Abstract
Lactobacillus gasseri LA39 and L. reuteri LA6 isolated from feces of the same human infant were found to produce similar cyclic bacteriocins (named gassericin A and reutericin 6, respectively) that cannot be distinguished by molecular weights or primary amino acid sequences. However, reutericin 6 has a narrower spectrum than gassericin A. In this study, gassericin A inhibited the growth of L. reuteri LA6, but reutericin 6 did not inhibit the growth of L. gasseri LA39. Both bacteriocins caused potassium ion efflux from indicator cells and liposomes, but the amounts of efflux and patterns of action were different. Although circular dichroism spectra of purified bacteriocins revealed that both antibacterial peptides are composed mainly of α-helices, the spectra of the bacteriocins did not coincide. The results of d- and l-amino acid composition analysis showed that two residues and one residue of d-Ala were detected among 18 Ala residues of gassericin A and reutericin 6, respectively. These findings suggest that the different d-alanine contents of the bacteriocins may cause the differences in modes of action, amounts of potassium ion efflux, and secondary structures. This is the first report that characteristics of native bacteriocins produced by wild lactobacillus strains having the same structural genes are influenced by a difference in d-amino acid contents in the molecules.
Bacteriocins are antimicrobial proteins or protein complexes that act mainly against related bacterial species. These components from lactic acid bacteria (LAB) have been classified into four classes, I to IV (16, 25). Recently, many class II bacteriocins, small hydrophobic peptides that do not contain modified amino acids such as lanthiothine, have been reported, and they have been divided into the categories IIa, IIb, IIc, and IId (21, 25). These LAB bacteriocins and the producing strains isolated from humans and foods are expected to be food preservatives and probiotics for preventing the growth of harmful bacteria in food and the human intestine.
The Lactobacillus acidophilus group of LAB is widely used in fermented milk products, and the intake of these bacteria may have beneficial effects on human health (2, 20). This group of bacteria, including L. gasseri, which is found in the human intestine, has been classified on the basis of similarities in rRNA sequences and compositions of the cell walls into six subgroups (9, 11, 18). Many strains in this group have been shown to be bacteriocin producers (6, 12, 19, 23).
In our laboratory, more than 100 strains of L. acidophilus group LAB isolated from human feces were screened to identify bacteriocin-producing strains (10, 15, 33). The bacteriocin gassericin A, produced by L. gasseri LA39 isolated from feces of a 4-month-old human infant, had a broad spectrum and high activity. This bacteriocin, which is active against the food-borne pathogenic bacteria Listeria monocytogenes, Bacillus cereus, and Staphylococcus aureus (10), was purified and sequenced by cloning of its structural gene (gaaA). Determination of the complete primary chemical structure of gassericin A revealed that this bacteriocin has a cyclic structure linking at N- and C-terminal amino acids and is a class II bacteriocin consisting of 58 amino acid residues (IYWIADQFGIHLATGTARKLLDAMASGASLGTAFAAILGVTLPAWALAAAGALGATA; molecular weight, 5,652 as determined by mass spectrometry) (14).
The heterofermentative L. reuteri LA6, which was isolated from feces of the same human infant when the infant was 2 months of age, produced the bacteriocin reutericin 6 (32). The molecular weight, primary amino acid sequence, and cyclic structure of reutericin 6 were found to be identical to those of gassericin A. PCR amplification of the chromosome DNA of L. reuteri LA6 by using primers that had been designed on the basis of gaaA and sequencing revealed that L. reuteri LA6 has the sequence gaaA with no variations between those primers (14). However, reutericin 6 has weaker activity and a narrower spectrum than gassericin A (10, 32). In this study, we examined modes of action, secondary structures, and d- and l-amino acid compositions to elucidate the structural and functional differences between gassericin A and reutericin 6, bacteriocins that have identical primary structures.
MATERIALS AND METHODS
Bacterial strains and media.
The bacteriocin producers L. reuteri LA6 and L. gasseri LA39 were isolated in our laboratory from the feces of a human infant when the infant was 2 and 4 months age, respectively (14, 32, 33). An indicator strain for bacteriocin, L. delbrueckii subsp. bulgaricus JCM 1002T, was obtained from the Japan Collection of Microorganisms (Riken, Wako, Japan). These strains were propagated twice in lactobacillus MRS broth (Difco Laboratories, Detroit, Mich.) with a 1% (vol/vol) inoculum at 37°C for 18 h. MRS agar medium was prepared by the addition of 1.5% (wt/vol) agar (agar no. 1; Oxoid, Unipath Ltd., Basingstoke, United Kingdom), and MRS soft-agar medium was prepared with 0.7% (wt/vol) agar. A modified MRS broth (DOB-MRS broth) for purification of the bacteriocins was prepared; MRS broth components without Tween 80 and beef extract were dialyzed against water, and 0.1% (vol/vol) oleic acid was added to the diffusate.
Bacteriocin assay.
