Abstract
Background
Caenorhabditis elegans hermaphrodites are capable of producing hundreds of progeny. However, genetic and environmental factors can keep many animals from attaining their full reproductive potential. In these situations, efficient use of any functional gametes becomes more important for reproductive success. To learn about this aspect of C. elegans reproductive biology, we examined oocyte production and sperm utilization patterns in a unique collection of semi-fertile sperm function mutants.
Results
In the mutants examined here, broods can be very small but sperm induced high levels of ovulation. Ovulation rates reach maximum levels between the first and second day of adulthood. Ovulations rates remain high during the reproductive period and gradually decline with age. These results further demonstrate a decoupling of the ability of sperm to fertilize oocytes and induce ovulation. We also observe that in our semi-fertile mutants the peak of successful fertilization events precedes the bulk of oocyte production. Mixing populations of functional and nonfunctional sperm under conditions without sperm competition also shows that functional sperm are utilized efficiently. Although overall brood size can be similar for different mutant strains, slight differences in the pattern of sperm utilization in these strains can lead to significant differences in resource utilization and population growth.
Conclusions
This study represents the first detailed description of oocyte and progeny production patterns over the entire reproductive period for wild-type and fertility impaired strains of C. elegans. The phenotype of our mutants provide an ideal system for studying sperm utilization patterns since they only affect one major process, the ability to fertilize oocytes. In semi-fertile mutants, the nature of the reproductive process and/or specific molecular mechanisms ensures that any functional sperm are utilized quickly. Only a fraction of the sperm produced by our semi-sterile mutants are functional as opposed to every sperm having a low but equal chance of fertilizing an oocyte. In addition to the number of progeny produced, the pattern of progeny production can have an important influence on the dynamics of population growth.
Background
The C. elegans strain considered "wild type" – N2, exists primarily in the hermaphroditic condition, although males are produced spontaneously (due to chromosomal nondisjunction) at an extremely low frequency [1,2]. During its fourth (and final) larval stage, the reproductive system of the hermaphrodite produces its lifetime's supply of sperm [3]. As development progresses, the germline undergoes a switch, and for the remainder of the animal's reproductive life will produce only oocytes [4]. In this organism, it is not the supply of oocytes which limits the number of progeny that each hermaphrodite produces, but rather, the number of functional sperm that are made in the larval stage [4,5].
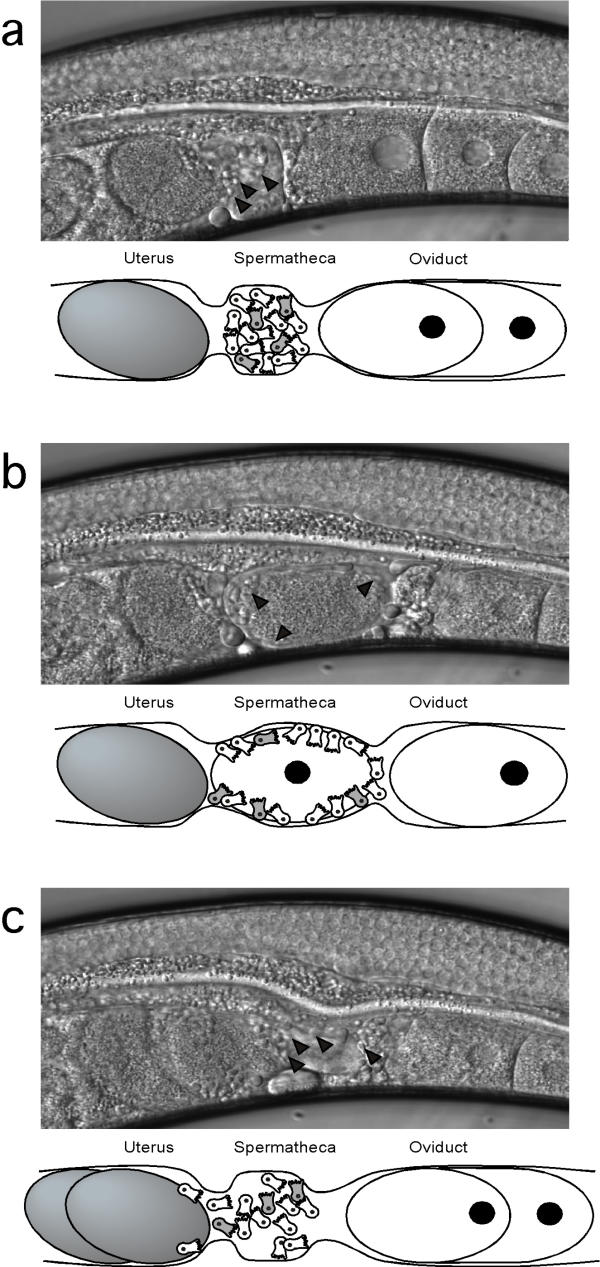
