Abstract
Spermatogenesis is a multistep process that generates millions of spermatozoa per day in mammals. A key to this process is the spermatogonial stem cell (SSC), which has the dual property of continually renewing and undergoing differentiation into a spermatogonial progenitor that expands and further differentiates. In this review, we will focus on how these proliferative and early differentiation steps in mammalian male germ cells are controlled by transcription factors. Most of the transcription factors that have so far been identified as promoting SSC self-renewal (BCL6B, BRACHYURY, ETV5, ID4, LHX1, and POU3F1) are upregulated by glial cell line-derived neurotrophic factor (GDNF). Since GDNF is crucial for promoting SSC self-renewal, this suggests that these transcription factors are responsible for coordinating the action of GDNF in SSCs. Other transcription factors that promote SSC self-renewal are expressed independently of GDNF (FOXO1, PLZF, POU5F1, and TAF4B) and thus may act in non-GDNF pathways to promote SSC cell growth or survival. Several transcription factors have been identified that promote spermatogonial differentiation (DMRT1, NGN3, SOHLH1, SOHLH2, SOX3, and STAT3); some of these may influence the decision of an SSC to commit to differentiate while others may promote later spermatogonial differentiation steps. Many of these transcription factors regulate each other and act on common targets, suggesting they integrate to form complex transcriptional networks in self-renewing and differentiating spermatogonia.
Keywords: Spermatogonial stem cell, Spermatogonia, Transcription factor, Self-renewal, Differentiation
1. Introduction
Spermatogenesis is a highly coordinated process requiring an orchestrated program of gene expression controlled by extrinsic and intrinsic factors. The extrinsic factors are derived from non-germ cells, including Sertoli cells, Leydig cells, and peritubular myoid cells within the testes; they trigger specific events in germ cells that dictate or influence spermatogenesis. The intrinsic factors are generated in germ cells and act within them. Among the intrinsic factors are transcription factors, which is the subject of this review.
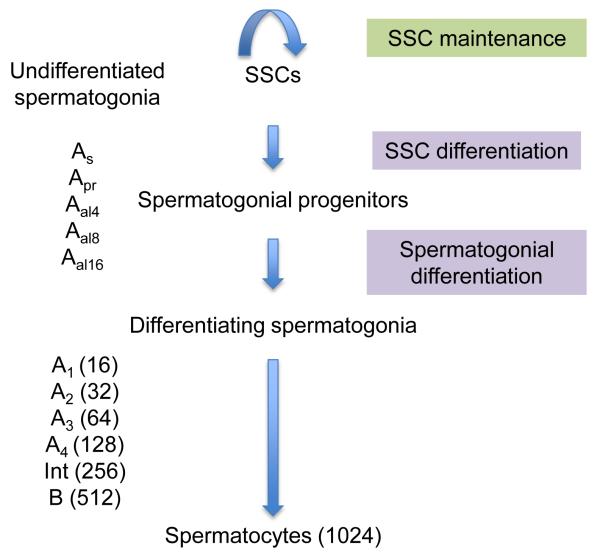
Spermatogenesis is one of the few developmental processes that occur in adults. In order for spermatogenesis to be maintained, it is essential that there are self-renewing cells in the testis. Specialized cells called spermatogonial stem cells (SSCs) serve in this capacity. In mammals, SSCs are located in a so-called “stem cell niche” in the basal compartment of the seminiferous epithelium, where SSCs proliferate to generate a clone of cells. The mitotic expansion of these clones is the foundation that sustains spermatogenesis. In mice, SSCs give rise to another type of undifferentiated spermatogonia called spermatogonial progenitors, which have a large proliferative capacity but are ultimately committed to differentiate. SSCs and spermatogonial progenitors are collectively called undifferentiated A-spermatogonia. Rather than being distinct cell types, SSCs and spermatogonial progenitors may represent distinct cellular states of a single cell type. After a series of cell divisions, these undifferentiated A-spermatogonia become differentiating A-spermatogonia, which, in turn, give rise to B-spermatogonia that further proliferate and differentiate into meiotic spermatocytes (Fig. 1). Upon completion of meiosis, spermatocytes convert into round spermatids and undergo a series of differentiation steps in the seminiferous epithelial tubule to become what is nearly a functional sperm: the elongated spermatid.
Figure 1. The three regulatory steps of spermatogonial development.
Not pictured is the fact that SSCs are a subset of undifferentiated As spermatogonia (and probably also Apr and Aal4 spermatogonia).
In this review, we will focus on mammalian transcription factors that are critical for SSC maintenance and spermatogonial differentiation (Fig. 1). Other recent reviews cover transcriptional regulators critical for other phases of mammalian spermatogenesis, including both meiotic and post-meiotic events [1–3].
2. SSCs
SSCs first arise a few days after birth in mice from non-dividing germ cells called gonocytes. SSCs are a subset of undifferentiated spermatogonia that are typically classified into being either A-single (As), A-paired (Apr), or A-aligned (Aal4, Aal8, Aal16, and in rare case, Aal32) spermatogonia (Fig. 1). This morphological classification is based on the unusual ability of A-spermatogonia to remain connected after mitosis by intercellular bridges created as a result of incomplete cytokinesis. The classical view has been that only As spermatogonia are SSCs, but it now appears that many Apr and even some of the shorter Aal spermatogonia have the potential to be SSCs [4,5].
Several protein markers, including PLZF, GFRα1, and ID4, have been shown to mark SSCs and other undifferentiated spermatogonia. Each differs in their specificity. Thus, while PLZF is expressed in all stages of undifferentiated spermatogonia [6,7], GFRα1 is mostly in As, Apr and Aal4 spermatogonia, and ID4 is specific for As spermatogonia [8]. To date, no marker has been identified as being exclusively expressed in SSCs [9]. Thus, the only reliable current means to unambiguously identify SSCs is to use an in vivo method: the germ cell transplantation assay [10]. The basis for this assay is that, by definition, SSCs are the only testicular germ cells that can colonize and initiate spermatogenesis. Thus, transplantation of a SSC (but not other cells) into a germ cell-free seminiferous tubule leads to the formation of a colony of descendent cells (after 2 to 3 months) that can be easily visualized. While it is not an entirely efficient assay (only ~10% of SSCs typically form a colony), in vivo transplantation allows one to compare the number of SSCs in different scenarios.
