Abstract
Background
Nanoparticles (NPs) have gained significance in medical fields due to their high surface-area-to-volume ratio. In this study, we synthesized NPs from a medicinally important plant – Plumbago zeylanica.
Materials and methods
Aqueous root extract of P. zeylanica (PZRE) was analyzed for the presence of flavonoids, sugars, and organic acids using high-performance thin-layer chromatography (HPTLC), gas chromatography-time of flight-mass spectrometry (GC-TOF-MS), and biochemical methods. The silver NPs (AgNPs), gold NPs (AuNPs), and bimetallic NPs (AgAuNPs) were synthesized from root extract and characterized using ultraviolet-visible spectra, X-ray diffraction (XRD), energy-dispersive spectrometry (EDS), transmission electron microscopy (TEM), and dynamic light scattering (DLS). The effects of these NPs on Acinetobacter baumannii, Staphylococcus aureus, and Escherichia coli biofilms were studied using quantitative biofilm inhibition and disruption assays, as well as using fluorescence, scanning electron microscopy, and atomic force microscopy.
Results
PZRE showed the presence of phenolics, such as plumbagin, and flavonoids, in addition to citric acid, sucrose, glucose, fructose, and starch, using HPTLC, GC-TOF-MS, and quantitative analysis. Bioreduction of silver nitrate (AgNO3) and chloroauric acid (HAuCl4) were confirmed at absorbances of 440 nm (AgNPs), 570 nm (AuNPs), and 540 nm (AgAuNPs), respectively. The maximum rate of synthesis at 50°C was achieved with 5 mM AgNO3 within 4.5 hours for AgNPs; and with 0.7 mM HAuCl4 within 5 hours for AuNPs. The synthesis of AgAuNPs, which completed within 90 minutes with 0.7 mM AgNO3 and HAuCl4, was found to be the fastest. Fourier-transform infrared spectroscopy confirmed bioreduction, while EDS and XRD patterns confirmed purity and the crystalline nature of the NPs, respectively. TEM micrographs and DLS showed about 60 nm monodispersed Ag nanospheres, 20–30 nm Au nanospheres adhering to form Au nanotriangles, and about 90 nm hexagonal blunt-ended AgAuNPs. These NPs also showed antimicrobial and antibiofilm activity against E. coli, A. baumannii, S. aureus, and a mixed culture of A. baumannii and S. aureus. AgNPs inhibited biofilm in the range of 96%–99% and AgAuNPs from 93% to 98% in single-culture biofilms. AuNPs also showed biofilm inhibition, with the highest of 98% in S. aureus. AgNPs also showed good biofilm disruption, with the highest of 88% in A. baumannii.
Conclusion
This is the first report on rapid and efficient synthesis of AgNPs, AuNPs and AgAuNPs from P. zeylanica and their effect on quantitative inhibition and disruption of bacterial biofilms.
Keywords: P. zeylanica, AgNPs, AuNPs, AgAuNPs, biofilm inhibition and disruption, GC-TOF-MS
Introduction
Nanomaterial synthesis and characterization have been an extensive area of research since the last decade. Biological synthesis of nanoparticles (NPs) is considered environmentally friendly, nontoxic, and cost-effective.1 Therefore, biological synthesis of NPs from various plant extracts2,3 and microorganisms like bacteria,4 yeasts,5 fungi,6 and algae7 have proven to be promising sources. However, plant extracts are preferred over microbial sources, as they do not require sterile conditions. Also, bioreduction of NPs using plant extracts is rapid, ecofriendly, biocompatible, and nontoxic.8,9
Silver has been exploited as antimicrobials from ancient period.10 With the evolution of nanomedicine as a study for treating infections, metallic silver in the form of NPs has regained its significance.11,12 Several bacteria have developed resistance against antibiotics, which has challenged the treatment of human infections.13–15 Therefore, silver NPs (AgNPs) as an antimicrobial agent seem to be beneficial compared to antibiotics.16 Moreover, few reports show synergistic enhancement of activity of antibiotics with AgNPs.17 There are a number of reports on green synthesis of gold NPs (AuNPs) by plants and microorganisms, and they are being exploited for research on drug delivery and cancer studies.18 However, as gold is not known to have an antimicrobial property, very few reports are available on the antimicrobial nature of AuNPs. It is of interest to some researchers whether AuNPs can inhibit biofilm formation and kill bacteria as they show surface bioconjugation.19 So far, syntheses of bimetallic NPs (AgAuNPs) from Diospyros kaki and Azadirachta indica extracts have been reported by Song and Kim20 and Shankar et al21 respectively, with an Au core and Ag shell observed in case of A. indica. However, research on their application as antimicrobial agents is still in its infancy.
Plumbago zeylanica, commonly known as agni (meaning fire) in Sanskrit, is an important medicinal plant used in ayurveda, an Indian system of traditional medicine.22 Ahmad and Beg23 reported on the antimicrobial activity of aqueous and alcoholic extracts from roots of P. zeylanica against Staphylococcus aureus, Pseudomonas aeruginosa, Proteus vulgaris, Bacillus subtilis, and Candida albicans. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) is a major flavonoid present in P. zeylanica, especially in roots, showing antifungal and antimicrobial properties.24,25 Plumbagin is also well known for its plasmid-curing property.26 Plumbagin has cured plasmid-encoding resistance to silver, cadmium, antimony, streptomycin, and ampicillin in Acinetobacter baumannii species.27
The phytochemical analysis of an aqueous root extract of P. zeylanica (PZRE) showed the presence of alkaloids, glycoside, reducing sugars, phenolics, tannins, lignin, saponins, and flavonoids.28 The majority of these compounds are known to act as reducing as well as stabilizing agents in the bioreduction of silver and gold salt.29–32 Moreover, there have been no reports on the synthesis of metal NPs by P. zeylanica to date. Therefore, we propose that P. zeylanica may synthesize novel metal NPs exhibiting antimicrobial and biofilm inhibition/disruption properties.
Materials and methods
Plant material and preparation of extract
P. zeylanica roots were collected from the natural landscape of Western Ghats, on Dr Homi Bhabha Road, Pune, Maharashtra, India. They were thoroughly washed with distilled water, shade-dried, chopped into thin slices, and ground to make fine powder. The aqueous extract was prepared by boiling 5 g of the powder in 100 mL sterile distilled water, followed by centrifugation at 8,000 rpm for 10 minutes. The supernatant of PZRE was filtered through a 0.45 μm membrane filter and stored at 4°C until further use.
Preparation of the PZRE samples for chromatographic analysis
PZRE was sequentially extracted along a polarity gradient using ethyl acetate and methanol for high-performance thin-layer chromatography (HPTLC; semiautomated Linomat 5; Camag, Muttenz, Switzerland) fingerprinting and gas chromatography-time of flight-mass spectrometry (GC-TOF-MS). The sequential extraction was done by suspending 5 mL of root extract in 5 mL of high-performance liquid chromatography-grade ethyl acetate, followed by stirring for 3 hours and centrifugation at 10,000 rpm at room temperature (RT). The supernatant was recovered, and 5 mL methanol was added to the pellet and redispersed. The solution was vigorously mixed by vortexing for 15 minutes, and then methanol extract was recovered. The ethyl acetate and methanol extracts were concentrated.
Phytochemical analysis of P. zeylanica
HPTLC analysis of PZRE was performed according to the protocol given in earlier studies.33,34 Plumbagin, quercetin, diosgenin, glucose, and fructose (Sigma-Aldrich, St Louis, MO, USA) were taken as standard for the analyses. Similarly, 5 μL of the ethyl acetate extract of PZRE was subjected to GC-TOF-MS (Pegasus 4D GCxGC-TOFMS system with Agilent 6890 gas chromatograph equipped with a dual-jet thermal modulator between primary and secondary columns and a Pegasus IV TOFMS as a detector; LECO, St Joseph, MI, USA). The program used was as follows: primary column, 5% phenyl polysilphenylene-siloxane HP-5MS capillary, 30 m ×0.32 mm, 0.25 μm; secondary column, 1.00 m ×0.10 mm ID ×0.10 μm of Rxi®-17 ms (Restek, Bellefonte, PA, USA) in GC oven (primary oven program, 100°C for 0.5 minute, followed by an increase up to 215°C at an increment of 20°C/minute, held for 0.5 minute, further to 270°C at 25°C/minute and held for 10 min; secondary oven program, rate and duration were identical to primary oven, target temperature was set at 30°C above primary oven). Sample injection: Gerstel PTV (Gerstel, Mülheim an der Ruhr, Germany) using solvent vent mode; modulator temperature +20°C. Carrier gas: helium at ramped pressure mode; transfer line 240°C. MS parameters: electron impact ionization 70 eV; ion source temperature 250°C; detector voltage 1,700 V; data-acquisition mass range m/z 50–500 at an acquisition rate of ten spectra; software ChromaTOF® 3.34 (LECO). Identification of components was based on comparison of their mass spectra with those of National Institute of Standards and Technology library spectra (v. 2.0).35 Citric acid, plumbagin, starch, and total reducing sugar content of PZRE were determined according to the methodology mentioned in the following sections.
