Abstract
Characterization of Salmonella enterica serovar Enteritidis was refined by incorporating new data from isolates obtained from avian sources, from the spleens of naturally infected mice, and from the United Kingdom into an existing lipopolysaccharide (LPS) O-chain compositional database. From least to greatest, the probability of avian isolates producing high-molecular-mass LPS O chain ranked as follows: pooled kidney, liver, and spleen; intestine; cecum; ovary and oviduct; albumen; yolk; and whole egg. Mouse isolates were most like avian intestinal samples, whereas United Kingdom isolates were most like those from the avian reproductive tract and egg. Non-reproductive tract organ isolates had significant loss of O chain. Isogenic isolates that varied in ability to make biofilm and to be orally invasive produced different O-chain structures at 25°C but not at 37°C. Hens infected at a 91:9 biofilm-positive/-negative colony phenotype ratio yielded only the negative phenotype from eggs. These results indicate that the environment within the hen applies stringent selection pressure on subpopulations of S. enterica serovar Enteritidis at certain points in the infection pathway that ends in egg contamination. The avian cecum, rather than the intestines, is the early interface between the environment and the host that supports emergence of subpopulation diversity. These results suggest that diet and other factors that alter cecal physiology should be investigated as a means to reduce egg contamination.
Salmonella enterica serovar Enteritidis is the leading cause of food-borne salmonellosis worldwide, in part because it is the only one of more than 2,000 serotypes that efficiently contaminates the hen egg and causes human illness (1, 48). S. enterica serovar Enteritidis resembles other pathogenic salmonellae with regard to known virulence mechanisms central to host cell invasion, survival, and growth in the host (7, 9-11, 15, 23, 36, 38, 46, 51, 52). It is important to determine differences between S. enterica serovar Enteritidis and other salmonellae because this information could help reduce egg contamination, specifically, compared to carcass contamination. The egg is produced, marketed, and used by the consumer differently from meat, which suggests that control strategies tailored to the egg industry are needed to realize reductions beyond those already achieved (5). Analysis of strain heterogeneity has established that egg-contaminating S. enterica serovar Enteritidis is predominantly clonal (25, 28, 31, 32, 35) but that it nonetheless generates substantial phenotypic variation that alters the incidence of egg contamination in infection models. Chemotyping of the lipopolysaccharide (LPS) O chain is a sensitive method of phenotypic analysis that combines stoichiometry with statistical analysis to produce clusters of data that correlate with LPS O-chain structure (41). Chemotyping has shown that S. enterica serovar Enteritidis efficiently produces high-molecular-mass (HMM) LPS O chain, whereas Salmonella enterica serovar Typhimurium does not. Chemotyping is rooted in analyses of LPS mutants. A strain of S. enterica serovar Enteritidis with mutations in the genes wzz and rol (encoding the regulator of O-chain length; originally referred to as the chain length determinant or cld) (3, 4) has been most useful for demarcating structural boundaries between isolates that produce low-molecular-mass (LMM) LPS O chain and those that produce HMM LPS O chain (21). Infection of hens with a high systemic dosage of a strain of S. enterica serovar Enteritidis that produces HMM LPS results in regression of the reproductive tract and dramatic loss of production, whereas birds infected with wzz mutant S. enterica serovar Enteritidis remain in production. Thus, HMM LPS is a molecule that influences avian reproductive tract biology in a remarkable way (40). The effect of HMM LPS at low dosage is to mitigate disease in hens, even though strains producing this molecule have cell surface properties that more closely resemble those of Salmonella enterica serovar Typhi, which causes significant illness in people (40, 43). Previous research has shown that the best source for isolates that efficiently produce HMM LPS is the egg (41). However, strains recovered from the egg are at the end point of selection within the chicken; thus, it is important to know whether control measures can be developed to decrease the emergence of subpopulations with reproductive tract tropism earlier in the infection pathway.
Another way in which clonal isolates of S. enterica serovar Enteritidis vary is in the ability to produce biofilm, which is an organic matrix composed of fimbriae, cellulose, flagella, and LPS O chain (2, 19, 45, 47). Some strains that are isogenic yield both biofilm-positive and -negative phenotypes (bf+ and bf−) in culture (19, 29), which is a desirable characteristic of cultures used to test the efficacies of vaccines (39). Once the phenotypes are separated into distinct subpopulations with stable culture characteristics, those cultures producing biofilm (also known as lacy, convoluted, radr) or HMM LPS exhibit enhanced oral invasiveness or an increase in egg contamination efficiency following systemic infection, respectively (19).
The major objective of this research was to expand the database on O-chain structure to better understand how subpopulations contribute to egg contamination at different sites within the infection pathway. For this purpose, S. enterica serovar Enteritidis was cultured from the intestine (pooled duodenum, jejunum, and ileum), paired ceca, organs (pooled liver, kidney, and spleen), reproductive tract tissue (ovary and oviduct), separated egg fractions (yolk and albumen), and whole egg obtained from experimentally infected hens. The LPS O chain was analyzed to determine the yields of rhamnose, which is the neutral sugar that reliably classifies serotype group D O chain as LMM or HMM LPS. Finally, standard bell curves were constructed, using fifth-polynomial curvilinear analysis, from average rhamnose yields and standard deviations (SDs) to visualize subpopulations. To further refine details of the infection pathway leading to egg contamination, LPS O chains from pairs of bf+ and bf− isolates from the United Kingdom were analyzed at permissive and nonpermissive temperatures of 25 and 37°C, respectively. Finally, three hen infection studies were conducted with two subpopulations that differed in their ability to make biofilm and HMM LPS but that were genetically indistinguishable using standardized ribotyping methodology. The combined outcome of these research approaches using stoichiometry, statistical analyses, and animal infection studies is the detection of subpopulation differences that help define the complexity of interactions between this important food-borne pathogen and its avian host.
MATERIALS AND METHODS
Source of isolates.
Tables 1 and 2 list all data that were used in statistical analyses by isolate accession number and source; some data originate with previously conducted investigations (41). The database used here includes 57 new isolates. The isolates were stored at −80°C in 20% glycerol or at 4°C as lyophilized cultures prior to analysis, which preserves phenotype stability for years in regard to the yield of rhamnose (41). United Kingdom isolates (Table 1) were characterized for phenotype upon initial isolation (T. Humphrey and K. Coles, unpublished data), and the phenotype was confirmed once the isolates were received at our laboratory. The term “wild type” is sometimes used in the European literature to describe strains that produce biofilm and that have a functional RpoS sigma factor, but the convention in the United States and in this report is to reserve the term for strains that are bf− and positive for RpoS.
TABLE 1.
