Abstract
Purpose
Seven genes involved in precursor mRNA (pre-mRNA) splicing have been implicated in autosomal dominant retinitis pigmentosa (adRP). We sought to detect mutations in all seven genes in Chinese families with RP, to characterize the relevant phenotypes, and to evaluate the prevalence of mutations in splicing genes in patients with adRP.
Methods
Six unrelated families from our adRP cohort (42 families) and two additional families with RP with uncertain inheritance mode were clinically characterized in the present study. Targeted sequence capture with next-generation massively parallel sequencing (NGS) was performed to screen mutations in 189 genes including all seven pre-mRNA splicing genes associated with adRP. Variants detected with NGS were filtered with bioinformatics analyses, validated with Sanger sequencing, and prioritized with pathogenicity analysis.
Results
Mutations in pre-mRNA splicing genes were identified in three individual families including one novel frameshift mutation in PRPF31 (p.Leu366fs*1) and two known mutations in SNRNP200 (p.Arg681His and p.Ser1087Leu). The patients carrying SNRNP200 p.R681H showed rapid disease progression, and the family carrying p.S1087L presented earlier onset ages and more severe phenotypes compared to another previously reported family with p.S1087L. In five other families, we identified mutations in other RP-related genes, including RP1 p. Ser781* (novel), RP2 p.Gln65* (novel) and p.Ile137del (novel), IMPDH1 p.Asp311Asn (recurrent), and RHO p.Pro347Leu (recurrent).
Conclusions
Mutations in splicing genes identified in the present and our previous study account for 9.5% in our adRP cohort, indicating the important role of pre-mRNA splicing deficiency in the etiology of adRP. Mutations in the same splicing gene, or even the same mutation, could correlate with different phenotypic severities, complicating the genotype–phenotype correlation and clinical prognosis.
Introduction
Retinitis pigmentosa (RP; OMIM 268000) is clinically characterized by bone spicule-like pigment deposits in the fundus with attenuated retinal vessels and waxy pallor of the optic disc, and retinal degenerations with reduced or absent responses on electroretinogram (ERG) [1]. On a cellular basis, RP predominantly affects rod photoreceptors and/or RPE cells, and likely involves cone photoreceptors at a later stage. As a consequence, patients with RP initially develop nyctalopia, followed by constriction of the visual fields (VFs) and severe visual impairment or even complete blindness eventually [2]. As the most common form of inherited retinal dystrophies (IRDs), RP affects 1 in 3,500 to 5,000 individuals worldwide [3-5].
RP exhibits significant genetic heterogeneity involving more than 50 loci thus far. Of note, the defects of precursor mRNA (pre-mRNA) splicing, a fundamental process required by most genes before transcription, have been implicated in the autosomal dominant form of RP (adRP). Among adRP-causing genes, seven genes are involved in the process of pre-mRNA splicing, including PRPF3, PRPF8, PRPF31, RP9, SNRNP200, PRPF6, and PRPF4 [6-11].
Pre-mRNA splicing is regulated by the spliceosome complex, a macromolecular machinery comprising five small nuclear ribonucleoproteins (snRNPs), U1, U2, U4, U6, and U5, as well as more than 100 non-snRNP-specific proteins [12]. The U5 snRNP bonds to the U4/U6 di-snRNP to assemble the U4/U6-U5 tri-snRNP, which acts as the catalytic component for splicing and is crucial for the pre-mRNA structural rearrangements [13,14]. All seven identified adRP-causing splicing genes encode either important components or regulators of U4/U6-U5 tri-snRNP complex, which is required for proper splicing of pre-mRNA in nearly all types of cells. However, the mechanism by which mutations in the seven ubiquitously expressed genes cause retinal specific phenotypes is not quite clear. Although functional studies on animal models are needed to further understand the mechanism, extensive genetic analysis with comprehensive clinical characterization of such patients would also be useful for better understanding the genotype–phenotype correlation, and directing the clinical prognosis, management, and gene replacement therapy.
Herein, we sought to detect mutations in the seven pre-mRNA splicing genes among eight Chinese families with RP to predict the functions, evaluate the prevalence, and analyze the associated phenotypes. As described previously, we used a targeted sequence capture with a next-generation massively parallel sequencing (NGS) approach to screen 189 genes associated with IRDs, including the seven splicing genes, for potential mutations [15]. Our current study revealed a frameshift mutation in PRPF31 and two missense mutations in SNRNP200 in three families with adRP, respectively. Taken together with our previous identification of a PRPF4 mutation, we have found that four (9.5%) out of our cohort of 42 families with adRP were associated with mutations in the pre-mRNA splicing genes. Interestingly, some splicing gene mutations identified here correlate with phenotypes that differ from those of previously reported families who carry mutations in the same gene, and therefore provide additional insights into the genotype–phenotype correlation of RP caused by pre-mRNA splicing deficiency. Mutations identified in other RP-causing genes in the present study were also summarized herein.