Total reducing sugar
To 50 μL of PZRE, we added the same volume of 80% phenol (volume/volume) and vortexed. To this mixture, 2 mL of sulfuric acid was added and incubated for 10 minutes at RT, followed by recording absorbance at 490 nm. Total reducing sugar content was estimated using a standard glucose curve.36
Total phenolic content
PZRE (0.125 mL) was added to 0.5 mL of deionized water. To this mixture, 0.125 mL Folin–Ciocalteu reagent was added and incubated for 5 minutes at RT. After incubation, 1.25 mL of 7% Na2CO3 solution was added, and the volume was made up to 3 mL with distilled water. This reaction mixture was then incubated for 90 minutes at RT, and absorbance was recorded at 760 nm. Total phenolic content was estimated using a standard gallic acid curve.37
Total flavonoid content
Half a milliliter of 2% AlCl3 in methanol was mixed with a 0.5 mL sample of PZRE and incubated for 10 minutes at RT. After 10 minutes, absorbance was recorded at 368 nm. The total flavonoid content was determined using a standard quercetin curve.37
Citric acid content
One milliliter of PZRE was evaporated in order to dry it completely, and reconstituted in 1 mL 5% trichloroacetic acid. Dropwise addition of 8 mL of anhydrous acetic anhydride was done, followed by incubation at 60°C for 10 minutes. The tubes were then stoppered. After 10 minutes, 1 mL of pyridine was added to each tube and restoppered. The tube was then incubated at 60°C for 40 minutes, followed by incubation in an ice-water bath for 5 minutes. Absorbance was recorded at 420 nm. The total citric acid content was determined from a standard citric acid curve.37
Starch content
P. zeylanica root powder was weighed, and 5 g of the powder was washed with 70% ethanol. The washes were repeated to ensure the removal of sugars until it did not develop color when treated with anthrone reagent. The residue was dried and boiled in 100 mL distilled water. Then 1 mL of PZRE was evaporated completely and reconstituted in 60% perchloric acid. To this, 4 mL of anthrone reagent was added and was boiled in a water bath for 8 minutes. The intensity of the dark green color was recorded at 630 nm, and the starch content was estimated by comparing the absorbance with the standard glucose curve.37
Plumbagin content
Plumbagin was extracted using acetone, chloroform, and heptane according to the method described by Israni et al.38 In a mortar and pestle, 5 g root powder was macerated using acetone (3×50 mL). The acetone extract was then evaporated to dryness at RT and 25 mL chloroform was added, followed by an equal volume of distilled water (3×) to remove water-soluble impurities. The chloroform extract was evaporated till a dark brown oily residue was obtained. This residue was treated with 10 mL of phosphoric acid (10%) for 30 minutes followed by extraction with 5 mL n-heptane. To 0.2 mL of this yellow solution, 10% alcoholic KOH was added, the volume was made up to 5 mL using absolute ethanol, and absorbance was recorded at 520 nm. The concentration of plumbagin was determined using a standard plumbagin curve.
Synthesis and characterization of nanoparticles
UV-visible spectroscopy and FTIR analysis
Synthesis and optimization studies of AgNPs and AuNPs were carried out according to the methods given earlier.3,37 AgAuNPs were prepared using 1 mM silver nitrate (AgNO3) and 1 mM chloroauric acid (HAuCl4) salt solutions in a 1:1 ratio. To 5 mL PZRE, 47.5 mL of HAuCl4 solution was added, followed by addition of 47.5 mL AgNO3. Synthesis of NPs was monitored by recording the spectra on an ultraviolet (UV)-visible spectrophotometer (SpectraMax M5®; Molecular Devices, Sunnyvale, CA, USA) operating at a resolution of 1 nm. Optimization studies for rate of NP synthesis were carried out at different temperatures ranging from 4°C to 50°C, and at varying concentrations from 0.1 mM to 5 mM. For AgAuNPs, the salt mixture of AgNO3 and HAuCl4 was mixed in the ratio of 1:1, with concentrations differing from 0.1 mM to 5 mM. AgNPs, AuNPs, and AgAuNPs were also chemically synthesized and characterized according to the protocol reported earlier.39 Dried PZRE before and after bioreduction was subjected to Fourier-transform infrared spectroscopy (FTIR; Affinity-1; Shimadzu, Tokyo, Japan) using the potassium bromide pellet technique, and was exposed to an IR source of 500–4,000 cm−1. For application purposes, the NPs were synthesized, followed by washing twice with double-distilled water in order to use for antimicrobial and biofilm assays.
EDS and XRD analysis
NPs were studied using energy-dispersive spectrometry (EDS; JSM 6360A analytical scanning electron microscope [SEM]; JEOL, Tokyo, Japan) at an energy range 0–20 keV for elemental confirmation. Phase formation of the bioreduced NPs was studied using X-ray diffraction (XRD; D8 Advanced; Bruker Optik, Ettlingen, Germany) with a CuKα (1.54 Å) source.
DLS measurement and TEM analysis
Freshly prepared 3 mL AgNPs, AuNPs, and AgAuNPs were taken in polystyrene cuvettes and subjected to particle-size analysis employing dynamic light scattering (DLS; Zetasizer Nano-2590; Malvern Instruments, Malvern, UK). Zeta-potential values of each sample were also recorded on the same instrument. The surface morphology and size of the NPs were determined using transmission electron microscopy (TEM; Tecnai™ G2 20 U-Twin; FEI, Hillsboro, OR, USA).
Antimicrobial assay
The effects of NPs synthesized using PZRE were tested against A. baumannii AIIMS 7, Escherichia coli NCIM (National Collection of Industrial Microorganisms) 2931 and S. aureus MTCC (Microbial Type Culture Collection) 3160 on Mueller–Hinton (MH) agar plates using a disk-diffusion assay. Different concentrations of freshly prepared AgNPs, AuNPs, and AgAuNPs were prepared from 2,048 μg to 0.25 μg/disk by the double-dilution method. After incubation at 37°C for 18 hours, the zones of inhibition were measured.40,17 Chemically synthesized NPs were used as controls. All assays were performed in triplicate.
Determination of biofilm inhibition and disruption
Biofilm-inhibition assays were performed by coincubation of 5 μL of culture (optical density adjusted to 0.05 at 600 nm) with AgNPs, AuNPs, and AgAuNPs (1,024 μg/well) in 200 μL MH broth for 24 hours at 37°C.40 In the case of biofilm-disruption assays, bacterial biofilm was allowed to form for 24 hours, followed by washing with sterile phosphate-buffered saline (PBS; NaCl 8 g/L, KCl 0.2 g/L, Na2HPO4 1.44 g/L, KH2PO4 0.24 g/L). Biofilm disruption was initiated by the addition of 1,024 μg/well of freshly synthesized NPs.41,42 Additionally, the effect of NPs was checked against a mixed-biofilm formation by A. baumannii and S. aureus. Biofilm formation in the absence of NPs served as a positive control. The extent of biofilm inhibition and disruption was determined using the crystal violet staining method as reported earlier.43 Chemically synthesized NPs were used as controls. Biofilm inhibition and disruption assays were carried out in triplicate.
Visualization of biofilm employing fluorescence microscopy, SEM, and AFM
The biofilm inhibition and disruption assays were carried out on grease-free, sterile glass slides with tenfold-higher volumes to make the final volume 2 mL in 12-well plates and kept at 37°C for incubation. The concentration of the NPs was 1,024 μg/well. After 24 hours, the glass slides were washed with sterile PBS. The glass slides were stained with acridine orange and observed under a Zeiss fluorescence microscope using 465–495 nm excitation filters (AxioScope A1; Carl Zeiss Meditec, Jena, Germany). Glass slides fixed with glutaraldehyde were dehydrated sequentially using ethyl alcohol and analyzed using SEM (JSM 6360A; JEOL). Slides washed with PBS and air-dried were used for image analysis at a resolution of 2×2 μm under semi-contact atomic force microscopy (AFM; NTEGRA, NT-MDT, Moscow, Russia).
Results
Phytochemical analysis of P. zeylanica
HPTLC fingerprints for PZRE showed the bands corresponding to the standard plumbagin and diosgenin in the methanolic and ethyl acetate fractions of PZRE (Table 1 and Figure S1). Similarly, GC-TOF-MS analysis was done. The compounds detected in GC-TOF-MS analysis of ethyl acetate extract of PZRE are enlisted in Table 2. Sucrose was observed to be a major component of the PZRE, at 22%. Plumbagin content was found to be 2% in the plant extract. Total reducing sugars, phenolic content, flavonoid content, starch, citric acid and plumbagin content are quantitated and given in Table 3.
Table 1.
High-performance thin-layer chromatography of the root extract of Plumbago zeylanica
| Compounds | Type of extract | Amount (%) |
|---|---|---|
| Plumbagin | Hydromethanolic | 0.002 |
| Methanolic | 1.6 | |
| Ethyl acetate | 11.2 | |
| Diosgenin | Hydromethanolic | – |
| Methanolic | 0.21 | |
| Ethyl acetate | 2.8 | |
| Quercetin | Hydromethanolic | 0.0005 |
| Methanolic | – | |
| Ethyl acetate | 0.4 | |
| Glucose | Hydromethanolic | 14.3 |
| Fructose | Hydromethanolic | 0.5 |
Note: All experiments were performed in triplicate, and standard deviations were negligible.
Abbreviation: –, not detected.
Table 2.
Gas chromatography-time of flight-mass spectrometry analysis of Plumbago zeylanica root extract (PZRE)
| Name | Formula | Quantity | Area % | RT (s) |
|---|---|---|---|---|
| Sucrose | C12H22O11 | 57 | 21.912 | 558.3 |
| 2-Furancarboxaldehyde, 5-(hydroxymethyl)- | C6H6O3 | 126 | 19.378 | 444.9 |
| D-Allose | C6H12O6 | 60 | 6.2624 | 587 |
| Triethylene glycol | C6H14O4 | 45 | 5.0681 | 440.1 |
| Spiro[2.3]hexan-5-one, 4,4-diethyl- | C10H16O | 110 | 3.9047 | 446 |
| Ethanol, 2-(2-aminoethoxy)- | C4H11NO2 | 45 | 3.8949 | 559.7 |
| 4-Chromanol | C9H10O2 | 121 | 2.9764 | 690.5 |
| Ethanol, 2,2′-[oxybis(2,1-ethanediyloxy)]bis- | C8H18O5 | 45 | 2.8263 | 568.9 |
| Benzothiazole, 2-(2-hydroxyethylthio)- | C9H9NOS2 | 167 | 2.8117 | 832.1 |
| N-Butyl-tert-butylamine | C8H19N | 114 | 2.7494 | 432.3 |
| Plumbagin | C11H8O3 | 188 | 2.4452 | 639 |
| Pentane, 2,3,3,4-tetramethyl- | C9H20 | 85 | 2.0557 | 430 |
| Phenol, 2,4-bis(1,1-dimethylethyl)- | C14H22O | 191 | 1.214 | 569.4 |
| Acetaldehyde, tetramer | C8H16O4 | 45 | 1.2032 | 877.2 |
| Ethanol, 2-[2-(2-ethoxyethoxy)ethoxy]- | C8H18O4 | 45 | 1.1105 | 456.6 |
| Benzoic acid | C7H6O2 | 122 | 1.003 | 417.1 |
| Ethanol, 2,2′-[oxybis(2,1-ethanediyloxy)]bis- | C8H18O5 | 45 | 0.84417 | 750.8 |
| 1,4-Benzenediol, 2-methoxy- | C7H8O3 | 125 | 0.78487 | 530.8 |
| Semioxamazide | C2H5N3O2 | 103 | 0.73226 | 454.8 |
| n-Hexadecanoic acid | C16H32O2 | 60 | 0.72788 | 814.7 |
| 2-Octanamine | C8H19N | 55 | 0.72463 | 438.4 |
| Tridecanoic acid, methyl ester | C14H28O2 | 74 | 0.71858 | 792.4 |
| Benzoic acid, 4-ethoxy-, ethyl ester | C11H14O3 | 121 | 0.65169 | 576.1 |
| Ethane, 1,1′-oxybis, 2-methoxy- | C6H14O3 | 45 | 0.59053 | 428.9 |
| Ethane, iodo- | C2H5I | 156 | 0.58063 | 640.3 |
| N,3-Diethyl-3-heptanamine | C11H25N | 142 | 0.5475 | 633.5 |
Abbreviations: RT, reaction time; s, seconds.