Description of United Kingdom S. enterica serovar Enteritidis isolates
| SEPRL accession no.a,b | United Kingdom accession no. | Concn of rhamnose (37°C) (μg/100 μg of LPS) | Concn of rhamnose (25°C) (μg/100 μg of LPS) | Biofilm formation (37°C, 25°C) | HMM LPS production (37°C, 25°C) |
|---|---|---|---|---|---|
| 21119 | 49680 | 16.9 | 9.2 | −, + | +, − |
| 21113 | 27655R | 12.8 | 10.3 | −, + | +, − |
| 21110 | 59907 | 9.6 | 7.4 | −, + | −, − |
| 21129 | LA5 | 5.1 | 9.5 | −, + | −, − |
| 21118 | E | 8.0 | 6.5 | −, + | −, − |
| 21120 | I | 7.4 | 9.7 | −, − | −, − |
| 21128 | EV53 | 9.4 | 15.7 | −, − | −, + |
| 21123 | EV54 | 11.2 | 10.3 | −, − | −, − |
| 21121 | C | 10.1 | 18.1 | −, − | −, + |
| 21114 | 27655S | 8.8 | 13.7 | −, − | −, + |
SEPRL, Southeast Poultry Research Laboratory, Athens, Georgia.
All data are new and have been added to the existing LPS O-chain database for further analysis.
TABLE 2.
Other S. enterica serovar Enteritidis isolatesb
| SEPRL accession no. | Source | Concn of rhamnose (μg/100 μg of LPS) |
|---|---|---|
| 99015 | Whole egg | 36.3 |
| 99016 | Whole egg | 20.5 |
| 99017 | Whole egg | 7.1 |
| 99017 | Whole egg | 7.8 |
| 99017 | Whole egg | 9.6 |
| 99018 | Whole egg | 6.7 |
| 98129 | Whole egg | 9.3 |
| 98131 | Whole egg | 11.1 |
| 21071a | Yolk | 13.4 |
| 21072a | Yolk | 8.2 |
| 21073a | Yolk | 8.8 |
| 21074a | Yolk | 11.6 |
| 21075a | Yolk | 15.6 |
| 21080a | Albumen | 10.6 |
| 21081a | Albumen | 9.5 |
| 21082a | Albumen | 11.6 |
| 21084a | Albumen | 10.1 |
| 21090a | Albumen | 11.7 |
| 98098 | Ovary/oviduct | 9.4 |
| 98099 | Ovary/oviduct | 8.8 |
| 98113 | Ovary/oviduct | 12.2 |
| 98114 | Ovary/oviduct | 9.7 |
| 98115a | Ovary/oviduct | 10.5 |
| 98100 | Pooled organ | 1.8 |
| 98101 | Pooled organ | 0.8 |
| 98102 | Pooled organ | 9.8 |
| 98103 | Pooled organ | 7.1 |
| 98117 | Pooled organ | 11.0 |
| 98118 | Pooled organ | 8.1 |
| 98107a | Ceca | 7.5 |
| 98108a | Ceca | 7.0 |
| 98109a | Ceca | 9.5 |
| 98110a | Ceca | 9.6 |
| 98111 | Ceca | 10.6 |
| 98112 | Ceca | 12.0 |
| 98104 | Intestine | 9.4 |
| 98105a | Intestine | 8.6 |
| 98106a | Intestine | 8.0 |
| 98123a | Intestine | 8.8 |
| 98124a | Intestine | 8.8 |
| 98125a | Intestine | 8.5 |
| 21101a | Intestine | 4.6 |
| 21102a | Intestine | 10.9 |
| 21103a | Intestine | 7.8 |
| 21104a | Intestine | 11.3 |
| 21105a | Intestine | 7.6 |
| 72a | wzz mutant | 11.0 |
| 96a | wzz mutant | 12.0 |
| 120a | wzz mutant | 11.6 |
| 229a | wzz mutant | 12.7 |
| 230a | wzz mutant | 9.8 |
| 231a | wzz mutant | 10.7 |
| 185a | wzz mutant | 7.3 |
| 186a | wzz mutant | 11.0 |
| 99000 | Mouse spleen | 10.4 |
| 99001 | Mouse spleen | 10.8 |
| 99002 | Mouse spleen | 10.0 |
| 99003 | Mouse spleen | 5.0 |
| 99005a | Mouse spleen | 4.1 |
| 99006 | Mouse spleen | 7.0 |
| 99007a | Mouse spleen | 3.9 |
| 99008 | Mouse spleen | 15.8 |
| 99009 | Mouse spleen | 14.8 |
| 99010 | Mouse spleen | 10.5 |
| 99011 | Mouse spleen | 9.5 |
| 99012 | Mouse spleen | 8.0 |
| 99013a | Mouse spleen | 8.6 |
| 99014a | Mouse spleen | 7.8 |
New data added to the database for further analysis.
Avian origin unless otherwise indicated.
Infection of hens.
Specific-pathogen-free mature leghorn hens between 25 and 55 weeks of age housed singly in layer cages were used to compare the abilities of biofilm-forming S. enterica serovar Enteritidis and wild-type strains to contaminate eggs. Contact infection was begun by contaminating the room by injecting 4 to 6 hens out of 24 per experimental group with 106 CFU of S. enterica serovar Enteritidis intravenously. The strains used to challenge hens, namely, SEPRL 22023, SEPRL 21000, and SEPRL 20127, are isogenic as determined by internationally accepted standards using two-enzyme ribotype analysis (35), and they are of phage type 13a (data not shown). The phenotypes of these three strains in regard to the ability to produce HMM LPS O chain (HMM) and biofilm were as follows: 22023, HMM+ bf+; 21000, HMM+ bf−; 20127, HMM− bf−. Strains 22023 and 21000 are wild-type strains that originated from the parental strain SE6 (12, 20). Strain 22023 was obtained as a third-passage liver-spleen isolate subsequent to infection of hens with the parental strain, SE6 (14). Strain 21000 derives from SE6, and it has been shown to produce HMM LPS (43). Strain 20127 is a wzz mutant derived from SE6 by insertion of a kanamycin cassette by transduction and homologous recombination. The number of hens injected intravenously was a function of room size, with one room being ∼50% of the size of the larger room. The birds were euthanatized 21 days postchallenge (not counting the day of infection).
Culture of eggs.
Eggs were collected for 21 days postinfection, labeled individually by bird and date, and stored for 7 days at room temperature prior to transfer to longer-term storage at 4°C. Egg contamination was classified as not detectable (no contamination detected), low incidence (<1% contaminated), or high incidence (>1% contaminated) as previously described (41). The culture of eggs that had been externally disinfected in Lugol's solution, either as whole contents or separated components, was previously described (13, 21, 40). Collection and culture of 1,000 eggs prior to infection did not detect any bacterial contamination. Culture from the rooms, prior to infection of the birds, using a methodology described previously, did not detect any environmental salmonellae (40).
Culture of S. enterica serovar Enteritidis from the intestinal tracts and ceca of hens.
Intestinal isolates were obtained from pooled 1-in. segments of the duodenal loop, the middle of the jejunum, and the ileum 1 in. proximal to the ileocecal junction and cultured as previously described (18). Cecal isolates were obtained by pooling 1 in. of the two terminal pouches of each bird. Briefly, samples were inoculated into cryptic soy broth for 48 h at 37°C. Culture (1 ml) was then transferred to Rappaport-Vassiliadis broth for incubation at 37°C for 24 h. Single colonies were obtained by passage of the Rappaport-Vassiliadis broth culture onto brilliant green (BG) agar (BBL) and streaking it for isolation. Determination of the serotype was initially made by reactivity with serovar group D1 O-antigen factor 9 (tyvelose). Confirmation of the strain genotype was done by ribotype analysis as previously described (35).