Methods
Patient recruitment and clinical evaluation
Six unrelated Chinese families affected with adRP and two families with RP with uncertain inheritance mode were recruited from multiple areas of China. The eight families (numbered families 01 to 08) included 24 presently affected individuals with conspicuous clinical hallmarks of RP, 14 unaffected siblings, and 14 spouses. Complete familial history and medical records were obtained from each family. Ophthalmic examinations were performed at local hospitals, including assessments of best-corrected visual acuities (BCVAs), slit-lamp examination, funduscopy, VF test, and ERG test. Peripheral blood samples drawn from median cubital veinwere collected from all recruited family members and 150 unrelated controls that were free of ocular diseases. We further saved all collected blood samples in -80°C before extracting genomic DNA. Genomic DNA was isolated from leukocytes as previously mentioned [16]. Genomic DNA was extracted from leukocytes. The study was prospectively approved and reviewed by local institutional ethics committee and adhered to the ARVO statement on human subjects and the tenets of the Declaration of Helsinki. Written informed consent was signed by all participants or their legal guardians.
Targeted gene capture
Design and validation of the sequence capture microarray were described previously [15]. Briefly, the microarray was designed and constructed on a 2.1-Mb NimbleGen sequence capture microarray platform (Roche NimbleGen, Madison, WI). A total of 179 genes associated with IRDs and ten candidate genes were selected from the RetNet database and relevant publications. This panel of genes includes the seven pre-mRNA splicing genes. Probes of 250–300 bases were designed accordingly to capture all exons, together with necessary flanking intronic sequences and 5′- and 3′-untranslated regions (UTRs) in these genes according to the human reference genome (NCBI build 37.1). The total targeted sequence was approximately 1.38 Mb.
Next-generation sequencing and bioinformatics analysis
Targeted sequence capture with NGS was performed on designated individuals of the families to identify potential disease-causing genetic variants. DNA template preparation, hybridization, and NGS were performed in collaboration with the Beijing Genome Institute (BGI, Shenzhen, China), as described previously [14]. Read alignments, calling of SNPs, and insertions and deletions (indels) were performed as previously described [15]. Briefly, sequencing reads were aligned to the NCBI human reference genome (NCBI build 37.1) using the Short Oligonucleotide Alignment Program (SOAP) to detect SNPs [16], and the Burrows-Wheeler Aligner (BWA) to detect indels as previously described [17]. Only mapped reads were selected for subsequent analysis, including coverage calculations, depth calculations, and genotype identification. All the identified variants were subsequently compared with the five SNP databases, including the dbSNP137, HapMap Project, 1000 Genome Project, YH database, and Exome Variant Server (EVS).
Mutation validation
We performed Sanger sequencing to validate variants, intrafamilial consegregation analysis, and prevalence testing in 150 unrelated controls. The standard protocol for Sanger sequencing has been described previously [18]. Information about the primers is available upon request.
In silico analyses
To estimate the evolutionary conservation of the mutated amino acids, we aligned the SNRNP200 orthologous protein sequences of the following species using the Vector NTI Advance 2011 software: human (Homo sapiens), gorilla (Gorilla gorilla), cow (Bos taurus), horse (Equus caballus); pig (Sus scrofa), chicken (Gallus gallus); mouse (Mus musculus), zebrafish (Danio rerio), and fruit fly (Drosophila melanogaster). In addition, three online prediction software tools, including Polymorphism Phenotyping (PolyPhen-2) [19] and SIFT Human Protein database [20], were used to predict the potential impact of the substitution.
Results
Analysis of variants detected with targeted next-generation sequencing
One patient was selected from each of the eight families for the targeted gene sequencing approach. The eight patients selected included Family 01-V:1, Family 02-II:2, Family 03-IV:3, Family 04-III:3, Family 05-II:1, Family 06-II:1, Family 07-II:1, and Family 08-II:5. Using NGS, we possessed an average call rate of more than 99.8% for the targeted bases with a mean depth of 76.37-fold. A total of 29,654 variants were initially identified in all 8 selected families and were subsequently filtered as following. First, 29,476 variants were found in the five SNP databases that included normal individuals, and were consequently filtered out. In the remaining 178 variants, we further removed synonymous variants and other non-coding variants (n = 158) and focused on non-synonymous (NS) variants, splice site (SS) variants, and short frame-shift coding indels, all of which were more likely to be pathogenic. Twenty coding variants were next confirmed with Sanger sequencing, and were further tested in all members from the corresponding family. Only nine variants were found to be cosegregated with phenotype in the corresponding families. Among them, one variant detected in Family 08 was further discarded due to the presence in 150 unrelated controls screened in this study. At this point, eight likely causative mutations were identified in the eight families, respectively.