Table 3.
Phytochemical analysis of Plumbago zeylanica root extract (PZRE)
| Sample | Phytochemicals (mg/mL) | |||||
|---|---|---|---|---|---|---|
| PZRE | Plumbagin | Total reducing sugars | Total phenolic content | Total flavonoid content | Starch | Citric acid |
|
| ||||||
| 0.12±0.04 | 1.25±0.04 | 0.73±0.02 | 0.95±0.05 | 0.93±0.02 | 3.38±0.01 | |
Note: The data is indicated as the means ± standard deviation, n=3.
Characterization of nanoparticles
Visual observation, UV-visible spectroscopy, and FTIR analysis
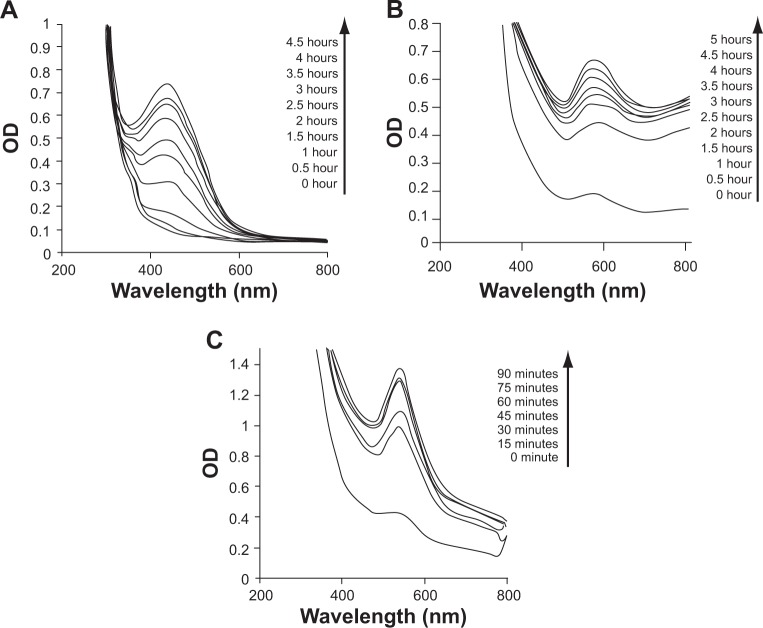
AgNPs, AuNPs, and AgAuNPs were bioreduced by PZRE. The yellowish-brown color of AgNPs appeared due to excitation of surface plasmon vibrations with absorbance maxima at 440 nm and a violet color of the AuNPs at 570 nm. A peak appeared at t=1 hour, with a steady increase till t=4.5 hours in AgNPs and t=5 hours in AuNPs (Figure 1A and B). AgAuNPs developed a dark-violet brownish color with absorbance maxima shifting towards 540 nm. A small peak appeared at t=0 minutes with the rate of synthesis increasing steadily till 90 minutes (Figure 1C).
Figure 1A–C.
Ultraviolet-visible spectra of nanoparticles synthesized from PZRE at 37°C as a function of time. (A) AgNPs from PZRE in 1 mM AgNO3; (B) AuNPs from PZRE in 1 mM HAuCl4; (C) AgAuNPs from PZRE in 1 mM AgNO3 and HAuCl4 solution in the ratio 1:1.
Abbreviations: HAuCl4, chloroauric acid; AgNO3, silver nitrate; PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AuNPs, gold NPs; AgAuNPs, bimetallic NPs; OD, optical density.
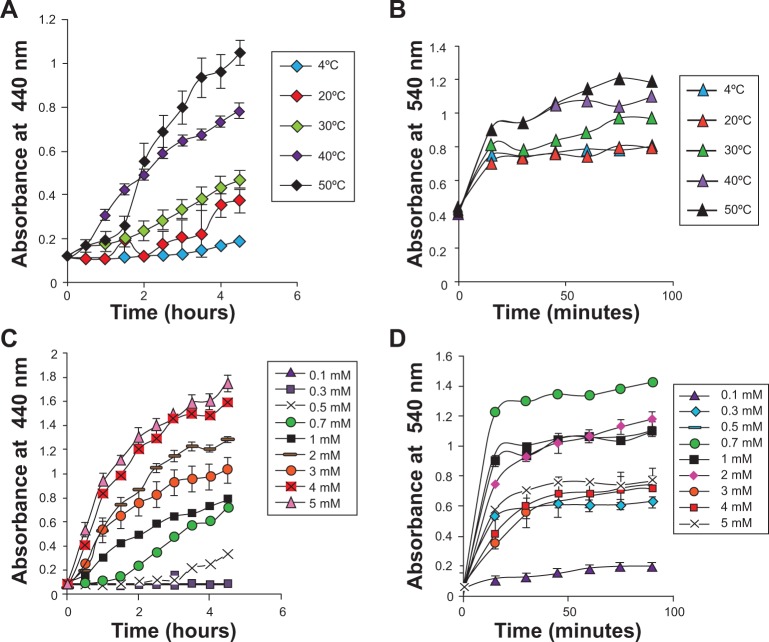
The effects of temperature on the rate of AgNPs and AgAuNPs synthesis showed a gradual increase in synthesis up to 50°C (Figure 2A and B). The study of the effect of salt concentration revealed an increase in the rate of AgNPs synthesis with an increase in concentration and maximum synthesis at 5 mM AgNO3 (Figure 2C). The rate of synthesis was highest in 0.7 mM and decreased further from 1 mM to 5 mM in AgAuNPs (Figure 2D). Similar results were observed in the case of AuNPs (Figure S2).
Figure 2A–D.
Bioreduction of AgNPs and AgAuNPs from PZRE at varying reaction temperatures and concentrations of the salts as a function of time. (A) Time course of AgNPs synthesis with varying reaction temperatures; (B) time course of AgAuNPs synthesis against reaction temperature; (C) time course of AgNPs formation obtained at different AgNO3 concentrations; (D) time course of AgAuNPs formation obtained at 1:1 AgNO3 and HAuCl4.
Abbreviations: HAuCl4, chloroauric acid; AgNO3, silver nitrate; PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AgAuNPs, bimetallic NPs.
The plant extracts before and after bioreduction were subjected to FTIR analysis. The sample of plant extract before bioreduction showed a peak at 3,300 cm−1, signifying the presence of hydroxyl groups (Figure 3). In samples after reduction, a peak at 3,300 cm−1 was not observed and the free hydroxyls were observed at 3,390 cm−1, indicating that hydroxyl groups present in polyphenols from P. zeylanica were responsible for the bioreduction of the AgNO3 and HAuCl4 solutions to form respective NPs. A significant peak at 1,741 cm−1of the symmetric stretch of C=O indicated the presence of aldehydes, ketones, esters, or carboxylic acids.
Figure 3A–D.
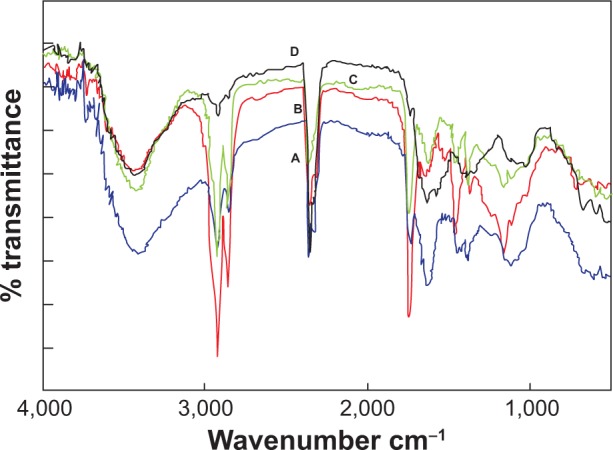
Fourier-transform infrared absorptive spectra before and after bioreduction of dried PZRE. (A) PZRE; (B) AgNPs; (C) AuNPs; (D) AgAuNPs.
Abbreviations: PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AuNPs, gold NPs; AgAuNPs, bimetallic NPs.
EDS, XRD, and DLS analysis
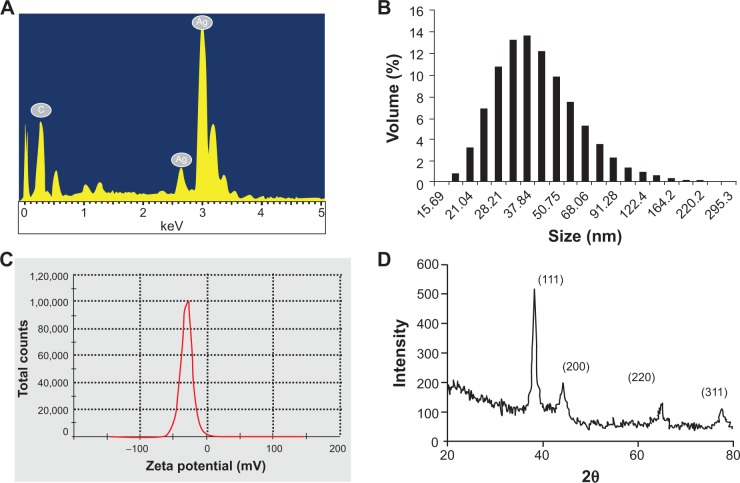
In EDS analysis, the characteristic peak at 3 keV confirmed the presence of AgNPs, whereas particle-size analysis employing DLS showed that the bioreduced AgNPs had an average size of 63.92 nm (Figure 4A and B). Stability of the particles was confirmed from the zeta-potential value, which was found to be −31, whereas phase formation was observed using XRD patterns. Phase formation was confirmed from characteristic peaks, such as (111), (200), (220), and (311). The data matched with the standard Joint Committee for Powder Diffraction Set, card 040783, confirming a face-centered cubic structure for the AgNPs (Figure 4C and D).