Culture of S. enterica serovar Enteritidis from the internal organs of hens.
The collection and culturing of S. enterica serovar Enteritidis from reproductive tract organs (ovary and oviduct) and other internal organs (kidney, liver, and spleen) was previously described (18). Briefly, 1 g each of liver and kidney tissue was collected and pooled with the entire spleen. Reproductive tract organs were collected (ovaries with mature ova removed and 1 in. each of the isthmus, the magnum, and the shell gland). Samples were pulverized in a stomacher and then cultured in brain heart infusion broth for 48 h at 37°C. Final isolation of suspect colonies was done on BG agar.
Extraction of LPS.
Stored isolates were cultured on BG agar and incubated for 16 h at 37°C. Colonies were inoculated into 500-ml Erlenmeyer flasks containing 500 ml of brain heart infusion (Difco) broth supplemented with 10 mM glucose and incubated for 24 h at 37°C or for 48 h at 25°C without shaking. The difference in incubation time was necessary to allow cultures grown at the lower temperature to reach equivalent stationary-phase optical densities at 600 nm of at least 0.85. The bacterial cells were pelleted by centrifugation at 10,000 × g (SLA1500 or SLA3000) for 30 min to 2 h at 4°C in preweighed sterile 250- or 500-ml polypropylene screw-cap bottles (Nalgene NNI 3141-0250 or NNI 3120-9500) with matched lids. The centrifugation time depended upon how well the cells pelleted. After the supernatant was decanted as much as possible, the wet weight of the cell pellet was recorded. The pellet was stored at 4°C overnight and then resuspended in 30 ml of sterile deionized distilled (DD) H2O, placed in a boiling water bath, and stirred vigorously for 30 min. The boiling water was replaced with ice, and the sample was stirred for another 90 min. After being balanced with DD H2O, the sample was centrifuged for 30 min at 10,000 × g and 4°C to pellet cell debris. The supernatant was transferred into a preweighed 250-ml polypropylene bottle and adjusted to 1% acetic acid by addition of concentrated stock. To precipitate crude polysaccharide, 60 ml of ethanol was added to the supernatant, followed by titration to a pH of 6.0 with ∼50 ml of 3 M sodium acetate (pH 5.2). The sample was incubated at −20°C for at least 16 h to facilitate precipitation, and the precipitate was pelleted the next day by centrifugation at 10,000 × g in a prechilled rotor. The supernatant was decanted, and the pellet was dried under nitrogen and then dissolved in 2 ml of nuclease buffer (21). Additional nuclease buffer (3.0 ml) was added if needed to facilitate solubilization. The tubes containing nuclease buffer were vortexed and centrifuged again to remove precipitate from the sides of the tubes. Nucleic acids were removed by incubating the sample with 2 μg of DNase/ml and 10 μg of RNase/ml for 16 h at 37°C as previously described (21). Proteinase K (500 μg) was added twice to the sample at 12-h intervals during incubation for 24 h at 42°C. After enzymatic digestion of contaminating proteins, the sample was centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was transferred to dialysis tubing with a 6,000- to 8,000-molecular-weight cutoff (Spectro-Por) and dialyzed in DD H2O for 3 days, with the water changed three times a day. After being frozen at −20°C for 16 h, the sample was lyophilized in a preweighed tube, and a final weight was recorded for the derivatized alditol acetates.
Preparation of alditol acetates for gas-chromatographic analysis.
To each LPS sample weighing between 500 μg and 1 mg, 20 μg of inositol was added as an internal sample. To hydrolyze the sample, 0.5 ml of 2 M trifluoroacetic acid was added and the sample was heated at 121°C for 2 h. The acid was removed by evaporation with nitrogen or a dry air stream (Pierce Reacti-ThermIII heating module). Traces of acid were removed by washing the sample twice with 250 μl of isopropanol and evaporating the sample back to dryness each time. To reduce the sample, the hydrolysate was dissolved in 0.25 ml of 1 M NH4OH solution containing 10 mg of sodium borodeuteride/ml, and sample was then incubated at ambient temperature for 1 h. Glacial acetic acid (1 or 2 drops) was titrated until bubbling ceased, and the sample was evaporated to dryness. Methanol-acetic acid (0.5 ml of a 9:1 [vol/vol] solution) was added four times, and the sample was evaporated to dryness each time. Methanol (0.5 ml) was added four times, and the sample was evaporated to dryness each time. To O acetylate the sample, 0.1 ml of acetic anhydride and 0.02 ml of 1-methyl imidazole were added. The sample was vortexed (cap on) and incubated at ambient temperature for 15 min. Distilled H2O (0.5 ml) was added, and the sample was incubated for 10 min. Chloroform (0.5 ml) was added and mixed with the sample by vortexing. The lower organic layer containing derivatized product was transferred to a new tube, and the sample was evaporated. Methanol (2 drops) was added, and the sample was evaporated again to assure dryness. The final residue was dissolved in 50 to 100 μl of methanol for analysis by gas-liquid chromatography.
Gas-chromatographic analysis.
The gas chromatograph was an HP6890 Plus series with an autoinjector, equipped for capillary columns and with a flame ionization detector. Samples were run on a 25-m silicone midpolar column (Quadex, catalog no. 007-17-25W-0.25F) at an initial temperature of 150°C for 2 min, which was increased at a rate of 4°C/min and held at 260°C for 10 min or until the heptose peak was present. Peak positions for O-chain sugars were determined by preequilibration of the column with a mix of the standard sugars inositol, rhamnose, fucose, arabinose, mannose, glucose, galactose, N-acetylglucosamine, and N-acetylgalactosamine at an average molarity of 27 ± 4.0 mM per sugar. All yields were normalized to heptose as previously described (21, 41, 43). Smooth isolates are classified as those that produce HMM and LMM LPS. Isolates with yields of at least 12 μg of rhamnose/100 μg of LPS produce ≥50% HMM LPS, whereas those with between 4 and 12 μg of rhamnose/100 μg of LPS produce more LMM than HMM LPS. Isolates that yield <4 μg of rhamnose/100 μg of LPS no longer react well with group D1 serotype reagents (also called somatic O antigen), and thus, they are described as having a rough phenotype.
Detection of biofilm phenotype.
To detect the biofilm phenotype, a culture was plated as a lawn at a concentration of 50 CFU per plate on BG agar (19). The plates were incubated for 16 h at 37°C and then incubated for an additional 48 h at 25 ± 2°C. The number of biofilm-producing colonies was recorded as a percentage of the total colonies counted, with a minimum of 100 colonies counted per culture. Biofilm production is both medium and temperature dependent, and other laboratory media that will support its formation have been described (29).
Statistical analysis.