Identification of mutations in pre-mRNA splicing genes in three families with clinical evaluation
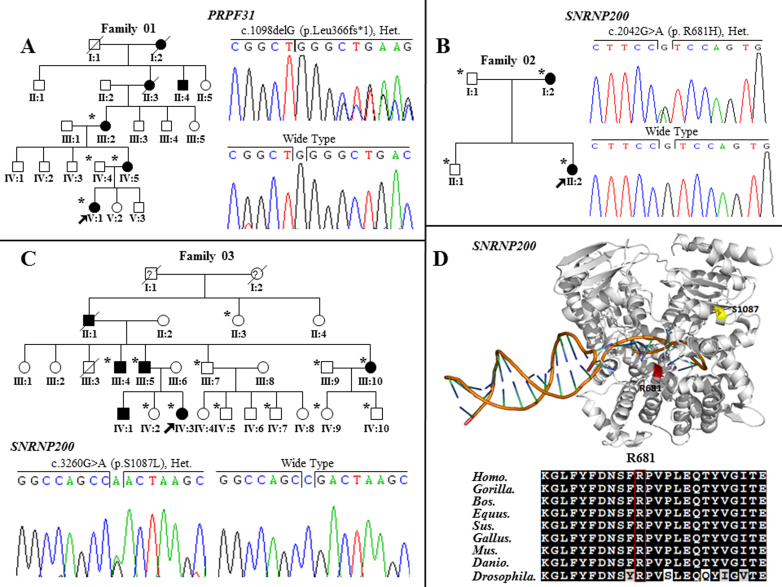
One novel putative mutation and two recurrent mutations were identified in the splicing genes in three individual families, respectively (Table 1). A novel frameshift variant, c.1098delG in exon 11 of PRPF31 (p.Leu366fs*1), was identified in Family 01 (Figure 1A) and was absent in 150 unrelated controls. The frameshift mutation in PRPF31 would likely generate a premature termination codon (PTC) that produce truncated proteins. Two recurrent mutations located in SNRNP200 were identified in families 02 (p.Arg681His) and 03 (p.Ser1087Leu) [10,21] (Figure 1B and 1C).
Table 1. Mutations identified in RP causative genes in this cohort.
| Family ID | Chr | Gene |
Mutation |
Status |
Bioinformatics Analysis |
Novel / Recurrent | |||
|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino Acid | Exon | SIFT | PolyPhen-2 | |||||
| 01 |
Chr19 |
PRPF31 |
c.1098delG |
p.Leu366fs*1 |
11 |
Het. |
NA |
NA |
Novel |
| 02 |
Chr2 |
SNRNP200 |
c.2042G>A |
p.Arg681His |
16 |
Het. |
D |
Probably D |
Recurrent |
| 03 |
Chr2 |
SNRNP200 |
c.3260C>T |
p.Ser1087Leu |
25 |
Het. |
D |
Probably D |
Reported |
| 04 |
Chr7 |
IMPDH1 |
c.931G>A |
p.Asp311Asn |
10 |
Het. |
D |
Possibly D |
Recurrent |
| 05 |
Chr8 |
RP1 |
c.2342C>G |
p.Ser781* |
4 |
Het. |
NA |
NA |
Novel |
| 06 |
Chr3 |
RHO |
c.1040C>T |
p.Pro347Leu |
5 |
Het. |
D |
Possibly D |
Recurrent |
| 07 |
ChrX |
RP2 |
c.409_411del |
p.Ile137del |
2 |
Hem. |
NA |
NA |
Novel |
| 08 | ChrX | RP2 | c.193C>T | p.Gln65* | 2 | Hem. | NA | NA | Novel |
Abbreviations: Het: heterozygous; Hom: homozygous; NA: not available; T: tolerated; D: damaging.
Figure 1.
Identification of mutations in pre-mRNA splicing related genes in the three corresponding families, conservation of SNRNP200 p.R681H and three-dimensional crystal of protein of SNRNP200. A: PRPF31 c.1098delG (p.Leu366fs*1) mutation in family 01. B: SNRNP200 c.2042G>A (p.R681H) mutation in family 02. C: SNRNP200 c.3260G>A (p.S1087L) mutation in family 03. D: SNRNP200 p.R681H and p.S1087 in the three-dimensional crystal of the protein of SNRNP200, which the SNRNP200 p.R681H was evolutionarily conserved (bottom). Men and women are symbolized by squares and circles, and slashed symbols denote that the subject is deceased. Affected and unaffected members are represented by filled and open symbols, respectively, while a question mark stands for an unclear physical condition. Arrows point to the probands, and asterisks indicate the subjects who participated in our study.
Three of the six traceable affected individuals in Family 01 were recruited in this study. All three patients were identified as carrying heterozygous PRPF31 p.Leu366fs*1. They developed night blindness within the first decade of their lives (Table 2), followed by progressive loss of central vision. The proband, Patient 01-V:1, developed visual impairment at age 5. Her fundus evaluation showed atrophy of the retina, attenuated retinal vessels, and bone spicule-like pigment deposits around the superior periphery of the left eye (Figure 2A). In addition, diffused loss of visual fields and diminished ERG responses were detected in both eyes.
