Abstract
We identified a polyketide synthase (PKS) gene, pksN, from a strain of Nectria haematococca by complementing a mutant unable to synthesize a red perithecial pigment. pksN encodes a 2,106-amino-acid polypeptide with conserved motifs characteristic of type I PKS enzymatic domains: β-ketoacyl synthase, acyltransferase, duplicated acyl carrier proteins, and thioesterase. The pksN product groups with the Aspergillus nidulans WA-type PKSs involved in conidial pigmentation and melanin, bikaverin, and aflatoxin biosynthetic pathways. Inactivation of pksN did not cause any visible change in fungal growth, asexual sporulation, or ascospore formation, suggesting that it is involved in a specific developmental function. We propose that pksN encodes a novel PKS required for the perithecial red pigment biosynthesis.
Polyketides comprise diverse natural products including antibiotics, pigments, and mycotoxins that are formed from small carbon precursor acids whose successive condensation is catalyzed by polyketide synthases (PKSs) (34). Filamentous fungi are prolific producers of polyketide metabolites of pharmacological and agricultural interest (4, 8, 13, 14, 28, 33, 42, 44, 46). These enzymes are quite diverse but belong to one of two basic types (23, 24). Modular PKS type I contains one or more large multifunctional protein subunits, while iterative PKS type II has only a few active sites on separate polypeptide chains. All identified fungal PKSs belong to modular type I, with a single polypeptide that contains up to eight domains for polyketide biosynthesis (21, 27). A typical fungal PKS consists of a linear succession of ketosynthetase (KS), acyltransferase (AT), dehydratase (DH), enoyl reductase (ER), ketoreductase (KR), acyl carrier protein (ACP), and thioesterase (TE) domains. In addition to the biochemically characterized enzymes, many other potential fungal PKSs have been identified by using degenerate-PCR approaches (5, 30, 36).
Over the past decade, several genes encoding fungal PKS enzymes involved in pigment production have been cloned and characterized. Most studies have focused on the biosynthesis of dark brown pigments, melanins (14, 35, 44), and green pigments (28, 33), because of their important role in fungal pathogenesis (29). Relatively little is known about the biosynthesis of red pigments, with the exception of the bikaverin produced by Gibberella fujikuroi (31). In Nectria haematococca, mutants in which the pigmentation of mycelia and perithecia is affected (i.e., mutants either show hyperproduction of red pigments or are albinos) have been obtained through mutagenesis (11, 40, 41). Extensive genetic analyses of such mutants have been conducted (11, 40). It has been established that at least six distinct loci control mycelial pigmentation. These pigmented metabolites are naphtoquinones (39), most of which have antibacterial, antifungal, insecticidal, and phytotoxic properties (26). The perithecial color phenotype appears to be governed by at least two genetically independent loci, of which one is marked by the I4 mutation (3). This mutation suppressed the red pigmentation of the perithecial wall but did not affect the color of the mycelium. This observation suggests that the I4 gene affected a function that was expressed in the sexual phase but not in the vegetative phase. Perithecial color mutants have been reported to occur in several ascomycete species including Hypomyces solani (19), Neurospora crassa (22), Magnaporthe grisea (9), and Gibberella fujikuroi (7), but the genes involved in such variation have not been cloned. A red pigment also accumulates in the ascospores of Aspergillus nidulans (6). This pigment is an anthraquinone, which suggests that it is a product of a PKS that has not yet been characterized.
Our objective in this study was to clone a gene involved in the biosynthesis of a perithecial red pigment in Nectria haematococca. Such genes might also be conserved in other fungi that produce red pigments during the sexual phase or play a role in the biosynthesis of other secondary metabolites.
MATERIALS AND METHODS
Fungal strains, media, and crosses.
All strains are derived from a homothallic species within the N. haematococca complex CBS 225.58 (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands). Based on DNA sequences of the 28S ribosomal DNA published in reference 17, we assigned this strain to clade 3 of the Fusarium solani complex established by O'Donnell (37) (data not shown).