The process of fertilization in C. elegans involves the dilation of the spermatheca, which allows the entry of the oocyte into this structure (Fig. 1). The spermatheca stores all of the sperm, which are amoeboid in nature. Sperm are required to induce high levels of oocyte maturation and ovulation [6,7]. Fertilization occurs within the spermatheca, and the zygote then enters the uterus. As the zygote moves into the uterus, it displaces many sperm from the spermatheca. These sperm must migrate back into the spermatheca in order to be utilized later. If fertilization was unsuccessful in the spermatheca, the unfertilized oocyte begins to endoreplicate its DNA and is eventually laid as such. There is no fertilization in the uterus. Thus, there is only a brief window of opportunity when the oocyte can be fertilized.
Figure 1.
Overview of the fertilization process in C. elegans (each panel consists of the Nomarski image and an explanatory cartoon). (a) shows an oocyte ready for ovulation into the spermatheca, where the sperm are stored. The mutants discussed in the paper produce a few fertilization-competent sperm (shown in the cartoons in grey) and mostly non-functional sperm (shown in white). In (b) the mature oocyte is ovulated into the spermatheca, which is the site of fertilization. Each oocyte will potentially contact all of the sperm, both functional and non-functional that are stored in the spermatheca simultaneously. After a short interval, the oocyte is pushed into the uterus (c) and any sperm that have also been displaced from the spermatheca into the uterus will make their way back into the spermatheca and await the next ovulation event. Although some of these displaced sperm might contact any unfertilized oocytes in the uterus, no fertilization occurs outside the spermatheca. In Nomarski images, examples of sperm are indicated by black arrowheads.
When hermaphrodites are mated to males, male sperm will outcompete the hermaphrodite's own sperm and will preferentially fertilize the oocytes [8]. This phenomenon is referred to as sperm competition, and in the nematode C. elegans specifically refers to the competition effects seen between male and hermaphrodite sperm. Unlike in other species, C. elegans exhibits no competition between sperm from different males [9]. Hermaphrodites mated to multiple males show no male precedence effects. The ability of male-derived sperm to outcompete hermaphrodite-derived sperm stems from intrinsic physiological differences between male and hermaphrodite sperm. It has been shown that male sperm are larger than hermaphrodite sperm, and that this size difference is important in the context of sperm competition in C. elegans [9-11]. Further, the ability of male sperm to outcompete hermaphrodite sperm has been shown to be independent of their ability to fertilize the oocyte [12].
A class of mutations has been defined that result in the production of sperm that are incapable of fertilizing the oocyte [5,13]. This is despite that fact that mutant sperm are produced in normal quantities, appear morphologically normal, activate to form crawling spermatozoa, localize correctly to the spermatheca and come in contact with oocytes [5,14]. Thus, these mutations seem to affect molecules that are required by the sperm specifically for fertilization and not for sperm development. These mutants are collectively called the spe-9 class, after the first of the corresponding genes to be cloned. While some alleles of these genes are completely sterile at all temperatures, others show temperature-sensitive sterility and still others show partial sterility (leaky sterile) at various culture conditions (Kadandale & Singson, unpublished observations).