Figure 4A–D.
Determination of purity, size, and nature of AgNPs synthesized by PZRE at 37°C. (A) Representative spot energy-dispersive spectrum; (B) histogram of size distribution; (C) zeta-potential analysis; (D) representative X-ray diffraction profile of thin film.
Abbreviations: PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles.
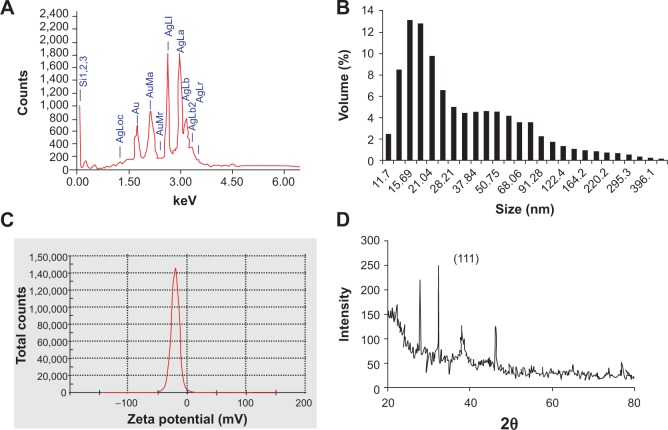
EDS of AgAuNPs showed that peaks were obtained at around 2 and 3 keV, confirming the presence of silver and gold (Figure 5A). The silver content was around 66%, and that of gold was 14%. The average size of the AgAuNPs was recorded to be 93.04 nm, with a zeta-potential value of −21 (Figure 5B and C). In XRD analysis, the position of the peak (38.9°C), located between the (111) reflections of Ag and Au, as seen in Figure 5D, represents the formation of alloy-like AgAuNPs. Similarly, characterization of AuNPs was carried out using the aforementioned techniques, and results are given in Figure S3.
Figure 5A–D.
Determination of purity, size, and nature of AgAuNPs synthesized by PZRE at 37°C. (A) Representative spot energy-dispersive spectrum; (B) Histogram of size distribution; (C) zeta-potential analysis; (D) representative X-ray diffraction profile of thin film.
Abbreviations: PZRE, Plumbago zeylanica root extract; AgAuNPs, bimetallic NPs.
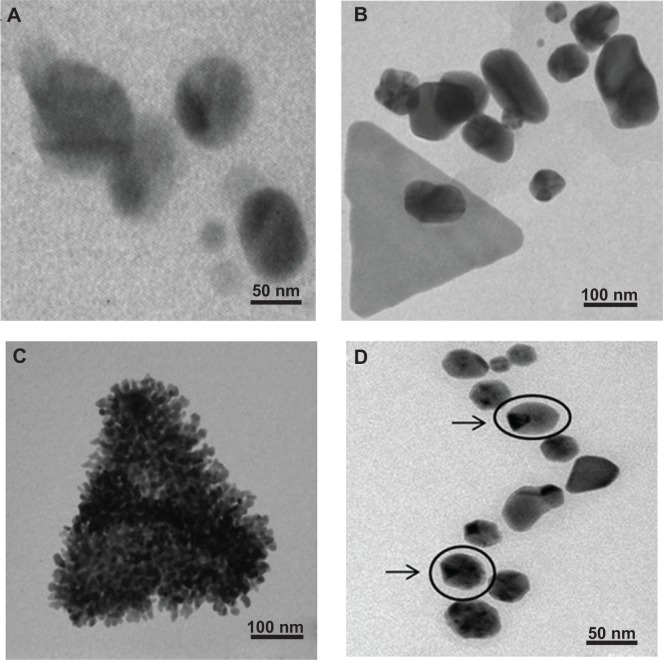
TEM analysis
The TEM micrograph showed spherically shaped AgNPs synthesized by PZRE (Figure 6A). The TEM micrographs revealed some unique features of AuNPs (Figure 6B and C). They were found to be anisotropic in nature, with spheres, triangles, and hexagons exhibiting an interesting and rare phenomenon of shape evolution. Figure 6B demonstrates an arrangement of gold nanospheres forming a gold nanotriangle. Furthermore, the shape evolution continued, with nanospheres adhering to the triangle, leading to the growth of nanotriangles with attached nanospheres (Figure 6C). In Figure 6D, the arrows indicate nanohexagons and nanospheres of AgAuNPs, but the majority of them were nanohexagons. A unique feature of AgAuNPs was their shape: blunt ended polygonal NPs. Chemically synthesized NPs were also characterized using EDS, XRD, TEM, and DLS (Figures S4–S6).
Figure 6A–D.
Characterization of nanoparticles synthesized by PZRE using transmission electron microscopy. (A) Spherical silver nanospheres; (B) shape evolution of gold nanotriangles; (C) assembly of gold nanospheres forming a gold nanotriangle; (D) arrows indicating blunt-ended AgAu nanopolygons.
Abbreviation: PZRE, Plumbago zeylanica root extract.
Antibacterial activity
The minimum inhibitory concentration (MIC) values of the NPs against Gram-negative and Gram-positive bacteria are given in Table 4. PZRE-mediated AgNPs showed effective antimicrobial activity compared to PZRE-mediated AuNPs and AgAuNPs, as well as the chemically synthesized AgNPs when used as a control. AgNPs exhibited a lower MIC value against E. coli (2 μg/disk) in comparison with A. baumannii and S. aureus strains (8 μg/disk). AuNPs showed the highest antimicrobial activity against S. aureus, with an MIC of 8 μg/disk, whereas chemically synthesized AuNPs did not show antimicrobial activity. Moreover, bioreduced AgAuNPs were found to show more effective antimicrobial activity compared to AgAuNPs synthesized chemically.
Table 4.
Minimum inhibitory concentration of silver nanoparticles (AgNPs), gold NPs (AuNPs) and bimetallic NPs (AgAuNPs) against bacterial cultures
| Name of the microorganism | Biologically synthesized
|
Chemically synthesized
|
||||
|---|---|---|---|---|---|---|
| AgNPs | AuNPs | AgAuNPs | AgNPs | AuNPs | AgAuNPs | |
| A. baumannii AIIMS 7 | 8 | 128 | 16 | 512 | – | 256 |
| E. coli NCIM 2931 | 2 | 256 | 4 | 512 | – | 32 |
| S. aureus aureus MTCC 3160 | 8 | 8 | 8 | 256 | – | 512 |
Note: All experiments were performed in triplicate and standard deviations were negligible.
Abbreviations: AIIMS, All India Institute of Medical Sciences; NCIM, National Collection of Industrial Microorganisms; MTCC, Microbial Type Culture Collection; –, No antimicrobial activity observed.
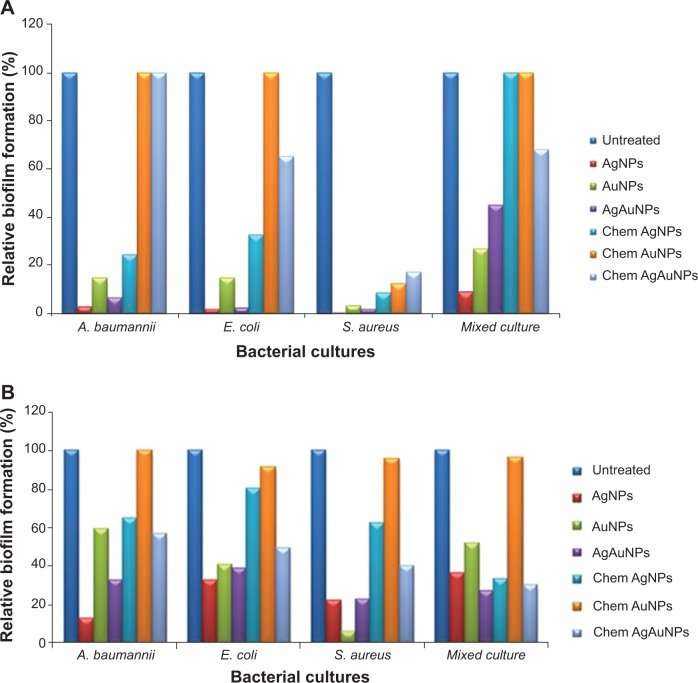
Biofilm inhibition and disruption using nanoparticles
As all three NP types – AgNPs, AuNPs, and AgAuNPs – showed superior antibacterial activity, these NPs were further studied for biofilm inhibition and biofilm disruption with chemically synthesized NPs as a control. Biofilm inhibition with NP concentration of 1,024 μg/well after 24 hours of coincubation is represented by the graph in Figure 7A. It was observed that AgNPs and AgAuNPs inhibited the biofilm in all the test-microorganism single cultures around 96%–99% and 93%–98%, respectively. This was interesting compared to the control AgNPs and AgAuNPs, which showed inhibition of 67%–91% and 0%–82%, respectively. In the case of mixed culture, chemically synthesized AgNPs did not inhibit biofilm formation, whereas AgNPs from PZRE showed 91% biofilm inhibition. AgAuNPs showed A. baumannii biofilm inhibition up to 94% and E. coli biofilm inhibition up to 98%, with the highest inhibition of S. aureus biofilm of 99%. Similar to control AgNPs, control AgAuNPs showed poor biofilm inhibition compared to biological AgAuNPs, whereas AuNPs from both biological and chemical sources showed poor biofilm inhibition and disruption, except AuNPs from PZRE, which showed 97% S. aureus biofilm inhibition.
Figure 7.
