A transcription factor, known as a positive regulator of nodule organogenesis in root cortex, acts as a negative regulator of epidermal infection events by rhizobia.
Abstract
Legume-rhizobium symbiosis occurs in specialized root organs called nodules. To establish the symbiosis, two major genetically controlled events, rhizobial infection and organogenesis, must occur. For a successful symbiosis, it is essential that the two phenomena proceed simultaneously in different root tissues. Although several symbiotic genes have been identified during genetic screenings of nonsymbiotic mutants, most of the mutants harbor defects in both infection and organogenesis pathways, leading to experimental difficulty in investigating the molecular genetic relationships between the pathways. In this study, we isolated a novel nonnodulation mutant, daphne, in Lotus japonicus that shows complete loss of nodulation but a dramatically increased numbers of infection threads. Characterization of the locus responsible for these phenotypes revealed a chromosomal translocation upstream of NODULE INCEPTION (NIN) in daphne. Genetic analysis using a known nin mutant revealed that daphne is a novel nin mutant allele. Although the daphne mutant showed reduced induction of NIN after rhizobial infection, the spatial expression pattern of NIN in epidermal cells was broader than that in the wild type. Overexpression of NIN strongly suppressed hyperinfection in daphne, and daphne phenotypes were partially rescued by cortical expression of NIN. These observations suggested that the daphne mutation enhanced the role of NIN in the infection pathway due to a specific loss of the role of NIN in nodule organogenesis. Based on these results, we provide evidence that the bifunctional transcription factor NIN negatively regulates infection but positively regulates nodule organogenesis during the course of the symbiosis.
Legumes develop a specialized symbiotic organ on their roots, the root nodule, in response to rhizobial infection. Benefiting from symbiotic nitrogen fixation by rhizobia in the nodule, plants can grow under nitrogen-limited conditions. The signaling pathways in nodule development are divided into two major events, rhizobial infection and organogenesis. For a successful symbiotic association, it is essential that the two phenomena proceed simultaneously in different root tissues (Crespi and Frugier, 2008; Madsen et al., 2010; Oldroyd, 2013). Rhizobial infection occurs in the epidermal cells of the root. Rhizobia penetrate the root tissues from curled root hair cells and progress toward the root cortex through an intracellular channel called the infection thread (IT; Vasse and Truchet, 1984; Gage, 2004; Jones et al., 2007; Fournier et al., 2008; Murray, 2011). In contrast, organogenesis begins with the reinitiation of cell division in the root cortex. Several nonnodulation or low-nodulation mutants have been identified by genetic mutant screening in the model legumes Lotus japonicus and Medicago truncatula. Those mutants are impaired in rhizobial infection processes at different steps, from earlier steps (root hair deformation and bacterial colonization; e.g. Lotus japonicus nod factor receptor5 (Ljnfr5)/Medicago truncatula nod factor perception (Mtnfp), Lotus japonicus symbiosis receptor kinase (Ljsymrk)/Medicago truncatula doesn't make infections2 (Mtdmi2), Lotus japonicus calcium- and calmodulin-dependent protein kinase (Ljccamk)/Medicago truncatula doesn't make infections3 (Mtdmi3)) to later steps (IT initiation, IT progression, and bacterial release; e.g. Lotus japonicus cyclops (Ljcyclops)/Medicago truncatula interacting protein of DMI3 (Mtipd3) and Lotus japonicus cerberus (Ljcerberus)/Medicago truncatula lumpy infections (Mtlin); Kouchi et al., 2010; Popp and Ott, 2011). The respective mutated genes are involved in each event. These genes show diverse spatial expression patterns in the epidermis, cortex, and nodule (Popp and Ott, 2011). This complexity has made it difficult to elucidate the molecular mechanism of the interrelationship between the two major signaling pathways of nodule development, occurring in both the epidermis and cortex at different developmental stages.
A few reports have recently focused on the cross talk or independence of these two pathways, using different approaches. By characterization of the phenotypes of various double mutant/transgenic plants harboring 14 individual infection-defective mutations and three spontaneous-nodule-formation mutations/transgenes, symbiotic genes in L. japonicus were categorized into four groups (Madsen et al., 2010): (1) genes for only infection, such as NAK-associated protein1 (NAP1), 121F-specific p53-inducible RNA (PIR1), and CERBERUS; (2) genes for organogenesis and indirectly for infection, such as SymRK, NUCLEOPORIN85, and POLLUX; (3) genes for both infection and organogenesis, such as NODULE INCEPTION (NIN), NODULATION SIGNALING PATHWAY1 (NSP1), and NSP2; and (4) genes for cross talk between infection and organogenesis, such as CCaMK and CYCLOPS. Another study of expression systems under the control of a tissue-specific promoter investigated the special contributions of MtNFP/LjNFR5 and MtDMI3/LjCCaMK to IT formation and nodule organogenesis in a tissue-autonomous manner (Rival et al., 2012). A third study of LjsymRK mutant alleles proposed different contributions of LjSYMRK to each pathway depending on different domains (Kosuta et al., 2011). These studies have shown that the SYMRK-CCaMK-CYCLOPS signaling cascade has two roles: rhizobial infection in the epidermis and nodule organogenesis in the root cortex. However, the tissue-specific roles or cellular interactions between the epidermal event and the inner tissue event of transcription factors functioning in later symbiotic signaling, such as NIN, NSP1, and NSP2, remain unclear.
Among the nonnodulation mutants, the nin mutant is defective in both IT initiation and nodule formation. It is believed that NIN, a transcription factor containing an RWP-RK domain which is named after a conserved motif, functions in both the infection and organogenesis pathways (Schauser et al., 1999; Borisov et al., 2003; Marsh et al., 2007). The expression pattern of the GUS reporter gene driven by the NIN promoter (ProNIN) indicates that epidermal expression a short time after inoculation is correlated with rhizobial infection, whereas expression in the root cortex at a later stage contributes to cell division (Heckmann et al., 2011; Kosuta et al., 2011; Popp and Ott, 2011). NIN transcription is highly induced only after rhizobial inoculation, and constitutive expression of NIN can lead to the ectopic division of cortical cells in the absence of rhizobia. These results indicate that NIN plays a central role in nodule organogenesis (Schauser et al., 1999; Tirichine et al., 2007; Suzaki et al., 2012; Soyano et al., 2013).
Cytokinin also plays an important role in nodule organogenesis, given that a loss-of-function mutation in the putative cytokinin receptors LOTUS HISTIDINE KINASE1 (LHK1) in L. japonicus and the knockdown of CYTOKININ RESPONSE1 in M. truncatula cause the low-nodulation phenotypes (Gonzalez-Rizzo et al., 2006; Murray et al., 2007). In contrast, the spontaneous nodule formation2 (snf2) mutant, which has a gain-of-function mutation in LHK1, yields a spontaneous nodulation phenotype in L. japonicus (Tirichine et al., 2007). Likewise, ectopic cortical cell division and NIN expression are induced by exogenous cytokinin application without rhizobial infection (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Heckmann et al., 2011). Cytokinin activates the cortical expression of NIN but does not induce the epidermal expression of NIN, suggesting that cytokinin activates only the organogenesis pathway and not the infection pathway mediated by NIN (Heckmann et al., 2011).
In this study, we identified a novel nin mutant allele, named daphne, which showed the interesting phenotypes of nonnodulation and hyper IT formation in L. japonicus. The mutant showed an altered expression pattern of NIN. In view of the relationship between the spatiotemporal expression pattern of NIN and the symbiotic phenotype of daphne, we propose a new cellular communication model controlled by NIN involving cross talk between infection and organogenesis for regulating rhizobial infection processes.