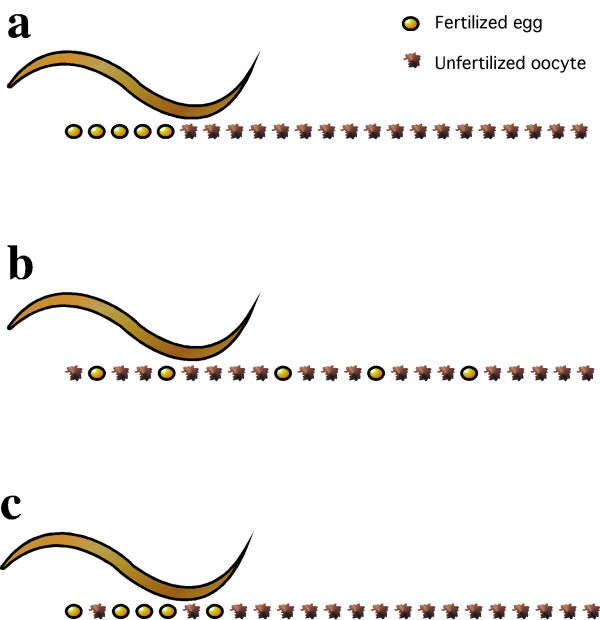
The nature of the C. elegans reproductive tract gives essentially every sperm in the spematheca the chance to contact the egg simultaneously (Fig. 1) [5,8]. This leads to extremely efficient use of sperm. In wild type animals, sperm are utilized at or very close to unit efficiency until sperm supplies are depleted [5,8,13]. In our collection of semifertile mutants, only a few successful fertilization events occur for each animal as opposed to the hundreds seen in wild type animals. Given that our mutants are only weakly fertile, we can hypothesize a number of possible sperm utilization patterns (Fig. 2). 1) All functional sperm are utilized immediately and with 100% efficiency (Fig. 2A). 2) Every sperm has a low chance of fertilizing a passing oocyte so the pattern of successful fertilization events is completely stochastic (Fig. 2B). The odds of a fertilization event would go up with an increase in the number of ovulations. Therefore the pattern of fertilization events would mirror that of ovulations. 3) Alternatively, sperm utilization patterns could be biased to one of the two patterns above but not a perfect match (Fig. 2C). Here we investigate patterns of ovulation and sperm utilization in our spe-9 class semisterile mutant collection where the majority of the sperm-egg interactions are unsuccessful.
Figure 2.
Expected patterns of fertilization in leaky fertilization-defective mutants of C. elegans. In cases where only a few functional sperm are made, we can expect completely efficient utilization of sperm (a), completely random fertilization patterns (b) or an intermediate, with a bias towards efficient utilization, due to issues such as sperm being swept into the uterus (c).
Results and discussion
Wild-type sperm utilization and ovulation patterns
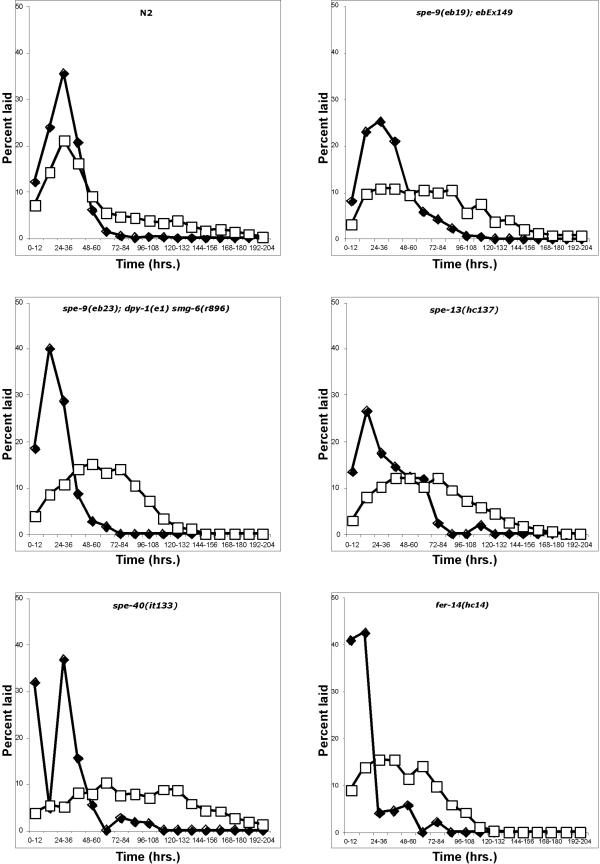
Wild-type (N2) progeny production under standard culture conditions (20°C, see Methods) peaks sharply 24–36 hours after the L4 larval stage (Fig. 3). The peak in ovulation events mirrors this pattern. Sperm are utilized very efficiently and almost 100% of the progeny are produced by 72 hours. When sperm are depleted, progeny production stops and oocyte production drops to basal levels. Eventually, as the worm ages, even basal oocyte production levels decrease. The initial lag before peak oocyte and progeny production corresponds to the time required to transition from the L4 larval stage (time = 0) to reproductively mature adults.