As an alternative to studying SSCs in vivo, in vitro SSC culture systems have been established. Two different SSC culturing methods that were established around the same time have been widely used. One method involves culturing so-called germline stem (GS) cells from neonatal (postnatal day-0 [P0] to P2) mouse testis [11]. In the other method, undifferentiated spermatogonia isolated from P6 to adult mice testes are enriched using the cell-surface marker, THY1, and then cultured [12]. Essential for the growth and maintenance of the SSCs in both GS and Thy1+ spermatogonial cell cultures is glial cell line-derived neurotrophic factor (GDNF). By using GDNF in combination with basic fibroblast growth factor (bFGF; also known as FGF2), both methods have successfully been used to culture and expand SSCs for >3 months without losing their stem cell activity, as assayed by the germ cell-transplantation assay [11,12]. Of note, these cultures harbor not only SSCs but also other spermatogonia, including spermatogonial progenitors. Therefore, it appears that these in vitro culture systems recapitulate what normally occurs in the stem cell niche in the testis in vivo: both the self-renewal and the differentiation of SSCs.
In vitro SSC culture systems afford considerable advantages over generating and characterizing SSC-mutant mice, both in terms of time and expense. By using small interfering RNAs (siRNAs) or short-hairpin RNAs (shRNAs) to knockdown the levels of specific factors of interest in cultured SSCs, followed by analysis using the transplantation assay, insights can be made as to the functions of such factors. Indeed, as described in the section below, there has been a blossom of studies using these in vitro systems to study the transcription factors involved in SSC self-renewal and differentiation.
3. SSC maintenance
There are ~2 × 104 SSCs in the adult mouse testis [13]. To maintain this number of SSCs, it is critical that an appropriate balance of self-renewal and differentiation occurs, including in response to environmental and genetic insults. If SSCs self-renew too frequently, they over accumulate, leading to defects in spermatogenesis. As an example of this, over-production of GDNF from Sertoli cells leads to an over-growth of SSCs, causing an arrest in early spermatogenesis [14]. Conversely, if there is an insufficient SSC self-renewal, such as in Gdnf-heterozygous mice, SSCs become “exhausted,” resulting in progressive germ cell loss [14].
Differentiation is accompanied by a cessation in proliferation in many cells types. In contrast, the differentiated cells derived from SSCs—spermatogonial progenitors—proliferate at a higher rate than SSCs [15]. This makes it challenging to distinguish between mechanisms controlling the proliferation of SSCs vs. spermatogonial progenitors, particularly given that these two cell types/cellular states cannot be unambiguously distinguished with known markers. Therefore, most of the SSC maintenance factors that have been defined have not been shown to increase the proliferation of SSCs specifically. This is not necessarily the case since SSC self-renewal is not only a reflection of SSC proliferation rate, but also: (i) SSC survival rate, and (ii) the proportion of self-renewing cell divisions relative to differentiating cell divisions. Molecules that promote SSC maintenance can act by altering the magnitude of any of these three mechanisms.
3.1. GDNF-inducible transcription factors important for SSC maintenance
To define genes potentially acting downstream of GDNF to promote SSC self-renewal, microarray analysis was employed to identify genes induced/upregulated by GDNF in SSCs [16]. This was accomplished by withdrawing GDNF for 18 hours and then providing cells with GDNF (or not) for a further 2, 4, or 8 hours. Genes defined as “GDNF-inducible” were those exhibiting decreased expression upon GDNF withdrawal and increased expression upon GDNF replacement. A subset of these genes was later shown to encode transcription factors that promote SSC maintenance, as described in this section.
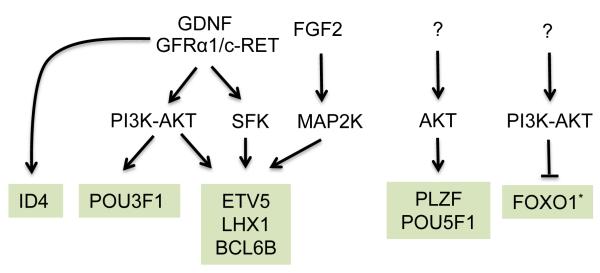
GDNF acts through two different signaling pathways to promote SSC self-renewal and induce target genes: the phosphatidylinositol 3-kinase (PI3K)/Ak strain thymoma (AKT)-dependent pathway [17] and the Src family kinase (SFK) pathway [18]. Some GDNF-inducible SSC self-renewal genes—including Etv5, Bcl6b, and Lhx1—are regulated by both of these pathways [18](Fig. 2 and Table 1).
Figure 2. Signaling pathway upstream of SSC maintenance factors.
Transcription factors that promote SSC maintenance are shown in green boxes. The signaling pathways depicted as upregulating these transcription factors act by increasing the expression of the genes encoding them. An asterisk denotes a case where the subcellular localization of a transcription factor is altered by a signaling pathway.
Table 1.
Summary of transcription factors important for spermatogonial maintenance and differentiation. (−) indicates negative regulation by a signaling pathway.
| Functions | Name | Upstream signaling pathway |
Knock-out1 |
In vitro siRNA/shRNA |
Positive targets |
Negative targets |
Microarray |
|---|---|---|---|---|---|---|---|
| SSC maintenance | ETV5 | GDNF-PI3K-AKT GDNF-SFK FGF2-MAP2K1 |
Null | siRNA |
Bcl6b*,
Lhx1*, T, miR21, cRet* |
siRNA, Thy1+ Spg culture |
|
|
|
|||||||
| BCL6B | GDNF-PI3K-AKT GDNF-SFK FGF2-MAP2K1 |
Null | siRNA | siRNA, Thy1+ Spg culture |
|||
|
|
|||||||
| LHX1 | GDNF-PI3K-AKT GDNF-SFK FGF2-MAP2K1 |
siRNA | |||||
|
|
|||||||
| POU3F1 | GDNF-PI3K-Akt | siRNA | siRNA, Thy1+ Spg culture |
||||
|
|
|||||||
| T | siRNA | ||||||
|
|
|||||||
| ID4 | GDNF | Null | siRNA | ||||
|
|
|||||||
| PLZF | AKT | Null | siRNA - no effect | Redd1 | cKit, Line1 |
In vivo KO spermatogonia |
|
|
|
|||||||
| POU5F1 | AKT | siRNA - no effect shRNA - SSC defect |
|||||
|
|
|||||||
| FOXO1 | PI3K-AKT (−) | Conditional (Vasa Cre) |
Lhx1*,
cRet* |
In vivo KO testes (P4) |
|||
|
|
|||||||
| TAF4B | Null |
In vivo KO testes (P8) |
|||||
|
| |||||||
| SSC/spermatogonial differentiation |
SOX3 | Null, Conditional (Vasa-Cre) |
Ngn3* | ||||
|
|
|||||||
| STAT3 | GDNF (−) | siRNA, shRNA | Ngn3 | ||||
|
|
|||||||
| NGN3 | GDNF (−) | siRNA, shRNA | |||||
|
|
|||||||
| SOHLH1, SOHLH2 |
RA | Null, Double null |
Ngn3,
Sox3, cKit |
Gfrα1,
cRet |
In vivo KO testes (P7) |
||
|
|
|||||||
| DMRT1 | Conditional (Ngn3-Cre) |
Sohlh1 | Stra8 | ChIP-chip (P9), In vivo KO testes (P9) |
|||
Only knockout studies that analyzed spermatogonial phenotypes are shown. Asterisks denote regulated genes that have not been tested as to whether they are direct targets or not. Spg, spermatogonia
ETV5
Ets-variant gene-5 (Etv5) encodes a member of the large ETS transcription factor family. Its role in spermatogenesis was first revealed by gene targeting studies conducted by Chen et al. (2005), who showed that loss of ETV5 in mice causes male infertility [19]. Remarkably, these mice became infertile during adulthood despite the fact that at 4 weeks of age—when the first wave of spermatogenesis is taking place—there was no obvious abnormality in their testes. Defects did not occur until the subsequent waves of spermatogenesis, when there was a dramatic loss of germ cells, including spermatogonia. By 10 weeks of age, most of the seminiferous tubule regions in Etv5-null male mice were completely devoid of germ cells. This phenotype is consistent with these mutant mice having a defect in SSC maintenance. The relatively normal phenotype at mid-postnatal development follows from the fact that the first wave of spermatogenesis is independent of SSCs [15]. Subsequent waves of spermatogenesis depend on SSCs, explaining the progressive decline in germ cells in the seminiferous tubules in Etv5-null male mice as they age.