Bacteriocin activity was assayed by the agar well diffusion method (33). Samples were mixed with 50 mM sterile sodium phosphate buffer (PB), pH 6.8, or a mixture of PB and 50% (vol/vol) 2-propanol (final concentration) and then diluted serially. MRS agar plates (4 mm deep) were overlaid with an MRS soft-agar indicator lawn prepared by the addition of a 1/10-diluted portion of an overnight culture. Wells of 5 mm in diameter were cut, and 65 μl of the sample was poured into each well. One unit of activity (AU) was arbitrarily defined as the reciprocal of the highest twofold dilution that inhibited the growth of the indicator strain.
Purification of bacteriocins.
After incubation at 37°C for 24 h (L. reuteri LA6) and 36 h (L. gasseri LA39) in DOB-MRS broth, culture supernatants (fraction A) were obtained by centrifugation (5,000 × g for 20 min at 4°C) and then filtered through filter paper (model no. 2; Toyo Roshi Kaisha, Ltd., Tokyo, Japan) at 4°C in order to separate oleic acid. The filtrate (fraction B) was dialyzed against water at 4°C. The dialysate (fraction C) was subjected to hydrophobic chromatography with a C4-butyl-Toyopearl column (3 by 9 cm; Tosoh, Tokyo, Japan) by stepwise elution with water and 50% (vol/vol) and 100% methanol, and then the permeate (eluate with water, fraction D) was subjected to reverse-phase chromatography with LiChroprep RP-8 resin (E. Merck, Darmstadt, Germany) by stepwise elution with water, 90% (vol/vol) acetonitrile, and 90% (vol/vol) 2-propanol. Each active fraction (eluate with 90% 2-propanol, fraction E) diluted with water was subjected to the repetition of reverse-phase chromatography with the same resin (E. Merck) by stepwise elution with water; 30, 60, and 90% acetonitrile, and 20, 40, 60, and 90% 2-propanol. Purified bacteriocins were obtained in a 60% 2-propanol fraction (fraction F) and stored at −20°C.
SDS-PAGE and in situ activity assay.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a 4.5% spacer gel and a 20% separating gel (17). After electrophoresis, the gel was stained with Coomassie brilliant blue R-250 (Fluka Chemie AG, Buchs, Switzerland). A protein marker kit with molecular weights of 2,350 to 46,000, 2,512 to 16,949, or 14,000 to 97,000 (Amersham International plc, Little Chalfont, United Kingdom) was used. For detection of bacteriocin activity on a gel, the other half of the gel was used for an in situ activity assay, as described by Daba et al. (7).
Analytical method.
The protein content of each sample was determined by using a micro bicinchoninic acid protein assay reagent kit (Pierce, Rockford, Ill.) with insulin (from bovine pancreas; Wako Pure Chemical Industries, Tokyo, Japan) as a standard peptide.
N-terminal amino acid sequencing.
The purified bacteriocins on a polyvinylidene difluoride (PVDF) membrane (Immobilon-PSQ; Millipore Corp., Bedford, Mass.) were subjected to N-terminal sequence analysis using an automatic protein sequencer (model 473A), an online phenylthiohydantoin analyzer (model 120A), and a data analysis system (model 610A; Applied Biosystems, Inc., Foster City, Calif.).
CD spectroscopy.
Circular dichroism (CD) spectra for the purified bacteriocins (fraction F) were recorded between 190 and 300 nm at room temperature by using a 1-mm-pathlength quartz cell on a JMS-700 spectropolarimeter (JASCO, Tokyo, Japan). Samples were scanned five times, and the data were averaged. A peptide-free control spectrum with 60% (vol/vol) 2-propanol was subtracted, providing the mean residue ellipticity of the peptide. The contents of α-helix, β-sheet, and unordered structures were estimated according to the report of Yang et al. (35).
Analysis of d- and l-amino acid compositions.
Purified bacteriocins (5 nM; fraction F) on a PVDF membrane (Immobilon-PSQ; Millipore Corp.) were hydrolyzed (110°C for 24 h) in 200 μl of 6 N HCl containing 0.2% (vol/vol) phenol. Amino acids from the PVDF membrane were extracted three times by using 100 μl of 0.1 N HCl-30% (vol/vol) methanol and then vacuum dried. The extracts were dissolved with 10 μl of 25 mM sodium borate buffer (pH 10.4), and 10 μl of a solution containing 260 mM N-acetyl-l-cysteine and 170 mM o-phthalaldehyde was added for reaction at 50°C for 30 s (26). Amino acid derivatives dissolved in 80 μl of solvent A (25 mM sodium acetate, pH 6.0) were analyzed by high-performance liquid chromatography using an L-6200 pump (Hitachi Co., Ltd., Tokyo, Japan) and a TSKgel ODS-80TsQA column (2 by 250 mm; Tosoh). Elution was done at 40°C with a linear gradient from 100/0 (solvent A/solvent B [90% {vol/vol} acetonitrile] ratio) to 30/70 (solvent A/solvent B ratio) for 50 min at a flow rate of 0.2 ml/min. Each peak was monitored by an F-1080 fluorescence detector (Hitachi Co., Ltd.) at an excitation wavelength of 340 nm and an emission wavelength of 440 nm, and the detector was linked to a D-5500 data station (Hitachi Co., Ltd.).
pH stability.
