targeted to subcellular regions through the addition of short targeting sequences. Such targeting allows determination of localized kinase activity occurring at the subcellular region being targeted. A variety of genetically encoded reporters have been targeted to subcellular locations such as the plasma membrane, Golgi, ER, mitochondria, and within the nucleus to reveal unique signatures of signaling at each location [5]. The subcellular targeting of the kinase activity reporter for PKD, DKAR (D Kinase Activity Reporter), is an example of how to measure localized kinase signaling.
Identification of an interaction of PKD via its PDZ-binding motif to the scaffold protein NHERF-1 (Na+/H+ exchanger regulatory factor-1) prompted studies to target DKAR to this protein complex and monitor signaling at a protein scaffold [17]. Addition of ten amino acids constituting a PDZ-binding motif to the C-terminus of DKAR (generating DKAR-PDZ) served to relocalize DKAR in a NHERF-1-dependent manner to the apical surface of the polarized epithelial cells, MDCK cells (Figure 2B). Thus, in cells where DKAR-PDZ is visibly localized to the NHERF scaffold, one can monitor PKD activity at NHERF, revealing highly enriched PKD signaling at this protein complex (Figure 3; [17]). Below, we describe how we utilized this DKAR-PDZ targeted to the NHERF protein scaffold to measure PKD signaling at this subcellular location in live cells.
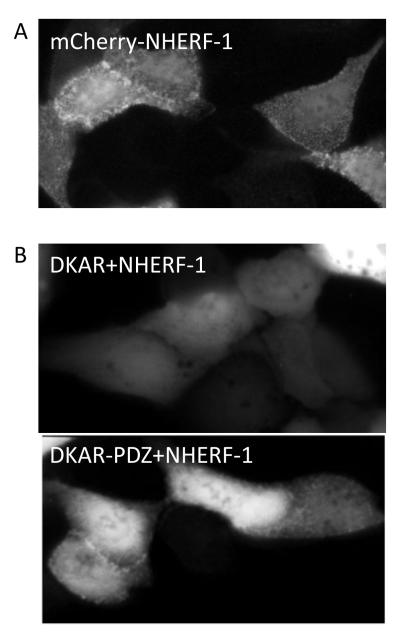
Figure 2.
Fluorescent images of MDCK cells. A. mCherry-tagged NHERF-1 localizes to NHERF complexes at the apical membrane of polarized epithelial cells (e.g. MDCK cells) and this presents in a punctate pattern. B. Untargeted DKAR is present throughout the cytosol and nucleus of the cell (top). Addition of the PDZ-targeting motif to DKAR (DKAR-PDZ) localizes it to the NHERF scaffold in NHERF-1-overexpressing MDCK cells (bottom); as not all cells display relocalization of DKAR-PDZ to the NHERF scaffold, care should be taken to select those that do.
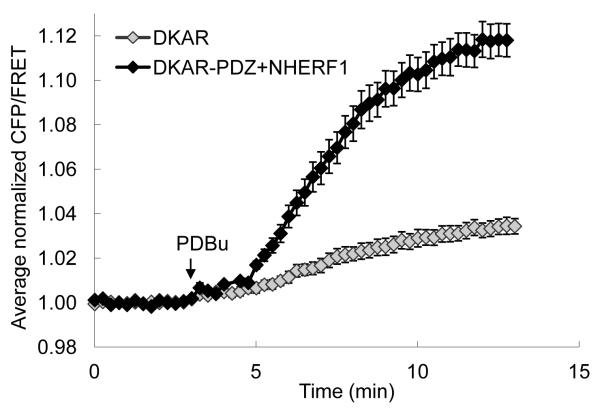
Figure 3.
Plot of the average normalized CFP/FRET ratio from DKAR or DKAR poised at NHERF-1 (DKAR-PDZ+NHERF1) following addition of PDBu. Adapted from (17).
2. Materials
2.1. Cell Culture
Madin Darby canine kidney (MDCK) cells (ATCC)
Dulbecco’s Modification of Eagle’s Medium/Ham’s F12 50/50 Mix (DMEM/F12) with 10% fetal bovine serum (FBS) (Cellgro)
35 mm, sterile, glass-bottom culture dishes, No. 1.0 (MatTek Corporation)
Effectene transfection reagent (QIAGEN)
DNA encoding DKAR, DKAR-PDZ, NHERF-1
2.2. Microscope Setup
Details listed here are specific to the experimental setup that our laboratory uses:
Axiovert 200M microscope (Zeiss) with Xenon lamp (XBO 75/2 OFR, Zeiss)
MicroMAX 512BFT CCD camera (Roper Scientific)
- Filters from Chroma Technology
- for cyan fluorescent protein (CFP): 420/20 nm excitation, 450 nm dichroic, 475/40 nm emission
- for yellow fluorescent protein (YFP): 495/10 nm excitation, 505 nm dichroic, 535/25 nm emission
- 10% neutral density filter (22000A ND filter 1.0)
Metafluor software (Molecular Devices) (see Note 1).