Evidence that Etv5-null male mice have a cell-autonomous defect in SSC maintenance was obtained from the germ cell transplantation assay. Tyagi et al. (2009) found that testicular cells from Etv5-null mice failed to colonize germ cell-free testes [20]. Further evidence that ETV5 promotes SSC maintenance in a cell autonomous manner was the finding that siRNA-mediated depletion of ETV5 from Thy1+ spermatogonial cell cultures resulted in reduced number of SSCs, as demonstrated by the transplantation assay [18]. Etv5-null spermatogonia exhibited reduced proliferation in vivo [20], suggesting that ETV5 promotes SSC maintenance by stimulating their proliferation. ETV5 may achieve this by increasing the receptivity of SSCs to GDNF since Etv5-null spermatogonia have reduced expression of the GDNF co-receptor, c-RET [20]. Coupled with the fact that GDNF induces Etv5 gene expression [16,21], this suggests the existence of a positive feedback loop revolving around both ETV5 and GDNF that leads to SSC self renewal (Fig. 3A).
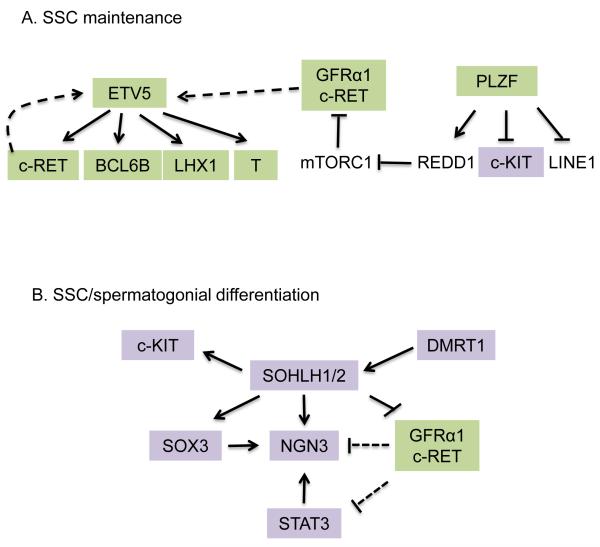
Figure 3. Transcriptional regulatory networks.
Green boxes denote factors that promote SSC maintenance. Purple boxes denote factors that promote SSC/spermatogonial differentiation. Known regulatory relationships are designated with solid arrows/lines; predicted regulatory relationships are denoted with dotted arrows/lines.
Microarray analysis of Thy1+ spermatogonial cell cultures depleted of ETV5 (by RNAi) revealed that ETV5 regulates (directly or indirectly) several genes important for SSC self-renewal, including Bcl6b, Lhx1, Cxcr4, and Brachyury (T) [22] (Fig. 3A, and see below). Evidence that ETV5 directly activates T transcription was the finding that ETV5 binds the T gene promoter, as shown by chromatin immunoprecipitation (ChIP) analysis [22]. Interestingly, a recent study suggested that ETV5 also directly activates the transcription of miR-21, a microRNA (miRNA) that promotes SSC maintenance in vitro [23].
Further evidence that ETV5 promotes SSC self-renewal is the finding that Etv5 gene expression is induced by FGF2, which itself is an important extrinsic factor that supports SSCs proliferation [24]. FGF2 acts through mitogen-activated protein kinase kinase 1 (MAP2K1) to transcriptionally activate Etv5, as well as its downstream genes, Bcl6b and Lhx1, in cultured GS cells (Fig. 2). ETV5 appears to be a major effector of the FGF2/MAP2K1 pathway in promoting SSC proliferation based on the finding that GS cells stably overexpressing ETV5 grow independently of FGF2 in vitro [24].
ETV5 was initially reported to be specifically expressed in Sertoli cells, which implied it acted in a non-cell autonomous manner to promote SSC self-renewal [19]. Subsequent studies demonstrated that ETV5 is also expressed in germ cells, but at a lower level than in Sertoli cells [18,20]. The current consensus view is that ETV5 functions in both cell types to maintain SSCs. The evidence that ETV5 functions cell-autonomously in germ cells is described above. The primary evidence that ETV5 acts through Sertoli cells to control SSC self-renewal comes from microarray analysis of primary Sertoli cells from Etv5-null vs. control mice [19]. This revealed that ETV5 upregulates the expression of several chemokine genes that have been implicated in regulating the stem cell niche [25,26]. One of these ETV5-upregulated genes—Ccl9—encodes a chemokine that facilitates Sertoli cell chemoattraction of undifferentiated spermatogonia through its receptor, CCR1 [27]. Another ETV5-upregulated gene is Cxcl12, which encodes a chemokine expressed by Sertoli cells that is required for SSC maintenance [28]. ETV5 also upregulates the receptor for CXCL12, which is present on SSCs and critical for SSC maintenance [22].