Figure 3.
Sperm utilization patterns in various strains of C. elegans. The strains tested are labeled on the graphs. Solid diamonds represent percentage of fertilized eggs laid and unfilled squares represent percentage of total ovulations at each time point.
Sperm utilization patterns in leaky spe-9 class mutants
We examined sperm utilization and ovulation patterns in mutant strains with low fertility at 20°C (spe-9, spe-13, spe-40 and fer-14) (Fig. 3). In the spe-9 class mutants, non-functional sperm remain in the spermatheca and continue to induce high levels of oocyte maturation and ovulation (Table 1) [5-7,14]. These levels of ovulation also drop off steadily as the animals age (Fig. 3). In all cases, we see that despite the fact that brood size in these mutants can be very low (Table 1), successful fertilizations occur early in the reproductive life of the animal (Fig. 3). Progeny production peaks at roughly the same time as wild-type. Furthermore, in these mutants, we also see that the peak of progeny production now precedes the peak in ovulations. Our data most closely resemble the scenario depicted in figure 2C with a very strong bias towards the pattern in figure 2A. Thus, there appears to be a robust mechanism that operates within the reproductive tract of the hermaphrodite that ensures that functional sperm are utilized quickly. In "real life" since many complicated factors are likely to come into play (e.g. the number of functional sperm temporarily displaced from the spermatheca by passing oocytes) we would not expect to see the perfectly efficient utilization pattern depicted in figure 2A. Finally, since we see this strong bias to early progeny production that does not mirror ovulation patterns, we conclude that these mutants produce two populations of sperm. One population are the few functional sperm and the other population are the non-functional sperm that will never be able to fertilize an oocyte.
Table 1.
Brood sizes of various mutants used in this study.
| Straina | ||||||
| N2 | spe-9(eb19); ebEx149 | spe-9(eb23); dpy-1, smg-6 | spe-13 | spe-40 | fer-14 | |
| brood size | 244 (35) | 136 (51) | 74 (22) | 9 (3) | 3 (2) | 4 (3) |
| total ovulations | 446 (57) | 325 (89) | 438 (56) | 474 (61) | 468 (75) | 388 (80) |
aAll data are for broods at 20°C. Numbers in brackets refer to standard deviation.