Lambda 10-2 filterwheel shutter controller (Sutter) (see Note 2).
40x/1.3 NA oil-immersion objective (Zeiss)
Immersion oil, Type DF (Cargille Labs)
2.3. Data Acquisition and Analysis
Hanks’ Balanced Salt Solution (HBSS, Cellgro) supplemented with 1 mM Ca2+ on the day of imaging
Phorbol-12,13-dibutyrate (PDBu, Millipore)
Gö 6976 (Millipore)
Excel (Microsoft) or equivalent
3. Methods
3.1. Cell Culture
MDCK cells are propagated in DMEM/F12 media containing 10% FBS; these cells adhere well to the uncoated, glass coverslip of the imaging dish. Two days prior to imaging, a confluent 10 cm dish of MDCK cells should be split 1:40 into a 35 mm imaging dish.
24 hours after plating cells into the 35 mm imaging dish (when they are at approximately 75% confluence), the MDCK cells should be transfected using the Effectene transfection reagent according to the manufacturer’s protocol. One microgram of DKAR DNA and 0.1 μg of NHERF-1 DNA should be transfected to attain proper relative expression levels for imaging the following day (see Note 3).
3.2. Data Acquisition
We perform all of our imaging experiments at room temperature.
On the day of imaging (one day post-transfection), turn on the lamp, microscope, filterwheel changer, and camera. Open the Metafluor software application.
Set up (or load) the imaging protocol. For our setup, this protocol consists of acquiring CFP (420/20 nm excitation, 450 nm dichroic, 475/40 nm emission, 200 ms exposure), FRET (420/20 nm excitation, 450 nm dichroic, 535/25 nm emission, 200 ms exposure) and YFP (495/10 nm excitation, 505 nm dichroic, 535/25 nm emission, 100 ms exposure) once every 15 seconds through the 40x objective and with a 10% neutral density filter in place. The CFP/FRET ratio is plotted to assess the progress of the experiment in real-time and the intensity of YFP is plotted as a measure of fluorophore photobleaching (see Notes 4 and 5).
Clean the 40x oil-immersion objective. Apply one drop of oil onto the objective.
Aspirate media from the cells, rinse cells in Hanks’ balanced salt solution containing 1 mM CaCl2 (HBSS/1 mM CaCl2), and replace with 2 ml HBSS/1 mM CaCl2 for imaging.
Place the imaging dish on the microscope stage and secure it (we use small pieces of modeling clay) to prevent subtle movements of the dish when adding drugs during the experiment.
Focus on the cells and identify those reflecting proper localization as well as optimal expression levels of reporter. For NHERF-overexpressing cells, proper localization of DKAR-PDZ consists of DKAR localized to the apical membrane of the MDCK cells as shown in the bottom panel of Figure 2B. In addition, optimal levels of DKAR are those in which the kinase activity reporter is expressed at levels within the range of cellular substrates (~1 μM) (see Note 6).
Acquire one series of images.
Select regions from the selected cells for analysis throughout the course of the experiment.
Subtract background levels estimated from areas with no cells (see Note 7).
Within Metafluor, save the log file that will contain data (intensities and ratios) from each region during the course of the experiment, and more importantly, save the images so that one can reanalyze the experiment once complete (see Note 8).
Begin the experiment, plotting both the FRET ratio (CFP/FRET) and YFP intensity (as a control for photobleaching) in real time. Continue acquiring until a stable baseline FRET ratio is established.
Addition of PDBu to stimulate PKD activity is performed by first removing 0.5-1 ml of the imaging buffer from the imaging dish during the experiment, resuspending the drug into this volume, and then adding it back drop-wise to the dish. A final concentration of 200 nM PDBu is used to activate PKD.
Pharmacologic inhibition of PKD activity, and thus reversal of the DKAR response, can be observed by addition of the inhibitor Gö 6976 (using the same method as described in step 12) at a final concentration of 500 nM (see Notes 9 and 10).
3.3. Data Analysis
Data within the saved log file can be opened from within Excel. The file will contain time as the first column, followed by four columns from each region analyzed: CFP intensity, FRET intensity, YFP intensity, CFP/FRET ratio calculation.
The baseline FRET ratio varies from cell to cell, thus, in order to compare the relative magnitude of response from the kinase activity reporter, the traces should be normalized to the average baseline FRET ratio from each region and then referenced about the time of drug addition. The averages of these normalized CFP/FRET ratios are then plotted with respect to time (Figure 3).