A practical feature of the stoichiometry associated with LPS structure is that shifts in clustering correlate with a more significant change in cell surface composition than is readily apparent by probability analysis, because LMM and HMM LPS O chains share local elements within a single repeat unit that nonetheless differ globally in mass. The wzz mutant of S. enterica serovar Enteritidis, which lacks the primary O-chain regulator of length, is used to help detect global changes in structure that differentiate cluster groups by providing boundaries (41). The mutant is also used to improve probability analysis by Student's t test, because the artificially elongated LMM LPS that it produces defines the upper limits for this structure. Rhamnose is a neutral sugar that is used to quantify structure for serovars of group B and D salmonellae, because it is one of four stoichiometric sugars in the O repeat unit that has no other known cellular source.
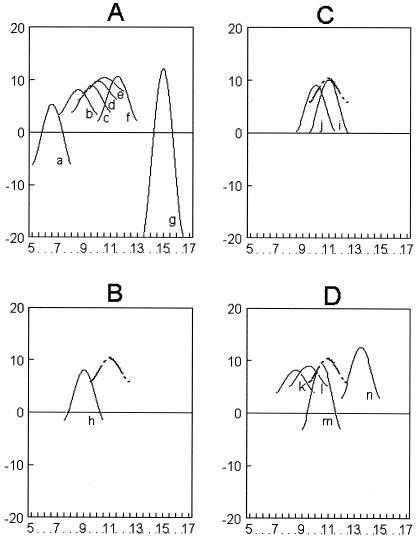
To visualize subpopulation differences, the average yield of rhamnose for each sample group was plotted to its numerical place value on the abscissa. Then, data points that were three SDs from the average, as well as the average itself, were used to generate a bell curve by application of the fifth-polynomial differential (Slidewrite version 6). For example, the average for the yolk group was plotted in the data field at 11.5 on the x axis (see Fig. 1A, curve f). To generate the curve, data of 2.17, 5.28, 8.39, 11.5, 8.39, 5.28, and 2.17 were generated by subtracting the SD of 3.11 μg of rhamnose/100 μg of LPS from the average of 11.5 μg of rhamnose/100 μg of LPS. A fifth-polynomial curve fit equation was applied to generate a standard bell curve to show the range of data that might be encountered for this one isolate in any particular evaluation. The area under the curve thus reflects the average of the data set, as well as the deviation that can be expected to occur within the subpopulation. Some data sets generated negative numbers due to a large SD, and these negative numbers were plotted as such. This method of analysis is used as an analytical tool to visualize bacterial subpopulation diversity according to LPS composition characteristics only. Tables 1 and 2 list all raw data used to generate the curves shown in Fig. 1. The bell curve for wzz mutant S. enterica serovar Enteritidis LPS is included in Fig. 1B, C, and D to facilitate comparison between groups; however, it is not included in Fig. 1A because it is obscured by curve e. Because structural detail shared between LMM and HMM LPS structures makes the application of probability analysis by Student's t test less sensitive to significant differences in structure, a high degree of statistical certainty that structures are different is assigned at a P value of <0.075, as determined by taking 50% of 0.150, which is the probability that wzz mutants and whole-egg isolates are similar (Table 3). Whole egg was used to define parameters of detection for HMM LPS, because it is the source that was previously shown to favor the production of HMM LPS (41).
FIG. 1.
Curvilinear analysis of Salmonella enteritidis subpopulation diversity. All curves were generated by application of fifth-polynomial analysis of the average yield of rhamnose ± 3 SDs as described in Materials and Methods. Curve set A: avian isolates, as recovered from organs (a) (pooled kidney, liver, and spleen), intestine (b) (pooled duodenum, jejunum, and ileum), ceca (c), ovary and oviduct (d), albumen (e), yolk (f), and whole egg (g). Cultures were grown at 37°C as otherwise described in the text. The control curve generated for wzz S. enteritidis, shown in sets B to D as a broken, line overlaps with albumen (e) in curve set A. Curve set B: mouse spleen isolates (h), compared to the curve for wzz S. enteritidis (broken line). Curve set C: Overall averages for United Kingdom isolates grown at 25°C (i) and United Kingdom isolates grown at 37°C (j). The broken line is the curve for wzz S. enteritidis. Curve set D: United Kingdom isolates, grown at the temperature indicated and divided by phenotype as follows: bf+, 25°C (k); bf−, 37°C (l); bf+, 37°C (m); bf−, 25°C (n). See Table 1 for accession and phenotype information for United Kingdom isolates. The broken line is the curve for wzz S. enteritidis.
TABLE 3.
Characterization of S. enterica serovar Enteritidis subpopulations by LPS chemotype
| Sourcec | Fig. 1 curve label | No. of isolates analyzed | % of isolates with phenotypeb:
|
Avg yield of rhamnose μg/100 μg of LPS (SD) | Student's t test P value
|
No. of subpopulations | |||
|---|---|---|---|---|---|---|---|---|---|
| HMM LPS | LMM LPS | Rough | Whole-egg comparisona | wzz mutant comparison | |||||
| Organ | a | 6 | 0 | 66.7 | 33.3 | 6.43 (4.21) | 0.056* | 0.010* | 2 |
| Intestine | b | 11 | 0 | 100.0 | 0 | 8.57 (1.77) | 0.031* | 0.007* | 1 |
| Ceca | c | 6 | 0 | 100.0 | 0 | 9.37 (1.88) | 0.126 | 0.082 | 1 |
| Ovary/oviduct | d | 5 | 0 | 100.0 | 0 | 10.12 (1.31) | 0.180 | 0.238 | 1 |
| Albumen | e | 5 | 0 | 100.0 | 0 | 10.70 (0.95) | 0.207 | 0.468 | 1 |
| Yolk | f | 5 | 20.0 | 80.0 | 0 | 11.52 (3.11) | 0.256 | 0.288 | 2 |
| Whole egg | g | 6 | 33.3 | 66.7 | 0 | 15.20 (11.51) | 0.229 | 2 | |
| Mouse spleen | h | 14 | 14.3 | 78.6 | 7.1 | 9.01 (3.53) | 0.039* | 0.101 | 3 |
| UK all, 25°C | i | 10 | 20.0 | 80.0 | 0 | 11.04 (3.67) | 0.152 | 0.424 | 2 |
| UK all, 37°C | j | 10 | 10.0 | 90.0 | 0 | 9.93 (3.22) | 0.095 | 0.258 | 2 |
| UK bf+, 25°C | k | 5 | 0 | 100.0 | 0 | 8.58 (1.57) | 0.120 | 0.019* | 1 |
| UK bf−, 37°C | l | 5 | 0 | 100.0 | 0 | 9.38 (1.42) | 0.149 | 0.075* | 1 |
| UK bf+, 37°C | m | 5 | 40.0 | 60.0 | 0 | 10.48 (4.54) | 0.209 | 0.436 | 2 |
| UK bf−, 25°C | n | 5 | 60.0 | 40.0 | 0 | 13.50 (3.56) | 0.382 | 0.042* | 2 |
| wzz mutant | 8 | 0 | 100.0 | 0 | 10.77 (1.65) | 0.150 | 1 | ||
Probability values of ≤0.075 indicate significant difference (*).