The purified bacteriocins (fraction F) in 50% (vol/vol) 2-propanol (0.5 ml) were adjusted to pH 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12 with 1 M NaOH or HCl and were stored at 4°C for 48 h. After adjustment of the solutions to pH 6.8 and the final volumes to 1 ml with 50% 2-propanol, the remaining bacteriocin activity was determined by the agar well diffusion method.
Mode of action.
The effects of the bacteriocins on indicator cells of L. delbrueckii subsp. bulgaricus JCM 1002T were examined as follows. An indicator strain was harvested by centrifugation of an overnight culture (5 ml) in MRS broth, washed twice in the same volume of sterile PB, and resuspended in 5 ml of PB or a mixture of PB and 3% (vol/vol) 2-propanol to be dispensed in aliquots (1 ml each). After addition of PB (1 ml) as blank or crude bacteriocins (fraction C; 1 ml; 120 AU) to each indicator cell suspension, cell viability, cell lysis (optical density), and bacteriocin activity remaining in the suspension were determined after 0, 60, and 120 min of incubation at 37°C. The cell population (number of CFU per milliliter) was measured by plating anaerobically onto MRS agar, and the optical density of 10-fold-diluted samples was determined spectrophotometrically at 650 nm. Amounts of the bacteriocin attached to the indicator cells were indirectly calculated as remainders from control (60 AU/ml) to bacteriocin activity in the cell suspension.
Quantification of ATP efflux from the indicator strain.
The ATP content of each sample tested was determined using an ATP assay reagent kit (Kikkoman Co. Ltd., Noda, Japan) based on the firefly luciferin-luciferase reaction (28). A solution of the indicator cells was prepared as follows. After incubation at 37°C for 18 h in MRS broth, the indicator cells were harvested by centrifugation (3,000 × g for 10 min at 20°C) and then washed three times with 50 mM imidazole-HCl buffer (pH 6.8). The washed cells were suspended in the imidazole-HCl buffer, and the suspension was adjusted to a 300-fold dilution of an 18-h culture.
The solution of indicator cells (4 ml) was mixed with 60% 2-propanol (100 μl) used as a control or each purified bacteriocin (fraction F; 100 μl; 5 AU). At intervals of 0, 60, and 120 min, the samples were withdrawn and then filter sterilized through a Millex-GV filter (0.22 μm pore size; Nihon Millipore, Yonezawa, Japan) in order to remove contaminated cells. The bioluminescence reagent (100 μl; Kikkoman) for the ATP assay was added to the filtrate (700 μl), and the bioluminescence was immediately measured at room temperature by using a Lumitester K-210 luminometer (Kikkoman). The ATP concentration was calibrated from the ATP standard, and total ATP in the indicator solution was determined after treatment with an ATP-releasing reagent (Kikkoman).
Quantification of potassium efflux from indicator cells.
The indicator solution was prepared as described above with some modifications. The harvested cells were washed three times with 50 mM imidazole-HCl buffer (pH 6.8), and the cells were then suspended in the imidazole-HCl buffer, and the suspension was adjusted to a sixfold dilution of an 18-h culture.
The indicator solution (4 ml) was mixed with 4 μl of CD222 potassium indicator (2 × 10−3 mM; Molecular Probes, Inc., Eugene, Oreg.) and then with 60% (vol/vol) 2-propanol (200 μl) as a control or each purified bacteriocin (fraction F; 200 μl; 250 AU). At intervals of 0, 60, and 120 min, the fluorescence was measured at an excitation wavelength of 396 nm and an emission wavelength of 480 nm by using a model PTL-396S fluorometer (Japan Spectroscopic Co., Hachioji). The potassium concentration was calibrated from the potassium chloride standard curve.
Quantification of potassium ion efflux from liposomes.
Large unilamellar vesicles composed of l-α-phosphatidylcholine (from soybeans; Wako) were prepared as follows. Lipid (l-α-phosphatidylcholine; 20 mg) dissolved in chloroform (200 μl) was dried under a stream of N2 gas in a siliconized microcentrifuge tube. After removal of residual solvent under a vacuum for 1 h, the dried lipid was resuspended in a solution (1.2 ml) containing 100 mM KCl and 1 mM CaCl2 (prepared at pH 5 by using dimethyl glutaric acid). The mixture was sonicated for 1 h to obtain large unilamellar vesicles and then freeze-thawed for two cycles for formation of multilamellar vesicles.
Lipid vesicles (70 μl) were added to 100 mM choline solution (14 ml) prepared at pH 5 by using HNO3, and the mixture was agitated slowly. After addition of 60% (vol/vol) 2-propanol (65 μl) as a control or each antibacterial substance in 60% 2-propanol (65 μl), the purified bacteriocins (fraction F; 0.3 μM; 60 AU/ml) and two ionophores, gramicidin D (0.98 μM; 120 AU/ml; Sigma Chemical Co., St. Louis, Mo.) and valinomycin (34.2 μM; 960 AU/ml; Sigma), were added to the lipid vesicle solution and the concentration of potassium in external solution was measured at 0, 30, 60, 90, and 120 min by using a 692 pH/ion meter (Metrohm, Herisau, Switzerland) with a potassium ion electrode (Metrohm).
Nucleotide sequence accession number.