Inhibition and disruption of bacterial biofilms by biologically and chemically synthesized nanoparticles at a concentration of 1,024 μg/well at 24 hours. (A) Biofilm inhibition; (B) biofilm disruption.
Abbreviations: A. baumannii, Acinetobacter baumannii; E. coli, Escherichia coli; S. aureus, Staphylococcus aureus; AgNPs, silver nanoparticles; AuNPs, gold NPs; AgAuNPs, bimetallic NPs; chem, chemically synthesized.
To elaborate more on the control of bacterial biofilms, a biofilm-disruption assay was carried out to check the effect of NPs on biofilm grown for 24 hours (Figure 7B). Bacterial biofilms were seen to be disrupted in presence of AgNPs up to 88% in A. baumannii, 67% in E. coli, 78% in S. aureus, and 64% in mixed biofilm, whereas chemically synthesized AgNPs showed poor disruption activity of about 19%–60%. AuNPs from PZRE showed lower biofilm disruption of around 40%, with a maximum of 95% in S. aureus, whereas chemically synthesized AuNPs did not show any biofilm-disruption activity. AgAuNPs also showed poor disruption of about 61%–77%, but this was superior to that of chemical AgAuNPs (40%–70%).
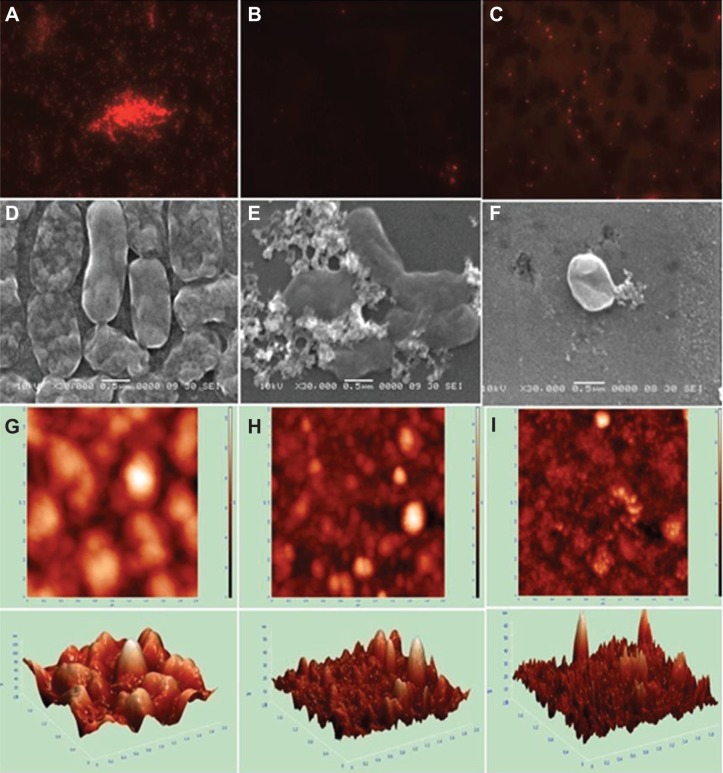
Visualization of biofilm using fluorescence microscopy, SEM, and AFM
The cells were seen in aggregates in the absence of NPs (Figure 8A) in images obtained under fluorescence microscopy at a resolution of 40×. In the presence of AgNPs, the biofilm as well as cells were absent (Figure 8B), whereas in Figure 8C, although biofilm was absent in the presence of AgAuNPs, the cells were seen to be isolated. In SEM images, A. baumannii cells and the biofilm formed, seen in Figure 8D, were intact. Due to the presence of intact biofilm, the cells were present in colonies, and no change in the morphology was observed. Coincubation of the bacterial cells with AgNPs led to a change in the cell morphology and disrupted the biofilm of A. baumannii (Figure 8E), whereas cells were seen to be lysed in the presence of AgAuNPs (Figure 8F).
Figure 8A–I.
Images of Acinetobacter baumannii biofilm formation and inhibition using fluorescence microscopy, scanning electron microscopy (SEM), and atomic force microscopy (AFM). Upper panel, fluorescence microscopy; middle panel, SEM; lower panel, AFM. (A,D,G) Biofilm formation; (B,E,H) biofilm inhibition with AgNPs; (C,F,I) biofilm inhibition with AgAuNPs.
Abbreviations: AgNPs, silver nanoparticles; AgAuNPs, bimetallic NPs.
AFM images in 3-D represent A. baumannii biofilm formation (Figure 8G) and inhibition in the presence of AgNPs and AgAuNPs (Figure 8H and I). The roughness of the glass slide containing the biofilm and NPs was calculated, which varied for different bacterial cultures. Table 5 presents the relative reduction in the roughness values of the biofilms when treated with AgNPs, AuNPs, and AgAuNPs. The roughness of the untreated biofilms was observed to be 16 nm, 5 nm, and 11 nm in A. baumannii, S. aureus, and E. coli, respectively. The percentage of relative roughness observed when the bacterial cultures were coincubated with biologically synthesized NPs was 12%–36% in A. baumannii and S. aureus and 40%–60% in E. coli. Chemically synthesized AgNPs (18%–45%) showed a reduction in roughness values, but were poor compared to biological AgNPs (12%–18%) except E. coli (20%). Chemically synthesized AuNPs and AgAuNPs showed lower reductions in roughness values of 80%–90% and 40%–60%, respectively, compared to biologically synthesized AuNPs (18%–60%) and AgAuNPs (25%–40%). Furthermore, it was also observed when fully formed biofilm was treated with AgNPs, roughness was reduced to 12% in A. baumannii and 50% in E. coli, which indicated that AgNPs also played a role in disrupting bacterial biofilms (Figure S7).
Table 5.
Roughness values of Acinetobacter baumannii, Escherichia coli, and Staphylococcus aureus biofilm in the presence and absence of nanoparticles, determined by atomic force microscopy
| Name of the microorganism | Roughness values (nm)
|
||||||
|---|---|---|---|---|---|---|---|
| Untreated | Biologically synthesized
|
Chemically synthesized
|
|||||
| AgNPs | AuNPs | AgAuNPs | AgNPs | AuNPs | AgAuNPs | ||
| A. baumannii | 16 | 2 | 5 | 4 | 3 | 13 | 10 |
| E. coli | 5 | 2 | 3 | 2 | 1 | 4 | 2 |
| S. aureus | 11 | 2 | 2 | 4 | 5 | 10 | 6 |
Note: All experiments were performed in triplicate and standard deviations were negligible.
Abbreviations: AgNPs, silver nanoparticles; AuNPs, gold NPs; AgAuNPs, bimetallic NPs.
Discussion
P. zeylanica is reported to be a rich source of phytochemicals and flavonoids, as observed in HPTLC fingerprinting and GC-TOF-MS profile.44 Higher concentrations of sucrose, glucose, and fructose determined by chromatographic techniques and quantitation assays may have stabilized the NPs synthesized from the root extract and may have controlled NP shapes, such as spherical31 and hexagonal,45 during the course of the synthesis. Starch concentration in PZRE was the same, whereas citric acid was found to be three times that of Dioscorea bulbifera.37 Starch stabilizes the NPs by capping them.46 Lee et al47 demonstrated that citrate can adsorb on NPs during their shaping and stabilize them. Therefore, owing to higher concentrations of citric acid in PZRE, NPs were better stabilized and evolved.
The 90 minutes required for the synthesis of AgAuNPs is the shortest reported for the biological synthesis of bimetallic AgAuNPs, in contrast to those reported from neem extract, which took 296 minutes for complete synthesis.21 The rate of synthesis was observed to be significantly dependent on the concentration of the salt solutions and temperature, which is well in agreement with the previous reports.48,49 The hydroxyl groups of polyphenols might be playing a significant role in bioreduction, as revealed by FTIR analysis.37
UV-visible spectroscopy indicating a shift from 570 nm to 540 nm in synthesis of AgAuNPs, as well as EDS analysis indicating a lower percentage of gold compared to silver, may be attributed to the Au core and Ag shell.21 The size of AgNPs synthesized by PZRE was comparable to earlier reports on Aloe vera50 and Cardiospermum halicacabum,51 as well as commercially available AgNPs.52 TEM micrographs of AuNPs showed a unique feature of shape evolution in the form of nanotriangles, in which adherence of gold nanospheres covering the whole surface area of gold nanotriangles was observed. However, Shankar et al53 reported on the Turkevich protocol for synthesis of gold nanotriangles and nanospheres, while Ghosh et al37 reported on anisotropic AuNPs from D. bulbifera. AgAuNPs were comprised of nanopolygons that were blunt-ended.54
We observed that chemically synthesized AgNPs showed poor antibacterial activity compared to biological AgNPs. It is important to note that AuNPs synthesized by PZRE showed antimicrobial activity. However, chemically synthesized AuNPs failed to show any activity. The reason for this may be the protein coating of NPs obtained from the biological extract.55
Biofilm formation by human pathogenic microbes causing severe chronic lung, urinary tract, and various nosocomial infections is attributed to lower penetration and activity of drugs, altered genotype of bacteria exposed to drugs, lower oxygen, and acidic pH.56–59 Therefore, research on biogenic NPs for control of bacterial biofilms is very promising. Various attempts have been made to control biofilms by biosurfactants, rhamnolipids, and enzyme-based detergents.60,61 Recently, Thawal et al62 demonstrated 80% disruption of biofilm by a phage from the family Podoviridae isolated from A. baumannii.
Control of bacterial biofilms using NPs like chitosan, zinc oxide, and silver are interesting, because of their high surface-to-volume ratio.42,63,64 Our results are in agreement with the report of Kalishwaralal et al65 where 50 nM of AgNPs significantly arrested P. aeruginosa biofilm formation without affecting cell viability, whereas 100 nM inhibited the growth of the organism itself and resulted in a 95%–98% decrease in biofilm. Also, AgNPs and AuNPs showed better biofilm inhibition than disruption; however, in the case of mixed cultures, AgAuNPs showed better biofilm disruption than biofilm inhibition. This shows the potential of AgAuNPs to disrupt mixed biofilms, which could be because of the synergistic effect of silver and gold components. Mixed biofilms, however, were inhibited and disrupted by AgNPs and AuNPs to a lesser extent compared to single bacterial biofilm, which might be due to more resistance to the penetrability of NPs challenging their potential for biofilm disruption.