Two-male mate experiments
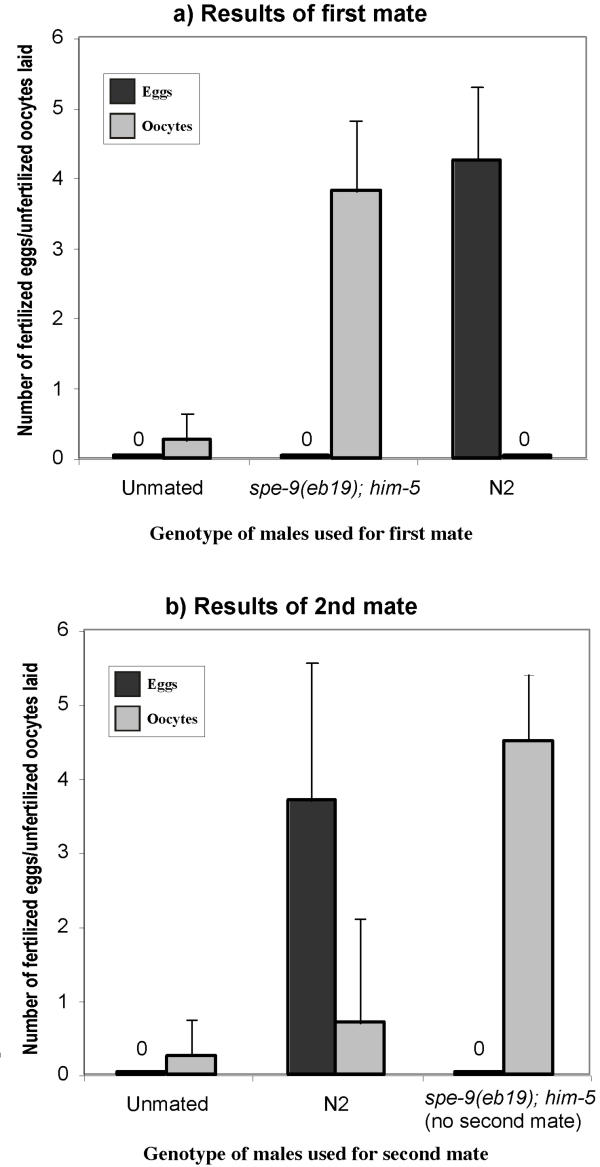
The results above suggest that there are two populations of sperm in our mutants (functional and non-functional). We wanted to see if we could introduce two populations of sperm into a feminized hermaphrodite (fem-1 mutants contain no endogenous sperm) and see the efficient utilization of a functional population of sperm without interference from a non-functional population. For this, we used a strategy involving the mating of the hermaphrodite to multiple males. Previous work has established that there are no sperm precedence effects when hermaphrodites are mated sequentially to multiple males. There also appear to be no male-male sperm competition effects in C. elegans [9,10]. We first introduced sperm from spe-9(eb19) males into fem-1(hc17) hermaphrodites by simply mating males to hermaphrodites. Since spe-9(eb19) is a null allele, all the sperm from such males are completely non-functional for fertilization [12,14]. An increase in the ovulation rates (Fig. 4a) of the fem-1(hc17) animals indicates successful sperm transfer, since ovulation is known to require a sperm-derived factor [7]. When such successfully mated fem-1(hc17) animals are subsequently mated to wild-type animals, we see an immediate shift towards laying fertilized eggs (Fig. 4b), as opposed to an equal number of eggs and unfertilised oocytes (or greater number of unfertilised oocytes). This shift is quite remarkable, considering the short time frame of this assay (see Methods). In animals that are not second-mated to wild type males, the ovulation rates are higher than the basal fem-1(hc17) level, indicating the persistence of the spe-9(eb19) sperm for the duration of the assay (Fig. 4b). In a similar experiment, it was found that reversing the order of the matings (i.e., N2 first, followed by spe-9(eb19) males) resulted in the production of only eggs, even after the second mate (results not shown).
Figure 4.
Sequential male mates to look at sperm utilization patterns in the absence of sperm competition. Unfertilized oocytes are represented by unfilled bars, the solid bars are fertilized eggs laid. Ovulation rates increase comparably after fem-1 hermaphrodites are mated to either spe-9 mutant males, or N2 controls (a). (b) shows results after hermaphrodites previously mated to spe-9 mutant males are re-mated to N2 males. As can be seen, there is an immediate shift towards utilization of the N2 male-derived sperm and the production of progeny. The animals not mated a second time to any males continue to show elevated ovulation rates (as compared to unmated controls), indicating the persistence of sperm for the duration of the experiment.