A mutant strain (a1 s1 I4 mod) (hereafter referred to as [a1s1I4mod]) was used as recipient in transformation experiments (16). The a1 and s1 mutations prevent the appearance of two morphological modifications called the “Anneau” and the “Secteur,” respectively (16, 43). The I4 mutant produces white, rather than red, perithecia and was obtained by treatment with N-methyl-N′-nitro-nitrosoguanidine (3). The mod mutation prevents ascospore formation within perithecia. The I4 and mod mutations are genetically linked (0.5% of recombination frequency) (3). A strain containing the a*58 mutation (hereafter referred to as [a*58]), which has altered “Anneau” expression (10), produced red perithecia and was used in some crosses.
General culture conditions and manipulations were as previously described (10). Strains were purified via subculture of microconidia and maintained at 26°C on potato dextrose agar made of potato broth (potatoes [200 g/liter] boiled for 1 h, peeled, and sliced) and glucose (20 g/liter) supplemented with agar (20 g/liter). Long-term stocks were stored as agar plugs under mineral oil at 12°C.
Crossing methods for the homothallic species Fusarium sp. strain CBS 225.58 have been described elsewhere (3, 40). When PR1, a transformant with a wild-type phenotype, was crossed with [a*58], which produces red fertile perithecia, a preliminary analysis of a sample of descendants from each perithecium collected on the line of contact was required to distinguish hybrids from parental perithecia (11). The progeny from two hybrid perithecia (250 ascospores from each perithecium) were analyzed for color marker segregation (3, 40), although these progeny may not all have originated from independent meiotic events.
DNA and RNA manipulation.
All nucleic acid manipulations were performed by standard methods (2). For Southern blot analysis, genomic DNA was extracted as described previously (32). The nucleotide sequence of the mutant allele I4 was determined from two independent amplicons obtained following PCR with oligonucleotide primers I4-187 (5′-GGCGCTTGAATGCTTGATAG-3′) and ip2393c (5′-TGGCCATCTTCTCGAATCGT-3′) by using the following program: 30 cycles of 1 min at 96°C, 30 s at 56°C, and 3 min at 72°C, followed by a final extension of 5 min at 72°C.
Transformation of N. haematococca.
Transformation was performed as previously described (16), with hygromycin B resistance as the selectable marker. For transformation with cosmids carrying the hygromycin resistance gene (hph), 10 μg of each cosmid pool was used. When cotransformation experiments were performed, usually with plasmid pBC-Hygro, 5 μg of the transforming plasmid was mixed with 5 μg of the DNA to be tested. Transformants were selected on PH8 medium (potato dextrose agar with 200 g of sucrose/liter, 3 g of agar/liter, and 8 mg of hygromycin B/liter) (Sigma-Aldrich).
Cosmid library screening.
We constructed an “XSG” gene library (16) by inserting genomic DNA from the N. haematococca wild-type strain partially digested with XhoI into the pMOCosX cosmid as previously described (38). We tested 20 cosmid pools in transformation experiments with [a1s1I4mod] as the recipient. An average of 400 hygromycin-resistant transformants per cosmid pool were cultured on PH8. After 7 days of growth, plates were exposed to daylight, and 15 days later they were examined for perithecial color. One cosmid pool, XSG11, led to the production of red perithecia. The complementing cosmid was identified by examining 20 subpools corresponding to each of the 12 columns and eight rows of the microtiter plate.
Subcloning of cosmid E12.
E12 was digested with BamHI, ClaI, EcoRI, HindIII, PstI, SalI, and XhoI. Pools of fragments larger than 1 kb were eluted from the agarose gel and used to cotransform the [a1s1I4mod] strain. Fragments of pools restoring the I4+ phenotype were cloned into pBluescript and then tested in the same way. pI4C3-2 was derived from E12 and carried a 4.5-kb ClaI fragment containing a truncated pksN gene.