RESULTS
Isolation of the daphne Mutant, Which Showed Nonnodulation and Dramatically Increased Infection of Rhizobia in L. japonicus
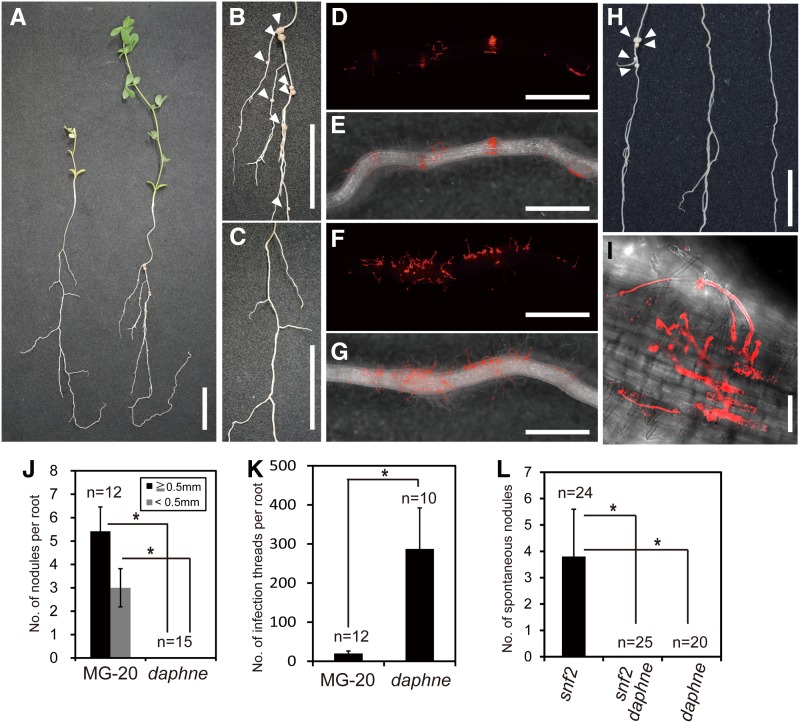
To date, several host genes necessary for nodule development have been identified (Madsen et al., 2010; Oldroyd, 2013). However, most nonnodulation mutants have defects in both the rhizobial infection and organogenesis pathways, so the molecular relationship between these two pathways and the mechanisms for controlling each pathway have remained obscure. To find new components involved in the infection or organogenesis pathways, we first screened ion-beam-mutagenized L. japonicus seeds of accession Miyakojima MG-20 (3,400 M1 lines) for the nonnodulation mutants. We next evaluated their infection ability. As a final step, we focused on a mutant, named daphne, displaying the novel phenotype of nonnodulation and hyperinfection. The daphne mutant was completely defective in nodule formation, being different from hyper infected1 (hit1), which was previously isolated as a hyperinfection mutant able to form a few nodules (Supplemental Fig. S1; Murray et al., 2007). In the daphne mutant, no nodules were observed even 28 d after inoculation (DAI; Fig. 1, A–C and I). daphne showed a typical nonnodulation phenotype, with pale-yellow leaves and growth delay under low-nitrogen conditions (Fig. 1A). However, in daphne, the number of ITs per root was dramatically increased, to 15-fold greater than that on the MG-20 wild type (Fig. 1, D–G and J). In the wild type, ITs tend to be formed in small restricted regions called susceptible zones (Vasse et al., 1993; Penmetsa and Cook, 1997; Krusell et al., 2002; Gage, 2004). On the other hand, ITs were observed on almost all regions of daphne roots. This extended rhizobial susceptibility has been observed previously in the nin mutant (Schauser et al., 1999). Additionally, IT elongation in daphne was visible in root hair but aborted and burst in the epidermal cell layer, and no cortical ITs were observed (Fig. 1I; Supplemental Fig. S2).
Figure 1.
Isolation of a novel nonnodulation mutant, daphne. A, Shoot and root phenotypes of the daphne mutant (left) and the Miyakojima MG-20 wild type (right) at 28 DAI. B, Nodulation phenotype of Miyakojima MG-20. Arrowheads indicate nodules. C, The nonnodulation phenotype of daphne. D to G, IT formation of the Miyakojima MG-20 root (D and E) and of the daphne root (F and G) following inoculation with M. loti MAFF303099 constitutively expressing DsRED. D and F, Red fluorescence images of roots. Linear red signals indicate ITs. E and G, Red fluorescence images and transmitted light images are merged. H, Spontaneous nodule formation in snf2 (left), the daphne snf2 double mutant (middle), and daphne (right). Arrowheads indicate spontaneous nodules. I, Confocal microscopic image of a daphne root. Z-stack series are shown in Supplemental Figure S2. Bars = 2 cm in A to C and H and 1 mm in D to G. J, Nodules were counted at 28 DAI with M. loti MAFF303099. K, The number of ITs per root was counted at 7 DAI with M. loti MAFF303099 constitutively expressing LacZ. L, Six weeks after germination, spontaneous nodules were counted without rhizobial infection under the no-nitrogen condition. Error bars indicate sd. *P < 0.05 by Student’s t test.
To identify the step of the organogenesis pathway that is blocked in daphne, we created the daphne snf2 double mutant and evaluated its ability for spontaneous nodule formation. snf2 has a gain-of-function mutation in the LHK1 gene, which encodes a cytokinin receptor protein. In this mutant, only genes downstream of cytokinin signaling are constitutively active. Accordingly, snf2 can form a nodule-like structure without rhizobial infection (Tirichine et al., 2007). The nonspontaneous nodule formation phenotype of the daphne snf2 double mutant indicates that the nonnodulation phenotype of daphne is caused by defects downstream of cytokinin signaling (Fig. 1, H and K).
Identification of the daphne Mutation by Map-Based Cloning and Inverse PCR
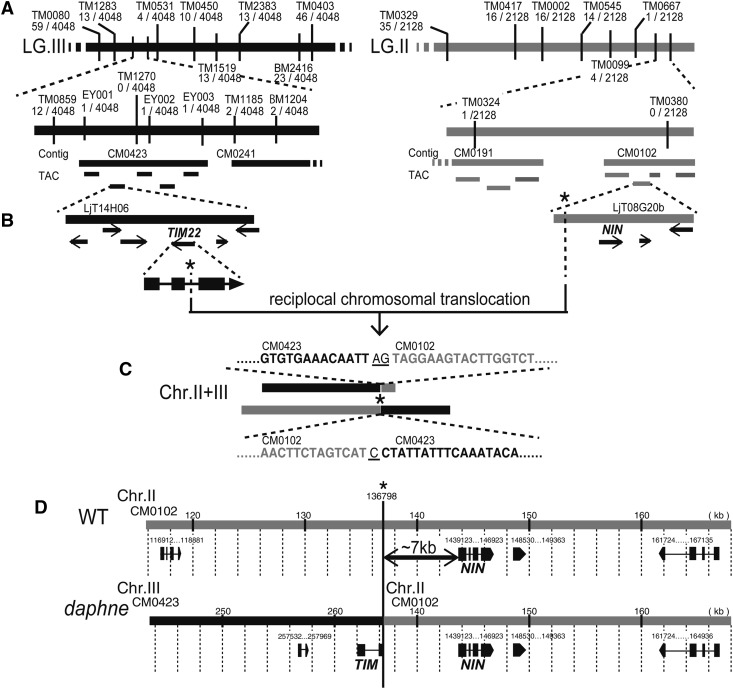
We roughly identified two loci on chromosomes II and III linked to the nonnodulation phenotype of daphne using a small F2 mapping population by map-based cloning (http://www.kazusa.or.jp/lotus/; Sandal et al., 2006; Supplemental Fig. S3). This result was apparently inconsistent with the observation that the F2 population segregated in an approximately 3:1 ratio, indicating that daphne is a recessive mutant. (Supplemental Table S1). We further explored the locus using a large F2 population with markers on linkage group III, and the translocation fusion point was identified by reverse transcription (RT)-PCR (Fig. 2; Supplemental Fig. S4). We finally detected the fused sequences, which originated from chromosomes II and III in the daphne genome, by inverse PCR. This finding suggested that the ion beam irradiation had induced a reciprocal chromosomal translocation. The translocation points lie in the second intron of the Translocase of the inner membrane (TIM) gene (chr3.CM0423.360.r2.d) on chromosome III and in an intergenic region (IGR) on chromosome II. The IGR sequence on the IGR lies approximately 7 kb upstream of NIN, which is known to be an essential gene for nodule development (Schauser et al., 1999). No mutation in the NIN coding region was detected in the daphne genome.