BCL6B
Bcl6b—a paralog of the well-studied Bcl6 tumor suppressor gene—encodes a zinc finger-containing transcriptional repressor that is known to enhance immune responses [29]. The first hint that Bcl6b might also have a role in SSCs was the discovery that it is highly expressed in rat germ cells enriched for SSC activity [30]. Its potential role in SSCs was reinforced when it was found that Bcl6b was among the suite of genes induced by GDNF in Thy1+ spermatogonial cell cultures [16]. Empirical evidence for a role in SSCs was the finding by Oatley et al. (2006) that Bcl6b-null mice exhibit the “progressive germ cell loss” phenotype, described above, indicative of a SSC maintenance defect [16]. To further test whether BCL6B promotes SSC self-renewal, Thy1+ spermatogonial cell cultures were transiently depleted of BCL6B (using siRNAs) and cultured for 7 days to allow the SSCs to undergo approximately one cell division. Germ cell transplantation analysis demonstrated that these BCL6B-depleted cell cultures had a dramatic loss (by 88%) of SSCs. BCL6B-depleted Thy1+ spermatogonial cell cultures exhibited a ~2-fold increase in the frequency of apoptotic cells, providing evidence that BCL6B promotes both SSC self-renewal and survival [16]. BCL6B appears to act downstream of the FGF2/MAP2K1 signaling pathway to promote the proliferation of SSCs (Fig. 2), based on the findings that (i) FGF2 induces Bcl6b expression (probably through ETV5) and (ii) GS cells stably overexpressing BCL6B proliferate independently of FGF2 [24].
Microarray analysis conducted by Wu et al. (2011) on BCL6B-depleted Thy1+ spermatogonial cells identified many BCL6B-positively regulated genes encoding proliferation regulators that are candidates to mediate the proproliferative function of BCL6B [22]. Also enriched were genes encoding proteins involved in cell-to-cell signaling and interactions, raising the possibility that BCL6B promotes SSC maintenance through regulating cell-cell interactions in the stem cell niche. Surprisingly, there was little overlap between the genes regulated by BCL6B and ETV5, despite the fact that Bcl6b is a downstream target gene of ETV5 [22] (Table 1). Thus, even though these two transcriptional regulators both promote SSCs maintenance, they may act by largely different means [22].
LHX1
LHX1 is a LIM-subclass homeobox transcription factor that has roles in embryonic development, including head formation and kidney tubular morphogenesis [31,32]. It was first implicated as having a role in SSCs when it was shown to be induced by GDNF in Thy1+ spermatogonial cell cultures [16]. RNAi-mediated knockdown of LHX1 in these cell cultures reduced their ability to form colonies in the in vivo germ-cell transplantation assay, demonstrating that LHX1 promotes SSC maintenance in vitro [18]. Like Bcl6b and Etv5, Lhx1 expression is induced by FGF2/MAP2K1 signaling [24]. However, unlike Bcl6b and Etv5, Lhx1 does not appear to be a major mediator of FGF2 action, as GS cells stably overexpressing LHX1 do not proliferate independently of FGF2 [24].
POU3F1
POU3F1 (also known as OCT6, SCIP, and TST-1) is a POU-subclass homeobox transcription factor that is expressed in the testis as well as the brain. Pou3f1-mutant mice die shortly after birth with various defects in nervous system development, precluding an analysis of the role of POU3F1 in the testis [33,34]. Like Bcl6b, Pou3f1 was first recognized as a candidate to be involved in SSCs when it was shown to be highly expressed in rat germ cells enriched for SSC activity [30] and induced by the GDNF-PI3K-AKT pathway in mouse Thy1+ spermatogonial cell cultures [35]. Its functional role in SSCs was demonstrated by knockdown studies conducted by Wu et al. (2010) in the latter. Transient depletion of POU3F1 by RNAi reduced the number of SSCs (~by 5-fold) in these cultures (as assayed by germ cell transplantation), indicating that POU3F1 promotes SSC maintenance, at least in vitro. POU3F1 may act by increasing SSC survival since depletion of POU3F1 led to a significant increase in the frequency of apoptotic cells in these cell cultures. However, it is not known whether the increase in apoptosis occurred in SSCs or the spermatogonial progenitors in these cultures.
Wu et al. (2011) performed microarray analysis of POU3F1-depleted vs. control Thy1+ spermatogonial cell cultures to identify POU3F1-regulated genes [22]. Overrepresented categories among the genes upregulated by POU3F1 were “post-translational modifications,” “protein folding,” and “chemotaxis.” POU3F1-downregulated genes encoded proteins enriched for “apoptosis-related functions.” Few of the POU3F1-regulated genes were in common with those regulated by BCL6B or ETV5. This was somewhat unexpected given that all three of these transcription factors promote SSC maintenance and all three are induced by GDNF signaling. While more data is required before making firm conclusions, the existing data suggests that POU3F1 promotes SSC self-renewal through a regulatory network largely independent from that of BCL6B and ETV5.
BRACHYURY (T)
The T-box transcription factor, BRACHYURY (T), is best known for its roles in defining cell fate and differentiation in the mesoderm cell lineage [36]. Its role in SSCs was first suggested by the discovery that the T gene is positively regulated by the SSC self-renewal factors, ETV5 and GDNF, in Thy1+ spermatogonial cell cultures [22]. A role for T in SSCs was directly demonstrated by knockdown studies performed in these cell cultures by Wu et al. (2011) [22]. Depletion of T reduced the number of SSCs, as assessed by the germ cell transplantation assay, providing in vitro evidence that T promotes SSC maintenance. T depletion also reduced the total number of cells in these Thy1+ spermatogonial cell cultures, suggesting that T may also have effects on spermatogonial progenitors, which may dominate these cultures. It is not known how T promotes the maintenance of SSCs, but it is tempting to speculate it acts by increasing both their proliferation rate and survival, since T is known to have both of these roles in allantoic cells in the early embryo [37].
The gene targets of T in SSCs have not been identified. T may serve as a subordinate transcription factor acting downstream of ETV5, based on the finding that the T promoter has consensus ETS-family binding sites and interacts with ETV5 in intact cells, as assayed by ChIP analysis [22]. Interestingly, recent evidence suggests that T acts not only in SSCs, but also in primordial germ cells (PGCs). In particular, T was shown to promote PGC specification by activating expression of germ cell determinants in mice embryos [38]. Thus, T appears to be a multi-functional transcription factor that acts in undifferentiated germ cells in both the fetal and adult gonad. To our knowledge, T is the only transcription factor known to have roles in both PGC specification and SSC maintenance.
