Abstract
During an inventory of Phlebotomine sand flies carried out in Madagascar, we have identified some specimens showing morphological characters related to the subgenus Sintonius of the genus Sergentomyia. We started a molecular study based on cytochrome b mtDNA and on D1–D2 and D8 domains of the rDNA. The sampling includes all the Sergentomyia species available and also S. (Sergentomyia) schwetzi, S. (Parrotomyia) magna, and the following species belonging to the subgenus Sintonius: S. clydei, S. christophersi, S. affinis vorax, S. adleri and S. meilloni. The Sintonius subgenus (sensu Theodor) is paraphyletic. The Malagasy specimens morphologically Sintonius-like are never clustered with the continental Sintonius. We propose a new subgenus to include them: Trouilletomyia subg. nov. Due to the lack of mesanepisternal setae, the species huberti is removed from the genus Phlebotomus and we propose here a new combination: Sergentomyia huberti comb. nov. The male of S. huberti is pinpointed and described for the first time. Lastly, a new species for Science is described on one female: Sergentomyia (Trouilletomyia) boironis n. sp.
Introduction
The subgenus Sintonius of the genus Sergentomyia was created in 1931 [1] in an article about a questionable classification of the Phlebotomine sandflies opposed to that previously proposed [2]. Curiously, P. (Euphlebotomus) philippinensis, and the American species Lutzomyia gomezi and Nyssomyia intermedia were included in this group [1]. Nitzulescu’s classification has not obtained the approval of any subsequent authors [3]–[7] who have considered Sintonius as a subgenus of the genus Sergentomyia França and Parrot, 1920, including the species exhibiting a cibarial armature and annealed spermathecae (designed as “spermathèques crénelées” by Nitzulescu). Later, Theodor considered this subgenus as an artificial group that does not sufficiently take into account the structures of the pharynx and particularly of the male genitalia. Consequently, he provided a new definition of this group [6], [7]: scanty erect hairs on abdominal tergites, style with four spines (either all terminal or two terminal and two subterminal), hooked parameres, pointed aedeagus, segmented spermathecae, buccal cavity of varying forms, pharynx lampglass shaped with a few teeth posteriorly or with ridges only.
During the last decade, we carried out several sandfly inventories in many parts of Madagascar. They included several specimens of males and females sharing the characters defining the subgenus Sintonius. Taking into account the high level of endemism in Madagascar [8] especially within the phlebotomine sandflies [9]–[17] before including these specimens in the subgenus Sintonius, we carried out a study based on two ribosomal and one mitochondrial molecular markers. This study is not a phylogenetic analysis of the subgenus Sintonius due to a limited sampling, nor a phylogeny of the genus Sergentomyia. However it demonstrates that the Sintonius subgenus (sensu Theodor) is paraphyletic. It also permitted new insights in the Malagasy sand flies with i) the creation of a new subgenus in the genus Sergentomyia, ii) the identification of a new species and iii) the correction of the position of one Malagasy species wrongly placed in the genus Phlebotomus.
Materials and Methods
Ethics Statement
For insect collections, we obtained a license for collecting and transporting zoological material N° 154/10/MEF/SG/DGF/DCB.SAP/SLRSE. No endangered or protected species were collected in this study.
Sand Fly Sampling
In total, the molecular sampling encompasses 41 specimens of Sergentomyia from eight countries (Table 1). It includes seven species of Sintonius sensu Theodor, the Sergentomyia known from Madagascar and some other African species. They were collected using CDC miniature light traps (John W. Hock company, Gainesville, FL), ultraviolet miniature light traps, sticky traps, or Malaise traps. The traps usually run overnight from 5 p.m. to 8 a.m. the following morning.
Table 1. Sergentomyia processed.
| Genbank accession numbers | |||||||
| sample | subgenus | species | collection sites | Cytb | D1–D2 | D8 | |
| SEYCH2 | Sintonius | clydei | Seychelles | Aldabra | KC669784 | ||
| SEYCH10 | KC669792 | ||||||
| CLSN2 | Senegal | Mont Rolland | KC669759 | KJ721094 | |||
| CLSN1 | KC669758 | KJ721095 | |||||
| MEIL | meilloni | Namibia | KJ746895 | KJ721096 | KJ721129 | ||
| AFBK5 | affinis vorax | Burkina Faso | Ouagadougou | KJ746893 | KJ721097 | KJ721130 | |
| AFBK1 | KJ746894 | KJ721098 | KJ721131 | ||||
| XXBK3 | adleri | Burkina Faso | Ouagadougou | KJ746879 | KJ721099 | KJ721132 | |
| CHR351 | christophersi | Algeria | Tamanrasset | KJ746880 | KJ721100 | KJ721133 | |
| MADA 1 | Trouilletomyia subg. nov. | huberti comb. nov. | Madagascar | Bemaraha-Anjohikinakina | KJ746896 | KJ721134 | |
| MADA 98 | KJ746897 | KJ721101 | KJ721135 | ||||
| MADA 618 | KJ746881 | KJ721102 | KJ721136 | ||||
| MADA 612 | KJ746882 | KJ721103 | KJ721137 | ||||
| MADA 606 | KJ746883 | KJ721104 | KJ721138 | ||||
| MADA 605 | KJ746884 | KJ721105 | KJ721139 | ||||
| MADA 594 | Mahajanga-Anjohikely | KJ746885 | KJ721106 | KJ721140 | |||
| MADA 586 | KJ746886 | KJ721107 | KJ721141 | ||||
| MADA 579 | KJ746887 | KJ721108 | KJ721142 | ||||
| MADA 576 | KJ746888 | KJ721109 | KJ721143 | ||||
| MADA 34 | Namoroka | KJ746889 | KJ721110 | KJ721144 | |||
| MADA 33 | KJ746890 | KJ721111 | KJ721145 | ||||
| MADA 32 | KJ746891 | KJ721112 | KJ721146 | ||||
| MADA 30 | KJ721113 | KJ721134 | |||||
| MADA 146 | boironis n.sp. | Madagascar | Isalo | KJ746892 | KJ721102 | KJ721135 | |
| MADA 22 | ungrouped Sergentomyia | majungaensis | Madagascar | Namoroka | EF522778 | KJ721103 | KJ721136 |
| FR1 | Sergentomyia | minuta | France | Luberon | |||
| DBSN1 | dubia | Senegal | Mont Rolland | KJ746900 | KJ721114 | KJ721147 | |
| MADA 60 | Rondanomyia | goodmani | Madagascar | Ankarana | JQ434695 | KJ721115 | KJ721148 |
| MADA 162 | Ankiliefatra | JQ421004 | KJ721116 | KJ721149 | |||
| MADA 161 | KJ746898 | KJ721117 | KJ721150 | ||||
| COMO 6 | goodmani comorensis | Comoros | Grande Comore | JQ421017 | KJ721118 | KJ721151 | |
| COMO 16 | JQ421012 | KJ721119 | KJ721152 | ||||
| MADA 17 | Vattieromyia | namo | Madagascar | Namoroka | EU143775 | KJ721120 | KJ721153 |
| MADA 15 | EU143774 | KJ721121 | KJ721154 | ||||
| MADA 76 | anka | Madagascar | Ankarana | EU143784 | KJ721122 | KJ721155 | |
| MADA 69 | EU143779 | KJ721123 | KJ721156 | ||||
| MADA 66 | sclerosiphon | Madagascar | Ankarana | EU143778 | KJ721124 | KJ721157 | |
| MADA 65 | EU143777 | KJ721125 | KJ721158 | ||||
| COMO 11 | pessoni | Comoros | Grande Comore | JQ421020 | KJ721126 | KJ721159 | |
| COMO 10 | JQ421019 | KJ721127 | KJ721160 | ||||
| 46CAM | Parrotomyia | magna | Cameroon | Daoud Safari (N-W) | KJ746899 | KJ721128 | |
| SOU3 | magna | Sudan | KJ721161 | ||||
Moreover, several other specimens not processed for molecular biology and the P. huberti holotype and paratype have been examined.