4. Notes
Metafluor software drives the acquisition of the CFP, FRET and YFP channels as well as a real-time analysis of the user-defined FRET ratio (CFP/FRET for DKAR) during the course of the experiment. Thus, one can evaluate the progress of the experiment and acquire images until a steady baseline is reached before addition of drugs.
The Lambda 10-2 filter changer is driven through the imaging software; this changer controls the excitation, dichroic and emission filters individually.
0.1 μg of NHERF-1 DNA is transfected as this results in NHERF-1 expression levels that properly localize to the apical membrane in MDCK cells (see Figure 2A); transfection of too much DNA results in overexpression of NHERF-1, yielding significant levels of untargeted, non-localized NHERF.
DKAR is basally undergoing FRET (see Figure 1); therefore, we opt to plot the CFP/FRET ratio rather than the canonical FRET/CFP ratio to visualize PKD signaling.
Each imaging setup should be calibrated to control against fluorophore photobleaching; that is, for our setup, we determined that we can acquire a series of images (CFP, FRET, YFP) up to every 7 seconds through the 40x objective and a 10% neutral density filter with excitation exposure times of 200 ms for CFP and FRET and 100 ms for YFP with no observed bleaching.
We calibrated the brightness of the signal from the reporter such that its expression level is within the range of endogenous substrates (~1 μM) [5]. This is important as too high a concentration of exogenous reporter could buffer the endogenous enzymatic capacity of the kinase and thus alter the activity readout.
One can subtract background levels of the saved images for each channel during reanalysis of the images saved during the experiment.
Saving the images is critical; from these one can reanalyze the experiment by defining new regions or adjust for region movement if this was observed during the experiment.
Reversal of responses from kinase activity reporters using kinase-specific inhibitors (e.g. Gö 6976 for PKD) further corroborates that the response from the reporter is due to signaling by the kinase of interest.
A simple control for any experiment is to monitor FRET changes from the reporter construct in which the phosphoacceptor site (e.g. threonine in the example of DKAR) is mutated to a non-phosphorylatable residue (e.g. alanine). This reporter should not undergo a FRET change and thus serves as a negative control in experiments where there is concern that the reporter is physically relocalizing within the cell, or where the cell is changing morphology; such instances may appear as FRET changes as the local reporter concentration change impacts basal intermolecular FRET.
5. References
- 1.Toker A. The biology and biochemistry of diacylglycerol signalling. Meeting on molecular advances in diacylglycerol signalling. EMBO Rep. 2005;6(4):310–314. doi: 10.1038/sj.embor.7400378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez-Ruiloba L, Cabrera-Poch N, Rodriguez-Martinez M, Lopez-Menendez C, Jean-Mairet RM, Higuero AM, Iglesias T. Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J Biol Chem. 2006;281(27):18888–18900. doi: 10.1074/jbc.M603044200. [DOI] [PubMed] [Google Scholar]
- 3.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology. 2011;26(1):23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthews SA, Rozengurt E, Cantrell D. Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/Protein kinase Cmu. J Biol Chem. 1999;274(37):26543–26549. doi: 10.1074/jbc.274.37.26543. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel MT, Newton AC. Spatiotemporal Dynamics of Kinase Signaling Visualized by Targeted Reporters. Curr Protoc Chem Biol. 2009;1(1):17–18. doi: 10.1002/9780470559277.ch090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato M, Kawai Y, Umezawa Y. Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-regulated kinase in single living cells. Anal Chem. 2007;79(6):2570–2575. doi: 10.1021/ac062171d. [DOI] [PubMed] [Google Scholar]
- 7.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(26):15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98(26):14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437(7058):569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 10.Allen MD, Zhang J. Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun. 2006;348(2):716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki K, Sato M, Umezawa Y. Fluorescent indicators for Akt/protein kinase B and dynamics of Akt activity visualized in living cells. J Biol Chem. 2003;278(33):30945–30951. doi: 10.1074/jbc.M212167200. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J Biol Chem. 2005;280(7):5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X, Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Molecular biology of the cell. 2008;19(10):4366–4373. doi: 10.1091/mbc.E08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161(5):899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajimoto T, Sawamura S, Tohyama Y, Mori Y, Newton AC. Protein kinase C {delta}-specific activity reporter reveals agonist-evoked nuclear activity controlled by Src family of kinases. J Biol Chem. 2010;285(53):41896–41910. doi: 10.1074/jbc.M110.184028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunkel MT, Toker A, Tsien RY, Newton AC. Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J Biol Chem. 2007;282(9):6733–6742. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunkel MT, Garcia EL, Kajimoto T, Hall RA, Newton AC. The protein scaffold NHERF-1 controls the amplitude and duration of localized protein kinase D activity. J Biol Chem. 2009;284(36):24653–24661. doi: 10.1074/jbc.M109.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]