Food clearing assay
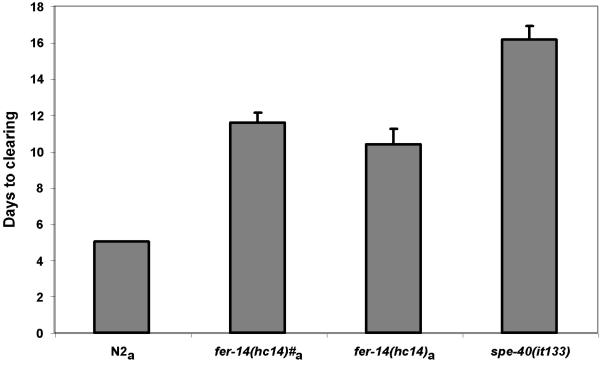
We have noted that some strains seem to display varying patterns of sperm utilization even though the bulk of their fertilization events occur early (Fig. 3, e.g. compare spe-40 to other strains). Therefore, we wished to determine if some of the variation in sperm utilization patterns seen in our strains could have an impact on population growth. Two of the mutants used in this study, spe-40 and fer-14 had similar brood sizes (See Table 1). The spe-40 strain seemed to have a more stochastic pattern of progeny production than other strains. We wanted to test whether this difference was biologically significant. For this experiment, we set up a standard food clearing assay [15,16] between fer-14 and spe-40. This assay has been previously shown to be a good measure of successful resource utilization and population growth [16]. Population growth is measured by the ability to clear a food resource from a culture dish. The spe-40 animals took 4–5 days longer to clear a spot of food than the fer-14 animals (Fig. 5), although their total brood sizes are not significantly different (p < 0.1). In order to confirm this, we set up the assay using only one fer-14 animal versus two spe-40 animals, and even in this case, the spe-40 animas still took significantly longer to clear the food (Fig. 5). We also observe a qualitative difference between plates cleared by spe-40 or fer-14. In the case of spe-40, the cleared plates have mostly older animals on them, while in the case of fer-14, the plates have some older worms, and many L1 and L2 animals that can potentially enter the dauer pathway. Worms in the L2 larval stage subjected to starvation, high population density, or high temperature can enter the dauer stage. This is a quiescent condition that is resistant to prolonged periods of harsh environmental conditions. When transported to more ambient surroundings, the worms can exit the dauer state and become reproductively viable adults [17]. Having more of a population capable of dauering when food resources are exhausted could lead to better long-term survival prospects for that population. The differences in food clearing cannot be attributed to developmental differences between the two strains, since the time they take to go from egg to adult to egg is not significantly different from each other (Table 2). Thus, differences in sperm utilization patterns are reflected in differential resource utilization times, and would appear to have biological relevance. It is not solely progeny number that influences the ability of a population to maximally utilize the available resources, but also the temporal pattern in which these progeny are produced.
Figure 5.
Food clearing assay. Worms of the indicated genotype were used to set up a food clearing assay. The "#" indicates plates where only one worm was used. In all other cases, two worms were used for the assay. spe-40(it133) takes a significantly longer time to clear a spot of food than fer-14(hc14) animals. p-Value is < 0.001 for worms marked with "a" (significantly different from the spe-40 animals, Student's t-Test).
Table 2.
Developmental time is the same for two spe-9-class mutants used in resource utilization assay.
| Straina | fer-14 (14) | spe-40 (17) |
| developmental timeb | 64.5 ± 0.6 | 64.6 ± 0.5 |
aNumbers in brackets refer to the number of worms tested. bNumber of hours taken for 50% of the eggs to develop into adults that begin laying eggs or unfertilized oocytes ± standard deviation.
In order to rapidly utilize functional sperm, it is reasonable to assume that the oocyte must have some means of distinguishing these, since it comes in contact, concurrently, with almost the whole population of sperm within the spermatheca (Fig. 1). The molecular components encoded by genes like spe-40 could influence the efficiency of this process. While our assays only measure quantitative differences in resource utilization, we can also imagine how such a method of discriminating between populations of sperm can also be used to produce qualitatively different offspring.
Conclusions
There appears to be a robust mechanism that operates within the reproductive tract of the C. elegans hermaphrodite that ensures that functional sperm are utilized quickly. Efficient sperm storage and presentation to the newly ovulated oocyte is likely to contribute to this efficiency and is an inherent feature of worm reproductive biology. In our mutants, only a fraction of the sperm produced by each worm are functional. Further, any functional sperm are used efficiently in all strains tested. However, small variations in this efficiency exits between strains. These small variations in sperm usage patterns can lead to significant differences in population growth, dynamics and resource utilization. The underlying molecular events and mechanism involved could have important influences on sexual selection and the reproductive success of animals in the wild.