Malagasy Study Sites
Captures have been carried out in the west and the south of Madagascar, which are subject to the trade winds with significant differences in rainfall and temperature explaining differences about climates.
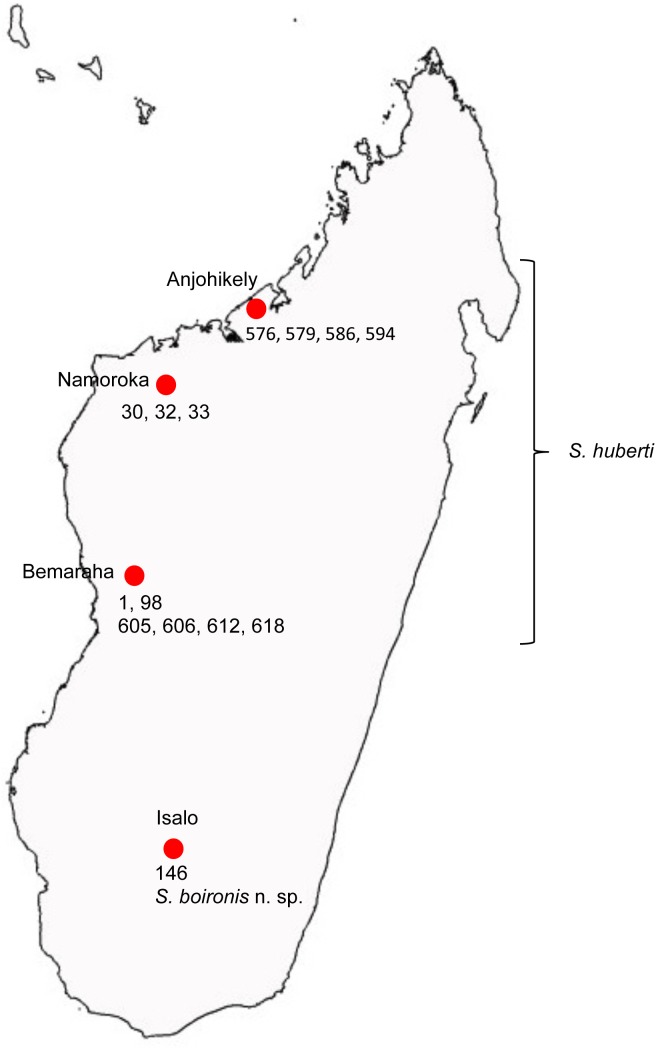
The four prospected localities where sandflies have been processed in the present study are detailed below and on figure 1.
Figure 1. Map of Madagascar Island with the prospected localities.
Anjohikely cave
This cave belongs to the Anjohibe caves complex, in the most southern part of the sandy tray Mahamavo, in the North-Western part of Madagascar at 80 km north of Mahajanga (15°33.7′S, 46°52.5′E, altitude: 100 m a.s.l.). The vegetation cover consists of “Mokoty” savannah with some patches of preserved forest on the limestone, building lapiaz. Sand flies were captured using a CDC light trap.
Anjohikinakina cave
It is located in the national park of Bemaraha, at 15.5 km north of Bekopaka in the western part of Madagascar (19°0′35.640″S, 44°46′3.720″E, altitude: 130 m a.s.l). The forest has dry deciduous type of limestone karst soil. The high plain of Bemaraha is a karst formation (Tsingy), located inside the reserve training pinnacles whose access is extremely difficult. Further north, can be found rolling hills interspersed with limestone rock formations. The river Manambolo limits the reserve to the south. Sand flies were captured using a CDC light trap.
Namoroka
This Special Reserve is located in the North-Western part of Madagascar in a dry deciduous forest on limestone karst ground. It is located within a large region from the Middle Jurassic limestone, similar to the Bemaraha’s Tsingy reserve located 250 km to the south. The limestone plateau that occupies the greater part of the reserve is in the form of a lapiaz. The south part of the Reserve is characterized by the presence of many sinkholes, some of which contain permanent pools. Sand flies have been collected using a Malaise trap near the Cave of Ambovonomby (16°28.2′S, 45°20.9′E, altitude: 200 m a.s.l).
Isalo
This National Park is located in the southern part of the centre of Madagascar. It is composed of Jurassic continental sandstone. The south and east of the massif consists of sandstone layers whose elements are size and resistance varies according to erosion. Sand flies were captured using a Malaise trap (22°37.60′S–45°21.49′E, altitude: 822 m a.s.l.).
Morphological Analysis
The sand flies collected were stored in 96% ethanol. The head and genitalia were cut off in a drop of ethanol, cleared in boiling Marc-André solution and mounted between microscope slide and cover slide for species identification directly in chloral gum or after dehydration in Canada balsam. To allow long-term preservation of the specimens previously mounted in chloral gum, they were remounted in Canada balsam after complete processing of washing and dehydration. The body related to the specimen was dried and stored in a vial at −20°C before DNA extraction. The specimens were observed under a BX50 microscope and measured using the Perfect Image software (Aries Company, Chatillon, France) and a video camera connected to the microscope. Drawings were made using the camera lucida.
Molecular Analysis
Genomic DNA was extracted from the thorax, wings, legs and abdomen of individual sand flies using the QIAmp DNA Mini Kit (Qiagen, Germany) following the manufacturer’s instructions, modified by crushing the sand fly tissues with a piston pellet (Treff, Switzerland), and using an elution volume of 50 to 200 µl [9].
All the mtDNA and rDNA amplifications were performed in a 50 µl volume using 5 µl of extracted DNA solution and 50 pmol of each of the primers. The PCR mix contained (final concentrations) 10 mM Tris HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.01% Triton X 100, 200 µM dNTP each base, and 1.25 units of 5 prime Taq polymerase (Eppendorf, Germany). The cycle begins with an initial denaturation step at 94°C for 3 min and finishes with a final extension at 68°C for 10 min. PCRs were done with the following temperature profiles.
A fragment of cytochrome b (Cyt b): 5 cycles with 30 sec 94°C, 40 sec 40°C, 1 min 68°C and 35 cycles with 30 sec 94°C, 30 sec 44°C, 1 min 68°C using the primers N1N-PDR: 5′-CAYATTCAACCWGAATGATA-3′ and C3B-PDR: 5′-GGTAYWTTGCCTCGAWTTCGWTATGA-3′ [18];
The D8 segment of the 28S rDNA: 40 cycles with 30 sec 94°C, 40 sec 48°C, 1 min 30 sec 68°C using the primers couple C’7: 5′-GTGCAGATCTTGGTGGTAG-3′ and D8E: 5′-GCTTTGTTTTAATTAAACAGT-3′;
The D1 and D2 fragments of the 28S rDNA: 30 cycles with 1 min 94°C, 1 min 58°C, 1 min 68°C using the primers couple C1’: 5′-ACCCGCTGAATTTAAGCAT-3′ and D2: 5′-TCCGTGTTTCAAGACGGG-3′ [19].
Amplicons were analyzed by electrophoresis in 1.5% agarose gel containing ethidium bromide. Direct sequencing in both directions was performed using the primers used for DNA amplification. The correction of sequences was done using Pregap and Gap softwares included in the Staden Package [20].
We used three different data sets for phylogenetic analyses: the rDNAs D1–D2 and D8, and the mtDNA cytochrome b.