Sequencing.
The insert in pI4C3-2 was entirely sequenced on both strands by using synthetic primers and the ABI PRISM Dye-terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.). The ends of each cloned ClaI fragment from cosmid E12 (pI4C3-1 to −5) were sequenced with plasmid-specific primers on an ABI373 automated DNA sequencer (Applied Biosystems).
Phylogenetic analysis.
Phylograms were inferred from concatenated ClustalX (41) protein multiple alignments of the AT and KS conserved domains. Neighbor-joining analyses were performed with Phylo_win (15) with the PAM (Point Accepted Mutation) distance matrix. One thousand bootstrap replicates were performed.
Nucleotide sequence accession numbers.
The nucleotide sequence of pksN was deposited in the GenBank and EMBL databases under accession number AY487573. The domain alignments and phylogenetic trees were deposited in the tree base database under accession number 26358.
RESULTS
Cloning of the I4+ gene by sib selection.
One cosmid pool, XSG11, transformed strain [a1s1I4mod] so that it produced red perithecia at a low frequency (3 of 250 transformants recovered). One transformant (PR1) had a wild-type phenotype, and two other transformants produced a mixture of red and white perithecia, suggesting instability of the transforming DNA. When the stable PR1 transformant was crossed with [a*58], which carries the I4+ allele, all of the progeny produced red perithecia. Thus, the complementing cosmid probably integrated at I4, suggesting that this cosmid carries the I4+ allele rather than a suppressor of the I4 mutation. In bidimensional subpooling experiments, only subpools E and 12 yielded transformants with red perithecia, thus identifying E12 as the complementing cosmid. When this cosmid was used to transform [a1s1I4mod], two types of transformants were recovered. A few (14%) transformants formed only red perithecia (stable transformants), while the remainder (86%) formed a mixture of red and white perithecia (unstable transformants), even when purified by subculturing single uninucleate conidia. pI4C3-2, a subclone of E12 carrying a 4.5-kb ClaI fragment, transformed strain [a1s1I4mod] so that it produced red perithecia at low frequency.
Identification and analysis of a polyketide synthase gene.
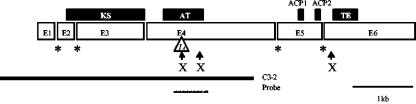
Analysis of the 4.5-kb fragment with the blastx algorithm (1) identified an open reading frame of 1,214 residues with a high degree of similarity to several type I PKS enzymes. The highest similarity was to the PKS encoded by the wA gene from A. nidulans (33). The 4.5-kb DNA fragment contains a 3′-truncated gene that we designated pksN. The complete pksN gene sequence was obtained from cosmid E12. Based on protein alignments and the presence of nucleotide sequences that closely matched the 5′ and 3′ end splice and internal consensus sequences (18), five putative introns ranging in size from 45 to 54 nucleotides were inferred (Fig. 1).
FIG. 1.
Diagram of the pksN gene showing six exons (E1 to E6) and five introns (intervals between E1 to E6). Conserved positions of introns relative to the wA gene are indicated by asterisks. Conserved amino acids at catalytic sites are indicated by boxes whose sizes correspond to the aligned domains KS, AT, ACP, and thioesterase (TE). The solid line indicates the length of the C3-2 fragment used in transformation experiments. The 0.6-kb PCR fragment (dashed line) was used as a probe in Southern blot experiments. X, XbaI site (the first one is absent in the I4 mutant).