Methods
Worm strains
fer-14(hc14), spe-13(hc137), and spe-40(it133) are all spe-9 class mutants that have not yet been cloned [5]. fem-1(hc17) has been described elsewhere [18]. Several alleles of spe-9 are not completely sterile [14]. The spe-9(eb23); dpy-1(e1) strain has a larger brood size (Kadandale & Singson, unpublished observations) when in the background of a smg-6 mutation. smg-6 is known to be required for mRNA surveillance and nonsense mediated decay [19-21]. The observation that the fertility of the eb23 allele increases in the smg background suggests that the semi-fertility of these mutants is due to read-through translation. spe-9(eb19); ebEx149 is a strain of worms that carry a transgene bearing the cosmid C17D12 and the dominant phenotypic marker, rol-6. Although C17D12 rescues spe-9 mutants, it has a truncated form of spe-9 that lacks the cytoplasmic tail and synthesizes less SPE-9 than can be detected by antibody staining (Zannoni and Singson, unpublished observations). Germline expression of transgenic extrachromosomal arrays is known to be poor [22,23], and this could explain why so little SPE-9 is made in these worms. The spe-9(eb19); him-5; ebEx149 strain carried the him-5 mutation. This mutation causes males to be produced at high frequency, but does not have any adverse effects on sperm [24]. At least 10 animals were used for all brood, ovulation and developmental time experiments (see below).
Sperm utilization patterns
Worms homozygous for the required mutation were grown up as a stock according to standard protocols and culture conditions (20°C see below) [1]. 20–30 young adult worms were picked to fresh plates and allowed to lay eggs for one hour. In cases where the homozygous stock did not produce sufficient progeny in one hour, heterozygotes were picked, and only their sterile progeny (which would be homozygous for the desired mutation) were used. The adult worms were then removed and the eggs were allowed to hatch and develop until they reached the L4 larval stage. At this time the worms were picked individually to fresh plates. The worms were transferred to fresh plates every twelve hours and the number of fertilized eggs and unfertilized oocytes that had been laid were counted for each worm. The first twelve-hour period in which either fertilized or unfertilized eggs were seen was taken as the 0–12 hour time point. Since L4 larvae were picked and had to still initiate zygote production, there was typically a short lag in ovulation rates for the 0–12 hour time point. The mutants used in this study were chosen since their semi-fertility was not completely temperature-sensitive, so that all brood sizes, sperm utilization patterns and total ovulation patterns could be checked at 20°C and compared relative to each other. Total ovulations were calculated as the sum of fertilized eggs and unfertilized oocytes laid at each time point.
Sequential two male mates
These were performed essentially as described [9]. In short, single fem-1(hc17) animals were picked to fresh plates and each was mated to four spe-9(eb19); him-5 males for three hours. The males were then removed and the fem-1 animals were allowed to lay eggs for one hour, after which they were transferred to fresh plates. Successful mating was scored by looking for an increase in the basal ovulation rate of fem-1 hermaphrodites. Half of these were not second-mated to any males, to check for persistence of sperm for the duration of the assay. The remaining animals were mated to four N2 males each for three hours. The males were removed and the hermaphrodites were allowed to lay eggs for one hour. The plates were checked for fertilized eggs or unfertilized oocytes.
Plate clearing assay
The plate clearing assay was modified from the method previously described [15]. Every plate was seeded with an equal amount of bacterial (E. coli – OP50) culture. Either two worms (spe-40(it133), fer-14(hc14) or N2) or one worm (fer-14) of each strain was picked to these plates as L4 animals and the time taken for each plate to be cleared of food was noted.
Developmental time
Heterozygous fer-14(hc14)/+ or spe-40(it133)/+ animals were picked and allowed to lay eggs for one hour. The adult worms were then picked off and the eggs allowed to hatch. Once they hatched, the young worms were transferred to individual plates and the time taken for them to lay their first egg/unfertilized oocyte was noted. Animals were scored as homozygous mutants or wild type (or heterozygotes) based on their fertility.