Consensus sequences were aligned by the Clustal W algorithm [21] from the BioEdit 4.8.10 sequence editor [22], and corrected manually.
Sequences Analysis
In the present study, the sequences have been analysed using Neighbor-Joining (NJ), maximum parsimony (MP) and maximum likelihood (ML) methods. Phlebotomus papatasi has been used as outgroup [19], [23]–[25].
Neighbor-Joining (NJ)
The NJ method [26] has been analysed by MEGA software version 5 [27]. Genetic distances were corrected according to the transition/transversion rate (Kimura’s two-parameter method). Bootstrap confidence values were calculated from 1,000 replications.
Maximum parsimony
Maximum parsimony (MP) analysis were performed using the branch and bound option of MEGA when possible or the heuristic search. The node support was assessed by bootstrapping over 100 replications.
Maximum likelihood
Sequence data were analysed by PhyML [28] based on maximum likelihood. The ML trees were constructed using the substitution models selected by MODELTEST program [29] with AIC: HKY85 [30] for D1–D2 and D8, and General Time Reversible [31] for cyt b.
Nomenclatural Acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature, and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix “http://zoobank.org/”. The LSID for this publication is: urn:lsid:zoobank.org:pub:D0300E46-E93C-4CEF-99DE-77D77E5FC442. The electronic edition of this work was published in a journal with an ISSN, and has been archived and is available from the following digital repositories PubMed Central, LOCKSS.
Results
Molecular Analysis
The sequences analysed in the present study have been deposited in Genbank as indicated in table 1. Despite several attempts, the direct sequencing of the S. clydei was not successfully performed for D8 and D1–D2 from the Seychelles specimens, like the sequencing of several markers of the specimens 1, 30 and 33, due to the small quantity of DNA extracts.
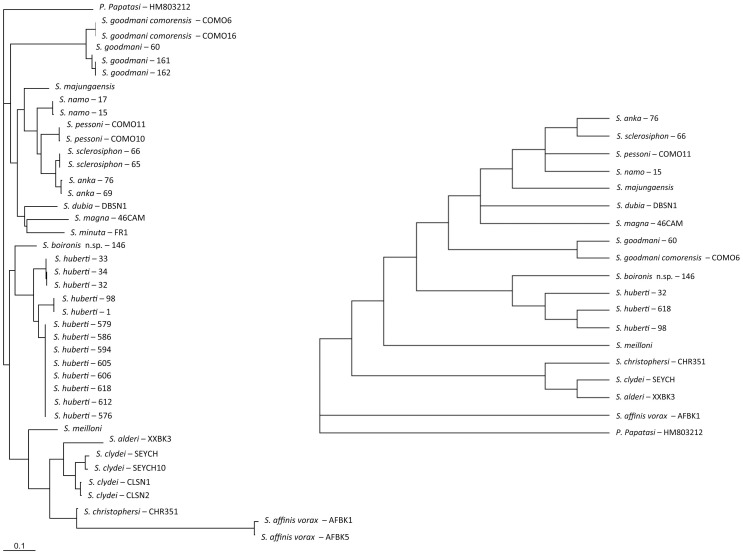
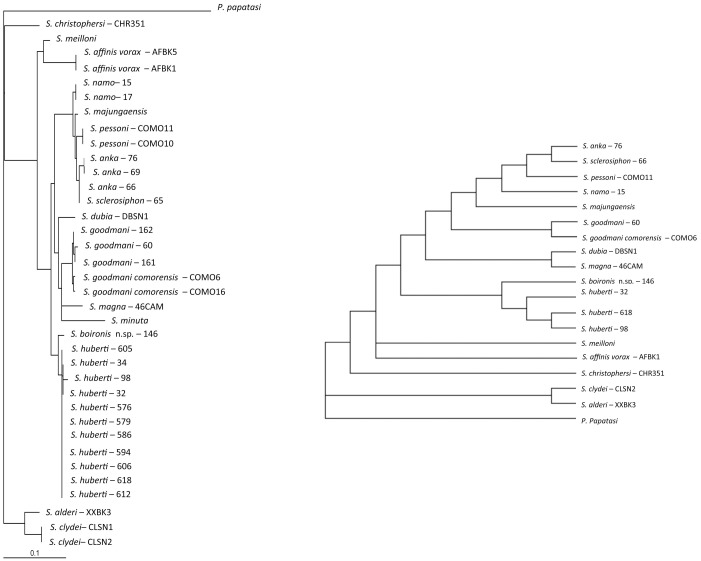
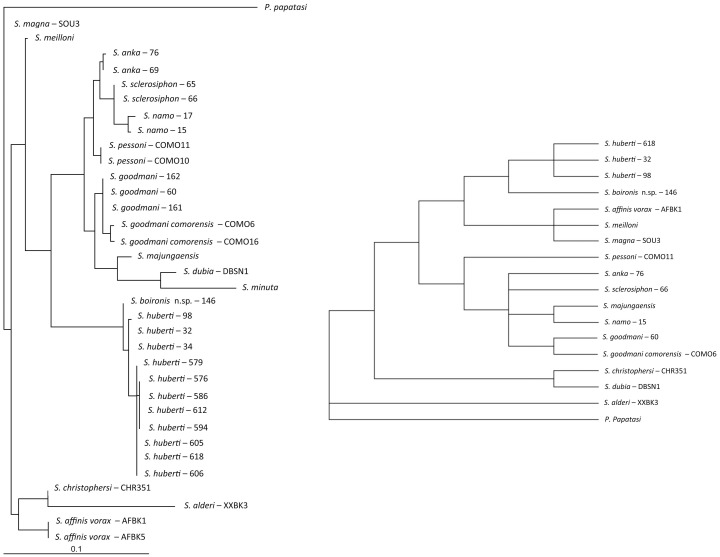
A high degree of homology is observed between the topologies obtained by ML and NJ analyses. Consequently, the NJ trees are not shown. The ML and MP trees related to the analysis of the sequences of D1–D2, D8 and cyt b are shown on figures 2, 3, and 4, respectively.
Figure 2. ML and MP trees based on cyt b sequences mtDNA.
Bootstrap values indicated have been obtained after 100 replications.
Figure 3. ML and MP trees based on D1–D2 sequences rDNA.
Bootstrap values indicated have been obtained after 100 replications.
Figure 4. ML and MP trees based on D8 sequences rDNA.
Bootstrap values indicated have been obtained after 100 replications.
All the markers and analyses isolate the Malagasy specimens exhibiting Sintonius morphological specimens from Anjohikinakina cave-Bemaraha (specimens 1, 98, 605, 606, 612, 618), from Anjohikely cave (specimens 576, 579, 586, 594), from Namoroka (specimens 30, 32, 33, 34) and from Isalo (specimen 146) in a clade not linked with the African Sintonius.
The females from Anjohikinakina cave and Bemaraha (holotype and topotypes) are morphologically similar to those from Anjohikely cave and from Namoroka. Sequences of males and females from these localities show 100% homology regarding D8 marker, a few differences for D1–D2 and cyt b sequences. Regarding the latter, differences are observed between localities, except for the specimens caught in the Bemaraha, divided in two populations. Consequently, we consider that these specimens belong to the same species. The 100% homology between the cyt b sequences of the male specimen number 98 and the female specimen number 1 (P. huberti holotype) allows us to describe the male of this species (see below).
The pairwise distance between the female specimen 146 from Isalo and the S. huberti populations (table 2) is high and is phylogeneticaly isolated (figs 2, 3 and 4). Moreover, this specimen 146 is morphologically very different from the females from Anjohikely, Namoroka and Bemaraha. All these data support the creation of a new species (see below).