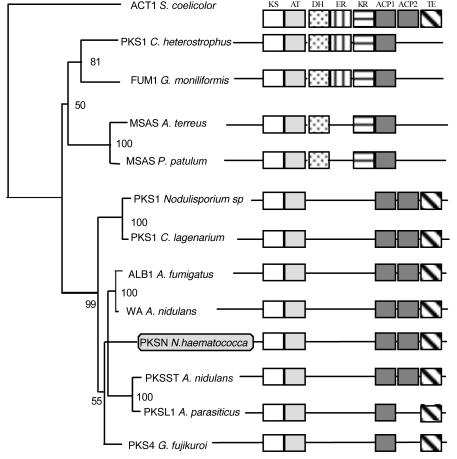
The predicted 2,106-amino-acid translation product of pksN contains five conserved functional domains (Fig. 1): a β-ketoacyl synthase domain (KS), an acyltransferase domain (AT), two acyl carrier protein motifs (ACP1 and ACP2), and a thioesterase domain (TE). Based on the organization of the functional domains, PKSN is of the WA type, which includes PKSs involved in pigment biosynthesis (14, 28, 33, 44) and in the aflatoxin/sterigmatocystin biosynthetic pathways (8, 13, 47). Phylogenetic analyses based on the KS (440 amino acids) and AT (240 amino acids) domains revealed similar evolutionary relationships. As shown on the tree obtained by combining domains AT and KS (Fig. 2), bootstrap values support the grouping of PKNS within the WA-type group.
FIG. 2.
Phylogenetic relationships of fungal PKSs inferred from the combined KS and AT domains. Corresponding organizations of the different functional domains within each fungal PKS are presented to the right. Domains are described in the top boxes, with the same abbreviations as for WA-type PKSs. Other domains are labeled as follows: dehydratase (DH), enoyl reductase (ER), and β-ketoacyl reductase (KR). Organisms and accession numbers (in parentheses): A. fumigatus ALB1 (X65866); A. nidulans wA (X65866); G. fujikuroi PKS4 (AF278141); Aspergillus parasiticus PKSL1 (L42765); N. haematococca PKSN (AY487573); Nodulisporium sp. PKS1 (AF151553); Colletotrichum lagenarium PKS1 (D83643); A. nidulans PKSST (L39121); Streptomyces coelicolor ACT1 (Q02059); C. heterostrophus PKS1 (U68040); G. moniliformis FUM5 (AF155773); Aspergillus terreus MSAS (U31329); Pittosporum patulum MSAS (X55776).
PKSN and perithecium pigmentation.
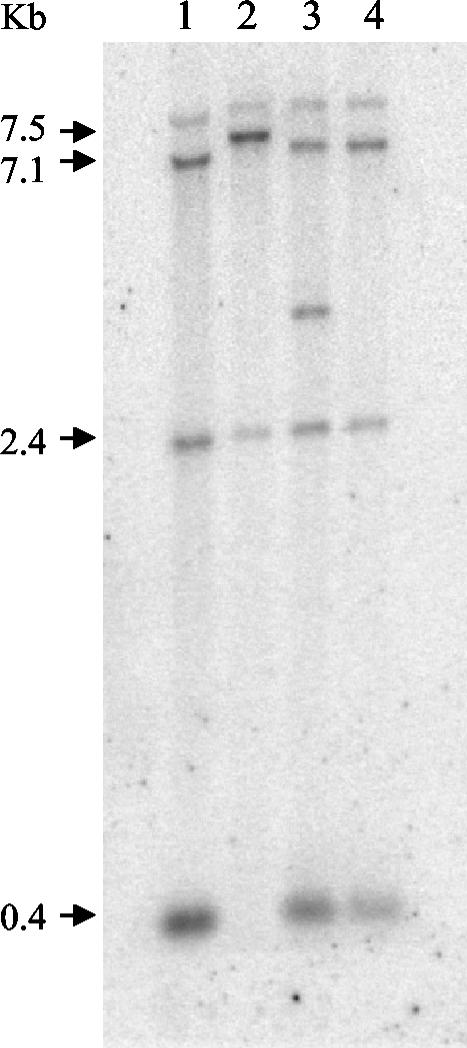
The I4 allele results from an 8-bp deletion at position 2422 downstream of the ATG codon. This mutation results in a stop codon 20 bp downstream of the mutation and thus encodes a truncated protein of 763 residues containing only the β-ketosynthase domain. To confirm that the I4 phenotype was not due to a secondary mutation, we cotransformed this strain with the pBC-Hygro plasmid and the I4C3-2 fragment, which lacks a portion of the 3′ region of pksN (Fig. 1). In the resulting analysis, some transformants were restored to the production of red perithecia and were presumed to result from an integration of the plasmid into the I4 locus. This hypothesis was tested by using a PCR approach based on the loss of an XbaI site associated with the deletion in the I4 allele. After amplification of a 1.9-kb fragment spanning the deletion and subsequent digestion with XbaI, we observed cleavage of the fragment in two C3-2 transformants with red perithecia and in the wild type but not in the [a1s1I4mod] strain (data not shown). The same two transformants were analyzed by Southern blotting (Fig. 3). The mutant lost a 0.4-kb band, and a 7.1-kb band was shifted to 7.5 kb. The two transformants had the same wild-type profile at the I4 locus, demonstrating that the red perithecial phenotype results from gene replacement. The additional hybridizing band in one transformant presumably results from an ectopic integration of another copy of the C3-2 fragment.