ID4
The inhibitor of DNA binding (ID) protein family has four members, all of which have the capacity to promote the undifferentiated cell state by virtue of their ability to inhibit basic helix-loop-helix (bHLH) transcription factors, most of which promote differentiation. ID proteins have a HLH domain, which allows them to heterodimerize with bHLH transcription factors, but they do not have a basic DNA-binding domain and thus they sequester bHLH transcription factors away from DNA. In addition to binding to bHLH transcription factors, ID proteins interact with cell cycle regulators, leading to promotion of the proliferative state characteristic of undifferentiated cells [39].
All four of the ID family members are expressed in the testes; albeit in different cell types [40]. ID4 is unique in being the only ID family member detectably expressed in A-spermatogonia [40]. It was recently shown by Oatley et al. (2011) that ID4 is exclusively expressed in As spermatogonia, based on using highly specific antibodies and Id4-GFP reporter transgenic mouse [8]. To our knowledge, ID4 is the only protein known to be expressed in As spermatogonia and not Apr or Aal spermatogonia. This is of considerable interest given that As spermatogonia are generally regarded as being more enriched for SSC activity than other A-spermatogonia [41].
Oatley et al. (2011) also obtained several lines of evidence indicating that ID4 is an SSC maintenance factor: First, like many other SSC maintenance factor genes, Id4 is induced by GDNF in Thy1+ spermatogonial cell cultures [8]. Second, siRNA-mediated knockdown of ID4 reduced the number of SSCs in these cultures, as shown by the germ cell transplantation assay [8]. Finally, Id4-mutant mice exhibit the “progressive germ cell loss” phenotype indicative of a SSC maintenance defect [8]. Together, these data support the notion that ID4 acts in the same manner in male germ cells as it does in other cell types: it promotes the undifferentiated, proliferative state. It will be interesting to identify the specific bHLH transcription factor(s) that ID4 represses to achieve this goal in SSCs.
3.2. GDNF-independent transcription factors important for SSC maintenance
SSC maintenance transcription factors expressed independently of the presence of GDNF are discussed in this section. Most of these transcription factors probably promote SSC maintenance by novel mechanisms that do not involve GDNF. However, there is evidence that at least one member of this class of transcription factors—PLZF—has roles in GDNF-dependent processes by virtue of its ability to regulate SSC responsiveness to GDNF.
PLZF
PLZF (otherwise known as ZBTB16 or ZFP145) is a transcriptional repressor that binds to DNA via its Kruppel-type zinc finger domains. It serves to repress transcription by recruiting histone deacetylases (HDACs) via its POZ domain [42]. PLZF was initially identified as the gene fused to the retinoic acid receptor-α gene as a result of chromosomal translocations in a subset of human acute promyeolocytic leukemia cases [43]. PLZF was later found to be essential for limb and axial skeleton patterning during embryogenesis [44]. Its role in the testis was revealed when Buaas et al. (2004) demonstrated that a nonsense mutation in the gene encoding Plzf was responsible for the male infertility phenotype of the luxoid (lu) mutant, which exhibits a progressive loss of germ cells [7]. Essentially the same phenotype was found in Plzf-null mice generated by conventional gene targeting methods by Costoya et al. (2004) [6]. The timing of the defect and the stages of the germ cell loss in a given section of seminiferous tubules were consistent with a SSC maintenance defect. As further evidence for this, Plzf-mutant germ cells failed to colonize germ cell-deficient recipient testes when transplanted. It is likely that PLZF functions cell autonomously to maintain SSCs since PLZF is expressed specifically in spermatogonia; indeed, PLZF is widely used as a marker of undifferentiated spermatogonia [9].
The exact mechanism by which PLZF maintains SSCs has not been elucidated. It seems unlikely that PLZF acts by promoting SSC survival, as its loss had little [6] or no significant effect [7] on germ cell apoptosis in early postnatal mice. This implies that, instead, PLZF serves to promote SSC self-renewal or SSC cell fate, but this remains to be directly determined.
To identify PLZF-regulated genes, Costoya et al. (2004) performed microarray analysis on Plzf-mutant vs. control spermatogonia isolated by FACS on the basis of the SSC marker α6-integrin [6]. This analysis revealed that PLZF regulates genes encoding proteins involved in “cell cycle control,” “cellular metabolism,” “the cytoskeleton,” and “gene expression.” With regard to the latter, PLZF regulates many genes encoding transcription factors and RNA-binding proteins, raising the possibility that PLZF is a master regulator on top of a hierarchy of transcriptional/post-transcriptional regulatory proteins in SSCs. Interestingly, the profile of genes regulated by PLZF appears to depend on cell type. In myeloid cells, PLZF inhibits many cell cycle genes, including cyclin A and c-myc, as well as the proliferation of these cells [45,46]. In contrast, these cell cycle genes are impervious to the repressive action of PLZF in spermatogonia, which may explain why PLZF does not repress the cell growth of undifferentiated germ cells.
Some direct targets of PLZF have been defined. One example is the c-Kit gene, which encodes a tyrosine kinase expressed by differentiating spermatogonia [47]. PLZF represses c-Kit transcription [48], which is consistent with the fact that c-KIT promotes spermatogonial differentiation, not self-renewal. A PLZF direct target positively regulated by PLZF is Redd1 [49], which encodes a natural inhibitor of mammalian target of rapamycin complex 1 (mTORC1). This PLZF-Redd1-mTORC1 circuit is functionally relevant for SSC maintenance since it is known that hyperactivity of mTORC1 leads to stem cell exhaustion through aberrant translation of downstream targets [50–52]. The activation of Redd1 transcription by PLZF is likely to be important for SSC maintenance since the repression of mTORC1 signaling by REDD1 is necessary for maximal expression of both components of the GDNF receptor—GFRα1 and c-RET—in SSCs [49]. Together, these data support a model in which PLZF operates in a molecular circuit that amplifies the responsiveness to GDNF signals as a means to maintain SSCs. The ability of PLZF to increase the sensitivity of SSCs to GDNF may also explain why deletion of PLZF in Thy1+ spermatogonial cell cultures (by RNAi) did not significantly affect the frequency of SSCs in these cultures [22]. This follows from the fact that Thy1+ spermatogonial cell cultures are normally supplemented with high levels of GDNF and thus they probably can grow normally even when expressing sub-optimal levels of factors involved in GDNF receptivity.