The sequence of the bacteriocin gene gaaA has been deposited in the DDBJ, EMBL, and GenBank databases under accession number AB007043.
RESULTS
Purification of bacteriocins.
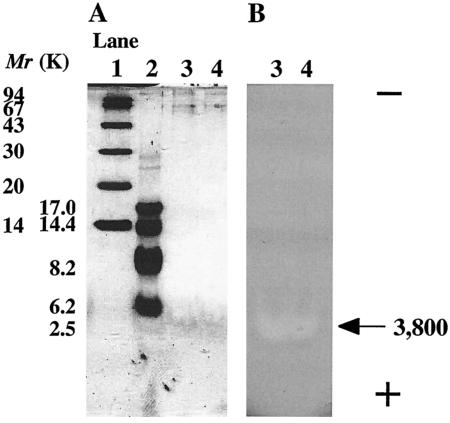
Gassericin A, a bacteriocin produced by L. gasseri LA39, and reutericin 6, a bacteriocin produced by L. reuteri LA6, were purified to homogeneity from DOB-MRS broth culture supernatant by reverse-phase chromatography. The average specific activities of both bacteriocins against an indicator strain (L. delbrueckii subsp. bulgaricus JCM 1002T) increased by about 4,000-fold with about 20% recovery after purification. The purity of both bacteriocins was analyzed by SDS-PAGE, in situ activity assay (single band holding bacteriocin activity) (Fig. 1), and N-terminal amino acid sequencing (no amino acid residues for cyclic structure were detected). Examination of both purified-bacteriocin activities against both producer strains by the agar well diffusion method showed that 16 ng of gassericin A inhibited the growth of L. reuteri LA6 but that over 960 ng of reutericin 6 did not inhibit the growth of L. gasseri LA39.
FIG. 1.
Results from SDS-PAGE (A) and in situ activity assay (B) of purified gassericin A and reutericin 6. Lanes 1 and 2, molecular weight (Mr) markers; lane 3, purified gassericin A; lane 4, purified reutericin 6. The arrow points to the location of the bacteriocins (Mr, 3,800). The plus and minus signs indicate the electric charge at the electrode of the SDS-PAGE.
pH stabilities of the two bacteriocins.
The pH stabilities (at 4°C for 48 h) of purified gassericin A and reutericin 6 (fraction F) were examined. Reutericin 6 lost 50 and 75% of activity at pH 3 and 2, respectively, but no decline in the activity level of gassericin A was observed at pH values in the range from 2 to 12.
Effects of bacteriocins on indicator cells.
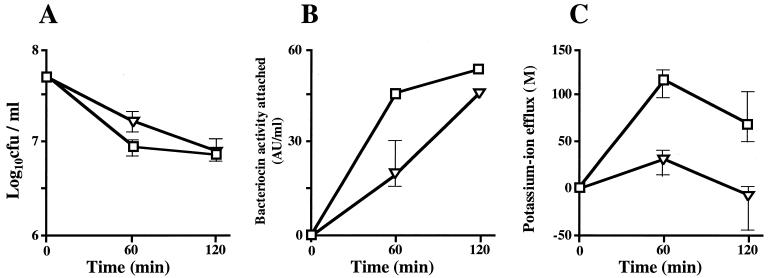
The effects of gassericin A and reutericin 6 on the viability and lysis of indicator cells of L. delbrueckii subsp. bulgaricus JCM 1002T and the amounts of the bacteriocins that attached to the cells were examined (Fig. 2A and B). The level of cell viability was decreased from 7.7 × 108 to 6.8 × 107 CFU/ml by the actions of both bacteriocins at the same levels of activity (60 AU/ml), but the times needed to reach the stationary cell number were different, 60 min (gassericin A) and 120 min (reutericin 6) (Fig. 1A). Optical density (ca. 0.48) at 650 nm was maintained after bacteriocin treatment (data not shown), indicating that no cell lysis was caused by either of the bacteriocins.
FIG. 2.
Effects of gassericin A and reutericin 6 on the indicator cells of L. delbrueckii subsp. bulgaricus JCM 1002T. Parameters tested were cell viability (A), amounts of bacteriocin activity attached to the cells (B), and changes in potassium ion efflux (C). (□), gassericin A; (▿), reutericin 6.
Figure 2B shows the amount of each bacteriocin activity that attached to the indicator cells (optical density of each solution, ca. 0.48). The level of activity of each bacteriocin detected in the cell suspension decreased with the advance of time, indicating that the levels of attachment of both bacteriocins increased. The activity level of reutericin 6 at 60 min was low in comparison with that of gassericin A, and the pattern of attachment was similar to the pattern of change in cell viability.
In these experiments, the same results were obtained with a mixed solution of PB and 1.5% (vol/vol) 2-propanol (final concentration). This finding indicates that there is no influence of 1.5% 2-propanol on the indicator strain, at least within 120 min.
Quantification of ATP and potassium ion efflux from indicator cells.
The amounts of ATP released from L. delbrueckii subsp. bulgaricus JCM 1002T by the actions of gassericin A and reutericin 6 were measured at 0, 60, and 120 min after the start of incubation. As determined by comparison with amounts of ATP in the control (60% 2-propanol) and total amounts of ATP after lysis, neither of the bacteriocins caused ATP efflux from the indicator cells after 120 min of incubation, at which time death of the indicator was observed (data not shown).