Figure 2.
Identification of the daphne mutation. A, Two genetic linkage maps of the regions of daphne loci in linkage group (LG) III (black, left) and linkage group II (gray, right). The newly developed marker (EY001-3) is shown in Supplemental Table S2. The number of recombination events (events/total chromosomes) is indicated. B, Physical maps of transformation-competent artificial chromosome (TAC) clone LjT14H06 (black, left) and TAC clone LjT08G20b (gray, right). Arrows indicate the annotations from miyakogusa.jp release 2.5 (http://www.kazusa.or.jp/lotus/; Sandal et al., 2006). C, Outline of chromosomal translocation between chromosome (Chr.) II and chromosome III, with sequences at the fusion point identified by inverse PCR amplified from the daphne genome. Black letters indicate the bases from CM0423 (chromosome III), and gray letters indicate the bases from CM0102 (chromosome II). Bases of unknown chromosomal origin are indicated by underlined letters. D, The locations of translocation fusion points in the contigs with gene annotations (exons shown as block boxes and introns shown as thin lines). Numbers on the ruler indicate the exact points (kb) in each contig. WT, Wild type. Asterisks indicate the reciprocal chromosomal translocation points of each locus.
daphne Is a Novel nin Mutant Allele, Different from the nin Null Mutant
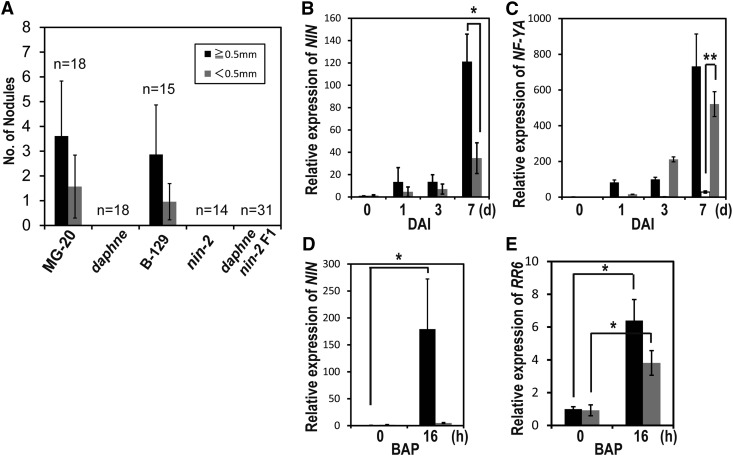
As described above, we determined two candidate loci responsible for the nonnodulation phenotype in daphne: TIM, on chromosome III, and NIN, on chromosome II. Although we introduced the coding sequence of TIM gene under the control of the ubiquitin promoter (ProLjUBQ; Maekawa et al., 2008) by hairy root transformation, the nonnodulation phenotype was not complemented (Supplemental Fig. S4). We next hypothesized that a translocation near the NIN locus causes the daphne phenotype, a notion supported by the previous characterization of a nin mutant that displays nonnodulation and no IT formation (Schauser et al., 1999). By crossing daphne (accession Miyakojima MG-20) and nin-2 (accession Gifu B-129), we tested whether daphne is a nin mutant allele. The success of crossing experiments using pollen of nin-2 was judged by the accumulation of anthocyanin, the dominant characteristic phenotype of Gifu B-129 (nin-2), in stems. All daphne × nin-2 F1 plants originating from three independent seed pods exhibited nonnodulation (Fig. 3A). These results suggested that daphne and nin-2 are allelic for the nonnodulation phenotype. Although the normal infection phenotype of daphne × wild-type F1 plants and the F2 segregation ratio indicate that both daphne phenotypes, nonnodulation and hyperinfection, are recessive (Supplemental Table S1), daphne × nin-2 F1 plants showed hyperinfection. We also observed that the excessive root hair deformation phenotype in daphne was similar to the phenotype of the nin mutant (Supplemental Fig. S5). For the subsequent analysis described below, we used nin-9 (Suzaki et al., 2012) as a canonical nin mutant because it has the same genetic background as daphne.
Figure 3.
daphne is a novel nin mutant allele. A, Allelism tests by crossing daphne and nin-2 mutants (Schauser et al., 1999). Nodules were counted at 14 DAI on each plant root. B to E, Quantitative real-time RT-PCR analysis of NIN (B and D), NF-YA (C), and RR6 (E) expression in the Miyakojima MG-20 wild type (black bars), in daphne (gray bars), and in nin-9 (white bars) noninoculated (0), at 1, 3, and 7 DAI (B and C), or dependent on cytokinin treatment (10−7 M benzylaminopurine [BAP]) for 16 h (D and E). Each cDNA was prepared from total RNA derived from whole root. Fold changes in expression are shown relative to roots at 0 DAI (B and C) or before cytokinin treatment (D and E). Error bars indicate sd of three biological replicates. *P < 0.05, **P < 0.01 by Student’s t test.
daphne Has Completely Lost the NIN Expression Induced by Cytokinin Application
NIN is a putative key transcription factor that plays a role in the infection and nodule organogenesis pathways. The expression level of NIN is strongly elevated in an inoculation-dependent manner (Schauser et al., 1999). Because the allelism test showed that daphne was a nin mutant allele, we next investigated the NIN expression pattern in daphne. Expression in whole roots was slightly induced by inoculation with Mesorhizobium loti. The transcript levels of NIN at earlier stages were almost identical in the wild type and daphne, whereas at 7 DAI, the level of NIN expression in daphne was almost one-third that in the wild type (Fig. 3B). The induction level of Nuclear transcription factor Y alpha (NF-YA), known as a downstream target of NIN (Soyano et al., 2013), also indicates that daphne partially retains the function of NIN, compared with almost no induction of NF-YA in a typical nin mutant, nin-9 (Fig. 3C).
We next evaluated the cytokinin-induced expression level of NIN, finding it to be completely absent in daphne (Fig. 3D). This loss is in good agreement with the finding that the snf2 mutation spontaneously activating the cytokinin signal does not rescue the nonnodulation phenotype of daphne (Fig. 1, H and K), because cytokinin is believed to induce only the organogenesis pathway and not the infection pathway (Heckmann et al., 2011). The cytokinin-induced expression of Lotus japonicus RESPONSE REGULATOR6 (RR6; Op den Camp et al., 2011) was detected even in daphne, suggesting that daphne retains the cytokinin responsiveness of genes other than NIN (Fig. 3E).