Phenotype designations (μg of rhamnose/100 μg of LPS): HMM, >12; LMM, >4, <12; rough, <4.
UK, United Kingdom.
RESULTS
Subpopulation characteristics of S. enterica serovar Enteritidis from different avian sources.
LPS compositional analysis of isolates generated the following rank order, from least to greatest, that describes the correlation between the source and recovery of HMM LPS (Fig. 1A): pooled organs (kidney, liver, and spleen) (curve a), pooled intestine (duodenum, jejunum, and ileum) (curve b), cecum (curve c), ovary-oviduct (curve d), albumen (curve e), yolk (curve f), and whole egg (curve g). The overall average for avian isolates, excluding those from the United Kingdom, was 9.98 ± 5.01 μg of rhamnose/100 μg of LPS. SDs from the different sources formed three clusters (Table 3), which were either <2, between 3 and 5, or >10 μg of rhamnose/100 μg of LPS.
Characteristics of homogeneous subpopulations of S. enterica serovar Enteritidis from avian sources.
SDs of <2 indicated that S. enterica serovar Enteritidis was as homogeneous as the wzz mutant; thus, intestine, ceca, reproductive tract organs (ovary and oviduct), and albumen sources yielded only one major LPS O-chain subpopulation (Fig. 1A, curves b, c, d, and e, respectively). When sources comprising homogeneous subpopulations were compared by probability analysis to the wzz mutant, with significance assigned at a P value of <0.075, P values of 0.01 (intestine), 0.08 (ceca), 0.24 (ovary-oviduct), and 0.47 (albumen) were obtained. These results indicated that the avian intestine is a highly selective environment for a homogeneous subpopulation of S. enterica serovar Enteritidis that produces an LMM LPS (Table 3). Compared to the whole-egg source, the high P value for albumen suggested that it was the most selective environment for subpopulation homogeneity associated with a gain in O chain (Table 3). Stringent selection pressure was not evident within ceca, which suggests that this is an early environment within the hen, following initial colonization of the intestine, which supports the emergence of strains more likely to produce HMM LPS (Fig. 1A). These findings strongly suggest that the microenvironment of the intestine selects for a different LPS structure than do those of the ceca, reproductive tract organs, and eggs.
Emergence of subpopulation diversity in avian organs and yolk.
Of the avian sources analyzed, organ (kidney, liver, and spleen) and yolk samples had intermediate SDs between 3 and 5 (Fig. 1A, curves a and f). However, these two sources applied very different selection pressures to S. enterica serovar Enteritidis, because the average rhamnose yield was significantly lower from organ than that from yolk, with a probability of 0.026 that the two populations were similar (Table 3). An SD range between 3 and 5 was found for sources that produced at least two out of the three LPS structures of interest, namely, rough, LMM, and HMM LPS (Table 3). A correlation of SD with emerging subpopulation diversity is further supported by the finding that the overall average of 9.98 ± 10 μg of rhamnose/100 μg of LPS, or two SD, includes all but 2 data points out of 46 analyzed (Table 2) (99015 and 99016 were outliers). The P values for organ and yolk isolates compared to the wzz mutant were 0.01 and 0.29, respectively. Thus, avian organs yielded two subpopulations, one producing LMM LPS O chain and one undergoing significant loss of O chain, whereas yolk subpopulations produced both LMM and HMM LPS (Table 3). The selection for loss of O chain by organs was more pronounced than that within intestines, because 33.3% of the isolates recovered from this source were clearly rough, whereas none of those from intestines were (Table 3).
The egg as a source of mixed selection pressures.
Whole-egg isolates generated a third class of SD, which measured 11.51 μg of rhamnose/100 μg of LPS (Table 3). This result was due to the inclusion of the two outlier data points, 99015 and 99016, in calculations (Tables 1 and 2). Without these data, the average rhamnose yield for whole-egg isolates was 8.6 ± 1.69 μg of rhamnose/100 μg of LPS. The curve generated without outlier values overlaid the curve generated from avian intestinal samples (Fig. 1A, curve b). These results suggest that whole egg is an environment that polarizes S. enterica serovar Enteritidis isolates into those that are similar to avian intestinal isolates and those that maximize HMM LPS production. The anatomical juxtaposition of two egg microenvironments that both support production of HMM LPS, namely, albumen as a highly selective environment and yolk as an environment that supports diversity, might contribute in some way to enhancement of virulence for humans. The only known association is that the causative agent of typhoid fever, namely, S. enterica serovar Typhi, produces HMM LPS; thus, selection pressure within the egg could inadvertently enhance the infectivity of S. enterica serovar Enteritidis for humans. Whole egg is not a growth site that would be typically encountered by S. enterica serovar Enteritidis except during food preparation, because the yolk and albumen are separated by a membrane. It is noteworthy that food storage practices that alter the integrity of the yolk membrane have been identified as a risk factor for infection in humans. Further research is in progress to investigate how different components of the egg alter the subpopulation biology of S. enterica serovar Enteritidis.
The mouse as a source of S. enterica serovar Enteritidis diversity in the henhouse environment.
Mouse spleen and avian intestinal sample groups had similar averages that indicated that selection pressures for both sample sources were biased toward loss of O chain (Table 3). However, the mouse spleen supported greater diversity than the avian intestine, as reflected by SDs of 3.53 and 1.77 μg of rhamnose/100 μg of LPS, respectively (Fig. 1B and A, curves h and b, respectively). It was the only source that yielded three subpopulations of S. enterica serovar Enteritidis (Table 3); thus, the spleen of the mouse polarizes subpopulations of S. enterica serovar Enteritidis, as did whole egg. These observations further support the concept that the mouse generates an unusual degree of S. enterica serovar Enteritidis subpopulation diversity compared to avian sources and that it has the potential to constantly seed the henhouse environment with orally invasive phenotypes (16). Understanding the role of the mouse in the infection pathway for S. enterica serovar Enteritidis is important, because this pest has been repeatedly identified as an on-farm risk factor associated with production of contaminated eggs.
Role of biofilm in the egg contamination process.
Strains showing the biofilm phenotype constitute a well-characterized subpopulation of S. enterica serovar Enteritidis that is often isolated from the mouse spleen. The proposed role of the biofilm-forming strain in egg contamination is that it is a helper for egg-contaminating strains, although it is itself poor at egg contamination (19). Recently, a general concept in epidemiology has been proposed that suggests that major changes in patterns of disease occur when the mucosal invasiveness of a pathogen increases (49). To explore whether this biology is relevant to the epidemic of human salmonellosis caused by S. enterica serovar Enteritidis, two analyses were undertaken to investigate subpopulation fluctuations of S. enterica serovar Enteritidis. The first approach was to analyze a group of biofilm-forming isolates from the United Kingdom for LPS O-chain microheterogeneity. This group of isolates is unique, because they have stable phenotypic characteristics and they are paired with isogenic variants that do not make biofilm. The other approach was to infect hens with a set of isogenic domestic isolates that differed in LPS characteristics and biofilm formation.