Figure 2C shows the amounts of potassium ion efflux from L. delbrueckii subsp. bulgaricus JCM 1002T as caused by the actions of gassericin A and reutericin 6 at 0, 60, and 120 min after the start of incubation. The amount of potassium ion efflux that occurred with 60% 2-propanol as a control was set as the 0 position. Both bacteriocins had caused potassium ion efflux from the indicator strain after 60 min of incubation, and the amount of efflux had decreased after 120 min of incubation. The potassium ion efflux caused by reutericin 6 was clearly less than that caused by gassericin A.
Quantification of potassium efflux from liposomes.
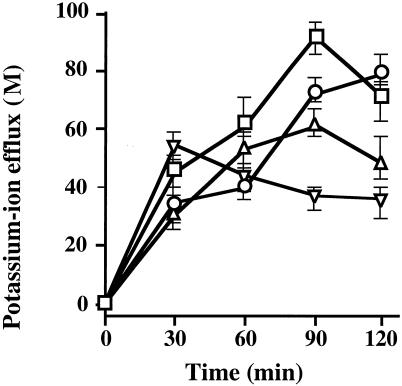
Fig. 3 shows the amounts of potassium ion efflux from liposomes, lipid vesicles composed of l-α-phosphatidylcholine, as caused by the action of ionophores and bacteriocins at 0, 30, 60, 90, and 120 min after the start of incubation. The amount of potassium ion efflux that occurred with 60% 2-propanol as a control was set at the 0 position. All antibacterial substances caused potassium ion efflux from liposomes, and the efflux pattern of gassericin A was similar to that of the ionophores tested. However, the amount of efflux caused by reutericin 6 peaked at 30 min, and the pattern of action of reutericin 6 was different from those of the ionophores and gassericin A.
FIG. 3.
Time courses of changes in amounts of potassium ion efflux from liposomes as caused by gassericin A and reutericin 6. The amounts of potassium ion efflux induced by the action of gramicidin D (○) (8 AU; 4.9 nM), valinomycin (▵) (64 AU; 170 nM), gassericin A (□) (4 AU; 1.5 nM), and reutericin 6 (▿) (4 AU; 1.5 nM) were plotted with that induced by 60% 2-propanol as the base. All experiments were performed at pH 5.
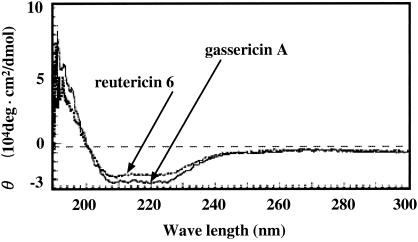
CD spectroscopy.
Fig. 4 shows CD spectra of gassericin A and reutericin 6 (fraction F) in 60% 2-propanol. A positive maximum near 190, negative maxima at 208 and 222 nm, and θ values indicate that both of the bacteriocins are peptides consisting mainly of α-helix structures. However, the spectroscopic patterns of the two bacteriocins did not coincide, and the absolute θ value of reutericin 6 was slightly lower than that of gassericin A.
FIG. 4.
CD spectra of purified gassericin A and reutericin 6.
Partial d- and l-amino acid composition analysis.
Gassericin A and reutericin 6 molecules are composed of 17 different amino acids, and all of the 58 amino acids in the bacteriocins were detected by chemical sequencing (14). The d- and l-amino acid compositions of both bacteriocins were analyzed for five kinds of amino acids as o-phthalaldehyde derivatives (Table 1). Two residues and one residue of d-Ala were detected among 18 Ala residues of gassericin A and reutericin 6, respectively, and no d-amino acids were detected among Asp, Tyr, Val, and Ile residues tested.
TABLE 1.
D- and L-amino acid compositions of gassericin A and reutericin 6
| Amino acid | No. of residues in:
|
|
|---|---|---|
| Gassericin A | Reutericin 6 | |
| d-Asp | NDa | ND |
| l-Asp | 2 | 2 |
| d-Tyr | ND | ND |
| l-Tyr | 1 | 1 |
| d-Ala | 2 | 1 |
| l-Ala | 16 | 17 |
| d-Val | ND | ND |
| l-Val | 1 | 1 |
| d-Ile | ND | ND |
| l-Ile | 4 | 4 |
ND, not detected.
DISCUSSION
L. gasseri LA39 and L. reuteri LA6 isolated from the same human infant were thought to both produce cyclic bacteriocins gassericin A and reutericin 6, which cannot be distinguished by molecular weights or primary amino acid sequences. In this study, gassericin A inhibited the growth of L. reuteri LA6, but reutericin 6, which has a narrower spectrum than gassericin A, did not inhibit the growth of L. gasseri LA39. Reutericin 6 lost less than 50% of its activity at a low acidic pH and showed a mode of action different from that of gassericin A. We further examined CD spectra and d- and l-amino acid compositions of gassericin A and reutericin 6 in order to determine the cause of the difference between these two bacteriocins.