FIG. 3.
Southern blot analysis of strains transformed with the C3-2 linear fragment. Genomic DNAs were digested by XbaI, electrophoresed on a 0.8% agarose gel, transferred to a nitrocellulose membrane, and hybridized with a 32P-labeled 663-bp PCR-amplified fragment of pksN spanning the two XbaI sites surrounding the I4 mutation (Fig. 1). Lane 1, [a*58] (I4+ allele); lane 2, [a1s1I4mod] (mutated allele); lanes 3 and 4, transformants E11 and E53, respectively, with restored I4+ perithecial red pigmentation.
Location of putative regulatory sites in the promoter of pksN.
The promoter region of pksN was examined for the presence of putative binding sites for fungal transcription factors. In pksN, we found two binding sites for AflR, the binuclear zinc cluster transcription factor required for gene expression in both the sterigmatocystin and aflatoxin pathways, three binding sites for AreA, a factor regulating genes involved in nitrogen metabolism, and three binding sites for PacC, which regulates genes based on ambient pH signals (Table 1). The AflR sites found in pksN are at the same distance (120 bp) from the translational initiation codon as those found for pksA, involved in aflatoxin biosynthesis in Aspergillus (12).
TABLE 1.
Locations of putative regulatory sites in the promoter of pksN
| Fungal transcription factor | Locations of binding sites (bp)a |
|---|---|
| AflR | −310, −143 |
| AreA | −599, −569, −354 |
| PacC | −161, −116, −96 |
Distance from the translational initiation codon.
DISCUSSION
We isolated a type I PKS gene (pksN) that participates in the biosynthesis of the red pigment present in the perithecial wall of N. haematococca. Prior to this study, the biosynthetic origin of the pigment in the perithecial wall was unknown. The demonstration that I4 encodes a PKS is consistent with the hypothesis that this red perithecial pigment also is a polyketide. Inactivation of the gene did not lead to significant changes with respect to hyphal growth, asexual spore formation, time course of perithecial formation, or sexual spore production. To our knowledge, this is the first report of the isolation and characterization of a fungal gene involved in the biosynthesis of a red pigment present in a specific cell type of the sexual stage. Although the precise function of this pigment has not yet been established, on the basis of what is known for other fungal spore pigments, particularly melanins (29), it probably protects the ascospores from environmental stresses such as UV irradiation and desiccation. It also could have an inhibitory effect on ascospore germination, since germination was never observed to occur inside red perithecia but does occur within the uncolored perithecia.