Table 2. Clinical manifestations of patients with identified mutations.
| Patient ID | Diagnosis | Gene | Mutation | Onset Age (year) | Age (years) /Sex |
BCVA (logMAR) |
Fundus Appearance |
ERG | VF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
O.D. |
O.S. |
||||||||||||||||
| O.D. | O.S. | MD | OD | AA | PD | MD | OD | AA | PD | ||||||||
| 01-V:1† |
ADRP |
PRPF31 |
p.Leu366fs*1 |
5 |
22/F |
0.5 |
0.6 |
Yes |
Waxy |
Yes |
Diffuse |
Yes |
Waxy |
Yes |
Diffuse |
Reduced |
Diffuse Loss |
| 02-II:2† |
ADRP |
SNRNP200 |
p.R681H |
6 |
22/F |
0.2 |
0.4 |
Yes |
Waxy |
Yes |
Intensive |
Yes |
Waxy |
Yes |
Intensive |
Reduced |
20 degrees |
| 03-IV:3† |
ADRP |
SNRNP200 |
p.S1087L |
7 |
22/F |
0.2 |
0.1 |
Yes |
Waxy |
Yes |
Diffuse |
Yes |
Waxy |
Yes |
Diffuse |
Reduced |
10 degrees |
| 04-III:3† |
ADRP |
IMPDH1 |
p.D311N |
10 |
65/M |
Light |
Light |
Yes |
Waxy |
Yes |
Intensive |
Yes |
Waxy |
Yes |
Intensive |
Reduced |
<5 degrees |
| 05-II:1† |
ADRP |
RP1 |
S781* |
8 |
58/M |
0.1 |
0.1 |
Yes |
Waxy |
Yes |
Intensive |
Yes |
Waxy |
Yes |
Diffuse |
Reduced |
10 degrees |
| 06-II:1† |
ADRP |
RHO |
p.P347L |
14 |
26/M |
0.2 |
0.3 |
Yes |
Waxy |
Yes |
Diffuse |
Yes |
Waxy |
Yes |
Diffuse |
Reduced |
20 degrees |
| 07-II:1† |
XLRP |
RP2 |
p.Ile137del |
8 |
23/M |
0.2 |
0.2 |
Yes |
Waxy |
Yes |
Diffuse |
Yes |
Waxy |
Yes |
Diffuse |
Reduced |
10 degrees |
| 08-II:5† | XLRP | RP2 | p.Q65* | 7 | 49/M | Light | Light | Yes | Waxy | Yes | Intensive | Yes | Waxy | Yes | Intensive | Reduced | <5 degrees |
Abbreviations: †, proband of each family; BCVA, best corrected visual acuity; logMAR, logarithm of the Minimum Angle of Resolution; O.D., right eye; O.S., left eye; MD, macular degeneration; OD, optic disc; AA, arterial attenuation; PD, pigment deposit; ERG, electroretinography; VF, visual field; NA, not available; NL, no light; F: female; M: male.
Figure 2.
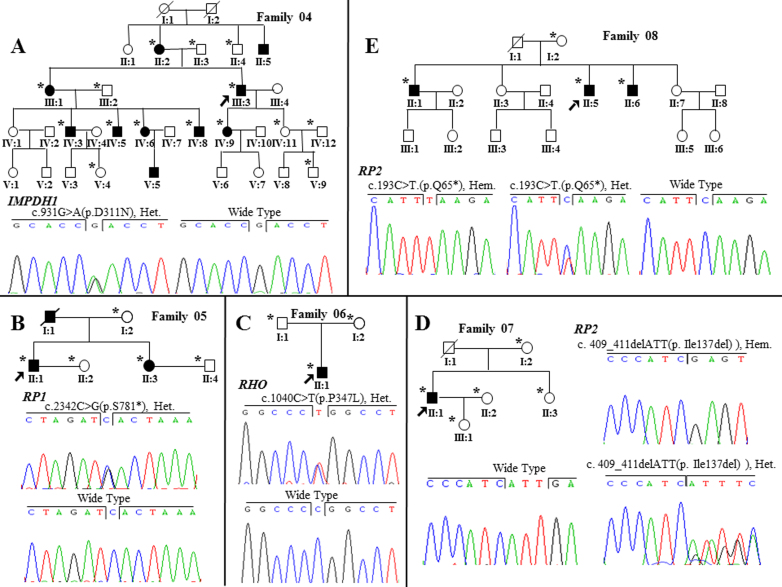
Identification of mutations in other retinitis pigmentosa–related genes in the five corresponding families. A: IMPDH1 c.823G>A (p.D275N) mutation in family 04. B: RP1 c.2342C>G (p. S781*) mutation in family 05. C: RHO c.1040C>T (p.P347L) mutation in family 06. D: RP2 c.409_411del (p.Ile137del) mutation in family 07. E: RP2 c.193C>T (p.Q65*) mutation in family 08. Men and women are symbolized by squares and circles, and slashed symbols denote that the subject is deceased. Affected and unaffected members are represented by filled and open symbols, respectively, while a question mark stands for an unclear physical condition. Arrows point to the probands, and asterisks indicate the subjects who participated in our study.