daphne Shows Broad Epidermal Expression Patterns of NIN
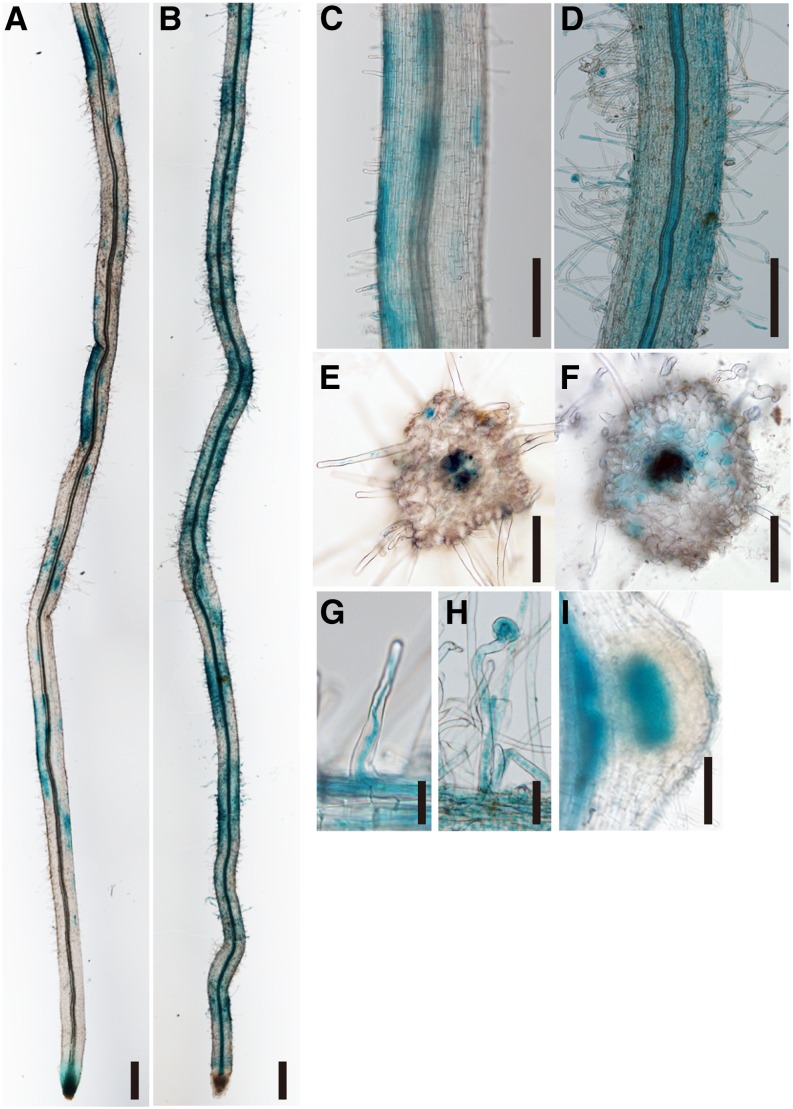
How is the lower expression of NIN in the daphne mutant implicated in the increased number of ITs? To identify the underlying mechanism of increased infection events, we investigated the spatial expression pattern of NIN. We cloned approximately 4 kb of ProNIN and approximately 2 kb of the NIN terminator (TerNIN) for promoter-GUS analysis. For 10 of 14 nin-9 plants, their IT formation was rescued by introducing the ProNIN::NIN::TerNIN construct. This indicated that the NIN promoter was sufficient for the function of NIN at least as involved in rhizobial infection (Table I; Supplemental Fig. S5). In the wild type, blue staining was restricted to several small epidermal regions of the root (Fig. 4, A, C, and E), as reported previously (Radutoiu et al., 2003; Kosuta et al., 2011). The inner cells of nodule primordia in the wild type were also stained, as observed previously (Fig. 4I; Heckmann et al., 2011). In contrast, we observed a broad range, almost the whole root area, of NIN promoter activity in the daphne root (Fig. 4, B, D, and F). The broader activity of ProNIN::GUS::TerNIN in daphne coincided with the region where excessive root hair curling and IT formation occur (Fig. 4, A–H).
Table I. Phenotypic effects of the expression of the NIN or NIN chimeric repressor in the MG-20 wild type, daphne, and nin.
J0571>>NIN rescued the nonnodulation phenotype. ProLjUBQ::NIN and J0571>>NIN inhibited hyperinfection in daphne. ProLjUBQ::NIN::SRDX showed the repression of both nodulation in MG-20 and infection in daphne. ProNIN::NIN::TerNIN rescued no infection phenotype of nin, yielding that the NIN promoter used in this study shows sufficient expression pattern at least for the infection pathway. Important results are highlighted in boldface.
| Plant Genotype | Transgene | Nodule |
abLRb |
IT |
|||||
|---|---|---|---|---|---|---|---|---|---|
| + | Lowa | − | ++c | Lowd | − | Total Plants | |||
| MG-20 | ProLjUBQ::GUS | 20 | 0 | 0 | 0 | 0 | 20 | 0 | 20 |
| ProLjUBQ::NIN | 19 | 0 | 0 | 10 | 0 | 19 | 0 | 19 | |
| ProLjUBQ::NIN::SRDX | 5 | 7 | 6 | 5 | 0 | 18 | 0 | 18 | |
| J0571>>mCherry-NLS | 20 | 0 | 0 | 0 | 0 | 20 | 0 | 20 | |
| J0571>>NIN | 20 | 0 | 0 | 3 | 0 | 20 | 0 | 20 | |
| daphne | ProLjUBQ::GUS | 0 | 0 | 18 | 0 | 18 | 0 | 0 | 18 |
| ProLjUBQ:NIN | 0 | 0 | 20 | 4 | 0 | 18 | 2 | 20 | |
| ProLjUBQ:NIN::SRDX | 0 | 0 | 19 | 2 | 3 | 14 | 2 | 19 | |
| J0571>>mCherry-NLS | 0 | 0 | 25 | 0 | 25 | 0 | 0 | 25 | |
| J0571>>NIN | 6 | 0 | 17 | 6 | 13 | 10 | 0 | 23 | |
| nin-9 | ProLjUBQ::GUS | 0 | 0 | 15 | 0 | 0 | 0 | 15 | 15 |
| ProLjUBQ:NIN | 0 | 0 | 14 | 5 | 0 | 0 | 6 | 6 | |
| ProNIN::NIN::TerNIN | 0 | 0 | 14 | 0 | 0 | 10 | 4 | 14 | |
| J0571>>NIN | 0 | 0 | 22 | 2 | 0 | 0 | 22 | 22 | |
Roots with small size and number of nodules (Supplemental Fig. S7).
Roots with aberrant lateral root such as enlarged tips and bumps (Supplemental Fig. S6).
Roots with the typical daphne phenotype, highly increased IT number.
Roots with the wild-type-like infection phenotype, normal IT number.
Figure 4.
Spatial expression analysis of the NIN gene. GUS staining images of Agrobacterium rhizogenes-mediated transformed roots with ProNIN::GUS::TerNIN at 7 DAI (with the M. loti MAFF303099 wild type) on the Miyakojima MG-20 wild type (A, C, E, and G) and daphne (B, D, F, and H). Blue staining was observed with transformed (GFP-positive) hairy roots in susceptible zones (A–F), including root hair cells (G and H) and an immature nodule in the wild type (I). Bars = 0.5 mm in A to D and 0.1 mm in E to I.
Overexpression of NIN Strongly Represses the Hyperinfection in daphne
The above result showed that daphne exerts broad ProNIN activity in the epidermis, indicating a broader susceptible zone for rhizobial infection than in the wild type. Based on these results, we hypothesized that NIN itself negatively regulates rhizobial infection. To address the negative function, we accordingly overexpressed NIN under the control of ProLjUBQ in daphne roots and observed the IT formation phenotype with M. loti expressing DsRED. Surprisingly, the hyperinfection phenotype of daphne was strongly suppressed in NIN-overexpressing transgenic roots, whereas GUS-overexpressing roots and nontransformed (GFP-negative) roots retained the excessive IT formation phenotype (Fig. 5, A–C and I–K; Table I). The hyperinfection phenotype of nontransformed roots indicates that the negative feedback regulation of IT formation is not long-distance signaling mediated by the shoot, in contrast to the regulation of the number of nodules (Okamoto et al., 2009). We also observed no normal nodules, but some lateral roots with enlarged tips and bumps, in ProLjUBQ::NIN transgenic roots in daphne (Table I; Supplemental Fig. S6; Suzaki et al., 2012; Soyano et al., 2013).
Figure 5.