Table 2. Male measurements.
| Bemaraha | Bemaraha | Anjohikely | Namoroka | ||||||||
| samples | 98 | 605, 606, 612, 618 | 523, 576, 579, 582, 586, 595 | 32 | 32 | ||||||
| average | min | max | average | min | max | average | |||||
| Head | |||||||||||
| AIII | 305.07 | 290.97 | 264.63 | 307.89 | 292.14 | 291.76 | 308.26 | 290.53 | 292.25 | 291.39 | |
| AIV | 138.18 | 132.27 | 119.58 | 142.10 | 133.03 | 123.54 | 142.10 | 137.48 | 136.97 | 137.22 | |
| AV | 145.14 | 138.46 | 124.83 | 151.81 | 140.06 | 130.26 | 151.81 | 145.79 | 141.09 | 143.44 | |
| L | 175.54 | 194.08 | 179.98 | 205.43 | 188.76 | 185.69 | 209.08 | 192.60 | 192.90 | 192.75 | |
| AIV+AV | 283.32 | 270.73 | 244.40 | 293.91 | 273.09 | 253.80 | 293.91 | 283.26 | 278.06 | 280.66 | |
| P1 | 36.14 | 44.20 | 39.30 | 49.31 | 42.24 | 35.44 | 51.83 | 34.09 | 36.36 | 35.23 | |
| P2 | 104.67 | 108.97 | 100.20 | 115.58 | 107.36 | 106.36 | 119.36 | 101.01 | 107.79 | 104.40 | |
| P3 | 143.24 | 162.02 | 144.92 | 170.91 | 155.27 | 142.47 | 170.91 | 128.79 | 136.00 | 132.39 | |
| P4 | 261.90 | 241.49 | 219.08 | 258.28 | 245.19 | 224.03 | 258.28 | 196.85 | 229.96 | 213.41 | |
| P5 | 361.08 | 400.50 | 367.41 | 450.10 | 394.77 | 411.00 | 450.10 | 257.44 | 389.14 | 323.29 | |
| Wings | |||||||||||
| Length | 1831.34 | 1822.98 | 1809.71 | 1851.39 | 1828.03 | 1824.49 | 1880.78 | 1890.00 | 1890.00 | ||
| Width | 408.81 | 464.80 | 443.77 | 525.51 | 478.03 | 470.56 | 525.51 | 404.66 | 404.66 | ||
| α | 364.94 | 369.35 | 325.53 | 402.84 | 365.91 | 366.73 | 402.84 | 368.36 | 368.36 | ||
| β | 293.91 | 396.00 | 380.76 | 425.81 | 400.85 | 380.76 | 445.54 | 412.35 | 412.35 | ||
| δ | 219.61 | 178.95 | 148.51 | 219.73 | 182.40 | 175.38 | 219.73 | 167.93 | 167.93 | ||
| γ | 364.02 | 308.22 | 279.69 | 334.97 | 307.63 | 287.56 | 338.31 | 317.55 | 317.55 | ||
| π | 113.06 | 161.03 | 124.81 | 183.81 | 156.55 | 124.96 | 182.11 | 192.90 | 192.90 | ||
| w/γ | 1.12 | 1.51 | 1.42 | 1.59 | 1.50 | 1.64 | 1.55 | 1.27 | 1.27 | ||
| Genitalia | |||||||||||
| Style | 100.16 | 116.87 | 113.97 | 119.46 | 112.62 | 101.11 | 117.10 | 96.29 | 103.14 | 99.72 | |
| Coxite length | 208.54 | 218.39 | 195.62 | 228.07 | 212.65 | 195.62 | 228.07 | 201.46 | 209.84 | 205.65 | |
| Paramere | 137.94 | 157.49 | 149.15 | 171.77 | 154.09 | 149.81 | 177.81 | 144.04 | 145.09 | 144.57 | |
| Aedeagus length | 66.70 | 79.31 | 69.25 | 89.46 | 76.18 | 79.01 | 94.46 | 71.20 | 67.41 | 69.31 | |
| Surstyles | 188.20 | 200.16 | 183.85 | 221.43 | 198.41 | 185.62 | 229.53 | 189.23 | 218.06 | 203.64 | |
| Genital filaments | ∼476.250 | 531.74 | 492.32 | 563.26 | 529.11 | 511.40 | 563.26 | 484.03 | 502.89 | 493.46 | |
| Genital pump | 117.65 | 101.27 | 131.07 | 116.66 | 108.04 | 131.07 | 120.09 | 120.09 | |||
Measurements in µm.
Morphological Analysis
The morphological examination of the Malagasy sandflies identified as Phlebotomus huberti (topotypes caught in 2012 and specimens from Namoroka) as well as the reexamination of the female holotype and the paratypes show a lack of mesanepisternal setae, despite the unexplained presence of a group of four mesanepisternal setae in the original description [10]. Consequently, the species huberti cannot belong to the genus Phlebotomus. We here propose a new combination: Sergentomyia huberti comb. nov. based on the existence of two synapomorphies i.e. the absence of pre-apical papilla on the fifth antennal segment and presence of an opened labial furca.
Linking Molecular and Morphological Analysis
The existence of annealed spermathecae and of small teeth on the cibarium is in agreement with the characters of inclusion in the subgenus Sintonius [1], [6], [7]. However, the molecular data do not support the inclusion of S. huberti comb. nov. in the subgenus Sintonius. The specimen 146 belongs to the sister species of S. huberti. Consequently, we propose a new subgenus including S. huberti comb. nov. and S. boironis n. sp.: Trouilletomyia subg. nov.
Description of Trouilletomyia subg. nov., depaquit and léger
According to the monophyly of the Malagasy species Sintonius-like, we have created for them the Trouilletomyia subg. nov. in the genus Sergentomyia, that we define by i) annealed spermathecae, ii) an armed cibarium in both sexes, iii) a remarkable pharyngeal armature with two types of teeth, like that observed in the subgenera Adlerius, Euphlebotomus or Anaphlebotomus pro parte (Asiatic species) of the genus Phlebotomus. To our knowledge, this pharyngeal armature had never been observed in the genus Sergentomyia.
Trouilletomyia Léger & Depaquit subg. Nov. urn:lsid:zoobank.org:act:E2091673-4A89-49BD-8AAE-3C41E5137C1F
Description of S. huberti comb. nov.
Genus Sergentomyia Rondani et Berté, in Rondani, 1840.
Subgenus Trouilletomyia subg. nov.
Species Sergentomyia huberti comb. nov.
The description is based on the specimen number 98.