LPS O-chain characteristics of S. enterica serovar Enteritidis isolates that vary in production of biofilm.
Chemotyping of all of the United Kingdom isolates at both 25 and 37°C yielded an average of 11.0 ± 3.67 and 9.93 ± 3.22 μg of rhamnose/100 μg of LPS, respectively. A P value of 0.245 indicated that similar structures were produced; in addition, curvilinear analysis showed that the two data sets overlapped substantially with each other and with data from the wzz mutant (Fig. 1C, curves i and j). The isolates were then divided according to biofilm phenotype (Table 1). Biofilm production is temperature dependent, and thus, analyses were made at the permissive temperature of 25°C and the nonpermissive one of 37°C. There was no significant difference between the two phenotype groups when isolates were grown at 37°C (Table 3), and curvilinear analysis showed only a modest offset between otherwise-overlapping curves (Fig. 1D, curves m and l). bf+ S. enterica serovar Enteritidis isolates yielded two similar subpopulations at 25 and 37°C, as indicated by a probability of 0.190. However, comparison of these averages to results for the wzz mutant suggests that growth of bf+ S. enterica serovar Enteritidis at 25°C decreased O-chain mass significantly, whereas growth at 37°C did not (P = 0.019 and 0.436, respectively) (Table 3). Curvilinear analysis shows the decrease in O-chain mass as a near-complete separation of curves (Fig. 1D, curves k and m). These results indicate that this group of biofilm-producing S. enterica serovar Enteritidis strains makes LMM LPS when grown at the permissive temperature of 25°C.
bf− S. enterica serovar Enteritidis grown at 25 and 37°C produced a significant difference in O-chain yields, with a probability of 0.029 that LPS O-chain mass was increased at 25°C (Fig. 1D, curves n and l). The probability that bf+ and bf−isolates produced different O-chain structures at 37°C was 0.341, whereas at 25°C it was 0.014. Therefore, the biofilm-permissive temperature of 25°C polarized LPS O-chain mass according to both the biofilm phenotype and the O-chain structure. The results suggest that growth conditions that support biofilm production are somehow similar to those in whole egg at 25°C, because both sources polarized subpopulations as defined by the generation of significantly different LPS O-chain structures. When the results from both phenotypes are considered, it appears that the environment is an important facilitator of egg contamination by providing signals that coordinate the physiologies of helper and effector subpopulations.
The hen as a selective host for reproductive tract-adapted S. enterica serovar Enteritidis.
To assess the degree to which selection pressure occurs in egg contamination following invasion from oral infection, three groups of hens were contact infected with one of three different isogenic strains of S. enterica serovar Enteritidis that varied in LPS O-chain structure and the ability to form biofilm. The first isolate had been recovered as a third-passage bird isolate from pooled liver and spleen (14). It had a bf+/bf− colony phenotype ratio of 91:9 and was capable of producing HMM LPS due to the presence of the wild type as the minority population; thus, the phenotype was HMM+ bf+. The second strain was the wzz mutant, which did not produce HMM LPS or biofilm but did produce LMM LPS (HMM− bf−) (21). In previous infection studies, this laboratory construct was consistently more virulent than field strains, and thus, it facilitates comparison of the results from these animal experiments to previous results. The third strain was wild-type S. enterica serovar Enteritidis, which produces HMM LPS but not biofilm (HMM+ bf−). Previous studies had demonstrated that it contaminated eggs at low incidence unless mixed with an orally invasive phenotype; in addition, it was capable of mitigating signs of disease in hens at low dosage and also demonstrated an ability to greatly alter the reproductive tract physiology of hens at high dosage (40).
Results were that 15 of 399 eggs (3.8%) from hens infected with the mixed phenotype (HMM+ bf+) were contaminated, 43 of 388 eggs (11.1%) from hens infected with wzz mutant S. enterica serovar Enteritidis (HMM− bf−) were contaminated, and 5 of 771 eggs (0.6%) from hens infected with wild-type S. enterica serovar Enteritidis (HMM+ bf−) were contaminated. Thus, wild-type S. enterica serovar Enteritidis mixed with the bf+ phenotype produced a high incidence of egg contamination (>1%), whereas the wild type alone correlated with a low incidence of egg contamination. In spite of the fact that the incidence of egg contamination was high following infection with a mixed-phenotype strain, 100% of the isolates recovered from eggs were bf−. This meant that an initial bf+/bf− ratio of 91:9 had changed to 0:100 by the time the isolates were recovered from the egg. These results strongly support the concept that a helper phenotype that is itself not recovered from eggs is an important part of the epidemiology associated with this pathogen. These results are in agreement with past egg contamination experiments, because the wzz mutant S. enterica serovar Enteritidis was again exceptionally virulent in these experiments, as it was in previous studies (40).
DISCUSSION
The environment of the hen intestine appears to act as an early clonal selector for S. enterica serovar Enteritidis strains that can initially colonize hens and subsequently develop reproductive tract tropism. In contrast, the mouse supplies invasive phenotypes within the henhouse that have time to respond to environmental conditions and to develop certain virulence properties most correlated with oral invasiveness prior to the infection of hens. If the theory is generally correct that emergent mucosal invasiveness is rate limiting for epidemic potential (49), then the coordinated emergence of an invasive subpopulation of S. enterica serovar Enteritidis capable of efficient oral infection with a parenterally adapted subpopulation that has tropism for the avian reproductive tract is a convergence of events that has had a disproportionate effect on human health. A need for coordination of subpopulations with specialized virulence functions provides a framework for understanding how S. enterica serovar Enteritidis can be prevalent in the poultry environment without resulting in egg contamination, why some vector hosts contribute more than others to high-incidence egg contamination, and why outbreaks occur sporadically (8, 34).
These results also suggest that S. enterica serovar Enteritidis differs from other paratyphoid salmonellae in having an alternative pathway of development that targets the avian reproductive tract. S. enterica serovar Enteritidis may be limited in its pathogenic potential without this alternative pathway, because it lost O chain in nonreproductive tract organs. This finding in part provides a plausible explanation for why this pathogen was of negligible incidence prior to its developing an association with eggs, because pathogenic paratyphoid salmonellae in general require LPS O chain for invasiveness to maintain complement resistance (22, 44). Recently, fepE has been proposed as a second regulator-of-length gene, and it too contributes to complement resistance (37); however, compositional analysis of this mutant is lacking, and it has not been determined if this mutant is growth limited in comparison to wild-type S. enterica serovar Enteritidis that grows to high cell density (17). S. enterica serovar Enteritidis may be unusual in its tendency to alter O-chain microheterogeneity, because it is not uncommon to find rough isolates resulting from spontaneous mutation of wzy, the O-chain polymerase gene (21). Further research is needed to determine if nonpathogenic salmonellae lose O-chain mass in non-reproductive tract organs as a general phenomenon, and conversely, to see if pathogenic serovars gain or maintain mass once organ invasion occurs. The presence of marginally rough phenotypes has been previously noted to compromise the accuracy of using immunodiagnostics to accurately classify Salmonella as rough or smooth (21).