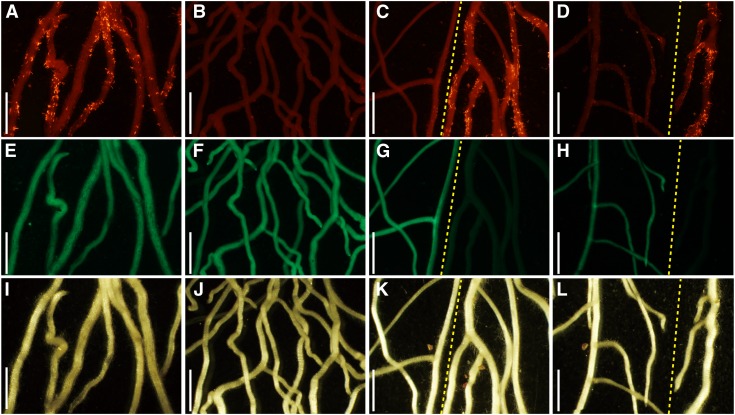
Ectopic expression of the NIN gene strongly suppresses excessive IT formation in daphne. Red fluorescence images (A–D), GFP fluorescence images (E–H), and transmitted light images (I–L) of A. rhizogenes-mediated transformed daphne roots are shown at 21 DAI. Roots were transformed with negative control vector ProLjUBQ::GUS (A, E, and I), ProLjUBQ::NIN (B, C, F, G, J, and K), or ProLjUBQ::NIN::SRDX (D, H, and L). ITs were observed by inoculating M. loti MAFF303099 constitutively expressing DsRED (A–D). GFP fluorescence showed transformed roots (E–H). Yellow dashed lines indicate the border between transformed and nontransformed roots. Bars = 2 cm.
Next, we confirmed the positive function of NIN in rhizobial infection in the daphne mutant background. We expressed a chimeric protein of NIN and the SRDX domain, a transcriptional repressor domain in the Arabidopsis (Arabidopsis thaliana) SUPERMAN repressor (Oshima et al., 2011). ProLjUBQ:NIN::SRDX dominantly repressed the target gene expression of NIN in the MG-20 wild type, causing a reduction in the number of nodules (Table I; Supplemental Fig. S7). In daphne, the ITs almost disappeared only in ProLjUBQ:NIN::SRDX-expressing transgenic roots (Fig. 5, D, H, and J; Table I), phenocopying the previously observed phenotype of a typical nin mutant (Schauser et al., 1999; Marsh et al., 2007). This suggests that daphne maintains the positive function of NIN in rhizobial infection, unlike a typical nin mutant.
These results indicate that NIN plays not only positive but also negative roles in IT formation, and daphne maintains the positive role but loses the negative role. In contrast to daphne, MG-20 wild-type plants formed a low number of ITs (less than 20 per root; Fig. 1J), which may account for the observation that ProLjUBQ:NIN apparently had no strong suppressive effects on IT number in the wild type (Table I).
Cortical But Not Epidermal Expression of NIN Was Specifically Lost in daphne
Both positive and negative roles of NIN in rhizobial infection have now been demonstrated. To further investigate the underlying mechanism, we hypothesized that the positive and negative actions of NIN are generated by epidermis and cortex, respectively, given that the lack of cytokinin-induced NIN in daphne results in an increase in the number of ITs and a typical nin mutant does not form ITs. We speculated that a less negative role of NIN in IT formation (cytokinin-induced NIN) resulted in excessive IT formation.
To address the tissue-specific activity of NIN, we attempted to express NIN using a cortex- and endodermis-specific enhancer isolated from Arabidopsis J0571 (Miyashima et al., 2011). The J0571 enhancer element was identified from the Arabidopsis GAL4-GFP enhancer-trap lines (http://www.plantsci.cam.ac.uk/haseloff; http://www.arabidopsis.org/abrc/haseloff.jsp). First, we tested the fluorescent marker (mCherry-with nuclear localization signal [NLS]) expressed by J0571 in hairy roots of L. japonicus. Although no marker expression was detected in the epidermis, signal was detected in the inner layers of the root, including cortex and endodermis (Supplemental Fig. S8), suggesting that the cortex- and endodermis-specific expression of J0571 is conserved in L. japonicus. Both the nonnodulation phenotype and excessive IT formation in daphne were partially rescued by J0571>>NIN (Fig. 6; Table1), confirming that the daphne phenotype was caused by the loss of NIN expression specifically in the cortex.
Figure 6.
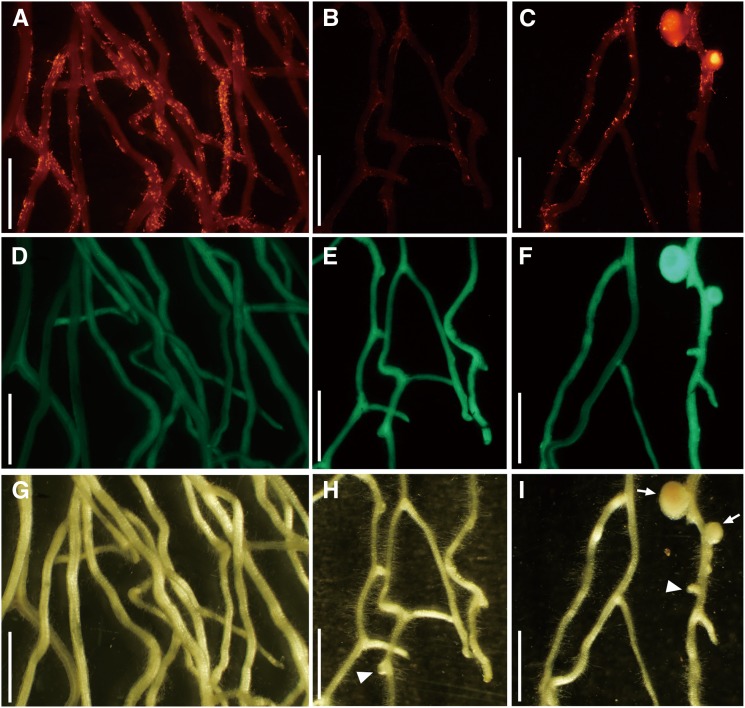
The nonnodulation phenotype in daphne is partially rescued by the cortical expression of NIN. Red fluorescence images (A–C), GFP fluorescence images (D–F), and transmitted light images (G–I) of A. rhizogenes-mediated transformed daphne roots are shown at 21 DAI. Roots were transformed with J0571>>NIN (A–I). Excessive ITs (A, D, and G), strongly suppressed ITs (B, E, and H), and nodules (C, F, and I) were observed by inoculating M. loti MAFF303099 constitutively expressing DsRED. GFP fluorescence showed transformed roots (D–F). Arrows and arrowheads indicate nodules and enlarged bumps, respectively. Bars = 2 cm.
DISCUSSION
NIN was first identified as a gene responsible for the nonnodulation phenotype in legumes (Schauser et al., 1999). Since then, it has been believed that NIN, a putative transcription factor, plays a positive role in nodule organogenesis and IT formation. Meanwhile, the possibility has been discussed that NIN also has a negative role in the rhizobial infection processes, based on the excessive root hair response or the expanded Early nodulin11 expression pattern in nin mutants and the lower expression of NIN in another hyper-IT mutant, hit1-1 (Schauser et al., 1999; Marsh et al., 2007; Murray et al., 2007). In this study, we identified the daphne mutant, a novel nin mutant allele displaying excessive IT formation as well as nonnodulation. The spatiotemporal expression patterns of the NIN gene in daphne provided new evidence of negative feedback regulation of the infection process mediated by NIN. In daphne, the level of NIN transcription from whole roots was less than that in the wild type. In contrast, the epidermal expression of NIN was broader than that in the wild type, indicating that the susceptible zone for rhizobial infection was enlarged in daphne. This increased susceptibility for infection could account for the excessive IT formation in daphne. Furthermore, although overexpression of NIN suppressed excessive infection, inner cell layer-specific expression of NIN rescued nodule formation in daphne. Based on these observations, we propose a negative feedback regulation of rhizobial infection mediated by NIN (Fig. 7). In this model, NIN plays two important roles, one in infection and the other in organogenesis. NIN functioning in infection is located in the epidermis in an earlier stage (in the susceptible zone) for proceeding with IT formation, whereas NIN functioning in organogenesis acts in a later stage. NIN functioning in organogenesis has not only a positive role in promoting cell division in the cortex but also a negative role in inhibiting rhizobial infection. In daphne, owing to the loss of expression of such NIN functioning in organogenesis, the root area for rhizobial infection becomes broader. Several reports have already suggested that genes downstream of cytokinin signaling or NIN itself are involved in preserving the balance of the nodule symbiosis (Murray et al., 2007; Mortier et al., 2010; Saur et al., 2011). Our study has experimentally confirmed one of those mechanisms, a negative role of NIN in rhizobial infection. Excessive root hair curling of a typical nin mutant also may be explained by our working model of NIN.