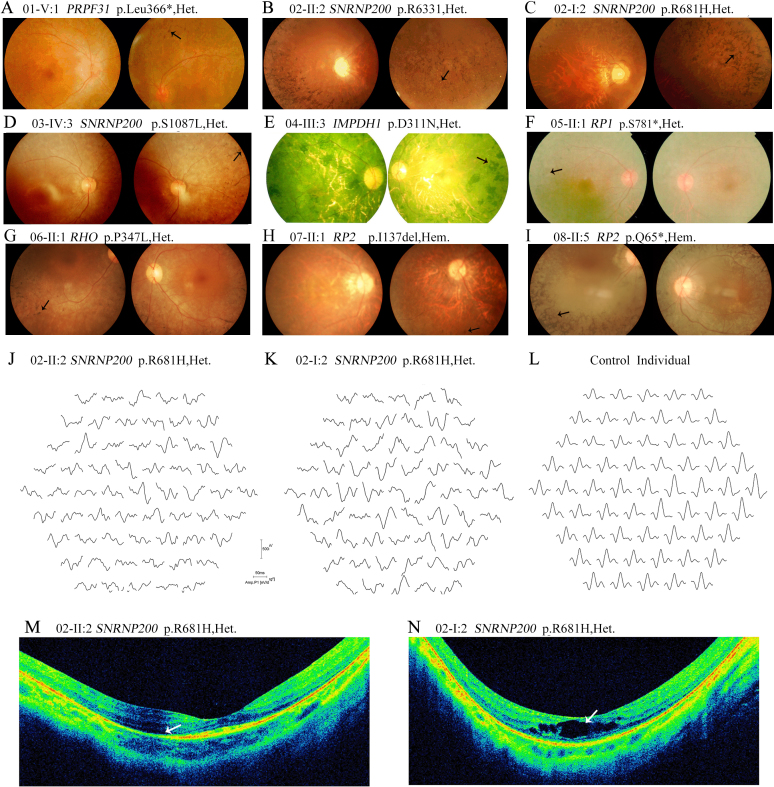
Two patients in Family 02, who carried a recurrent mutation in SNRNP200, p.Arg681His, showed early onset age of the disease, rapid disease progression, and severely decreased visual acuity in recent years. The proband (02-II:2), currently 22 years old, with high myopia, developed night blindness at age 6 (Table 2). Typical RP fundus and reduced ERG responses were also observed (Figure 2B,J). Retinal atrophy was revealed with optical coherence tomography (OCT) examination (Figure 2M). Her mother (02-I:2) also presented poor visual acuity, typical RP fundus with macular edema, and significantly reduced ERG responses (Figure 2C, K,N). Another recurrent mutation, SNRNP200 p.Ser1087Leu, was identified in all patients studied in Family 03. All patients in this family showed rapid and serious processing of RP with early onset of night blindness ranging from age 5 to 10. The proband, 03-IV:3, currently 22 years old, with diffuse bone spicule-like pigment deposits in the peripheral retina, has poor central vision, namely, 0.2/0.1, accompanying VF rapidly decreasing to 10 degrees (Figure 2D, Table 2). Detailed clinical characterization of this family has been described elsewhere [16].
Mutations identified in other retinitis pigmentosa–causing genes
In five other families, we identified five mutations in other RP-related genes, including two novel mutations and three recurrent mutations (Table 1). Two novel nonsense mutations, RP1 p. Ser781* and RP2 p.Gln65*, were cosegregated in families 05 and 08, and the novel frameshift mutation RP2 p.Ile137del was identified in Family 07 (Figure 3B,D,E). Two other recurrent mutations, IMPDH1 p.Asp311Asn [22] and RHO p.Pro347Leu [23], were identified in families 04 and 06, respectively (Figure 3A,C). All patients from these five families showed typical features of RP with atrophy of the retina, attenuated retinal vessels, and bone spicule-like pigment deposits (Figure 2E–I), and clinical information is summarized in Table 2.
Figure 3.
Clinical evaluations of patients from the eight families with mutations in retinitis pigmentosa genes. A: Patient 01-V:1 (age 22, carrying PRPF31 p.Leu366fs*1) has a waxy optic disc, attenuated retinal arterioles, and pigment deposits around the superior periphery of the left eye. B: Fundus examination of Patient 02-II:2 (age 22, carrying SNRNP200 p.R681H) shows numerous bone spicule-like pigments and crystalline forms were scattered in the peripheral fundus (arrow). C: The fundus of Patient 02-I:2 (age 51, carrying SNRNP200 p.R681H) shows intensive pigment deposits. D: Patient 03-IV:3 (age 22, carrying SNRNP200 p.S1087L) demonstrates diffuse bone spicule-like pigment deposits in the periphery retina of the right eye. E: Patient 04-III:3 (age 65, carrying IMPDH1 p.D311N) has intensive bone spicule-like pigment deposits, which severely impacted the central retina. F: The fundus of Patient 05-II:1 (age 58, carrying RP1 p. S781*) shows typical retinitis pigmentosa (RP) features with posterior subcapsular cataract. G: The fundus of Patient 06-II:1 (age 26, carrying RHO p.P347L) with typical RP features. H: Fundus examination of Patient 07-II:1 (age 23, carrying RP2 p.Ile137del) reveals typical features of RP fundus. I: Patient 08-II:5 (age 49, carrying RP2 p.Q65*) has typical fundus of RP. J: Multifocal electroretinogram (mfERG) test traces from the right eye of Patient 02-II:2 are shown. Responses of the peripheral retina are greatly reduced while responses within the central 10 degrees are slightly involved. K: MfERG of the right eye of Patient 02-I:2 demonstrates a global decrease in ERG responses. L: ERG responses of one unrelated control. M: Optical coherence tomography (OCT) examination of Patient 02-II:2 shows retinal atrophy. N: OCT examination of Patient 02-I:2 shows macular edema and retinal atrophy.