Recently, it was reported that PLZF regulates not only protein-coding genes, but also repetitive elements. Using two knock-in Plzf mouse models, Puszyk et al. (2013) demonstrated that PLZF inhibits both the transcription and transposition of L1 elements [53], the most abundant class of retrotransposons in humans [54]. PLZFOFF mice, which express a mutant version of PLZF that fails to bind DNA, were found to have hypomethylated L1 genomic loci and express high levels of L1 elements in the testis. In contrast, PLZFON mice, which express a form of PLZF that binds to DNA constitutively (i.e., independently of acetylation of its zinc finger motif by the HAT p300), had hypermethylated L1 loci and expressed only low levels of L1 elements in the testis. ChIP analysis provided evidence that PLZF directly represses L1 transcription by recruiting HDACs and DNA methyltransferases to L1 loci. The ability of PLZF to dampen retrotransposon activity in the testis is likely to have important implications for spermatogenesis given that it is well established that uncontrolled retrotransposition can result in detrimental effects that lead to loss of germ cells [55].
POU5F1
POU5F1 (otherwise known as OCT4) is a POU-subclass homeobox transcription factor that is essential for the establishment and maintenance of a variety of pluripotent stem cell populations [56]. A well-established example of its pluripotency activity is the ability of POU5F1, in combination with other factors, to reprogram somatic cells into pluripotent stem cells [57–59]. The specific roles of POU5F1 in the germ cell lineage is less well understood, but POU5F1 is expressed in a developmental pattern that is consistent with its having functions in immature and undifferentiated germ cells. At mid-gestation (after embryonic day 9.5 [E9.5]), the major site of POU5F1 expression in mouse embryos is in primordial germ cells (PGCs) [60,61]. POU5F1 expression persists in gonocytes, the precursors of SSCs that are most abundant at birth, as well as in undifferentiated spermatogonia after birth [7,62].
To examine what functions POU5F1 might have in SSCs, Dann et al. (2008) stably knocked down POU5F1 expression in GS cell cultures [63]. They found that POU5F1-depleted GS cell cultures had reduced numbers of cells with SSC activity, as measured by the germ cell transplantation assay. Depletion of POU5F1 in the GS cell cultures also reduced both their proliferation rate and survival, based on BrdU incorporation and Annexin V analysis, respectively. This suggested that POU5F1 maintains SSCs by promoting both their survival and proliferation, but these effects may instead be engendered on the spermatogoial progenitors that are in GS cell cultures.
In apparent contradiction to the results of Dann et al. (2008), Wu et al. (2010) found that transient knockdown of POU5F1 did not significantly reduce SSC numbers in Thy1+ spermatogonial cell cultures, as measured by the transplantation assay [35]. The apparent discrepancy between these two studies may be because they used different approaches. Dann et al. (2008) stably reduced POU5F1 level using a shRNA/lentivirus approach, while Wu et al. (2010) transiently depleted POU5F1 using a siRNA approach. The transient knockdown approach would specifically address the role of POU5F1 in SSC maintenance in vitro, while the stable knockdown approach would detect the role of POU5F1 either in vitro or in vivo. It is possible that POU5F1 is required for SSC maintenance in vivo, not in vitro, because there are high levels of growth factors present in the latter. By analogy, transient depletion of the SSC maintenance factor, PLZF, in Thy1+ spermatogonial cell cultures did not measurably affect the growth or survival of SSCs in these cultures, whereas stable loss of PLZF virtually eliminated SSCs, as measured by germ cell transplantation (see above). Of note, Dann et al. (2008) and Wu et al. (2010) also used different cell systems for their studies. The former study used GS cells (derived from DBA mice) cultured in serum-containing media, while the latter used Thy1+ spermatogonial cell cultures (derived from C57BL/6 mice) cultured in serum-free media [35,63]. The origin of the cells, the culture conditions and media, and the genetic background could all influence the function of POU5F1 in SSCs. Indeed, it is known that genetic background affects SSC self-renewal [64].
Like Plzf, Pou5f1 is expressed independently of GDNF in SSCs. A candidate to function in its place is the TGFβ superfamily secretory protein— NODAL—that has been shown to serve as an autocrine factor to positively regulate Pou5f1 expression in the spermatogonial cell line C18-4 [65]. It is unlikely that NODAL acts in an autoregulatory manner in SSCs in vivo since the Nodal gene is not detectably expressed in spermatogonia [35], but it may be secreted by Sertoli cells (or other somatic cells in the testes) to provide NODAL to SSCs.
FOXO1
FOXO1 is a member of the forkhead transcription factor family that has a variety of functions, including regulation of glucose metabolism, insulin signaling, control of cellular growth, and stem cell homeostasis [66,67]. FOXO1 was initially suspected of having a role in SSCs by virtue of its high expression in rat germ cells enriched for SSC activity [30]. To elucidate its functional role, Goertz et al. (2011) generated conditional knockout (cKO) mice lacking FOXO1 in germ cells (using Vasa-Cre mice, which express Cre recombinase specifically in germ cells as early as ~E15 [68]). These cKO mice exhibited a progressive age-dependent decline in spermatogenesis, suggesting they have a SSC maintenance defect. In addition, Goertz et al. (2011) found that adult Foxo1-cKO mice had seminiferous tubules with phenotypes suggesting other defects, including spermatogonial arrest.
Microarray analysis of P4 testes from Foxo1-cKO mice revealed that FOXO1 directly or indirectly regulates several SSC maintenance-promoting genes, including Lhx1 and c-Ret [69]. In addition, FOXO1 regulates stem cell marker genes (Gata2, Dppa4, and Sall4) and key regulators of spermatogenic progression (e.g., Egr2, Egr4 and Tex19). Of note, many of these genes encode transcription factors, raising the possibility that FOXO1 is a master regulator of a GDNF-independent transcriptional network in spermatogonia.