This novel PKS provides an opportunity to investigate the biosynthesis of red pigments accumulating in a specific cell type, e.g., ascospores or cleistothecia, in other fungi and the relationship between spore pigments and other secondary metabolites. The presence in the promoter of pksN of putative regulatory sites for three transcription factors, the aflatoxin pathway-specific regulatory protein, AflR, and the global regulatory proteins, AreA and PacC, also found in the promoter of pksA, involved in aflatoxin biosynthesis (12), is very intriguing. This finding suggests that the expressions of pksN and pksA are influenced by the same factors. In addition, the fact that the red pigment of A. nidulans ascospores is structurally similar to norsolonic acid, the first stable intermediate in the biosynthesis of aflatoxins (6), raises interesting questions about an evolutionary relationship between spore pigments and mycotoxins.
In addition, the results reported here show that phylogenetic analysis of revised alignments of fungal PKSs clarified the relationship between them. For example, the phylogenetic position of PKS1 from C. heterostrophus (45), which produces the T toxin, was not clearly established on the basis of the alignment of 240 amino acids of the KS domain (5). When the analyzed region is increased to 440 residues and the FUM1 gene product from G. moniliformis is included, both of these genes cluster with the 6-methylsalicylic synthase (MSAS)-type PKSs. Clustering with either AT, KS, or both domains appears to be correlated with the presence of the dehydratase and ketoreductase domains. Increasing the size of the aligned AT region from 50 to 240 amino acids also identified more highly conserved sequence regions. Therefore the AT and KS domains could be used together to develop degenerate-PCR approaches to efficiently screen for PKS genes.
By sequencing the genomic region surrounding the pksN gene, we also detected other open reading frames with predicted P-450 monooxygenase and O-methyl transferase activities. These enzymatic activities are associated with a PKS in several mycotoxin-biosynthetic pathways (20, 25) and could also be involved in the biosynthesis of the red pigment. Inactivation experiments are in progress in order to determine if these genes are coregulated and part of a new gene cluster.
Acknowledgments
We thank Olivier Lespinet and Marie Dufresne for helpful comments on the manuscript.
S. Graziani was supported by a fellowship from CNRS. This work benefited from grants from the CNRS (UMR 8621).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Babai-Ahary, A., M. J. Daboussi-Bareyre, and D. Parisot. 1982. Isolation and genetic analysis of self-sterility and perithecial pigmentation mutants in the homothallic isolate of Nectria haematococca. Can. J. Bot. 60:79-84. [Google Scholar]
- 4.Beck, J., S. Ripka, A. Siegner, E. Schiltz, and E. Schweizer. 1990. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 192:487-498. [DOI] [PubMed] [Google Scholar]
- 5.Bingle, L. E., T. J. Simpson, and C. M. Lazarus. 1999. Ketosynthase domain probes identify two subclasses of fungal polyketide synthase genes. Fungal Genet. Biol. 26:209-223. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. W., and J. J. Salvo. 1994. Isolation and characterization of sexual spore pigments from Aspergillus nidulans. Appl. Environ. Microbiol. 60:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaisrisook, C., and J. F. Leslie. 1990. A maternally expressed nuclear gene controlling perithecial pigmentation in Gibberella fujikuroi (Fusarium moniliforme). J. Hered. 81:189-192. [Google Scholar]
- 8.Chang, P. K., J. W. Cary, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B biosynthesis. Mol. Gen. Genet. 248:270-277. [DOI] [PubMed] [Google Scholar]