Figure 7.
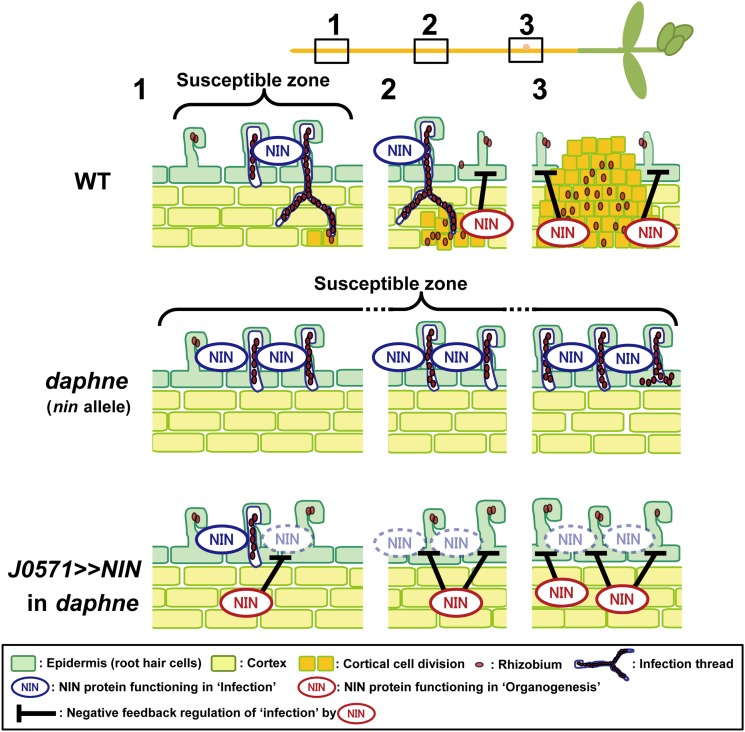
A model of the inhibition of rhizobial infection processes mediated by NIN. In the wild type (WT), NIN functions in both rhizobial infection (blue, in the epidermis) and organogenesis (red, in the cortex). In the earlier stage (1), NIN (blue) is predominant, but in the later stage (3), the proportion of NIN (red) has increased with nodule development. It is assumed that a potential negative correlation between the organogenesis and infection pathways (black bars) regulates the amount of infection and restricts the region of rhizobial susceptibility. In the daphne mutant, NIN functions only in infection (blue) and not in organogenesis (red), resulting in no activation of cortical cell division. The loss of NIN (red) enhances rhizobial infection. When we induced NIN expression in cortex in daphne, infection was strongly suppressed, owing to the loss of a negative feedback loop (black bars) mediated by NIN (red). The numbers indicate the developmental stages of nodulation. “NIN” surrounded by a dashed line indicates reduced gene expression or protein function of NIN, induced by the negative feedback loop described above (black bars).
How can two different biological events, infection and organogenesis, be controlled by the same transcription factor, NIN? How can NIN act both positively and negatively in a stage- or tissue-dependent manner during nodule organogenesis? Recent reports in Arabidopsis indicate that a transcription factor may act as a bifunctional transcription factor and mediate a wide variety of biological events. WUSCHEL acts as both a repressor and an activator in a domain-dependent manner (Ikeda et al., 2009). In our study, however, a NIN chimeric repressor can repress IT formation in the epidermis (Fig. 5; Table I), so that NIN functions only as an activator in both infection and organogenesis. If NIN functions as a repressor of IT formation, the NIN chimeric repressor should not repress IT formation. Another possibility is that different actions of NIN are dependent on tissue- or stage-specific downstream targets, including cotranscriptional regulators. The putative cortex- or late nodule-specific downstream factors might suppress the function of NIN for IT formation transcriptionally or posttranscriptionally. This potential mechanism is supported by several studies. Studies of LEAFY and Activator protein1 transcription factors indicate that a single transcription factor can bind to different groups of targets by interacting with individual cis-regions or cofactors following the developmental stages (Gregis et al., 2008; Liu and Mara, 2010). Moreover, several reports of cross talk in defense signaling propose a potential mechanism by which single transcription factors, such as WRKYs and TGAs (bZIP-type transcription factor), play both positive roles in the salicylic acid-dependent pathway and negative roles in the jasmonic acid-dependent pathway (Li et al., 2004; Gao et al., 2011; Van der Does et al., 2013).
We next discuss the candidates for cofactors or downstream target genes of NIN. In terms of the cell proliferation activity of NIN, a positive role for organogenesis, the contribution of NSP2 and NF-YA/YB has been demonstrated (Soyano et al., 2013). For the negative regulation of infection, ethylene-responsive factors are strong candidates, given that both of the hyperinfection mutants, Mtsickle and Medicago truncatula ethylene response factor required for nodule differentiation (Mtefd), harbor mutations in ethylene-signaling molecules. Although a few hyperinfection mutants, Ljhit1-1, Mtsickle, and Mtefd (Penmetsa and Cook, 1997; Murray et al., 2007; Vernié et al., 2008), suggest the existence of regulatory mechanisms for controlling rhizobial infection processes, such regulation is far less well known than that of the number of nodules (van Brussel et al., 2002; Oka-Kira and Kawaguchi, 2006; Kouchi et al., 2010). Furthermore, although Ljhit1-1 exhibits low nodulation, Mtsickle and Mtefd exhibit increased numbers of nodules, leaving mysterious the putative cross talk between the infection and organogenesis pathways. According to our model, the phenotype of Ljhit1-1 is caused by the lower expression of NIN functioning in organogenesis and inhibiting infection, similar to daphne. In contrast, in Mtsickle and Mtefd, NIN may be more highly expressed, but inhibitory mechanisms of infection mediated by NIN may be lost. Our study indicates that NIN may switch between positive and negative influence on rhizobial infection in different tissues or nodule developmental stages. NIN could be controlling the balance between infection and organogenesis. Future study of daphne will shed new light on the cofactors or downstream target genes of NIN that differ between those two pathways.
Although NIN is a key transcriptional factor in nodule development, the functional NIN promoter region necessary for nodule organogenesis has not yet been elucidated. Only IT formation, and not nodule formation, was rescued in nin-9 by the introduction of ProNIN(∼4kb)::NIN::TerNIN (Table I; Supplemental Fig. S5). In this study, we identified a novel mutant allele of nin, daphne, whose genome was changed approximately 7 kb upstream of NIN by chromosomal translocation (Fig. 2). These results raise at least two possibilities. One is that a cis-regulatory element necessary for the organogenesis pathway, including the cytokinin response element, has been lost from the upstream region of NIN in the daphne genome. In other words, a 7-kb segment of the NIN promoter region is sufficient for the function of NIN in the infection pathway. The other possibility is that epigenetic alteration leads to a different NIN expression pattern in daphne. Furthermore, ProUBQ::NIN induced aberrant roots including bumps in the wild type, daphne, and nin-9 (Supplemental Fig. S6), but we failed to rescue IT formation in nin-9 by the introduction of ProUBQ::NIN (Table I; Supplemental Fig. S5). This implies that the induction mechanism of NIN transcript is more complex than so far anticipated; spatial and temporal expression of NIN may need to be strictly controlled in order to achieve its functions in both infection and organogenesis pathways. The elucidation of the mechanism remains an important challenge.