The CD spectra of the purified bacteriocins revealed that both antibacterial peptides are composed mainly of α-helices and are nearer the α/β type than the α+β type as determined by the absolute θ values. However, the spectra of the two bacteriocins did not completely coincide, indicating that the secondary structures of the two bacteriocins are slightly different. The d- and l-amino acid composition was analyzed for five amino acids (Asp, Tyr, Ala, Val, and Ile) among 17 amino acids in the molecules. It was clarified that the difference between both bacteriocins was in only one d-Ala residue in the molecule. These data suggest that the different d-alanine contents of gassericin A and reutericin 6 may cause the differences in antibacterial spectra, modes of action, amounts of potassium ion efflux, and secondary structures.
Identification of Asp, Glu, Ala, Ser, Pro, Cys, Phe, Leu, Val, and Tyr d-amino acids in various antimicrobial substances such as bacitracin A, vernamycin B, polymyxin, tyrocidine, gramicidin, actinomycin, and valinomycin has been reported (5). In bacteriocins, a d-amino acid in lactocin S (lantibiotic; class I) produced by L. sake strain L45 has been reported (29), and it has been suggested that d-Ala residues are converted during maturation from original l-Ser cordons. As for gassericin A and reutericin 6, the cordons on the structural gene (gaaA) encoding Ser residues correspond to the amino acid sequence decided by chemical sequencing. This suggests that d-Ala residues in the two bacteriocins are produced by another mechanism.
Many peptides in peptidoglycan of the bacterial cell wall contain d-Ala and d-Glu residues, which are constructed by d-amino acid transaminase (27, 30). Bacterial racemases for Ala, Arg, Glu, Phe, and Pro have been reported (1, 3). In 1998, Nagata et al. investigated the d-amino acid contents in peptides and proteins extracted from organisms ranging from bacteria to eukaryotes, and they reported that d-Glu and d-Asp were detected at high frequencies (24). Notably, 12 and 10% of all d- and l-Ala residues were found to be d-Ala in the gram-positive Staphylococcus epidermidis and Streptococcus pyogenes bacteria tested, respectively, and Bacillus sp. strain YN-1 contained d-Glu at a frequency of 22%. Although gassericin A and reutericin 6 have no Glu residues in the molecules and no d-Asp was detected in this study, those findings suggest that the d- and l-amino acid composition analysis of both bacteriocins remains to be performed for Arg (one residue in the molecule), Leu (eight residues), Phe (two residues), Pro (one residue), and Ser (two residues).
As one of the major action mechanisms, hydrophobic and low-molecular-weight bacteriocins cause efflux of low-molecular-weight components such as ATP and cations by pore formation in the cytoplasmic membrane, resulting in depolarization of the cytoplasmic membrane to cause death of the indicator cell (21). No ATP efflux from the indicator cells of L. delbrueckii subsp. bulgaricus JCM 1002T was caused by gassericin A and reutericin 6, the activity levels of which were sufficient to inhibit the growth of the indicator cells, suggesting that neither of the bacteriocins causes lysis of cells or leakage of substances of molecular weights of more than 507 (ATP). Gramicidin D, which is composed of normal chain peptides A, B, and C, is an ionophore that forms pores in the cell membrane and transports monocations through α-helix structures more rapidly than valinomycin, a potassium cation carrier, across the cell membrane (34). In this study, the peptide concentration of gramicidin D (8 AU; 4.9 nM) that inhibited the growth of L. delbrueckii subsp. bulgaricus JCM 1002T cells and caused efflux of potassium ions from lipid vesicles was lower than that of valinomycin (64 AU; 170 nM). Moreover, gassericin A and reutericin 6 were more effective (1.6-fold) in specific activity than gramicidin D. These findings indicate that both bacteriocins act in the cytoplasmic membrane of a target cell and cause the death of the cell by efflux of potassium ions.
The two bacteriocins prepared by the same purification steps had similar activity levels and peptide concentrations for inhibition of the growth of indicator cells, but they induced different amounts and patterns of potassium ion efflux from the cells and liposomes. Although the pattern of the mode of action of gassericin A corresponded with the pattern of potassium ion efflux, the patterns of the mode of action and potassium ion efflux for reutericin 6 were different for the indicator cells and liposomes. These results suggest that reutericin 6, which rapidly causes potassium ion efflux from liposomes, is influenced by surface substances on the indicator cells which may cause the time lag on the mode of action and the low efflux pattern.
The purified gassericin A and reutericin 6 (fractions E and F) were highly hydrophobic peptides and did not dissolve in aqueous solution. Although 2-propanol was used to dissolve both bacteriocins in the test solutions, there were no differences in the modes of action within 120 min and no effects on the measurement of bacteriocin activity by using the agar well diffusion method. However, in microtiter plate assays (15), L. delbrueckii subsp. bulgaricus JCM 1002T as the indicator did not grow in a medium solution containing 1.5% 2-propanol after incubation for 24 h at 37°C (data not shown), indicating that 2-propanol caused serious damage to the indicator. In this study, we showed the functional differences of the two bacteriocins under the same conditions and found no direct elucidations of 2-propanol effects such as weakening of the cell walls of the indicator and conformational changes of the bacteriocins. The influences of 2-propanol remain to be identified in addition to other concerns for safe application of bacteriocins in aqueous environments such as those in food and in vivo.