Male (Fig. 5)
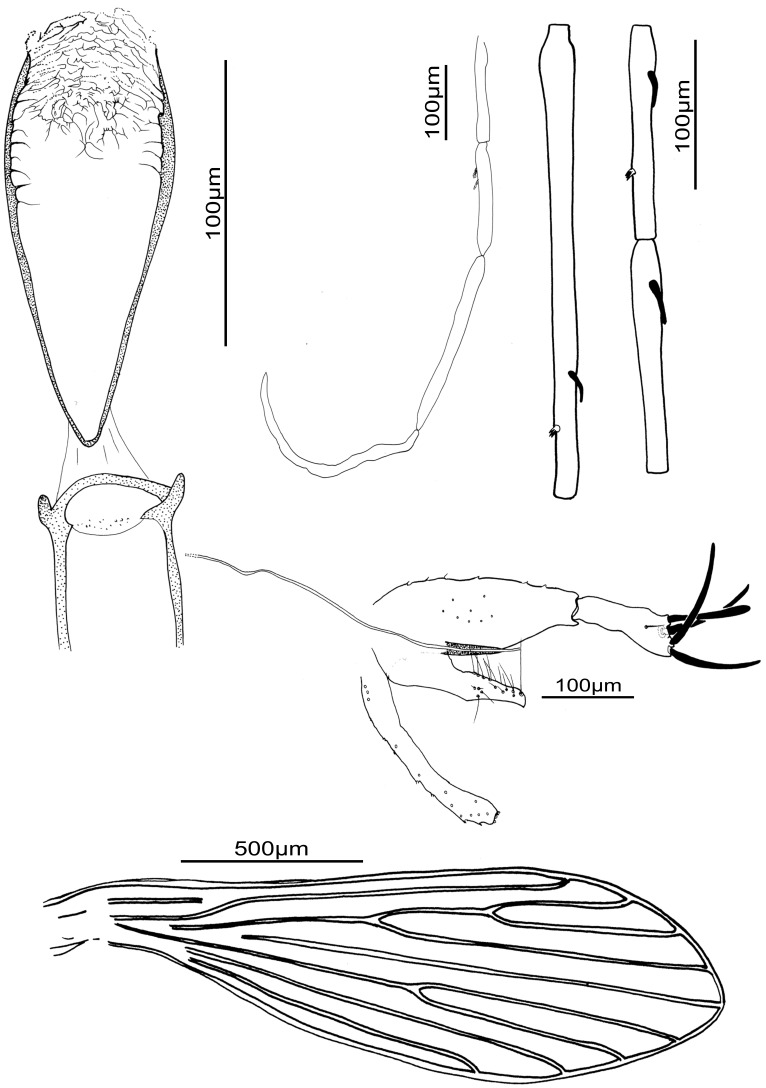
Figure 5. Sergentomyia (Trouilletomyia) huberti comb. nov. male from Bemaraha.
A: Cibarium and pharynx. B: Palp. C: Antennal segments III, IV and V. D: Genitalia. E: Wing.
*Head
Inter-ocular suture: incomplete.
Cibarial armature with some discrete denticles.
Pharynx quite narrow, with a discrete armature composed of very small aligned teeth, forming ripples. Some well developed lateral teeth.
Palpal formula: 1, 2, 3, 4, 5. A few Newstead’s scales on the third palpal segment.
Antennal formula: 1/III-XV with short ascoids. AIII = 305 µm more than AIV + AV.
Labrum = 176 µm. AIII/L = 1.73.
*Thorax
No setae on the mesanepisternum.
Wing: length = 1771 µm, width = 408 µm, α = 365 µm, β293 µm = , δ = 220 µm, γ = 364 µm, π = 113 µm.
Width/γ ratio = 1.1.
*Genital Armature
Coxite 209 µm long, with about ten internal setae implanted in its middle.
Style 100 µm long, narrow, with two terminal and two subterminal spines.
Single paramere, hooked at the top.
Surstyles 188 µm long.
Aedeagus: length = 67 µm, straight, regularly tapering toward the distal end.
Genital filaments: length = 476 µm, isodiametric.
The description of female [10] remains valid, but without setae on the mesanepisternum.
Description of S. boironis n. sp. Randrianambinintsoa & Depaquit
Genus Sergentomyia Rondani et Berté, in Rondani, 1840.
Subgenus Trouilletomyia subg. nov.
Species Sergentomyia boironis n. sp.
Sergentomyia boironis Randrianambinintsoa & Depaquit sp. nov. urn:lsid:zoobank.org:act: 734CFF03-B1DD-448C-92CF-1059D21D1BBC.
It is based on the specimen 146.
Female (Fig. 6)
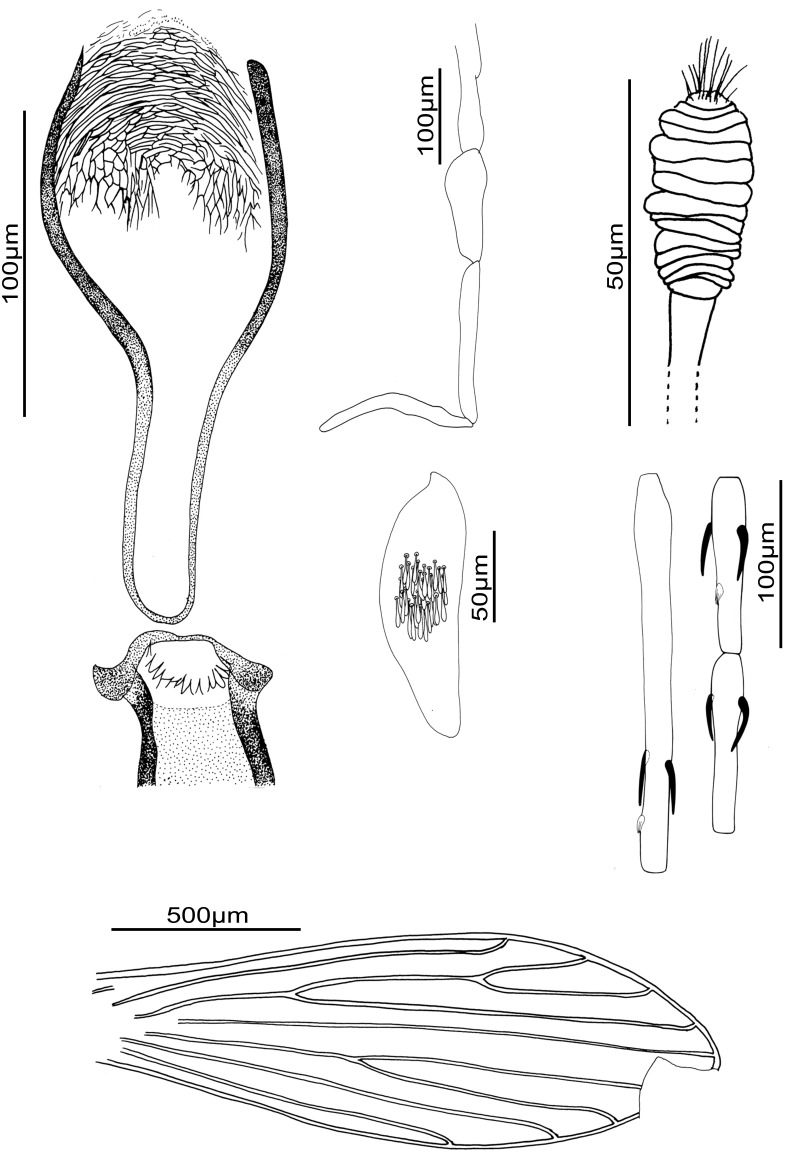
Figure 6. Sergentomyia (Trouilletomyia) boironis n. sp. female.
A: Cibarium and pharynx. B: Palp. C: 3rd palpal segment showing Newstead’s scales. D: Spermathecal body. E: Antennal segments III, IV and V. F: Wing.
Holotype.
*Head
Interocular suture incomplete.
Cibarial armature with fifteen pointed teeth directed backward, along an arc.
Pharyngeal armature Adlerius or Euphlebotomus-like, formed of two kinds of teeth: the posterior ones formed of several concentric ranges and the anterior ones long and oriented forward.
Palpal formula: 1, 2, 3, 4, 5. About thirty Newstead’s scales club-like in a patch on mesal face of the third segment.
Antennal formula: 2/III-XV. Short ascoids. A3 = 231 µm, longer than A4 ( = 103 µm) + A5 ( = 105 µm). Antennae lost during the remounting.
Labrum = 216 µm. A3/L = 1.07.
*Thorax
No setae on the mesanepisternum.
Wing: length about 1900 µm, width = 510 µm, β = 501 µm, δ = 79 µm, γ = 342 µm, π = 139 µm.
Width/γ ratio = 1.49.
*Spermathecae: annealed body formed by 15 rings. Rounded head, slightly invaginated in the last ring. Observation of ducts not possible.