The biological meaning of controlling the susceptibility of rhizobia has not been established. At least under our experimental conditions, daphne exhibited no difference in plant growth between the noninoculated and inoculated conditions (Supplemental Fig. S9). IT formation may be a less energy-consuming process than nodule formation or nitrogen fixation (van Brussel et al., 2002; Oka-Kira and Kawaguchi, 2006; Kouchi et al., 2010). Alternatively, it is possible that plants may need to avoid excessive bacterial infection even during an interaction with symbiotic bacteria. This may be related to a common mechanism for the establishment of plant-symbiont and plant-pathogen interactions (Vasse et al., 1993; Bozsó et al., 2009; Soto et al., 2009; Nakagawa et al., 2011).
Our study identified a novel nin mutant allele, daphne. We demonstrated that NIN, known to date as a positive factor for IT formation and nodule organogenesis, has a negative role in rhizobial infection processes. The multiple functions of the transcription factor NIN will afford an opportunity to investigate potential cross talk between infection in the epidermis and cell division in the cortex during the course of the establishment of nodule symbiosis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The daphne mutant was isolated by screening M2 progeny derived from Lotus japonicus Miyakojima MG-20 wild-type seeds mutagenized by irradiation with a carbon ion beam (C5+). The details of the ion beam irradiation have been reported previously (Oka-Kira et al., 2005; Magori et al., 2009; Yoshida et al., 2010). Seeds were sown in sterilized vermiculite soaked in autoclaved vermiculite supplied with Broughton and Dilworth solution (Broughton and Dilworth, 1971) containing 0.5 mm KNO3 with or without Mesorhizobium loti MAFF 303099 under a 16-h-light/8-h-dark cycle. Cytokinin treatment was applied by incubation of seedlings in vermiculite supplied with Broughton and Dilworth solution containing 10–7 m benzylaminopurine for 16 h. ITs were observed or counted after inoculation of M. loti rhizobia constitutively expressing LacZ (Yoshida et al., 2010) or DsRED. nin-2 was kindly provided by Jens Stougaard (Schauser et al., 1999) and was used for the allelism test. nin-9 (Suzaki et al., 2012) was used for gene expression analysis and complementation testing.
Microscopic Observation
Bright-field and fluorescence images were viewed with an SZX12/16 stereomicroscope or a BX50 microscope (Olympus). Images were acquired with a DP Controller (Olympus). Confocal images were viewed with an A1 confocal laser-scanning microscope (Nikon) and NIS Elements (Nikon).
Quantification of ITs
At 5 d after germination, plants were inoculated with M. loti constitutively expressing LacZ. At 7 DAI, roots were stained for β-galactosidase activity. ITs on all parts of the root were counted using the microscope (BX50; Olympus).
Map-Based Cloning and Inverse PCR
The daphne locus was mapped using F2 progeny of daphne and Gifu B-129. Two loci located on linkage groups II and III were identified using 52 F2 plants. Fine-mapping was performed in 2,048 F2 plants. The newly developed genetic markers in this study are shown in Supplemental Table S2. The deleted region located on CM0423 (chromosome III) was identified. daphne genomic DNA was extracted with the DNeasy Plant Mini Kit (Qiagen) and digested with EcoO109I, ApoI, and EcoRI. The digested DNA fragments were self-ligated with T4 DNA ligase (TaKaRa). Then, using inverse PCR analysis with two sets of primers designed on sequences in CM0423, the fused sequences originating from CM0423 (chromosome III) and CM0102 (chromosome II; Fig. 2) were detected. The primers used in inverse PCR analysis are shown in Supplemental Table S3.
Expression Analysis
Total RNA was isolated from each plant tissue using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was prepared using the QuantiTect Reverse Transcription Kit (Qiagen). Real-time RT-PCR was performed using the ABI Prism 7000 (Applied Biosystems) with THUNDERBIRD SYBR qPCR Mix (Toyobo) or with the QuanTitect SYBR Green RT-PCR Kit (Qiagen), according to each manufacturer’s protocol. The expression of ubiquitin or elongation factor-1α was used as the reference. The primers used in expression analysis are shown in Supplemental Table S3. Data are means ± sd of three biological and three technical replicates.
Plant Transformation
The recombinant plasmids were introduced into Agrobacterium rhizogenes strain AR1193 and were transformed into roots of L. japonicus by a hairy root transformation method described previously (http://www.bio-protocol.org/wenzhang.aspx?id=795; Okamoto et al., 2009).
Cloning of NIN Promoter Constructs and Promoter-GUS Assay
A 1.7-kb Gateway cassette (GW) fragment was excised from the DR5 (for auxin-inducible reporter gene)::GFP-NLS construct (Suzaki et al., 2012) and inserted into the BamHI site of pCAMBIA1300-GFP, named pCAMBIA1300-GW-GFP. Next, the GFP in the vector was removed using XhoI, and PCR-amplified GFP-LjLTI6b (Suzaki et al., 2012) was inserted into the XhoI site to create a new binary vector, pCAMBIA1300-GW-GFP-LjLTI6b. Then, using two sets of primers for the amplification of approximately 4 kb of ProNIN and approximately 2 kb of TerNIN, two fragments were cloned into pCAMBIA1300-GW-GFP-LjLTI6b. In the final step, GUS in pDONR221 (Invitrogen), which was provided by Detlef Weigel, and the NIN cDNA in pENTR/D-TOPO (Suzaki et al., 2012) were inserted between ProNIN and TerNIN by an LR recombination reaction (Invitrogen). A transfer DNA construct expressing ProNIN::GUS::TerNIN was transformed into MG-20 and daphne. GFP fluorescence was checked as a marker for transformation. Transformed roots were inoculated with M. loti MAFF303099. At 7 DAI, a GUS staining procedure was performed as described previously (Jefferson et al., 1987).
Analysis of the IT Phenotype with Overexpressing NIN or a Chimeric Repressor of NIN
NIN cDNA without a stop codon in pENTR/D-TOPO was generated from NIN cDNA in pENTR/D-TOPO (Suzaki et al., 2012) by site-directed mutagenesis with primers (Supplemental Table S3). A GW::SRDX fragment was amplified from the pDEST-BCKH plasmid (Oshima et al., 2011) and inserted between the KpnI and AscI sites of pUB-GFP (Maekawa et al., 2008), named pUB-GW-SRDX-GFP. NIN cDNA in pENTR/D-TOPO (Suzaki et al., 2012) and NIN cDNA without a stop codon were inserted into the GW sites of pUB-GW-GFP (Maekawa et al., 2008) and pUB-GW-SRDX-GFP, respectively, with the LR recombination reaction (Invitrogen). As a control, GUS in pDONR221 (Invitrogen) was inserted into the GW site of pUB-GW-GFP by the LR recombination reaction (Invitrogen). daphne plants were treated with M. loti MAFF303099 constitutively expressing DsRED. At 14 DAI, ITs were observed.
Analysis of Tissue Specificity Using a Cortex- and Endodermis-Specific Expression System
GAL4-VP16 (for herpes simplex virus protein) and the nopaline synthase (NOS) terminator sequence with flanking genomic regions were amplified by PCR from an enhancer trap line, J0571, in Arabidopsis (Arabidopsis thaliana; http://www.plantsci.cam.ac.uk/Haseloff/; Miyashima et al., 2011) and cloned into the HindIII site of pGWB501:5xUAS (Goh et al., 2012) using an In-Fusion HD cloning kit (TaKaRa), named pGWB501:5xUAS-J0571. Next, the GW::SRDX::TerNOS fragment was excised from pUB-GW-SRDX-GFP by KpnI and ScaI double digestion and inserted into pCAMBIA1300-GFP, named pCAMBIA1300-GW-SRDX-GFP. A J0571-GAL4-VP16-TerNOS-5xUAS-35S minimal promoter was amplified by PCR from the template plasmid pGWB501:5xUAS-J0571 and inserted into the KpnI site of pCAMBIA1300-GW-GFP or pCAMBIA1300-GW-SRDX-GFP, named pCAMBIA1300-J0571-GW-GFP or pCAMBIA1300-J0571-GW-SRDX-GFP, respectively. In the final step, NIN and mCherry-NLS coding sequence (Suzaki et al., 2012) or NIN and mKusabira Orange2 (mKO2) coding sequence without a stop codon were inserted into the GW site of pCAMBIA1300-J0571-GW-GFP or pCAMBIA1300-J0571-GW-SRDX-GFP by the LR recombination reaction (Invitrogen). As a control, mKO2 without a stop codon was amplified from the plasmid including mKO2 (Medical and Biological Laboratories; Sakaue-Sawano et al., 2008) by PCR and cloned into pENTR/D-TOPO vector using a TOPO cloning kit (Invitrogen). Primers used for these constructs are listed in Supplemental Table S3. At 21 DAI, nodules and ITs were observed in transformed hairy roots.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The phenotype of nodule formation in hit1-1 mutant.