The novelty of the study lies in the fact that PZRE-synthesized NPs were found to be antimicrobial against A. baumannii and able to control its biofilm. Although A. baumannii is found in the human skin microbiome, it is also considered to be a dreadful nosocomial pathogen, due to its high resistance to antibiotics and metal salts leading to severe mortality (17%–62%) among immunocompromised patients because of bacteremia.26,58,66–72 Deformity in A. baumannii cells treated with AgNPs, as observed in fluorescence and SEM micrographs, might be due to enhanced penetrability of AgNPs within the biofilm, unlike metallic silver.15,40,42 This can be a powerful strategy for sensitizing A. baumannii with high metal resistance.
This is the first report of its kind where inhibition as well as disruption of bacterial biofilms have been demonstrated employing AFM after treatment with NPs synthesized by PZRE. Significant reduction of roughness confirmed the efficacy of NPs in biofilm inhibition and disruption. This proved to be advantageous, owing to semi-contact mode, better resolution, biofilms observed in their own physiological condition with no addition of chemicals, direct contact with the surface with minimum use of lenses and electron beams, and deduction of roughness values based upon variation in height and texture.73,74
Conclusion
This is the first report of rapid and efficient synthesis of AgNPs, AuNPs, and AgAuNPs by P. zeylanica. All the three types of NPs had unique features: being spherical in case of AgNPs, revealing shape evolution in case of AuNPs, and showing blunt-ended hexagonal nanostructures in AgAuNPs. These diverse properties were obtained due to phenolics, flavonoids, sugars, starch, and citric acid present in PZRE. Bioreduced NPs were able to inhibit to significant levels, particularly in the case of E. coli and S. aureus. S. aureus biofilm was inhibited and disrupted significantly by AuNPs, which shows that AuNPs can be more effective against Gram-positive bacteria than Gram-negative bacteria. Therefore, the NPs synthesized, particularly AgNPs from P. zeylanica, proved to be very efficient in the control of bacterial biofilms.
Supplementary materials
Table of contents
| Serial number | Contents | Page number |
|---|---|---|
| 1. | Supplementary figure 1 HPTLC fingerprint of PZRE. (A) PZRE ethyl acetate extract, and (B) standard mixture of plumbagin (P), diosgenin (D), quercetin (Q) and catechin (C). | 2650 |
| 2. | Supplementary figure 2 Bioreduction of AuNPs from PZRE at varying reaction temperatures and concentrations of the salts as a function of time. (A) Time course of AuNPs synthesis against reaction temperature, and (B) time course of AuNPs formation at different HAuCl4 concentration. | 2650 |
| 3. | Supplementary figure 3 Determination of purity, size and nature of the AuNPs synthesized by PZRE at 37°C. (A) Representative spot energy-dispersive spectrum, (B) histogram of size distribution, (C) zeta potential analysis, and (D) representative X-ray diffraction profile of thin film. | 2651 |
| 4. | Supplementary figure 4 Determination of purity, size and nature of the AgNPs synthesized using 1% trisodium citrate. (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution. | 2651 |
| 5. | Supplementary figure 5 Determination of purity, size and nature of the AuNPs synthesized using 1% trisodium citrate. (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution. | 2652 |
| 6. | Supplementary figure 6 Determination of purity, size and nature of the AgAuNPs synthesized using 1% trisodium citrate. (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution. | 2652 |
| 7. | Supplementary figure 7 AFM micrographs for biofilm disruption. (A) A. baumannii biofilm disruption using AgNPs using AFM, and (B) amplitude mode of AFM image. | 2653 |
| 8. | Materials and methods for cytotoxicity assay | 2653 |
| 9. | Supplementary figure 8 Cytotoxicity of the nanoparticles synthesized from PZRE and 1% trisodim citrate determined using MCF7 cells showing no inhibition upto the concentration of 0.5 mg/well. | 2653 |
Abbreviations: HPTLC, high-performance thin-layer chromatography; PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AuNPs, gold NPs; HAuCl4, chloroauric acid; AgAuNPs, bimetallic NPs; AFM, atomic force microscopy.
HPTLC fingerprint of PZRE.
Notes: (A) PZRE ethyl acetate extract, and (B) standard mixture of plumbagin (P), diosgenin (D), quercetin (Q) and catechin (C).
Abbreviations: HPTLC, high-performance thin-layer chromatography; PZRE, Plumbago zeylanica root extract.
Bioreduction of AuNPs from PZRE at varying reaction temperatures and concentrations of the salts as a function of time.
Notes: (A) Time course of AuNPs synthesis against reaction temperature, and (B) time course of AuNPs formation at different HAuCl4 concentration.
Abbreviations: PZRE, Plumbago zeylanica root extract; AuNPs, gold nanoparticles; HAuCl4, chloroauric acid; h, hours.
Determination of purity, size and nature of the AuNPs synthesized by PZRE at 37°C.
Notes: (A) Representative spot energy-dispersive spectrum, (B) histogram of size distribution, (C) zeta potential analysis, and (D) representative X-ray diffraction profile of thin film.
Abbreviations: PZRE, Plumbago zeylanica root extract; AuNPs, gold nanoparticles.
Determination of purity, size and nature of the AgNPs synthesized using 1% trisodium citrate.
Notes: (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution.
Abbreviation: AgNPs, silver nanoparticles.
Determination of purity, size and nature of the AuNPs synthesized using 1% trisodium citrate.
Notes: (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution.
Abbreviation: AuNPs, gold nanoparticles.
Determination of purity, size and nature of the AgAuNPs synthesized using 1% trisodium citrate.
Notes: (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution.
Abbreviation: AgAuNPs, bimetallic nanoparticles.
AFM micrographs for biofilm disruption.
Notes: (A) A. baumannii biofilm disruption using AgNPs using AFM, and (B) amplitude mode of AFM image.
Abbreviations: AgNPs, silver nanoparticles; AFM, atomic force microscopy.
Materials and methods for cytotoxicity assay
MCF7 (human breast adenocarcinoma), was obtained from National Center for Cell Sciences, India and cultured in a humidified atmosphere (37°C, 5% CO2) in Roswell Park Memorial Institute1640 medium (Life Technologies, Waltham, MA, USA) supplemented with 10% fetal bovine serum.1 Nanoparticles were evaluated for cytotoxicity against MCF7 cancer cell lines. The compounds were evaluated in vitro at a various concentration. MTT colorimetric assay was used to determine growth inhibition. 100 μL of cell suspension (2×104 cells) were plated in 96-well plates and allowed to attach for 24 hours. Cells were exposed in triplicate wells to AgNPs, AuNPs and AgAuNPs both synthesized chemically as well as by PZRE for 48 hours. After 48 hours, 20 μL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) solution (5 mg/mL,) was added to each well. After 4 hours of incubation the formazan precipitate was dissolved in 100 μL of solubilization buffer (10% SDS in 0.1 N HCl) and kept overnight for complete dissolution. The absorbance at 576 nm was determined on a 96 well microplate reader (SpectraMax M5®, Molecular Devices Corporation, Sunnyvale, CA). The absorbance values were used to calculate % inhibition.
Results
Cytotoxicity of the nanoparticles synthesized from PZRE and 1% trisodim citrate determined using MCF7 cells showing no inhibition upto the concentration of 0.5 mg/mL.
Abbreviations: PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AuNPs, gold NPs; AgAuNPs, bimetallic NPs; Chem, chemically synthesized.