Discussion
To date, seven pre-mRNA splicing genes, including PRPF3, PRPF6, PRPF8, PRPF31, RP9, SNRNP200, and PRPF4, have been identified as adRP-causing genes. Proteins encoded by the seven genes are essential for pre-mRNA splicing in all cell types. However, mutations in these genes have been implicated only in retinal degeneration, indicating their tissue specificity. Expression of PRPF3, PRPF31, and PRPF8 has been consistently reported as significantly higher in the murine retina compared to other metabolically active tissues [24].
In this study, a cohort compromising six families affected with adRP and two families with uncertain inheritance mode were investigated for potential disease-causing mutations. Three mutations in two adRP-causing splicing genes were identified in three individual families in this study. Taken together with our previous identification of a PRPF4 mutation [11], four mutations accounted for 9.5% of the mutations in the pre-mRNA splicing genes in our cohort of 42 families with adRP. Although the number of the families studied herein was not large enough, this is the first report to provide etiological prevalence for mutations in all seven adRP-causing splicing genes among Chinese patients with adRP using NGS. To lend credit to the etiological prevalence, particular attention has been paid to sample selection and the approaches used. All families were recruited from different areas across China, which reflected well-distributed geographic origins. Mutations were identified with comprehensive genetic analyses comprising targeted sequencing of nearly all known genes associated with IRDs, optimized bioinformatic analysis of all variants detected, cosegregation analysis within corresponding families, and the prevalence test in 150 unrelated normal controls. Conservational analysis and in silico analyses were also conducted to predict the pathogenicity of the missense mutations. With these efforts, our results strongly support that the three variants detected in the splicing genes are disease-causing mutations rather than rare polymorphisms.
The protein encoded by PRPF31 is involved in maintaining the stability of the U4/U6-U5 tri-snRNP complex [25,26]. To date, more than 50 mutations in this gene, including missense substitutions, splice-site mutations, large deletions, and genomic rearrangement encompassing PRPF31, have been reported to cause RP [27-29]. In the present study, we identified a novel frameshift mutation, PRPF31 p.Leu366fs*1, which is predicted to generate a PTC, and may result in a truncated protein lacking 132 amino acids from the C-terminal. It has long been debated whether such mutations will eventually result in “null alleles” due to nonsense-mediated mRNA decay (NMD) [30] or a truncated protein, overexpression of which was shown to impair pre-mRNA splicing in vitro [31]. Although the existence of such truncated proteins has never been proved in vivo, compelling evidence has shown that PTC-causing mutations in PRPF31 led to RP via haploinsufficiency due to inactivation of transcripts or protein synthesis, which is consistent with our results [30]. The transcript of PRPF31 p.Leu366fs*1 was predicted to generate NMD. The patients carrying this mutation can be considered functional hemizygotes and exhibit consistent and severe phenotypes with relatively early onset of night blindness and diffused involvement of the retina, which are similar to those of a previously reported family that carries a frameshift deletion in PRPF31 [27].
SNRNP200 encodes hBrr2, an essential protein for U4/U6 unwinding [10]. To date, ten missense mutations in SNRNP200 have been implicated in adRP among different populations, which points out the important role of SNRNP200 in the disease etiology of retinitis pigmentosa [10,15,21,32]. In the present cohort, two recurrent mutations, p.Arg681His and p.Ser1087Leu, were identified in two families with adRP. The Arg681 residue is located in the first DExD-helicase domain, more specifically, in the boundaries between two helicases, the first ATP (ATP)-binding and the first C-terminal (CTER) domain. The Ser1087 residue is located in the ratchet helix of the first Sec63 domain of hBrr2. However, the two domains are located in this first Hel308-like module, which is essential for U4/U6 unwinding. The p.Arg681His residue impairs hBrr2 helicase/ATPase activity, leading to prematurely unwinding of U4/U6 or premature disassembly of the spliceosome, and thus to defects in spliceosome catalysis [10,33]. As for the p.Ser1087Leu mutation, its homolog mutation in budding yeast was shown to significantly inhibit unwinding of U4/U6 [10]. Accordingly, in this study, Family 03, who carried p.Ser1087Leu, presented a more severely affected retina and visual function compared with patients who harbor p.Arg681His.
Both residues are highly conserved among multiple species through evolution. Here, only two affected members in Family 02 carrying SNRNP200 p.Arg681His were identified. This mutation has been previously identified only in a Caucasian patient with sporadic RP. No clinical information about that individual is available. In the present study, this mutation was found to correlate with a panel of severe phenotypes. Patients carrying this mutation manifest severe phenotypes, including early disease onset age and rapid disease progression. That indicates the potentially hazardous nature of the mutation. Family 03, who carry SNRNP200 p.Ser1087Leu, presented complete penetrance with an earlier onset age of the disease (age ranging from 5 to 10 years) compared with another Chinese family who carry the same mutation [18,32]. In addition, the present family showed more severe phenotypes, including rapid decreases in VF and poor central vision.