Supplemental Figure S2. Z-stack image series of same area of daphne root.
Supplemental Figure S3. The two loci responsible for nonnodulation of daphne by rough mapping.
Supplemental Figure S4. The result of complementation tests by the constitutive expression of TIM.
Supplemental Figure S5. Complementation of IT formation phenotype of nin-9.
Supplemental Figure S6. Enlarged bumps and tips were induced by overexpressing NIN in both MG-20 and daphne.
Supplemental Figure S7. Nodule organogenesis was decreased in ProLjUBQ::NIN::SRDX-expressing roots.
Supplemental Figure S8. The expression pattern of marker gene under the control of J0571 enhancer.
Supplemental Figure S9. The plant growth difference between noninoculated and inoculated conditions.
Supplemental Table S1. The segregation ratio of mapping population and backcrossing population.
Supplemental Table S2. The newly developed genetic markers list used for fine mapping in this study.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Jens Stougaard for the mutant seeds of nin-2, Krzyztof Szczyglowski for the mutant seeds of hit1-1, Detlef Weigel for the GUS construct, Makoto Hyashi for M. loti MAFF303099 expressing DsRED, Shusei Sato for the genomic data of L. japonicus, Masaru Ohme-Takagi for the SRDX construct, Masanao Sato and Kiyoshi Tatematsu for valuable comments, the Functional Genomics Facility and the Spectrography and Bioimaging Facility of the National Institute for Basic Biology Core Research Facilities and the Model Plant Facilities of the National Institute for Basic Biology Bioresource Center for technical support, and Enago (www.enago.jp) for the English language review.
Glossary
- IT
infection thread
- DAI
days after inoculation
- RT
reverse transcription
- IGR
intergenic region
- cDNA
complementary DNA
Footnotes
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grants-in-Aid for Scientific Research nos. 23012038 to T.S. and 22128006 to M.K.) and the Yoshida Scholarship Foundation (to E.Y.).
The online version of this article contains Web-only data.
References
- Borisov AY, Madsen LH, Tsyganov VE, Umehara Y, Voroshilova VA, Batagov AO, Sandal N, Mortensen A, Schauser L, Ellis N, et al. (2003) The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol 131: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozsó Z, Maunoury N, Szatmari A, Mergaert P, Ott PG, Zsíros LR, Szabó E, Kondorosi E, Klement Z. (2009) Transcriptome analysis of a bacterially induced basal and hypersensitive response of Medicago truncatula. Plant Mol Biol 70: 627–646 [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Dilworth MJ. (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi M, Frugier F. (2008) De novo organ formation from differentiated cells: root nodule organogenesis. Sci Signal 1: re11. [DOI] [PubMed] [Google Scholar]
- Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG. (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148: 1985–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ. (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao QM, Venugopal S, Navarre D, Kachroo A. (2011) Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155: 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H. (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139: 883–893 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. (2008) AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J 56: 891–902 [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, Madsen LH, Jurkiewicz A, Nielsen MW, Tirichine L, Stougaard J. (2011) Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol Plant Microbe Interact 24: 1385–1395 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2009) Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5: 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S, Held M, Hossain MS, Morieri G, Macgillivary A, Johansen C, Antolín-Llovera M, Parniske M, Oldroyd GED, Downie AJ, et al. (2011) Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J 67: 929–940 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M. (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F, et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZC, Mara C. (2010) Regulatory mechanisms for floral homeotic gene expression. Semin Cell Dev Biol 21: 80–86 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kusakabe M, Shimoda Y, Sato S, Tabata S, Murooka Y, Hayashi M. (2008) Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol Plant Microbe Interact 21: 375–382 [DOI] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M. (2009) Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol Plant Microbe Interact 22: 259–268 [DOI] [PubMed] [Google Scholar]
- Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GED. (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K. (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313 [DOI] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153: 222–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kaku H, Shimoda Y, Sugiyama A, Shimamura M, Takanashi K, Yazaki K, Aoki T, Shibuya N, Kouchi H. (2011) From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J 65: 169–180 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Kawaguchi M. (2006) Long-distance signaling to control root nodule number. Curr Opin Plant Biol 9: 496–502 [DOI] [PubMed] [Google Scholar]
- Oka-Kira E, Tateno K, Miura K, Haga T, Hayashi M, Harada K, Sato S, Tabata S, Shikazono N, Tanaka A, et al. (2005) klavier (klv), a novel hypernodulation mutant of Lotus japonicus affected in vascular tissue organization and floral induction. Plant J 44: 505–515 [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. (2009) Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol 50: 67–77 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263 [DOI] [PubMed] [Google Scholar]
- Op den Camp RH, De Mita S, Lillo A, Cao Q, Limpens E, Bisseling T, Geurts R. (2011) A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiol 157: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima Y, Mitsuda N, Nakata M, Nakagawa T, Nagaya S, Kato K, Ohme-Takagi M. (2011) Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol 28: 201–210 [Google Scholar]
- Penmetsa RV, Cook DR. (1997) A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Popp C, Ott T. (2011) Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol 14: 458–467 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Rival P, de Billy F, Bono JJ, Gough C, Rosenberg C, Bensmihen S. (2012) Epidermal and cortical roles of NFP and DMI3 in coordinating early steps of nodulation in Medicago truncatula. Development 139: 3383–3391 [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Ohtawa K, Hama H, Kawano M, Ogawa M, Miyawaki A. (2008) Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem Biol 15: 1243–1248 [DOI] [PubMed] [Google Scholar]
- Sandal N, Petersen TR, Murray J, Umehara Y, Karas B, Yano K, Kumagai H, Yoshikawa M, Saito K, Hayashi M, et al. (2006) Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol Plant Microbe Interact 19: 80–91 [DOI] [PubMed] [Google Scholar]
- Saur IML, Oakes M, Djordjevic MA, Imin N. (2011) Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytol 190: 865–874 [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. (1999) A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Soto MJ, Domínguez-Ferreras A, Pérez-Mendoza D, Sanjuán J, Olivares J. (2009) Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol 11: 381–388 [DOI] [PubMed] [Google Scholar]
- Soyano T, Kouchi H, Hirota A, Hayashi M. (2013) Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. (2012) Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J. (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- van Brussel AAN, Tak T, Boot KJM, Kijne JW. (2002) Autoregulation of root nodule formation: signals of both symbiotic partners studied in a split-root system of Vicia sativa subsp. nigra. Mol Plant Microbe Interact 15: 341–349 [DOI] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse J, Debilly F, Truchet G. (1993) Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J 4: 555–566 [Google Scholar]
- Vasse JM, Truchet GL. (1984) The Rhizobium-legume symbiosis: observation of root infection by bright-field microscopy after staining with methylene blue. Planta 161: 487–489 [DOI] [PubMed] [Google Scholar]
- Vernié T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P. (2008) EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20: 2696–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida C, Funayama-Noguchi S, Kawaguchi M. (2010) plenty, a novel hypernodulation mutant in Lotus japonicus. Plant Cell Physiol 51: 1425–1435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.