Type-locality : Isalo National Park, Madagascar: 22°37.60′S–45°21.49′E, altitude: 822 m a.s.l.
The holotype has been deposited in the department of entomology of the Muséum National d’Histoire Naturelle, Paris.
The male remains unknown.
Derivatio Nominum
The subgenus Trouilletomyia subg. nov. is dedicated to our colleague Jean Trouillet.
The species Sergentomyia boironis n. sp. is dedicated to our colleague Pascal Boireau.
To meet the criteria of availability, the authors Randrianambinintsoa & Depaquit are responsible of the name Sergentomyia boironis n. sp. and the authors Depaquit, Léger & Randrianambinintsoa are responsible of the name Trouilletomyia subg. nov. and should be cited as the sole authority of these taxa, according to the Article 50(1) of the lnternational Code of Zoological Nomenclature, 4th edition, 2000.
Discussion
Within the Phlebotominae of the Old World, the genus Sergentomyia França & Parrot, 1920 appears to be a catch fall group, including all the Old World species excluded from all other genera (Phlebotomus, Idiophlebotomus, Chinius, Spelaeophlebotomus, Grassomyia, Parvidens, Spelaeomyia and Demeillonius) [24], [25], [32]. Species of the genus Sergentomyia share the following characters: a mesanepisternum without setae, abdominal tergites 2–6 carrying usually all or most recumbent hairs, an usual 1/III–XV antennal formula in the males and 2/III–XV in the females with some exceptions, a cibarium with an armature of teeth and/or denticled more developed in females than in males (beyond exceptions), a single paramere, a style with four terminal spines (or often 2 terminal and 2 subterminal) and an accessory spine.
The genus Sergentomyia is regularly mentioned as probable vector of leishmaniases [33], [34] and arboviruses [35], [36], it is important that the systematics of this group is well assessed.Currently, mainly based on the spermathecal morphology, the genus Sergentomyia is divided in seven subgenera: Sergentomyia França & Parrot, 1920; Neophlebotomus França & Parrot, 1920; Sintonius Nitzulescu, 1931; Parrotomyia Theodor, 1948; Rondanomyia Theodor, 1948; Capensomyia Davidson, and; Vattieromyia, Depaquit, Léger & Robert, 2008. However many species remain unclassified at subgeneric level.
The species S. huberti and S. boironis n. sp. show the characters of inclusion in the subgenus Sintonius [1], [6], [7]: i) scanty erect hairs on the abdominal segments II to VI, ii) annealed spermathecae, iii) a pointed aedeagus and iv) a style with two terminal and two subterminal spines. However, the markers and molecular analyses show that S. huberti and S. boironis n. sp. never cluster with the Sintonius included in the present study. Moreover, the latter group is paraphyletic. Regarding the different analyses, it appears the consistency index of cyt b analysis is low (0.41) whereas those calculated from the ribosomal markers D1–D2 and D8 are higher (0.75 and 0.72), respectively. Consequently, the ribosomal markers are more reliable than cyt b. This mitochondrial marker includes many homoplasic characters and the cyt b trees could be considered as doubtful. According to D1–D2 (figure 2), the MP and ML analyses show S. clydei as the sister species of S. adleri. The relationships between S. meilloni and S. affinis vorax are not resolved by MP and the phylogenetic position of S. christophersi differs. Regarding the D8 rDNA domain, the positions of the Sintonius differ according to MP and ML trees, especially the position of S. affinis vorax and S. meilloni. The paraphyly of the subgenus Sintonius is proven but a more extensive study comparing morphological and molecular analyses is needed in order to revise the group. In our opinion, the main morphological traits characterising the subgenus Sintonius could be symplesiomorphies. For example, many species of Phlebotomus or American sandflies have annealed spermathecae. These data did not allow us to include S. huberti and S. boironis n.sp. in the subgenus Sintonius.