However, several common issues in this study likely influence the prevalence of mutations in adRP-related splicing genes. The NGS technique currently cannot detect large deletions, insertions, and copy number variations. Some potential pathogenic non-coding variants may also be removed during our filtering process. Therefore, our current data cannot exclude the seven splicing genes as disease-causing genes in the remaining families. Similarly, for the three families with identified mutations in splicing genes, we cannot completely rule out the possibility that variants other than the identified ones may contribute partially or fully to the disease etiology. These limitations call for necessary follow-up analysis in all cases. Additionally, mutations in PRPF8 and PRPF31 could result in all or no symptoms, meaning mutation carriers could develop either complete phenotypes or no phenotypes [27,34,35]. This unique feature leads to the hypothesis that RP with uncertain inheritance mode may also be caused by mutations in the seven genes. Thus, a prevalence test of mutations in the seven genes in patients with RP other than adRP is needed.
In conclusion, our study expanded the spectrum of mutations in genes associated with RP and further emphasized the role of pre-mRNA splicing defects in the etiology of adRP. The novel genotype–phenotype correlations presented in our study are helpful for directing clinical prognosis, management, and genetic counseling.
Acknowledgments
The authors thank all the patients and their family members for their participation. This study was supported by National key basic research program of china (973 program No.2013CB967500); National Natural Science Foundation of China (No. 81222009, 81170856, and 81170867); Thousand Youth Talents Program of China (to Zhao C.); Jiangsu Outstanding Young Investigator Program (No.BK2012046); Jiangsu Province’s Key Provincial Talents Program (No. RC201149); The Fundamental Research Funds of the State Key Laboratory of Ophthalmology (No. 2013KF02); The Fundamental Research Funds.Supported by Jiangsu Province’s Key Discipline. The sponsor or funding organization had no role in the design or conduct of this research.
References
- 1.Bird AC. Retinal photoreceptor dystrophies LI. Edward Jackson Memorial Lecture. Am J Ophthalmol. 1995;119:543–62. doi: 10.1016/s0002-9394(14)70212-0. [DOI] [PubMed] [Google Scholar]
- 2.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 3.Carrillo J, Martinez P, Solera J, Moratilla C, Gonzalez A, Manguan-Garcia C, Aymerich M, Canal L, Del Campo M, Dapena JL, Escoda L, Garcia-Sagredo JM, Martin-Sala S, Rives S, Sevilla J, Sastre L, Perona R. High resolution melting analysis for the identification of novel mutations in DKC1 and TERT genes in patients with dyskeratosis congenita. Blood Cells Mol Dis. 2012;49:140–6. doi: 10.1016/j.bcmd.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Chizzolini M, Galan A, Milan E, Sebastiani A, Costagliola C, Parmeggiani F. Good epidemiologic practice in retinitis pigmentosa: from phenotyping to biobanking. Curr Genomics. 2011;12:260–6. doi: 10.2174/138920211795860071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collin RW, Safieh C, Littink KW, Shalev SA, Garzozi HJ, Rizel L, Abbasi AH, Cremers FP, den Hollander AI, Klevering BJ, Ben-Yosef T. Mutations in C2ORF71 cause autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2010;86:783–8. doi: 10.1016/j.ajhg.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maita H, Kitaura H, Ariga H, Iguchi-Ariga SM. Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp Cell Res. 2005;302:61–8. doi: 10.1016/j.yexcr.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Tanackovic G, Ransijn A, Ayuso C, Harper S, Berson EL, Rivolta C. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2011;88:643–9. doi: 10.1016/j.ajhg.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKie AB, McHale JC, Keen TJ, Tarttelin EE, Goliath R, van Lith-Verhoeven JJ, Greenberg J, Ramesar RS, Hoyng CB, Cremers FP, Mackey DA, Bhattacharya SS, Bird AC, Markham AF, Inglehearn CF. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet. 2001;10:1555–62. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 9.Vithana EN, Abu-Safieh L, Allen MJ, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer ND, Willis C, Moore AT, Bird AC, Hunt DM, Bhattacharya SS. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell. 2001;8:375–81. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhao C, Bellur DL, Lu S, Zhao F, Grassi MA, Bowne SJ, Sullivan LS, Daiger SP, Chen LJ, Pang CP, Zhao K, Staley JP, Larsson C. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85:617–27. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Liu Y, Sheng X, Tam PO, Zhao K, Chen X, Rong W, Liu Y, Liu X, Pan X, Chen LJ, Zhao Q, Vollrath D, Pang CP, Zhao C. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2014;23:2926–39. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- 12.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–57. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkas MH, Grant GR, Pierce EA. Transcriptome analyses to investigate the pathogenesis of RNA splicing factor retinitis pigmentosa. Adv Exp Med Biol. 2012;723:519–25. doi: 10.1007/978-1-4614-0631-0_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beggs JD, Teigelkamp S, Newman AJ. The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J Cell Sci Suppl. 1995;19:101–5. doi: 10.1242/jcs.1995.supplement_19.15. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Lai TY, Chiang SW, Tam PO, Liu DT, Chan CK, Pang CP, Zhao C, Chen LJ. Contribution of SNRNP200 sequence variations to retinitis pigmentosa. Eye (Lond) 2013;27:1204–13. doi: 10.1038/eye.2013.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao C, Lu S, Zhou X, Zhang X, Zhao K, Larsson C. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum Genet. 2006;119:617–23. doi: 10.1007/s00439-006-0168-3. [DOI] [PubMed] [Google Scholar]
- 19.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 21.Benaglio P, McGee TL, Capelli LP, Harper S, Berson EL, Rivolta C. Next generation sequencing of pooled samples reveals new SNRNP200 mutations associated with retinitis pigmentosa. Hum Mutat. 2011;32:E2246–58. doi: 10.1002/humu.21485. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan LS, Bowne SJ, Reeves MJ, Blain D, Goetz K, Ndifor V, Vitez S, Wang X, Tumminia SJ, Daiger SP. Prevalence of mutations in eyeGENE probands with a diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013;54:6255–61. doi: 10.1167/iovs.13-12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Millá E, Maseras M, Martínez-Gimeno M, Gamundi MJ, Assaf H, Esmerado C, Carballo M. Genetic and molecular characterization of 148 patients with autosomal dominant retinitis pigmentosa(ADRP). Arch Soc Esp Oftalmol. 2002;77:481–4. [PubMed] [Google Scholar]
- 24.Cao H, Wu J, Lam S, Duan R, Newnham C, Molday RS, Graziotto JJ, Pierce EA, Hu J. Temporal and tissue specific regulation of RP-associated splicing factor genes PRPF3, PRPF31 and PRPC8–implications in the pathogenesis of RP. PLoS ONE. 2011;6:e15860. doi: 10.1371/journal.pone.0015860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova OV, Makarov EM, Liu S, Vornlocher HP, Luhrmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 2002;21:1148–57. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–9. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini S, Robinson PN, Singh JR, Vanita V. A novel 7 bp deletion in PRPF31 associated with autosomal dominant retinitis pigmentosa with incomplete penetrance in an Indian family. Exp Eye Res. 2012;104:82–8. doi: 10.1016/j.exer.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Rose AM, Mukhopadhyay R, Webster AR, Bhattacharya SS, Waseem NH. A 112 kb deletion in chromosome 19q13.42 leads to retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2011;52:6597–603. doi: 10.1167/iovs.11-7861. [DOI] [PubMed] [Google Scholar]
- 29.Köhn L, Bowne SJ. S.S. L, S.P. Daiger, M.S. Burstedt, K. Kadzhaev, O. Sandgren, I. Golovleva. Breakpoint characterization of a novel approximately 59 kb genomic deletion on 19q13.42 in autosomal-dominant retinitis pigmentosa with incomplete penetrance. Eur J Hum Genet. 2009;17:651–5. doi: 10.1038/ejhg.2008.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rio Frio T, Wade NM, Ransijn A, Berson EL, Beckmann JS, Rivolta C. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J Clin Invest. 2008;118:1519–31. doi: 10.1172/JCI34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan L, Kawada M, Havlioglu N, Tang H, Wu JY. Mutations in PRPF31 inhibit pre-mRNA splicing of rhodopsin gene and cause apoptosis of retinal cells. J Neurosci. 2005;25:748–57. doi: 10.1523/JNEUROSCI.2399-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T, Jin X, Zhang X, Yuan H, Cheng J, Lee J, Zhang B, Zhang M, Wu J, Wang L, Tian G, Wang W. A Novel Missense SNRNP200 Mutation Associated with Autosomal Dominant Retinitis Pigmentosa in a Chinese Family. PLoS ONE. 2012;7:e45464. doi: 10.1371/journal.pone.0045464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benaglio P, McGee TL, Leonardo P. Capelli1,3, ShyanaHarper, Eliot L. Berson, and Carlo Rivolta. Next generation sequencing of pooled samples reveals new SNRNP200 mutations associated with retinitis pigmentosa. Hum Mutat. 2011;32:E2246–58. doi: 10.1002/humu.21485. [DOI] [PubMed] [Google Scholar]
- 34.Maubaret CG, Vaclavik V, Mukhopadhyay R, Waseem NH, Churchill A, Holder GE, Moore AT, Bhattacharya SS, Webster AR. Autosomal dominant retinitis pigmentosa with intrafamilial variability and incomplete penetrance in two families carrying mutations in PRPF8. Invest Ophthalmol Vis Sci. 2011;52:9304–9. doi: 10.1167/iovs.11-8372. [DOI] [PubMed] [Google Scholar]
- 35.Audo I, Bujakowska K, Mohand-Said S, Lancelot ME, Moskova-Doumanova V, Waseem NH, Antonio A, Sahel JA, Bhattacharya SS, Zeitz C. Prevalence and novelty of PRPF31 mutations in French autosomal dominant rod-cone dystrophy patients and a review of published reports. BMC Med Genet. 2010;11:145. doi: 10.1186/1471-2350-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]