Authors' contributions
PK conducted all of the experiments reported in this manuscript. AS was responsible for some of the original strain constructions and observations leading to this study. AS was the primary investigator and coordinated the research efforts. The manuscript was written by PK and AS.
Acknowledgments
Acknowledgements
We would like to thank Chi-hua Chiu, Jody Hey, Monica Driscoll, Mohamed Noor and Iris Dihlmann for critical comments on the manuscript. We thank members of the lab for helpful discussions. The Caenorhabditis Genetics Center provided some nematode strains, and it is funded by the National Institutes of Health National Center for Research Resources (NCRR). Work in the lab was supported by a grants to A.W.S. from the National Institutes of Health (R01 GM63089-01), the National Science Foundation (IBN-0000182), and a Charles & Johanna Busch Biomedical Fund. P.K. was supported by a Charles & Johanna Busch fellowship in molecular biology.
Contributor Information
Pavan Kadandale, Email: kadandale@waksman.rutgers.edu.
Andrew Singson, Email: singson@waksman.rutgers.edu.
References
- Brenner Sydney. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Barbara J. Sex determination and X chromosome dosage compensation. In: Donald L Riddle, Thomas Blumenthal, Barbara J Meyer and James R Priess, editor. C Elegans II. Cold Spring Harbor, Cold Spring Harbor Laboratory; 1997. pp. 209–240. [PubMed] [Google Scholar]
- Ward Samuel, Miwa Johji. Characterization of temperature-sensitive, fertilization-defective mutants of the nematode Caenorhabditis elegans. Genetics. 1978;88:285–303. doi: 10.1093/genetics/88.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Singson A. Every sperm is sacred: fertilization in Caenorhabditis elegans. Dev Biol. 2001;230:101–109. doi: 10.1006/dbio.2000.0118. [DOI] [PubMed] [Google Scholar]
- Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17:187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Ward Samuel, Carrel John S. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- LaMunyon Craig W., Ward Samuel. Larger sperm outcompete smaller sperm in the nematode Caenorhabditis elegans. Proceedings of the Royal Society, Series B. 1998;265:1997–2002. doi: 10.1098/rspb.1998.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Sperm precedence in a hermaphroditic nematode Caenorhabditis elegans is due to competitive superiority of male sperm. Experientia. 1995;51:817–823. doi: 10.1007/BF01922436. [DOI] [PubMed] [Google Scholar]
- LaMunyon CW, Ward S. Evolution of sperm size in nematodes: sperm competition favours larger sperm. Proceedings of the Royal Society, Series B. 1999;266:263–267. doi: 10.1098/rspb.1999.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Hill KL, L'Hernault SW. Sperm competition in the absence of fertilization in Caenorhabditis elegans. Genetics. 1999;152:201–208. doi: 10.1093/genetics/152.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault Steven W., Shakes Diane C., Ward Samuel. Developmental genetics of chromosome I spermatogenesis-defective mutants in the nematode Caenorhabditis elegans. Genetics. 1988;120:435–452. doi: 10.1093/genetics/120.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson Andrew, Mercer Kristina B., L'Hernault Steven W. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell. 1998;93:71–79. doi: 10.1016/S0092-8674(00)81147-2. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Barnes TM. More is not better: brood size and population growth in a self-fertilizing nematode. Proc R Soc Lond B Biol Sci. 1991;246:19–24. doi: 10.1098/rspb.1991.0119. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Kuwabara PE, Corneliussen B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr Biol. 2000;10:1615–1618. doi: 10.1016/S0960-9822(00)00867-8. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Albert PS. Genetic and environmental regulation of dauer larva development. In: Priess JR, editor. C elegans II. Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. [PubMed] [Google Scholar]
- Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali BM, Anderson P. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol Gen Genet. 1998;260:176–184. doi: 10.1007/s004380050883. [DOI] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-F. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault SW, Roberts TM. Cell biology of nematode sperm. Methods Cell Biol. 1995;48:273–301. doi: 10.1016/s0091-679x(08)61392-8. [DOI] [PubMed] [Google Scholar]