Ambient temperature is a regulatory signal for LPS O-chain structure, just as it is for biofilm formation. However, it is probable that there are other regulatory signals that coordinate the emergence of the two phenotypes. Some signals may favor the outgrowth of one phenotype over another. In the United Kingdom, the bf+ phenotype is considered the wild type, whereas the negative phenotype is the wild type in the United States (T. Humphrey, K. Coles, and J. Guard-Petter, personal communication). Considering that egg contamination in Europe has a higher incidence than in the United States, this reported difference between the continents may give a first indication of a shift in phenotype that presages high-incidence egg contamination. Both S. enterica serovar Enteritidis and S. enterica serovar Typhimurium produce biofilm, whereas only S. enterica serovar Enteritidis efficiently produces HMM LPS. These findings suggest that HMM LPS and avian reproductive tract tropism differentiate the cell membrane and the biology of S. enterica serovar Enteritidis from those of other paratyphoid salmonellae, whereas biofilm formation does not.
The paired ceca of poultry appear to be an early interface between environment and host that correlates with emerging subpopulation diversification. Certain anatomical aspects of the digestive tracts of farm animals distinguish their abilities to efficiently convert fibrous plant materials to energy. In brief, all farm animals have some anatomical site that facilitates the conversion of feedstuffs into volatile fatty acids, which are then converted into glucose for metabolic processes that support efficient production of food and fiber. Poultry have paired ceca, the horse has a single cecum and a fermenting large intestine, and the true ruminants have the four-compartment stomach. Without these structures and an associated enhanced ability to digest a range of feedstuffs, it is unlikely that modern farm animals could reach the level of production that they have. The pig is the farm animal with a monogastric digestive tract that most closely resembles that of humans, but even it has a well-developed cecum compared to the vestigial human organ (the appendix). If gut physiology is an important reason why salmonellae persistently colonize farm animals, then the cecal physiology of the hen may play a specific role in supporting the emergence of S. enterica serovar Enteritidis with tropism for the reproductive tract. There is some molecular evidence that the cecum supports the growth and colonization of subpopulations of Salmonella that vary in LPS O-chain composition, whereas the intestine favors smooth strains (33). Gut physiology can be manipulated, and many types of biological controls and diets have been designed with the idea of excluding pathogenic salmonellae from the gut (6, 24, 26, 27, 30, 42, 50). Refinement of these approaches to suppress the emergence of subpopulations of S. enterica serovar Enteritidis that have tropism for the reproductive tract of the hen might further reduce the number of contaminated eggs that reach the market. Differences in nutrient sources, molting practices, and other dietary management practices among continents and regions could have a substantial impact on subpopulation dynamics. In regard to S. enterica serovar Enteritidis, additional research on how avian cecal biology alters subpopulation diversification will be required in order to develop application from theory.
Acknowledgments
This work was made possible by the technical contributions of Joyce Jacks.
Funding was provided by USDA CRIS grant 6612-32000-042 and Intervet CRADA grant 58-3K95-0-819, which provided salary support for C. Morales. The United Kingdom component of this work was funded by the Health Protection Agency.
REFERENCES
- 1.Anonymous. 1988. Salmonella enteritidis phage type 4: chicken and egg. Lancet ii:720-722. [PubMed]
- 2.Austin, J. W., G. Sanders, W. W. Kay, and S. K. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295-301. [DOI] [PubMed] [Google Scholar]
- 3.Bastin, D. A., G. Stevenson, P. K. Brown, A. Haase, and P. R. Reeves. 1993. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol. Microbiol. 7:725-734. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor, R. A., G. E. Haraguchi, R. A. Hull, and S. I. Hull. 1991. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 173:5699-5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogan, T. A., and T. J. Humphrey. 2003. The rise and fall of Salmonella Enteritidis in the UK. J. Appl. Microbiol. 94:114-119. [DOI] [PubMed] [Google Scholar]
- 6.Corrier, D. E., B. Hargis, A. Hinton, Jr., D. Lindsey, D. Caldwell, J. Manning, and J. DeLoach. 1991. Effect of anaerobic cecal microflora and dietary lactose on colonization resistance of layer chicks to invasive Salmonella enteritidis. Avian Dis. 35:337-343. [PubMed] [Google Scholar]
- 7.Cotter, P. A., and V. J. DiRita. 2000. Bacterial virulence gene regulation: an evolutionary perspective. Annu. Rev. Microbiol. 54:519-565. [DOI] [PubMed] [Google Scholar]
- 8.Davies, R. H., and M. Breslin. 2003. Persistence of Salmonella enteritidis phage type 4 in the environment and arthropod vectors on an empty free-range chicken farm. Environ. Microbiol. 5:79-84. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 10.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay, B. B., and J. H. Brumell. 2000. Salmonella interactions with host cells: in vitro to in vivo. Philos. Trans. R Soc. Lond. B Biol. Sci. 355:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gast, R. K., and C. W. Beard. 1990. Production of Salmonella enteritidis-contaminated eggs by experimentally infected hens. Avian Dis. 34:438-446. [PubMed] [Google Scholar]
- 13.Gast, R. K., J. Guard-Petter, and P. S. Holt. 2002. Characteristics of Salmonella enteritidis contamination in eggs after oral, aerosol, and intravenous inoculation of laying hens. Avian Dis. 46:629-635. [DOI] [PubMed] [Google Scholar]
- 14.Gast, R. K., J. Guard-Petter, and P. S. Holt. 2003. Effect of prior serial in vivo passage on the frequency of Salmonella enteritidis contamination in eggs from experimentally infected laying hens. Avian Dis. 47:633-639. [DOI] [PubMed] [Google Scholar]
- 15.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guard-Petter, J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421-430. [DOI] [PubMed] [Google Scholar]
- 17.Guard-Petter, J. 1998. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl. Environ. Microbiol. 64:2166-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guard-Petter, J., D. J. Henzler, M. M. Rahman, and R. W. Carlson. 1997. On-farm monitoring of mouse-invasive Salmonella enterica serovar Enteritidis and a model for its association with the production of contaminated eggs. Appl. Environ. Microbiol. 63:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guard-Petter, J., L. H. Keller, M. M. Rahman, R. W. Carlson, and S. Silvers. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol. Infect. 117:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guard-Petter, J., B. Lakshmi, R. Carlson, and K. Ingram. 1995. Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method. Appl. Environ. Microbiol. 61:2845-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guard-Petter, J., C. T. Parker, K. Asokan, and R. W. Carlson. 1999. Clinical and veterinary isolates of Salmonella enterica serovar Enteritidis defective in lipopolysaccharide O-chain polymerization. Appl. Environ. Microbiol. 65:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulig, P. A. 1996. Pathogenesis of systemic disease, p. 2774-2787. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella. Cellular and molecular biology, vol. 2. ASM Press, Washington, D.C.