Reference
- 1.Pawar VU, Ghosh S, Chopade BA, Shinde VS. Design and synthesis of harzialactone analogues: Promising anticancer agents. Bioorg Med Chem Lett. 2010;20(24):7243–7245. doi: 10.1016/j.bmcl.2010.10.100. [DOI] [PubMed] [Google Scholar]
Acknowledgments
GS acknowledges the financial support for this work from the Institute of Bioinformatics and Biotechnology, University of Pune, Pune, India. SG thanks the Council of Scientific and Industrial Research (CSIR, Government of India) for a senior research fellowship (09/137[0516]/2012-EMR-I). Part of this work was supported by UPE Phase I and II, awarded to the University of Pune by the University Grant Commission (UGC), New Delhi, India. We are thankful to Dr Charegaonkar and Mr Naidu, Anchrome Enterprises (I) Pvt Ltd, Mulund, Mumbai, India for HPTLC analysis. We also acknowledge Professor SI Patil, Head of the Department of Physics, University of Pune for use of the TEM facility; and Purkayashta P, Khanka V, Banerjee S, and Koya R from India LECO instruments Pvt. Ltd. (Mumbai, Maharashtra, India) for GC-TOF-MS identification.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Tran QH, Nguyen VQ, Le A. Silver nanoparticles: synthesis, properties, toxicology, applications and perspectives. Adv Nat Sci Nanosci Nanotechnol. 2013;4(3):033001. [Google Scholar]
- 2.Elavazhagan T, Arunachalam K. Memecylon edule leaf extract mediated green synthesis of silver and gold nanoparticles. Int J Nanomedicine. 2011;6:1265–1278. doi: 10.2147/IJN.S18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh S, Patil S, Ahire M, et al. Synthesis of gold nanoanisotrops using Dioscorea bulbifera tuber extract. J Nanomater. 2011;2011:354793. [Google Scholar]
- 4.He S, Zhirui G, Zhang Y, Zhang S, Wang J, Gu N. Biosynthesis of gold nanoparticles using the bacteria Rhodopseudomonas capsulata. Mater Lett. 2007;61(18):3984–3987. [Google Scholar]
- 5.Mourato A, Gadanho M, Lino AR, Tenreiro R. Biosynthesis of crystalline silver and gold nanoparticles by extremophilic yeasts. Bioinorg Chem Appl. 2011;2011:546074. doi: 10.1155/2011/546074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mukherjee P, Ahmad A, Mandal D, et al. Fungus-mediated synthesis of silver nanoparticles and their immobilization in the mycelial matrix: a novel biological approach to nanoparticle synthesis. Nano Lett. 2001;1(10):515–519. [Google Scholar]
- 7.Govindaraju K, Kiruthuga V, Kumar VG, Singaravelu G. Extracellular synthesis of silver nanoparticles by a marine alga, Sargassum wightii Grevilli and their antibacterial effects. J Nanosci Nanotechnol. 2009;9(9):5497–5501. doi: 10.1166/jnn.2009.1199. [DOI] [PubMed] [Google Scholar]
- 8.Chopade NB, Ghosh S, More P, et al. Biogenic copper nanoparticles as novel antidiabetic and antioxidant in nanomedicine; Poster presented at: Second International Translational Nanomedicine Conference; July 26–28, 2013; Boston, MA. [Google Scholar]
- 9.Khan M, Khan M, Adil SF, et al. Green synthesis of silver nanoparticles mediated by Pulicaria glutinosa extract. Int J Nanomedicine. 2013:81507–81516. doi: 10.2147/IJN.S43309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo KJ, Hye CK, Ki WK, Sook S, So HK, Yong HP. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl Env Microbiol. 2008;74(7):2171–2178. doi: 10.1128/AEM.02001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, Du Z, Lv H, et al. Green synthesis of silver nanoparticles by Chrysanthemum morifolium Ramat. Extract and their application in clinical ultrasound gel. Int J Nanomedicine. 2013;8:1809–1815. doi: 10.2147/IJN.S43289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27(1):76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Shakibaie MR, Dhakephalkar P, Kapadnis BP, Chopade BA. Removal of silver from photographic wastewater effluent using Acinetobacter baumannii BL54. Can J Microbiol. 1999;45(12):995–1000. [PubMed] [Google Scholar]
- 14.Waters AE, Contente-Cuomo T, Buchhagen J, et al. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin Infect Dis. 2011;52(10):1227–1230. doi: 10.1093/cid/cir181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhakephalkar PK, Chopade BA. High levels of multiple metal resistance and its correlation to antibiotic resistance in environmental isolates of Acinetobacter. Biometals. 1994;7(1):67–74. doi: 10.1007/BF00205197. [DOI] [PubMed] [Google Scholar]
- 16.Chopra I. The increasing use of silver-based products as antimicrobial agents: a useful development or a cause for concern? J Antimicrob Chemother. 2007;59(4):587–590. doi: 10.1093/jac/dkm006. [DOI] [PubMed] [Google Scholar]
- 17.Singh R, Wagh P, Wadhwani S, et al. Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics. Int J Nanomedicine. 2013;8:4277–4290. doi: 10.2147/IJN.S48913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shebdadkar U, Singh R, Wadhawani S, Chopade BA. Microbial synthesis of gold nanoparticles: current status and future prospects. Adv Colloid Interface Sci. 2014 Jan 2; doi: 10.1016/j.cis.2013.12.011. Epub. [DOI] [PubMed] [Google Scholar]
- 19.Glomm WR. Functionalized gold nanoparticles for applications in bionanotechnology. J Dispers Sci Technol. 2005;26(3):389–414. [Google Scholar]
- 20.Song JY, Kim BS. Biological synthesis of bimetallic Au/Ag nanoparticles using persimmon (Diopyros kaki) leaf extract. Korean J Chem Eng. 2008;25(4):808–811. [Google Scholar]
- 21.Shankar SS, Rai A, Ahmad A, Sastry M. Rapid synthesis of Au, Ag, and bimetallic Au core-Ag shell nanoparticles using neem (Azadirachta indica) leaf broth. J Colloid Interface Sci. 2004;275(2):496–502. doi: 10.1016/j.jcis.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Patwardhan RB, Dhakephalkar P, Chopade BA. Medicinal importance of chitraka plant: a review. Shrushti Health Bull. 2005;4:4–5. [Google Scholar]
- 23.Ahmad I, Beg AZ. Antimicrobial and phytochemical studies on 45 Indian medicinal plants against multi-drug resistant human pathogens. J Ethnopharmacol. 2001;74(2):113–123. doi: 10.1016/s0378-8741(00)00335-4. [DOI] [PubMed] [Google Scholar]
- 24.van der Vijver LM. Distribution of plumbagin in the mplumbaginaceae. Phytochemistry. 1972;11(11):3247–3248. [Google Scholar]
- 25.de Paiva SR, Figueiredo M, Aragão TV, Kaplan MA. Antimicrobial activity in vitro of plumbagin isolated from Plumbago species. Mem Inst Oswaldo Cruz. 2003;98(7):959–961. doi: 10.1590/s0074-02762003000700017. [DOI] [PubMed] [Google Scholar]
- 26.Shakibaie MR, Dhakephalkar PK, Kapadnis BP, Salayaghe G, Chopade BA. Plasmid mediated silver and antibiotic resistance in Acinetobacter baumannii BL54. Iran J Med Sci. 1998;23(12):30–36. [Google Scholar]
- 27.Deshpande LM, Chopade BA. Plasmid mediated silver resistance in Acinetobacter baumannii BL54. Biometals. 1994;7(1):49–56. doi: 10.1007/BF00205194. [DOI] [PubMed] [Google Scholar]
- 28.Dhale DA, Markandeya SK. Antimicrobial and phytochemical screening of Plumbago zeylanica Linn. (Plumbaginaceae) leaf. J Exp Sci. 2011;2(3):4–6. [Google Scholar]
- 29.Asharani PV, Mun GLK, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3(2):279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 30.Egorova EM, Revina AA. Synthesis of metallic nanoparticles in reverse micelles in the presence of quercetin. Colloids Surf A Physicochem Eng Asp. 2000;168(1):87–96. [Google Scholar]
- 31.Panigrahi S, Kundu S, Ghosh SK, Nath S, Pal T. General method of synthesis for metal nanoparticles. J Nanopart Res. 2004;6(4):411–414. [Google Scholar]
- 32.Zhao X, Shi Y, Wang T, Cai Y, Jiang G. Preparation of silica-magnetite nanoparticle mixed hemimicelle sorbents for extraction of several typical phenolic compounds from environmental water samples. J Chromatogr A. 2008;1188(2):140–147. doi: 10.1016/j.chroma.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 33.Chothani DL, Patel MB, Mishra SH. HPTLC fingerprint profile and isolation of marker compound of Ruellia tuberosa. Chromatogr Res Int. 2012;2012:180103. [Google Scholar]
- 34.Skalska-Kamińska A, Matysik G, Wójciak-Kosior M, Donica H, Sowa I. Thin-layer chromatography of sugars in plant material. Curr Issue Pharm Med Sci. 2009;22(4):17–24. [Google Scholar]
- 35.Parveen I, Moorby JM, Fraser MD, Allison GG, Kopka J. Application of GCMS-TOF metabolite profiling techniques to the analysis of heathland plant diets of sheep. J Agric Food Chem. 2007;55(4):1129–1138. doi: 10.1021/jf062995w. [DOI] [PubMed] [Google Scholar]
- 36.Dubois M, Gilles DA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for the determination of sugars and related substances. Anal Chem. 1956;28(3):350–356. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S, Patil S, Ahire M, et al. Synthesis of silver nanoparticles using Dioscorea bulbifera tuber extract and evaluation of its synergistic potential in combination with antimicrobial agents. Int J Nanomedicine. 2012;7:483–496. doi: 10.2147/IJN.S24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Israni SA, Kapadia NS, Lahiri SK, Yadav G, Shah MB. An UV-visible spectrophotometric method for the estimation of plumbagin. Int J Chem Tech Res. 2010;2(2):856–859. [Google Scholar]
- 39.Augustine R, Rajarathinam R. Synthesis and characterization of silver nanoparticles and its immobilization on alginate coated sutures for the prevention of surgical wound infections and the in vitro release studies. Int J Nano Dimens. 2012;2(3):205–212. [Google Scholar]
- 40.Deshpande LM, Kapadnis, Chopade BA. Metal resistance in Acinetobacter and its relation to β-lactamase production. Biometals. 1993;6(1):55–59. doi: 10.1007/BF00154233. [DOI] [PubMed] [Google Scholar]
- 41.Sahu PK, Iyer PS, Oak A, Pardeshi K, Chopade BA. Characterization of eDNA from the clinical strain Acinetobacter baumannii AIIMS 7 and its role in biofilm formation. Scientific World Journal. 2012;2012:973436. doi: 10.1100/2012/973436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaidhani SV, Singh R, Singh D, et al. Biofilm disruption activity of silver nanoparticles synthesized by Acinetobacter calcoaceticus PUCM 1005. Mater Lett. 2013;108:324–327. [Google Scholar]
- 43.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–179. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 44.Kishore N, Mishra BB, Tiwari V, Tripathi V. An account of phytochemicals from Plumbago zeylanica (Family: Plumbaginaceae): a natural gift to human being. Chron Young Sci. 2012;3(3):178–198. [Google Scholar]
- 45.Swarnavalli GCJ, Joseph V, Kannappan V, Roopsingh D. A simple approach to the synthesis of hexagonal-shaped silver nanoplates. J Nanomater. 2011;2011:825637. [Google Scholar]
- 46.Gao XH, Wei LQ, Wang J, Xu BS. Green synthesis of starch-stabilized silver nanoparticles and their antibacterial properties. Adv Mater Res. 2011:236–238. 1945–1948. [Google Scholar]
- 47.Lee GP, Bignell LJ, Romeo TC, et al. The citrate-mediated shape evolution of transforming photomorphic silver nanoparticles. Chem Commun. 2010;46(41):7807–7809. doi: 10.1039/c0cc02580a. [DOI] [PubMed] [Google Scholar]
- 48.Jiang XC, Chen WM, Chen CY, Xiong SX, Yu AB. Role of temperature in the growth of silver nanoparticles through a synergetic reduction approach. Nanoscale Res Lett. 2011;6(1):32. doi: 10.1007/s11671-010-9780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zabetakis K, Ghann WE, Kumar S, Daniel M. Effect of high gold salt concentrations on the size and polydispersity of gold nanoparticles prepared by an extended Turkevich-Frens method. Gold Bull. 2012;45(4):203–211. [Google Scholar]
- 50.Chandran SP, Chaudhary M, Pasricha R, Ahmad A, Sastry M. Synthesis of gold nanotriangles and silver nanoparticles using Aloe vera plant extract. Biotechnol Prog. 2006;22(2):577–583. doi: 10.1021/bp0501423. [DOI] [PubMed] [Google Scholar]
- 51.Mitra B, Vishnudas D, Sant SB, Annamalai A. Green-synthesis and characterization of silver nanoparticles by aqueous leaf extracts of Cardiospermum helicacabum leaves. Drug Invent Today. 2012;4(2):340–344. [Google Scholar]
- 52.Cytodiagnostics [website on the Internet] Burlington, ON, Canada: Cytodiagnostics; [Accessed March 2, 2013]. Available from: http://www.cytodiagnostics.com. [Google Scholar]
- 53.Shankar SS, Bhargava S, Sastry M. Synthesis of gold nanospheres and nanotriangles by the Turkevich approach. J Nanosci Nanotechnol. 2005;5(10):1721–1725. doi: 10.1166/jnn.2005.192. [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee P, Nandi AK. Bimetallic Au(core)-Ag(shell) nanoparticles from interfacial redox process using poly(o-methoxyaniline) J Colloid Interface Sci. 2010;344(1):30–36. doi: 10.1016/j.jcis.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 55.Klein J. Probing the interactions of proteins and nanoparticles. Proc Natl Acad Sci U S A. 2007;104(7):2029–2030. doi: 10.1073/pnas.0611610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costertan JW, Montataro L, Arciola R. Biofilm in implant infections: its production and regulation. Int J Artif Organs. 2005;28(11):1062–1068. doi: 10.1177/039139880502801103. [DOI] [PubMed] [Google Scholar]
- 57.Pour NK, Dusane DH, Dhakephalkar PK, Zamin FR, Zinjarde SS, Chopade BA. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol Med Microbiol. 2011;62(3):328–338. doi: 10.1111/j.1574-695X.2011.00818.x. [DOI] [PubMed] [Google Scholar]
- 58.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 59.National Institutes of Health Targeted research on oral microbial biofilms. 1998. [Accessed April 3, 2013]. Available from: http://grants.nih.gov/grants/guide/rfa-files/RFA-DE-98-006.html.