FOXO1 is predominantly in the cytoplasm of gonocytes and it shifts to the nucleus in spermatogonia [69]. While the functional significance of this developmentally regulated shift in sub-cellular location is not known, a likely possibility is that it restricts FOXO1 transcriptional activity to spermatogonia. In support of this notion, studies in non-germ cells provided evidence that the subcellular location of FOXO1 controls its activity. In particular, phosphorylation of FOXO1 by AKT prevents FOXO1 from accumulating in the nucleus of various cell types (including cerebellar neurons) by retaining it in the cytoplasm, rendering it incapable of regulating downstream gene targets [70,71]. Given the fact that PI3K-AKT pathway is important for SSCs maintenance (Fig. 2), Goertz et al. (2011) tested whether AKT signaling also controls FOXO1 localization and function in germ cells. In support of this possibility, they found that germ cell-specific loss of the AKT repressor, PTEN, caused FOXO1 to be localized in the cytoplasm of spermatogonia. Consistent with this cytoplasmically localized FOXO1 being non-functional, these cKO mice exhibited an SSC maintenance defect phenotype; i.e., an age-dependent increase of seminiferous tubules devoid of germ cells [69]. As a reciprocal test of this hypothesis, mice mutant for the positive regulator of PI3K-AKT signaling—PDK1—were tested. Its loss led to AKT hypophosphorylation and accumulation of FOXO1 in the nuclei of gonocytes, a cell type that normally has FOXO1 in the cytoplasm. The testes of these Pdk1-cKO mice had an overabundance of proliferative spermatogonia and largely lacked advanced germ cells [69]. This suggests that PI3K-AKT signaling is required to prevent FOXO1 from entering the nuclei of gonocytes and activating a cell proliferation program precociously. Of note, this “pro-differentiation” role of AKT pathway is at apparent odds with the fact that PI3KAKT pathway promotes SSC self-renewal by virtue of its being a crucial signaling pathway downstream of GDNF in SSCs [17]. Whether the PI3K-AKT pathway promotes proliferation or differentiation may depend on the presence of stage-specific factors that influence this decision.
TAF4B
The RNA polymerase II pre-initiation complex is composed of several factors, including transcription factor IID (TFIID). TFIID is itself comprised of several proteins—called TBP-associated factors (TAFs)—some of which have tissue-specific roles, including in the reproductive tract [72–75]. One of the TAFs with tissue-specific roles is a paralog of the Taf4 gene—Taf4b—that was initially shown to function in granulosa cells in the ovary where it promotes folliculogenesis [76]. TAF4B was later found to also function in the testis. Falender et al. (2005) found that Taf4b-null males were fertile when young, but subsequently exhibited an age-dependent progressive loss of germ cells [77]. By 11 weeks of age, they had very low sperm counts and were infertile. This age-dependent defect in spermatogenesis suggested that TAF4B has roles in SSC self-renewal. Further supporting this possibility was the finding that Taf4b-null mice had reduced spermatogonial proliferation. However, the timing of this proliferative defect (P3) suggested that it could also be caused by a malfunction in non-SSC proliferation or a delay in the differentiation of gonocytes (which are non-dividing) into spermatogonia. Indeed, it is clear that TAF4B has at least one SSC-independent male gonadal function based on the finding that Taf4b-null mice have less male germ cells than normal even at a time point (P2) when SSCs are yet to form.
To examine whether TAF4B acts in a cell autonomous or non-autonomous manner in germ cells, Falender et al. (2005) used the germ cell transplantation assay [77]. They found that transplanting wild-type germ cells into Taf4b-deficient testes supported normal spermatogenesis. This indicated that TAF4B is not required in testicular somatic cells to drive spermatogenesis and it implied that, instead, TAF4B functions in a cell autonomous manner in germ cells. In the future, it will be critical to define precisely what roles TAFB has in germ cells, including in SSCs.
4. SSC and spermatogonial differentiation
This section considers transcription factors that have been shown to promote the differentiation of SSCs and/or later stage A-spermatogonia. The transcription factors that drive the differentiation of SSCs into spermatogonial progenitors are important because they influence the balance between SSC self-renewal and differentiation (Fig. 1). Hence, they contribute to determining both the number of stem cells in the testes and the number of sperm that are ultimately produced from the testes. The identification of such transcription factors is challenging because the SSC-to-spermatogonial progenitor differentiation step appears to be reversible and because SSCs and spermatogonial progenitor largely share the same markers [5,78,79].
The second type of transcription factors that we will consider in this section is those that promote the conversion of spermatogonial progenitors into differentiating A-spermatogonia, an event that occurs after spermatogonial progenitors undergo several rounds of mitosis (Fig. 1). This retinoic acid-dependent differentiation step occurs at stage VIII of the seminiferous epithelial cycle and is characterized by the acquisition of c-KIT, the receptor for Kit ligand (Kitl; also known as SCF) that is critical for spermatogonial differentiation to occur [80–82].
SOX3
SOX3 is a member of the SRY-related high mobility group (SOX) transcription factor family that functions in cell fate determination [83]. While most commonly studied with regard to its function in embryonic brain development and cognition, SOX3 is also important for spermatogenesis [84]. However, elucidation of the spermatogenic role of SOX3 has been clouded by ambiguity with regard to its expression pattern and the effect of its loss in different genetic backgrounds.
The first study to report the effects of targeted mutation of Sox3—Weiss et al. (2003)—examined mice with a 129/svj genetic background [85]. Sox3-null mice in this genetic background had reduced testes size and displayed heterogeneous defects in seminiferous tubules, including some tubule regions lacking all germ cells and others harboring multiple layers of pachytene spermatocytes with a cluster of round and elongated spermatids in the lumen, detached from the epithelium. The authors interpreted these germ cells defects as being secondary to a Sertoli cell defect since their immunohistochemical analysis suggested that SOX3 was present in Sertoli cell nuclei. Consistent with this notion, they found that the seminiferous tubules from these mutant mice had large vacuoles in the epithelium, as well as disrupted basement membranes.
Rizzoti et al. (2004) examined the effect of loss of SOX3 in a mixed, mostly outbred (MF1), genetic background [84]. Unlike the mice in the 129/svj genetic background, the Sox3-null mice in this outbred genetic background were fertile and exhibited less severe seminiferous tubule defects; albeit the qualitative effects were similar. By careful morphological analysis, these authors found that SOX3 is not expressed in Sertoli cell nuclei, but rather in a subset of spermatogonia. This raised possibility that SOX3 has a cell autonomous role in germ cells. However, these authors also observed pituitary gland defects in Sox3-null mice, which led them to speculate that disruption of normal endocrine function could be responsible for the spermatogenic defects in Sox3-mutant mice. This was of considerable interest given that studies in humans have shown that SOX3 mutations are associated with X-linked hypopituitarism [86]. It will be important in the future to determine whether SOX3 has roles in the hypothalamic-pituitary-gonadal axis.
A third report—Raverot et al. (2005)—elected to examine the effect of loss of SOX3 in a C57BL/6 genetic background since this genetic background often sensitizes mice to testes defects caused by genetic perturbations [87]. Unlike the mixed outbred background, the C57BL/6 genetic background did not support fertility when SOX3 was knocked out, even though the levels of endocrines (FSH, LH, and testosterone) were normal [88]. Histological analysis revealed a variety of germ cell defects in the seminiferous tubules of adult Sox3-mutant mice in this genetic background, including some regions with only spermatogonia, some with reduced numbers of germ cells at all stages, and some that appeared to completely lack germ cells. Elongated spermatids and mature sperm were largely absent, suggesting a block in germ cell development. Analysis of postnatal development revealed no defects at P7, but by P14 there was an abnormal accumulation of undifferentiated spermatogonia, which suggested that these mice have a defect in spermatogonial differentiation. In support of this, there were few spermatocytes at P21, which normally are abundant at this time point.