Although bacteriocins with the same primary structures produced by different bacterial species have been reported for other LAB species such as Pediococcus acidilactici and L. plantarum (pediocin AcH) (8, 22) and L. curvatus and L. sake (curvacin A-sakacinA) (4, 31), there have been no reports on the different characteristics of native primary identical bacteriocins such as gassericin A and reutericin 6. This is the first report that characteristics of native bacteriocins produced by wild lactobacillus strains having the same structural genes are influenced by a difference in d-amino acid contents in the molecules. In the case of heterologous expression of gassericin A and reutericin 6 in Escherichia coli, the specific activity of the noncyclic bacteriocin expressed with free N- and C-termini was weaker than those of native bacteriocins (13) and no d-Ala residues were contained in the expressed molecule (data not shown). The attention to and check for characteristics and the d- and l-amino acid analysis of peptides and proteins after expression using identified genes and those from wild microorganisms having the same genes would be worthwhile and needed in coming periods. The development of an automatic d- and l-amino acid sequencer will make it easier to check the structures and the functions of the molecules in future.
Acknowledgments
We thank Kazuaki Akasaka, Graduate School of Life Sciences, Tohoku University, for his assistance in CD spectroscopy analysis and Shigehiro Obata, Kishida Chemical Co. Ltd. (Mita, Japan), for his assistance in d- and l-amino acid composition analysis.
This work was partially supported by the NIG Cooperative Research Program (2003-30) of the National Institute of Genetics and by a grant-in-aid for young scientists to Y. Kawai (no. 12760083) from the Japanese Ministry of Education, Science, and Culture.
REFERENCES
- 1.Adams, E. 1972. Amino acid racemases and epimerases, p. 479-507. In P. D. Boyer (ed.), Enzymes, 3rd ed., vol. 6. Academic Press, New York, N.Y.
- 2.Anderson, J. W., and S. E. Gilliland. 1999. Effect of fermented milk (yogurt) containing Lactobacillus acidophilus L1 on serum cholesterol in hypercholesterolemic humans. J. Am. Coll. Nutr. 18:43-50. [DOI] [PubMed] [Google Scholar]
- 3.Ashiuchi, M., K. Tani, K. Soda, and H. Misono. 1998. Properties of glutamate racemase from Bacillus subtilis IFO 3336 producing poly-gamma-glutamate. J. Biochem. (Tokyo) 123:1156-1163. [DOI] [PubMed] [Google Scholar]
- 4.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 177:2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodanszky, M., and D. Perlman. 1969. Peptide antibiotics. Science 163:352-358. [DOI] [PubMed] [Google Scholar]
- 6.Contreras, B. G., L. De Vuyst, B. Devreese, K. Busanyova, J. Raymaeckers, F. Bosman, E. Sablon, and E. J. Vandamme. 1997. Isolation, purification, and amino acid sequence of lactobin A, one of the two bacteriocins produced by Lactobacillus amylovorus LMG P-13139. Appl. Environ. Microbiol. 63:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daba, H., S. Pandian, J. F. Gosselin, R. E. Simard, J. Huang, and C. Lacroix. 1991. Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl. Environ. Microbiol. 57:3450-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ennahar, S., D. Aoude-Werner, O. Sorokine, A. Van Dorsselaer, F. Bringel, J. C. Hubert, and C. Hasselmann. 1996. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl. Environ. Microbiol. 62:4381-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujisawa, T., Y. Benno, T. Yaeshima, and T. Mitsuoka. 1992. Taxonomic study of Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al., 1980) with the type strain of Lactobacillus amylovorus (Nakamura, 1981). Int. J. Syst. Bacteriol. 32:487-491. [DOI] [PubMed] [Google Scholar]
- 10.Itoh, T., Y. Fujimoto, Y. Kawai, T. Toba, and T. Saito. 1995. Inhibition of food-borne pathogenic bacteria by bacteriocins from Lactobacillus gasseri. Lett. Appl. Microbiol. 21:137-141. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. L., C. F. Phelps, C. S. Cummin, J. London, and F. Gasser. 1980. Taxonomy of the Lactobacillus acidophilus group. Int. J. Syst. Bacteriol. 30:53-68. [Google Scholar]
- 12.Kanatani, K., M. Oshimura, and K. Sano. 1995. Isolation and characterization of acidocin A and cloning of the bacteriocin gene from Lactobacillus acidophilus. Appl. Environ. Microbiol. 61:1061-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai, Y., K. Arakawa, A. Itoh, B. Saitoh, Y. Ishii, J. Nishimura, H. Kitazawa, T. Itoh, and T. Saito. 2003. Heterologous expression of gassericin A, a bacteriocin produced by Lactobacillus gasseri LA39. Anim. Sci. J. 74:45-51. [Google Scholar]