- 60.Rodrigues L, van der Mei HC, Teixeira JA, Oliveira R. Biosurfactant from Lactococcus lactis 53 inhibits microbial adhesion on silicone rubber. Appl Microbiol Biotechnol. 2004;66(3):306–311. doi: 10.1007/s00253-004-1674-7. [DOI] [PubMed] [Google Scholar]
- 61.Augustine N, Ali-Vehmas T, Atroshi F. Assesment of enzymatic cleaning agents and disinfectants against bacterial biofilms. J Pharm Pharm Sci. 2004;7(1):55–64. [PubMed] [Google Scholar]
- 62.Thawal ND, Yele AB, Sahu PK, Chopade BA. Effect of a novel podophage AB7-IBB2; Poster presented at: Second International Translational Nanomedicine Conference; July 26–28, 2013; Boston, MA. [Google Scholar]
- 63.Chopade BA, Ghosh S, Jagtap S, et al. Dioscorea bulbifera mediated synthesis of AgNPs exhibiting novel antibiofilm and antileishmanial activity; International Translational Nanomedicine: ITNANO; July 26–28, 2013; Northeastern University, Boston, USA. Abstract 59. [Google Scholar]
- 64.Shrestha A, Shi Z, Neoh KG, Kishen A. Nanoparticulates for antibiofilm treatment and effect of aging on its antibacterial activity. Basic Res Technol. 2010;36(6):1030–1035. doi: 10.1016/j.joen.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 65.Kalishwaralal K, BaraManiKanth S, Pandian SR, Deepak V, Gurunathan S. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf B Biointerfaces. 2010;79(2):340–344. doi: 10.1016/j.colsurfb.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 66.Chopade BA, Patwardhan RB, Dakephalkar PK. Acinetobacter infections in India: genetic and molecular biology studies and some approaches to the problem. In: Kumar Sushil, Sen AK., editors. Tropical Diseases: Molecular Biology and Control strategies. New Delhi: Council of Scientific and Industrial Research; 1994. pp. 704–716. [Google Scholar]
- 67.Patil JR, Jog NR, Chopade BA. Isolation and characterization of Acinetobacter from upper respiratory tract of healthy humans and demonstration of lectin activity. Indian J Med Microbiol. 2001;19(1):30–35. [Google Scholar]
- 68.Pardesi KR, Yavankar SP, Chopade BA. Plasmid distribution and antimicrobial susceptibility patterns of Acinetobacter genospecies from healthy skin of a tribal population in western India. Indian J Med Res. 2007;125(1):79–88. [PubMed] [Google Scholar]
- 69.Patil JR, Copade BA. Studies on bioemulsifier production by Acinetobacter strains isolated from healthy human skin. J Appl Microbiol. 2000;91(2):290–298. doi: 10.1046/j.1365-2672.2001.01382.x. [DOI] [PubMed] [Google Scholar]
- 70.Robenshtok E, Paul M, Leibovici L, et al. The significance of Acinetobacter baumannii bacteraemia compared with Klebsiella pneumoniae bacteraemia: risk factors and outcomes. J Hosp Infect. 2006;64(3):282–287. doi: 10.1016/j.jhin.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 71.Sunenshine RH, Wright MO, Maragakis LL, et al. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg Infect Dis. 2007;13(1):97–103. doi: 10.3201/eid1301.060716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect Dis. 2010;10(196) doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oh YJ, Jo W, Yang Y, Park S. Influence of culture conditions on Escherichia coli O157: H7 biofilm formation by atomic force microscopy. Ultramicroscopy. 2007;107(10–11):869–874. doi: 10.1016/j.ultramic.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 74.Dufrene YF. Using nanotechniques to explore microbial techniques. Nat Rev Microbiol. 2004;2(6):451–460. doi: 10.1038/nrmicro905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table of contents
| Serial number | Contents | Page number |
|---|---|---|
| 1. | Supplementary figure 1 HPTLC fingerprint of PZRE. (A) PZRE ethyl acetate extract, and (B) standard mixture of plumbagin (P), diosgenin (D), quercetin (Q) and catechin (C). | 2650 |
| 2. | Supplementary figure 2 Bioreduction of AuNPs from PZRE at varying reaction temperatures and concentrations of the salts as a function of time. (A) Time course of AuNPs synthesis against reaction temperature, and (B) time course of AuNPs formation at different HAuCl4 concentration. | 2650 |
| 3. | Supplementary figure 3 Determination of purity, size and nature of the AuNPs synthesized by PZRE at 37°C. (A) Representative spot energy-dispersive spectrum, (B) histogram of size distribution, (C) zeta potential analysis, and (D) representative X-ray diffraction profile of thin film. | 2651 |
| 4. | Supplementary figure 4 Determination of purity, size and nature of the AgNPs synthesized using 1% trisodium citrate. (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution. | 2651 |
| 5. | Supplementary figure 5 Determination of purity, size and nature of the AuNPs synthesized using 1% trisodium citrate. (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution. | 2652 |
| 6. | Supplementary figure 6 Determination of purity, size and nature of the AgAuNPs synthesized using 1% trisodium citrate. (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution. | 2652 |
| 7. | Supplementary figure 7 AFM micrographs for biofilm disruption. (A) A. baumannii biofilm disruption using AgNPs using AFM, and (B) amplitude mode of AFM image. | 2653 |
| 8. | Materials and methods for cytotoxicity assay | 2653 |
| 9. | Supplementary figure 8 Cytotoxicity of the nanoparticles synthesized from PZRE and 1% trisodim citrate determined using MCF7 cells showing no inhibition upto the concentration of 0.5 mg/well. | 2653 |
Abbreviations: HPTLC, high-performance thin-layer chromatography; PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AuNPs, gold NPs; HAuCl4, chloroauric acid; AgAuNPs, bimetallic NPs; AFM, atomic force microscopy.
HPTLC fingerprint of PZRE.
Notes: (A) PZRE ethyl acetate extract, and (B) standard mixture of plumbagin (P), diosgenin (D), quercetin (Q) and catechin (C).
Abbreviations: HPTLC, high-performance thin-layer chromatography; PZRE, Plumbago zeylanica root extract.
Bioreduction of AuNPs from PZRE at varying reaction temperatures and concentrations of the salts as a function of time.
Notes: (A) Time course of AuNPs synthesis against reaction temperature, and (B) time course of AuNPs formation at different HAuCl4 concentration.
Abbreviations: PZRE, Plumbago zeylanica root extract; AuNPs, gold nanoparticles; HAuCl4, chloroauric acid; h, hours.
Determination of purity, size and nature of the AuNPs synthesized by PZRE at 37°C.
Notes: (A) Representative spot energy-dispersive spectrum, (B) histogram of size distribution, (C) zeta potential analysis, and (D) representative X-ray diffraction profile of thin film.
Abbreviations: PZRE, Plumbago zeylanica root extract; AuNPs, gold nanoparticles.
Determination of purity, size and nature of the AgNPs synthesized using 1% trisodium citrate.
Notes: (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution.
Abbreviation: AgNPs, silver nanoparticles.
Determination of purity, size and nature of the AuNPs synthesized using 1% trisodium citrate.
Notes: (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution.
Abbreviation: AuNPs, gold nanoparticles.
Determination of purity, size and nature of the AgAuNPs synthesized using 1% trisodium citrate.
Notes: (A) Representative X-ray diffraction profile of thin film, (B) representative spot energy-dispersive spectrum, (C) transmission electron micrograph, and (D) histogram of size distribution.
Abbreviation: AgAuNPs, bimetallic nanoparticles.
AFM micrographs for biofilm disruption.
Notes: (A) A. baumannii biofilm disruption using AgNPs using AFM, and (B) amplitude mode of AFM image.
Abbreviations: AgNPs, silver nanoparticles; AFM, atomic force microscopy.
Cytotoxicity of the nanoparticles synthesized from PZRE and 1% trisodim citrate determined using MCF7 cells showing no inhibition upto the concentration of 0.5 mg/mL.
Abbreviations: PZRE, Plumbago zeylanica root extract; AgNPs, silver nanoparticles; AuNPs, gold NPs; AgAuNPs, bimetallic NPs; Chem, chemically synthesized.