The specific value of S. boironis n. sp. is supported by both morphological and molecular data. Its cibarial armature is very different from that of S. huberti and we note the head of the spermathecae is rounded and included in the most distal ring, differing from that of S. huberti and Sintonius. Moreover, phylogenetical analyses show S. boironis n. sp. is the sister species of S. huberti. The pairwise distances are high between S. boironis n. sp and S. huberti: 9.5 to 12.8% for cyt b and 12 to 19% for D1–D2 (table 2). The low values (0.3 to 0.5%) observed for D8 are explained by the MEGA algorithm not taking into account the positions including indels, which support the variability between S. boironis n. sp. and S. huberti.
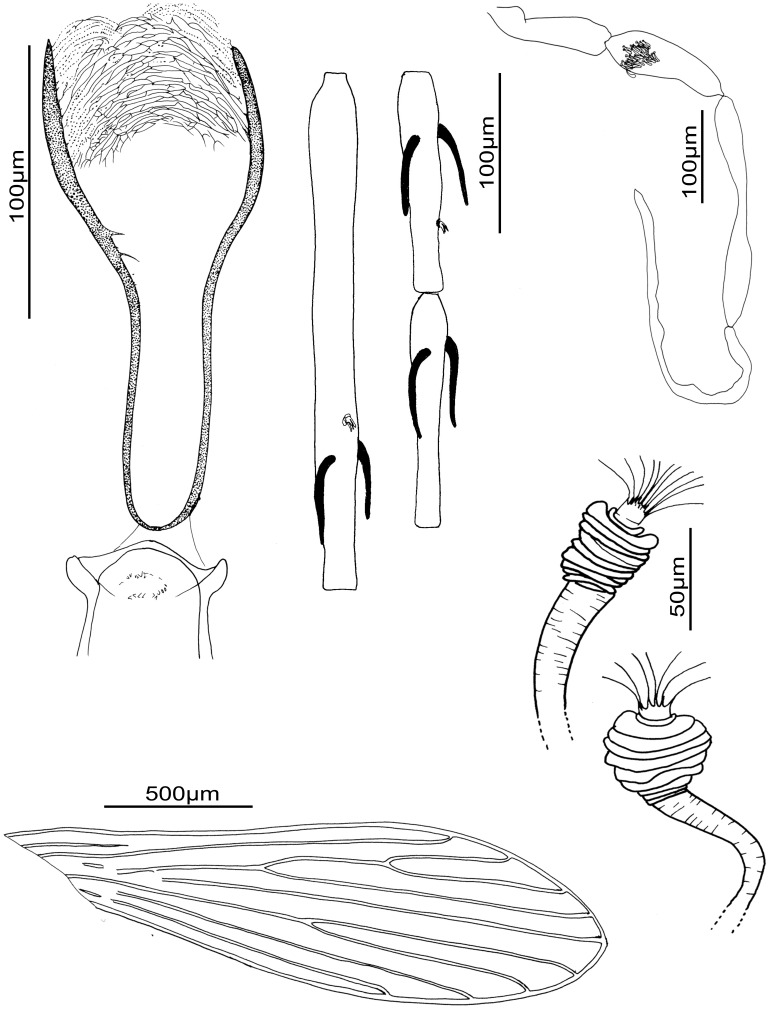
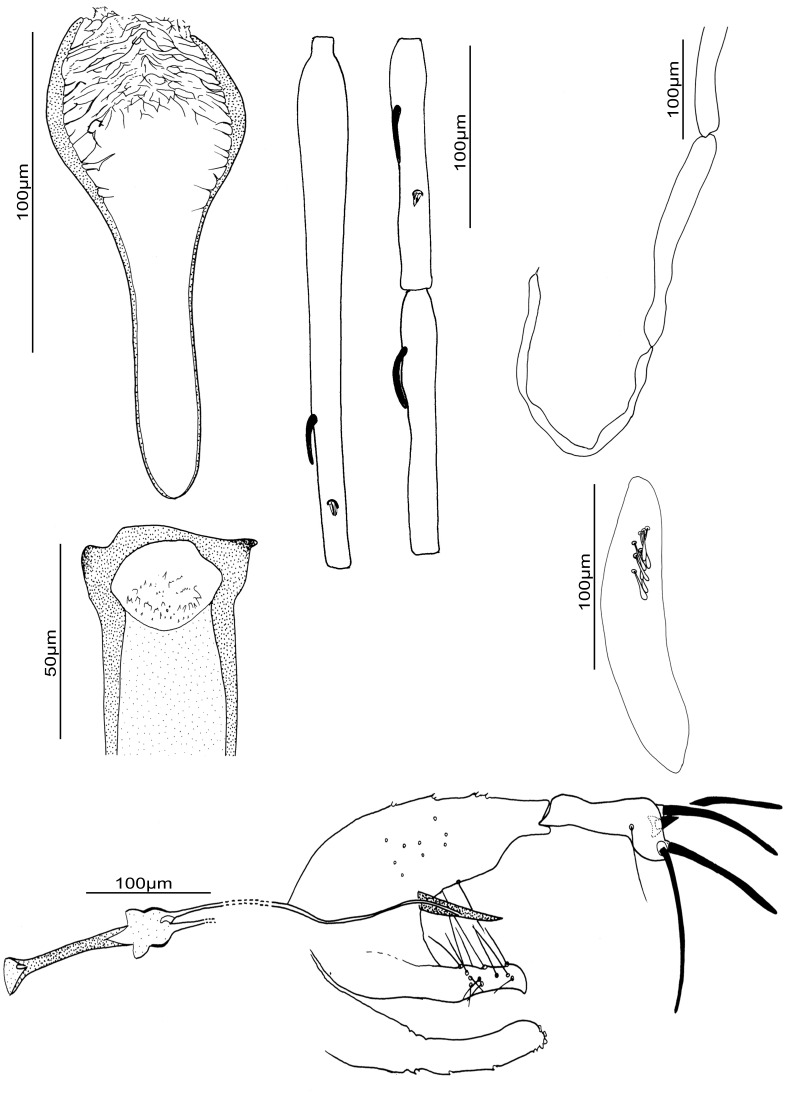
We observe important molecular cyt b variability between the different populations of S. huberti comb. nov. However there is not enough material available from different populations and genders in order to assess if they belong to cryptic species or not. The observation of specimens from Namoroka shows little differences in females (figure 7) and males (figure 8) especially regarding the higher number of cibarial teeth. However, these observations do not seem significant enough at the present time to justify the creation of a new species. The morphometric descriptive statistics (table 3) do not emphasise significant differences between these populations. Despite the existence of two sympatric mitochondrial populations in the Anjohikinakina cave-Bemaraha (specimens 1 and 98 on the one hand, and specimens 605, 606, 612 and 618 on the other hand) will encourage new investigations related to this species.
Figure 7. Sergentomyia (Trouilletomyia) huberti comb. nov. female from Namoroka.
A: Cibarium and pharynx. B: Antennal segments III, IV and V. C: Palp. D. Spermathecae. E: Wing.
Figure 8. Sergentomyia (Trouilletomyia) huberti comb. nov. male from Namoroka.
A: Cibarium. B: Pharynx. C: Antennal segments III, IV and V. D: Palp. E: 3rd palpal segment showing Newstead’s scales. E: Genitalia.
Table 3. Pairwise genetic distances (%) between and within taxa.
| Cyt b | (1) | (2) | (3) | (4) | (5) |
| Bemaraha (no type) (1) | |||||
| Anjohikely (2) | 0.000 | ||||
| Mada 1 et 98 (3) | 0.011 | 0.011 | |||
| Namoroka (4) | 0.012 | 0.012 | 0.013 | ||
| Isalo (5) | 0.016 | 0.016 | 0.019 | 0.018 | |
| D1–D2 | (1) | (2) | (3) | (4) | (5) |
| Bemaraha (no type) (1) | |||||
| Anjohikely (2) | 0.000 | ||||
| Mada 98 (3) | 0.007 | 0.007 | |||
| Namoroka (4) | 0.001 | 0.001 | 0.006 | ||
| Isalo (5) | 0.012 | 0.012 | 0.019 | 0.013 | |
| D8 | (1) | (2) | (3) | (4) | (5) |
| Bemaraha (no type) (1) | |||||
| Anjohikely (2) | 0.001 | ||||
| Mada 98 (3) | 0.002 | 0.002 | |||
| Namoroka (4) | 0.002 | 0.002 | 0.000 | ||
| Isalo (5) | 0.005 | 0.005 | 0.003 | 0.003 |
Identification key of the Sergentomyia and Grassomyia from Madagascar:
Females:
1- capsulated spermathecae 2
non capsulated spermathecae 3
2- spherical spermathecal capsule Grassomyia
spermathecae narrower in the median part S. (Vattieromyia)
3- smooth spermathecae S. majungaensis
segmented spermathecae 4
4- completely segmented spermathecae; well developed pharyngeal armature with two kinds of teeth 5
partially segmented spermathecae; discrete pharyngeal armature S. goodmani
5- cibarial armature of about 15 well developed pointed teeth, oriented backward, along a curved line S. boironis n. sp
cibarial armature with discrete teeth or denticle S. huberti comb. nov.
Males:
1- one ascoid on the third antennal segment (AIII) 2
absence of ascoid on the third antennal segment (AIII) 3
2- antennal formula 1/III-XII S. goodmani
antennal formula 1/III-XV S. huberti
3- AIII shorter than 200 µm Grassomyia
AIII longer than 200 µm 4
4- genital filaments shorter than 250 µm S. majungaensis
genital filaments longer than 250 µm S. (Vattieromyia).
Supporting Information
Neighbor-Joining tree based on cytochrome b sequences mtDNA. Bootstrap values indicated have been obtained after 1,000 replicates.
(TIF)
Neighbor-Joining tree based on D1-D2 sequences rDNA. Bootstrap values indicated have been obtained after 1,000 replicates.
(TIF)
Neighbor-Joining tree based on D8 sequences rDNA. Bootstrap values indicated have been obtained after 1,000 replicates.
(TIF)
Acknowledgments
The authors thank Sylvette Gobert for proofreading the manuscript and Steve Goodman (Field Museum of Natural History of Chicago, NGO Vahatra) for logistical support in the field. They are grateful to the Ministry of Forests of Madagascar and Madagascar National Park for providing a research permit.