- 9.Crawford, M. S., F. G. Chumley, C. G. Weaver, and B. Valent. 1986. Characterization of the heterokaryotic and vegetative diploid phases of Magnaporthe grisea. Genetics 114:1111-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daboussi-Bareyre, M. J. 1980. Heterokaryosis in Nectria haematococca: complementation between mutants affecting the expression of two differentiated states. J. Gen. Microbiol. 116:425-433. [Google Scholar]
- 11.Daboussi-Bareyre, M. J., D. Laillier-Rousseau, and D. Parisot. 1979. Contrôle génétique de deux états différenciés de Nectria haematococca. Can. J. Bot. 57:1161-1173. [Google Scholar]
- 12.Ehrlich, K. C., B. G. Montalbano, J. W. Cary, and P. J. Cotty. 2002. Promoter elements in the aflatoxin pathway polyketide synthase gene. Biochim. Biophys. Acta 1576:171-175. [DOI] [PubMed] [Google Scholar]
- 13.Feng, G. H., and T. J. Leonard. 1995. Characterization of the polyketide synthase gene (pksL1) required for aflatoxin biosynthesis in Aspergillus parasiticus. J. Bacteriol. 177:6246-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulton, T. R., N. Ibrahim, M. C. Losada, D. Grzegorski, and J. S. Tkacz. 1999. A melanin polyketide synthase (PKS) gene from Nodulisporium sp. that shows homology to the pks1 gene of Colletotrichum lagenarium. Mol. Gen. Genet. 262:714-720. [DOI] [PubMed] [Google Scholar]
- 15.Galtier, N., M. Gouy, and C. Gautier. 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Applic. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 16.Graziani, S. 2002. Caractérisation de gènes impliqués dans le déterminisme de deux modifications contagieuses chez Nectria haematococca. Ph.D. thesis. Université Paris 11, Orsay, France.
- 17.Guadet, J., J. Julien, J. F. Lafay, and Y. Brygoo. 1989. Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Mol. Biol. Evol. 6:227-242. [DOI] [PubMed] [Google Scholar]
- 18.Gurr, S. J., S. E. Unkles, and J. R. Kinghorn. 1987. The structure and organization of nuclear genes in filamentous fungi, p. 93-139. In J. R. Kinghorn (ed.), Gene structure in eukaryotic microbes. IRL Press, Oxford, England.
- 19.Hansen, H. N., and W. C. Snyder. 1943. The dual phenomenon and sex in Hypomyces solani f. sp. cucurbitae. Am. J. Bot. 30:419-422. [Google Scholar]
- 20.Hohn, T. M., A. E. Desjardins, and S. P. McCormick. 1995. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 248:95-102. [DOI] [PubMed] [Google Scholar]
- 21.Hopwood, D. A., and D. H. Sherman. 1990. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24:37-66. [DOI] [PubMed] [Google Scholar]
- 22.Howe, H. B. J., and T. E. Johnson. 1976. Phenotypic diversity among alleles at the per-1 locus of Neurospora crassa. Genetics. 82:595-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchinson, C. R. 1999. Microbial polyketide synthases: more and more prolific. Proc. Natl. Acad. Sci. USA 96:3336-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kealey, J. T., L. Liu, D. V. Santi, M. C. Betlach, and P. J. Barr. 1998. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. USA 95:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 26.Kurobane, I., L. C. Vining, A. G. McInnes, and N. N. Gerber. 1980. Metabolites of Fusarium solani related to dihydrofusarubin. J. Antibiot. 39:205-214. [DOI] [PubMed] [Google Scholar]
- 27.Lal, R., R. Kumari, H. Kaur, R. Khanna, N. Dhingra, and D. Tuteja. 2000. Regulation and manipulation of the gene clusters encoding type-I PKSs. Trends Biotechnol. 18:264-274. [DOI] [PubMed] [Google Scholar]
- 28.Langfelder, K., B. Jahn, H. Gehringer, A. Schmidt, G. Wanner, and A. A. Brakhage. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187:79-89. [DOI] [PubMed] [Google Scholar]