- 14.Kawai, Y., Y. Ishii, K. Uemura, H. Kitazawa, T. Saito, and T. Itoh. 2001. Lactobacillus reuteri LA6 and Lactobacillus gasseri LA39 isolated from feces of the same human infant produce identical cyclic bacteriocin. Food Microbiol. 18:407-415. [Google Scholar]
- 15.Kawai, Y., T. Saito, J. Uemura, and T. Itoh. 1997. Rapid detection method for bacteriocin and distribution of bacteriocin-producing strains in Lactobacillus acidophilus group lactic acid bacteria isolated from human feces. Biosci. Biotechnol. Biochem. 61:179-182. [DOI] [PubMed] [Google Scholar]
- 16.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lauer, E., C. Helming, and O. Kandler. 1980. Heterogeneity of the species Lactobacillus acidophilus (Moro) Hansen and Mocquot as revealed by biochemical characteristics and DNA-DNA hybridization. Zentbl. Bakteriol. Mikrobiol. Hyg. Abt. Orig. C 1:150-168. [Google Scholar]
- 19.Leer, R. J., J. M. van der Vossen, M. van Giezen, J. M. van Noort, and P. H. Pouwels. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629-1635. [DOI] [PubMed] [Google Scholar]
- 20.Lievin-Le, M. V., R. Amsellem, A. L. Servin, and M. H. Coconnier. 2002. Lactobacillus acidophilus (strain LB) from the resident adult human gastrointestinal microflora exerts activity against brush border damage promoted by a diarrhoeagenic Escherichia coli in human enterocyte-like cells. Gut 50:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moll, G. N., W. Konings, and A. Driessen. 1999. Bactreriocins: mechanism of membrane insertion and pore formation. Antonie Leewenhoek 76:185-198. [PubMed] [Google Scholar]
- 22.Motlagh, A. M., A. K. Bhunia, F. Szostek, T. R. Hansen, M. C. Johnson, and B. Ray. 1992. Nucleotide and amino acid sequence of pap-gene (pediocin AcH production) in Pediococcus acidilactici H. Lett. Appl. Microbiol. 15:45-48. [DOI] [PubMed] [Google Scholar]
- 23.Muriana, P. M., and T. R. Klaenhammer. 1991. Cloning, phenotypic expression, and DNA sequence of the gene for lactacin F, an antimicrobial peptide produced by Lactobacillus spp. J. Bacteriol. 173:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagata, Y., T. Fujiwara, K. Kawaguchi-Nagata, Y. Fukumori, and T. Yamanaka. 1998. Occurrence of peptidyl d-amino acids in soluble fractions of several eubacteria, archaea and eukaryotes. Biochim. Biophys. Acta 1379:76-82. [DOI] [PubMed] [Google Scholar]
- 25.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 26.Nimura, N., and T. Kinoshita. 1986. O-Phthalaldehyde-N-acetyl-l-cysteine as a chiral derivatization reagent for liquid chromatographic optical resolution of amino acid enantiomers and its application to conventional amino acid analysis. J. Chromatogr. 352:169-177. [Google Scholar]
- 27.Peisach, D., D. M. Chipman, P. W. Van Ophem, J. M. Manning, and D. Ringe. 1998. Crystallographic study of steps along the reaction pathway of d-amino acid aminotransferase. Biochemistry 37:4958-4967. [DOI] [PubMed] [Google Scholar]
- 28.Sakakibara, T., S. Murakami, N. Hattori, M. Nakajima, and K. Imai. 1997. Enzymatic treatment to eliminate the extracellular ATP for improving the detectability of bacterial intracellular ATP. Anal. Biochem. 250:157-161. [DOI] [PubMed] [Google Scholar]
- 29.Skaugen, M., J. Nissen-Meyer, G. Jung, S. Stevanovic, K. Sletten, C. Inger, M. Abildgaard, and I. F. Nes. 1994. In vivo conversion of l-serine to d-alanine in a ribosomally synthesized polypeptide. J. Biol. Chem. 269:27183-27185. [PubMed] [Google Scholar]
- 30.Tanizawa, K., S. Asano, Y. Masu, S. Kuramitsu, H. Kagamiyama, H. Tanaka, and K. Soda. 1989. The primary structure of thermostable d-amino acid aminotransferase from a thermophilic Bacillus species and its correlation with l-amino acid aminotransferases. J. Biol. Chem. 264:2450-2454. [PubMed] [Google Scholar]
- 31.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1993. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch. Microbiol. 160:279-283. [DOI] [PubMed] [Google Scholar]
- 32.Toba, T., S. K. Samant, E. Yoshioka, and T. Itoh. 1991. Reutericin 6, a new bacteriocin produced by Lactobacillus reuteri LA6. Lett. Appl. Microbiol. 13:281-286. [Google Scholar]
- 33.Toba, T., E. Yoshioka, and T. Itoh. 1991. Potential of Lactobacillus gasseri isolated from infant faeces to produce bacteriocin. Lett. Appl. Microbiol. 12:228-231. [Google Scholar]
- 34.Wallance, B. A. 1998. Recent advances in the high-resolution structures of bacterial channels: gramicidin A. J. Struct. Biol. 121:123-141. [DOI] [PubMed] [Google Scholar]
- 35.Yang, J. T., C. S. Wu, and H. M. Martinez. 1986. Calculation of protein conformation from circular dichroism. Methods Enzymol. 130:208-269. [DOI] [PubMed] [Google Scholar]