Funding Statement
The fieldwork received funding from the Institute of Research for Development, Institut Pasteur de Madagascar, and Volkswagen Stiftung. Laboratory work received funding from: (i) the Program of “Institut Français de la biodiversité/CNRS/AIRD Biodiversité dans les îles de l’Océan Indien'” itself included in the Regional Project “Insectes vecteurs (phlébotomes et moustiques) dans les îles de l’Océan Indien: Madagascar, Seychelles et Comores”, the GDRI “Biodiversité et Développement Durable à Madagascar”; (ii) “Université de Reims Champagne – Ardenne”; and (iii) the Robert S. McNamara Fellowship Program of the World Bank. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nitzulescu V (1931) Essai de classification des phlébotomes. Annales de Parasitologie 9: 271–275. [Google Scholar]
- 2. França C, Parrot L (1920) Introduction à l'étude systématique des Diptères du genre Phlebotomus. . Bull SocPathol Exot 13: 695–708. [Google Scholar]
- 3.Abonnenc E (1972) Les phlébotomes de la région éthiopienne (Diptera, Psychodidae). Cah ORSTOM, Sér Ent Méd Parasitol 55: 239 p. [Google Scholar]
- 4. Lewis DJ (1978) The phlebotomine sandflies (Diptera: Psychodidae) of the Oriental Region. Bulletin of the British Museum (Natural History), Entomology Series 37: 217–343. [Google Scholar]
- 5.Perfiliev P (1968) Fauna of USSR. Diptera. Phlebotomidae (sandflies). THEODOR O, editor. Jérusalem: Israel program for scientific translations. 363 p. [Google Scholar]
- 6. Theodor O (1948) Classification of the old world species of the subfamily Phlebotominae (Diptera : Psychodidae). Bull Ent Res 39: 85–118. [DOI] [PubMed] [Google Scholar]
- 7.Theodor O (1958) 9c. Psychodidae-Phlebotominae. Stuttgart: Erwin lindner. 1–55 p. [Google Scholar]
- 8. Goodman S, Benstead J (2005) Updated estimates of biotic diversity and endemism for Madagascar. Oryx 39: 73–77. [Google Scholar]
- 9. Depaquit J, Léger N, Ferté H, Robert V (2004) Les phlébotomes de Madagascar (Diptera: Psychodidae). II. Description de la femelle de Phlebotomus (Anaphlebotomus) fertei Depaquit, Léger & Robert, 2002; description du mâle et redescription de la femelle de Phlebotomus (Anaphlebotomus) berentiensis (Léger & Rodhain, 1978) comb. nov. Parasite 11: 201–209. [DOI] [PubMed] [Google Scholar]
- 10. Depaquit J, Léger N, Robert V (2002) Première mention de Phlebotomus à Madagascar (Diptera: Psychodidae). Description de Phlebotomus (Anaphlebotomus) fertei n. sp. et de Phlebotomus (Anaphlebotomus) huberti n. sp. Parasite 9: 325–331. [DOI] [PubMed] [Google Scholar]
- 11. Depaquit J, Léger N, Robert V (2004) Les Phlébotomes de Madagascar (Diptera: Psychodidae). III–Description de Phlebotomus (Anaphlebotomus) fontenillei n. sp. Parasite 11: 261–265. [DOI] [PubMed] [Google Scholar]
- 12. Depaquit J, Léger N, Robert V (2007) Les phlébotomes de Madagascar (Diptera: Psychodidae). V-. Description de Sergentomyia majungaensis n. sp. Parasite 14: 219–223. [DOI] [PubMed] [Google Scholar]
- 13. Depaquit J, Léger N, Robert V (2008) Les phlébotomes de Madagascar (Diptera: Psychodidae). VI–Un sous-genre nouveau (Vattieromyia) avec trois espèces nouvelles: Sergentomyia (V.) sclerosiphon, S. (V.) namo et S. (V.) anka . Parasite 15: 15–26. [DOI] [PubMed] [Google Scholar]
- 14. Léger N, Depaquit J, Robert V (2005) Les phlébotomes de Madagascar (Diptera: Psychodidae). IV. Description de Sergentomyia (Rondanomyia) goodmani n. sp. Rétablissement du sous-genre Rondanomyia Theodor. Parasite 12: 51–57. [DOI] [PubMed] [Google Scholar]
- 15. Léger N, Rodhain F (1978) Sergentomyia berentiensis n. sp. (Diptera, Psychodidae). Description à partir d'un exemplaire femelle récolté à Madagascar. Bull Soc Path Ex 71: 476–479. [PubMed] [Google Scholar]
- 16. Randrianambinintsoa FJ, Depaquit J (2013) Phlebotomine sand flies from Madagascar (Diptera: Psychodidae). VIII - Phlebotomus (Anaphlebotomus) vincenti n. sp. Bull Soc Pathol Exot 106: 206–211. [DOI] [PubMed] [Google Scholar]
- 17. Randrianambinintsoa FJ, Leger N, Robert V, Depaquit J (2013) Phlebotomine sand flies from Madagascar (Diptera: Psychodidae). VII. An identification key for Phlebotomus with the description of Phlebotomus (Anaphlebotomus) vaomalalae n. sp. Parasite 20: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esseghir S, Ready PD, Killick-Kendrick R, Ben-Ismail R (1997) Mitochondrial haplotypes and geographical vicariance of Phlebotomus vectors of Leishmania major . Insect Molecular Biology 6: 211–225. [DOI] [PubMed] [Google Scholar]
- 19. Depaquit J, Perrotey S, Lecointre G, Tillier A, Tillier S, et al. (1998) Systématique moléculaire des Phlebotominae: étude pilote. Paraphylie du genre Phlebotomus . C R Acad Sci III 321: 849–855. [DOI] [PubMed] [Google Scholar]
- 20. Bonfield J, Staden R (1996) Experiment files and their application during large-scale sequencing projects. DNA Seq 6: 109–117. [DOI] [PubMed] [Google Scholar]
- 21. Thompson J, Higgins D, Gibson T (1994) Clustal W: improving the sensivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice.. Nucleic Acid Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hall T (1999) Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 4: 95–98. [Google Scholar]
- 23. Aransay AM, Scoulica E, Tselentis Y, Ready PD (2000) Phylogenetic relationships of phlebotomine sandflies inferred from small subunit nuclear ribosomal DNA. Insect Molecular Biology 9: 157–168. [DOI] [PubMed] [Google Scholar]
- 24. Rispail P, Léger N (1998) Numerical taxonomy of Old World Phlebotominae (Diptera : Psychodidae). 1. Considerations of morphological characters in the genus Phlebotomus Rondani et Berté 1840. Mem Inst Oswaldo Cruz 93: 773–785. [DOI] [PubMed] [Google Scholar]
- 25. Rispail P, Léger N (1998) Numerical taxonomy of Old World Phlebotominae (Diptera : Psychodidae). 2. Restatement of classfication upon subgeneric morphological characters. Mem Inst Oswaldo Cruz 93: 787–793. [DOI] [PubMed] [Google Scholar]
- 26. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 27. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Posada D, Crandall K (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 30. Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape spliting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174. [DOI] [PubMed] [Google Scholar]
- 31. Tavare S (1986) Some probabilistic and statistical problems on the analysis of DNA sequences. Lectures in Mathematics in the Life Sciences 17: 57–86. [Google Scholar]
- 32. Abonnenc E, Léger N (1976) Sur une classification rationnelle des Diptères Phlebotomidae. Cah ORSTOM, Sér Ent Méd Parasitol 14: 69–78. [Google Scholar]
- 33. Berdjane-Brouk Z, Kone AK, Djimde AA, Charrel RN, Ravel C, et al. (2012) First detection of Leishmania major DNA in Sergentomyia (Spelaeomyia) darlingi from cutaneous leishmaniasis foci in Mali. PLoS One 7: e28266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Senghor MW, Faye MN, Faye B, Diarra K, Elguero E, et al. (2011) Ecology of phlebotomine sand flies in the rural community of Mont Rolland (Thies region, Senegal): area of transmission of canine leishmaniasis. PLoS One 6: e14773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Charrel RN, Izri A, Temmam S, de Lamballerie X, Parola P (2006) Toscana virus RNA in Sergentomyia minuta files. Emerg Infect Dis 12: 1299–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Depaquit J, Grandadam M, Fouque F, Andry PE, Peyrefitte C (2010) Arthropod-borne viruses transmitted by Phlebotomine sandflies in Europe: a review. Euro Surveill 15: 19507. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-Joining tree based on cytochrome b sequences mtDNA. Bootstrap values indicated have been obtained after 1,000 replicates.
(TIF)
Neighbor-Joining tree based on D1-D2 sequences rDNA. Bootstrap values indicated have been obtained after 1,000 replicates.
(TIF)
Neighbor-Joining tree based on D8 sequences rDNA. Bootstrap values indicated have been obtained after 1,000 replicates.
(TIF)