- 23.Gunn, J. S. 2001. Bacterial modification of LPS and resistance to antimicrobial peptides. J. Endotoxin Res. 7:57-62. [PubMed] [Google Scholar]
- 24.Heres, L., B. Engel, F. van Knapen, M. C. de Jong, J. A. Wagenaar, and H. A. Urlings. 2003. Fermented liquid feed reduces susceptibility of broilers for Salmonella enteritidis. Poult. Sci. 82:603-611. [DOI] [PubMed] [Google Scholar]
- 25.Hilton, A. C., and C. W. Penn. 1998. Comparison of ribotyping and arbitrarily primed PCR for molecular typing of Salmonella enterica and relationships between strains on the basis of these molecular markers. J. Appl. Microbiol. 85:933-940. [DOI] [PubMed] [Google Scholar]
- 26.Hinton, A., Jr., D. E. Corrier, G. E. Spates, J. O. Norman, R. L. Ziprin, R. C. Beier, and J. R. DeLoach. 1990. Biological control of Salmonella typhimurium in young chickens. Avian Dis. 34:626-633. [PubMed] [Google Scholar]
- 27.Hinton, A., Jr., D. E. Corrier, R. L. Ziprin, G. E. Spates, and J. R. DeLoach. 1991. Comparison of the efficacy of cultures of cecal anaerobes as inocula to reduce Salmonella typhimurium colonization in chicks with or without dietary lactose. Poult. Sci. 70:67-73. [DOI] [PubMed] [Google Scholar]
- 28.Hudson, C. R., M. Garcia, R. K. Gast, and J. J. Maurer. 2001. Determination of close genetic relatedness of the major Salmonella enteritidis phage types by pulsed-field gel electrophoresis and DNA sequence analysis of several Salmonella virulence genes. Avian Dis. 45:875-886. [PubMed] [Google Scholar]
- 29.Humphrey, T. J., A. Williams, K. McAlpine, M. S. Lever, J. Guard-Petter, and J. M. Cox. 1996. Isolates of Salmonella enterica Enteritidis PT4 with enhanced heat and acid tolerance are more virulent in mice and more invasive in chickens. Epidemiol. Infect. 117:79-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubena, L. F., R. H. Bailey, J. A. Byrd, C. R. Young, D. E. Corrier, L. H. Stanker, and G. E. Rottinghaust. 2001. Cecal volatile fatty acids and broiler chick susceptibility to Salmonella typhimurium colonization as affected by aflatoxins and T-2 toxin. Poult. Sci. 80:411-417. [DOI] [PubMed] [Google Scholar]
- 31.Laconcha, I., D. L. Baggesen, A. Rementeria, and J. Garaizar. 2000. Genotypic characterisation by PFGE of Salmonella enterica serotype Enteritidis phage types 1, 4, 6, and 8 isolated from animal and human sources in three European countries. Vet. Microbiol. 75:155-165. [DOI] [PubMed] [Google Scholar]
- 32.Landeras, E., M. A. Usera, C. Calderon, and M. C. Mendoza. 1997. Usefulness of phage typing and “two-way ribotyping” to differentiate Salmonella enteritidis strains. Microbiologia 13:471-480. [PubMed] [Google Scholar]
- 33.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 64:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liebana, E., L. Garcia-Migura, C. Clouting, F. A. Clifton-Hadley, M. Breslin, and R. H. Davies. 2003. Molecular fingerprinting evidence of the contribution of wildlife vectors in the maintenance of Salmonella Enteritidis infection in layer farms. J. Appl. Microbiol. 94:1024-1029. [DOI] [PubMed] [Google Scholar]
- 35.Liebana, E., L. Garcia-Migura, J. Guard-Petter, S. W. McDowell, S. Rankin, H. M. Opitz, F. A. Clifton-Hadley, and R. H. Davies. 2002. Salmonella enterica serotype Enteritidis phage types 4, 7, 6, 8, 13a, 29 and 34: a comparative analysis of genomic fingerprints from geographically distant isolates. J. Appl. Microbiol. 92:196-209. [DOI] [PubMed] [Google Scholar]
- 36.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 38.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259-274. [DOI] [PubMed] [Google Scholar]
- 39.Parker, C., K. Asokan, and J. Guard-Petter. 2001. Egg contamination by Salmonella serovar enteritidis following vaccination with Delta-aroA Salmonella serovar typhimurium. FEMS Microbiol. Lett. 195:73-78. [DOI] [PubMed] [Google Scholar]
- 40.Parker, C. T., B. Harmon, and J. Guard-Petter. 2002. Mitigation of avian reproductive tract function by Salmonella enteritidis producing high-molecular-mass lipopolysaccharide. Environ. Microbiol. 4:538-545. [DOI] [PubMed] [Google Scholar]
- 41.Parker, C. T., E. Liebana, D. J. Henzler, and J. Guard-Petter. 2001. Lipopolysaccharide O-chain microheterogeneity of Salmonella serotypes Enteritidis and Typhimurium. Environ. Microbiol. 3:332-342. [DOI] [PubMed] [Google Scholar]
- 42.Que, J. U., S. W. Casey, and D. J. Hentges. 1986. Factors responsible for increased susceptibility of mice to intestinal colonization after treatment with streptomycin. Infect. Immun. 53:116-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman, M. M., J. Guard-Petter, and R. W. Carlson. 1997. A virulent isolate of Salmonella enteritidis produces a Salmonella typhi-like lipopolysaccharide. J. Bacteriol. 179:2126-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roantree, R. J. 1967. Salmonella O antigens and virulence. Annu. Rev. Microbiol. 21:443-466. [DOI] [PubMed] [Google Scholar]
- 45.Romling, U., and M. Rohde. 1999. Flagella modulate the multicellular behavior of Salmonella typhimurium on the community level. FEMS Microbiol. Lett. 180:91-102. [DOI] [PubMed] [Google Scholar]
- 46.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 47.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 48.St Louis, M. E., D. L. Morse, M. E. Potter, T. M. DeMelfi, J. J. Guzewich, R. V. Tauxe, and P. A. Blake. 1988. The emergence of grade A eggs as a major source of Salmonella enteritidis infections. New implications for the control of salmonellosis. JAMA 259:2103-2107. [PubMed] [Google Scholar]
- 49.Su, C., D. Evans, R. H. Cole, J. C. Kissinger, J. W. Ajioka, and L. D. Sibley. 2003. Recent expansion of toxoplasma through enhanced oral transmission. Science 299:414-416. [DOI] [PubMed] [Google Scholar]
- 50.Tellez, G., C. E. Dean, D. E. Corrier, J. R. Deloach, L. Jaeger, and B. M. Hargis. 1993. Effect of dietary lactose on cecal morphology, pH, organic acids, and Salmonella enteritidis organ invasion in Leghorn chicks. Poult. Sci. 72:636-642. [DOI] [PubMed] [Google Scholar]
- 51.Tsolis, R. M., R. A. Kingsley, S. M. Townsend, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261-274. [PubMed] [Google Scholar]
- 52.Zhou, D., and J. Galan. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3:1293-1298. [DOI] [PubMed] [Google Scholar]