- 29.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38:143-158. [DOI] [PubMed] [Google Scholar]
- 30.Lee, T., S. H. Yun, K. T. Hodge, R. A. Humber, S. B. Krasnoff, G. B. Turgeon, O. C. Yoder, and D. M. Gibson. 2001. Polyketide synthase genes in insect- and nematode-associated fungi. Appl. Microbiol. Biotechnol. 56:181-187. [DOI] [PubMed] [Google Scholar]
- 31.Linnemannstöns, P., J. Schulte, M. del Mar Prado, R. H. Proctor, J. Avalos, and B. Tudzynski. 2002. The polyketide synthase gene pks4 from Gibberella fujikuroi encodes a key enzyme in the biosynthesis of the red pigment bikaverin. Fungal Genet. Biol. 37:134-148. [DOI] [PubMed] [Google Scholar]
- 32.Malardier, L., M. J. Daboussi, J. Julien, F. Roussel, C. Scazzocchio, and Y. Brygoo. 1989. Cloning of the nitrate reductase gene (niaD) of Aspergillus nidulans and its use for transformation of Fusarium oxysporum. Gene 78:147-156. [DOI] [PubMed] [Google Scholar]
- 33.Mayorga, M. E., and W. E. Timberlake. 1992. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235:205-212. [DOI] [PubMed] [Google Scholar]
- 34.Metz, J. G., P. Roessler, D. Facciotti, C. Levering, F. Dittrich, M. Lassner, R. Valentine, K. Lardizabal, F. Domergue, A. Yamada, K. Yazawa, V. Knauf, and J. Browse. 2001. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290-293. [DOI] [PubMed] [Google Scholar]
- 35.Money, N. P. 1997. Mechanism linking cellular pigmentation and pathogenicity in the rice blast disease. Fungal Genet. Biol. 22:151-152. [DOI] [PubMed] [Google Scholar]
- 36.Nicholson, T. P., B. A. Rudd, M. Dawson, C. M. Lazarus, T. J. Simpson, and R. J. Cox. 2001. Design and utility of oligonucleotide gene probes for fungal polyketide synthases. Chem. Biol. 8:157-178. [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell, K. 2000. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia 92:919-938. [Google Scholar]
- 38.Orbach, M. J. 1994. A cosmid with a HgR marker for fungal library construction and screening. Gene 150:159-162. [DOI] [PubMed] [Google Scholar]
- 39.Parisot, D., M. Devys, and M. Barbier. 1990. Naphtoquinone pigments related to fusarubin from the fungus Fusarium solani. Microbios 64:31-47. [PubMed] [Google Scholar]
- 40.Parisot, D., M. Maugin, and C. Gerlinger. 1984. Genes controlling pigmentation in Nectria haematococca. J. Gen. Microbiol. 130:1543-1555. [Google Scholar]
- 41.Parisot, D., M. Maugin, and C. Gerlinger. 1981. Genetic and epigenetic factors involved in the excretion of naphtaquinone pigments into the culture medium by Nectria haematococca. J. Gen. Microbiol. 126:443-457. [Google Scholar]
- 42.Proctor, R. H., A. E. Desjardins, R. D. Plattner, and T. M. Hohn. 1999. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi mating population A. Fungal Genet. Biol. 27:100-112. [DOI] [PubMed] [Google Scholar]
- 43.Silar, P., and M. J. Daboussi. 1999. Non-conventional infectious elements in filamentous fungi. Trends Genet. 15:141-145. [DOI] [PubMed] [Google Scholar]
- 44.Takano, Y., Y. Kubo, K. Shimizu, K. Mise, T. Okuno, and I. Furusawa. 1995. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol. Gen. Genet. 249:162-167. [DOI] [PubMed] [Google Scholar]
- 45.Yang, G., M. S. Rose, B. G. Turgeon, and O. C. Yoder. 1996. A polyketide synthase is required for fungal virulence and production of the polyketide T-toxin. Plant Cell 8:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, J., P. K. Chang, J. W. Cary, M. Wright, D. Bhatnagar, T. E. Cleveland, G. A. Payne, and J. E. Linz. 1995. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 61:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, J. H., and T. J. Leonard. 1995. Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J. Bacteriol. 177:4792-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]