Raverot et al. (2005) confirmed Rizzoti et al. (2004)’s finding that SOX3 is exclusively expressed in undifferentiated spermatogonia, thereby further supporting the notion SOX3 promotes spermatogonial differentiation in a cell autonomous manner [88]. To definitively address this, Laronda et al. (2011) conditionally knocked out Sox3 in germ cells (using Vasa-Cre mice, which express CRE recombinase in germ cells at ~E15 [68]). They found that these Sox3-cKO mice had spermatogenic defects that were very similar to those in ubiquitous Sox3-null mice; e.g., accumulation of spermatogonia and reduced spermatocytes [89]. This clearly demonstrated that SOX3 functions cell-autonomously in germ cells. To assess the underlying mechanism, these investigators used flow cytometry to quantify the number of undifferentiated (c-Ret+, c-Kit−) and differentiating (c-Ret−, c-Kit+) spermatogonia. They found that the number of differentiating spermatogonia was much lower in Sox3-cKO testes compared to control testes, clearly demonstrated that SOX3 promotes spermatogonial differentiation.
Whether SOX3 also promotes SSC differentiation is not known. In support of this possibility, the expression pattern of SOX3 overlaps with that of NGN3, an SSC differentiation factor ([90] and see below). Indeed, Raverot et al. (2005) obtained evidence that SOX3 stimulates Ngn3 expression [88]. Further evidence that SOX3 promotes SSC differentiation is the finding it suppresses the expression of the SSC self-renewal factor gene Pou5f1 [88].
STAT3
STAT3 belongs to the signal transducers and activators of transcription (STAT) family. It is best known for its role in mouse ES cells, where it acts downstream of leukemia inhibitor factor to maintain these stem cells in an undifferentiated cell state [56,91]. In the testis, STAT3 is expressed in gonocytes and spermatogonia [92]. The activated (phosphorylated) form of STAT3 is expressed in Thy1+ spermatogonial cell cultures, suggesting that STAT3 is functional in these cells [93]. Consistent with STAT3 serving to promote SSC differentiation, withdrawal of the SSC self-renewal promoting factor, GDNF, led to increased levels of phosphorylated STAT3 (without a change in total STAT3 level) [90]. To directly examine its function in spermatogonia, Oatley et al. (2010) used three independent approaches to inhibit the level/function of STAT3 in Thy1+ spermatogonial cell cultures: (i) siRNA-mediated knockdown, (ii) incubation with a cell-permeable STAT3-inhibitor peptide, and (iii) incubation with a JAK2 inhibitor that represses STAT3 phosphorylation. All three treatments increased the number of SSCs in these cultures, as measured using the germ cell transplantation assay [93]. Together with the finding that these treatments did not measurably affect cell proliferation or cell survival in vitro, this suggested that SSC differentiation was inhibited, resulting in an accumulation of SSCs. To directly test this, Oatley et al. (2010) used a lentivirus expressing an shRNA against Stat3 to stably depleted STAT3 in these cell cultures. Unlike control cells, the STAT3-depleted cells failed to re-establish full spermatogenesis in recipient mice testes. Instead they gave rise to patches of As, Apr, and Aal spermatogonia, indicative of a defect in SSC differentiation.
A role for STAT3 in promoting SSC differentiation was unexpected given that STAT3 has the opposite effect in mouse ES cells and germline stem cells in Drosophila melanogaster: it promotes their self-renewal [56,91,94]. A likely explanation is that STAT3’s function depends on its cellular context. Thus, in pluripotent cells, STAT3 regulates a specific repertoire of genes—including Klf4, Utf1, and Myc—that together serve to promote the maintenance of that cellular state [95]. In contrast, undifferentiated cells already committed to the germ cell lineage may respond to STAT3 by regulating a set of genes that, instead, promote their differentiation. While genome-wide studies have not been conducted to identify the specific targets of STAT3 in germ cells, one direct target that has been identified by a candidate approach is the Ngn3 gene [90]. As discussed next, this gene encodes a transcription factor that promotes SSC differentiation, suggesting the existence of a STAT3-NGN3 circuit critical for SSC differentiation (Fig. 3B).
NGN3
The bHLH family member, NGN3, is best known for its role in promoting the development of endocrine cells in both the pancreas and the intestine [96,97]. In the testis, NGN3 is expressed in a subset of undifferentiated spermatogonia, as demonstrated by in situ hybridization and immunostaining [86,97]. As another means to test this, Nakagawa et al. (2007) used reporter mice that contain a stop cassette in between two loxP sites that prevents downstream LacZ expression unless excised out by Cre-mediated recombination. Crossing these reporter mice with tamoxifen-inducible Ngn3-Cre ER mice permitted the fate of NGN3+ cells to be followed by pulse-chase analysis [5]. Using this approach, these authors demonstrated that most NGN3-lineage spermatogonia advance to subsequent stages of spermatogenesis and leave the testes within 3 months. This provided strong support for the idea that NGN3+ germ cells are spermatogonial progenitors committed to undergo differentiation.
To determine the role of NGN3 in undifferentiated spermatogonia, the same in vitro and in vivo approaches used to analyze STAT3 function [93] were employed by Kaucher et al. (2012) [90]. These investigators found that transient depletion of NGN3 (using siRNAs) in Thy1+ spermatogonial cell cultures triggered an increase in the number of SSCs without measurably increasing cell proliferation, suggesting that the SSCs accumulate because of a block in SSC differentiation. As further evidence for this, they found that stable depletion of NGN3 (using a shRNA/lentivirus approach) blocked the ability of SSCs in Thy1+ spermatogonial cell cultures to regenerate full spermatogenesis after transplantation. Instead, there were patches of As, Apr, and Aal spermatogonia, indicating a block in SSC differentiation [90]. Together, these results indicate that, like STAT3, NGN3 promotes SSC differentiation. This effect of NGN3 on undifferentiated germ cells is reminiscent of its role in endocrine progenitors, where NGN3 promotes their differentiation into mature endocrine cells [99].
While the molecular circuitry downstream of NGN3 that drives SSC differentiation is unknown, NGN3 is a good candidate to be a central hub in the circuitry driving SSC differentiation. This follows from the finding that the Ngn3 is regulated by all the other known SSC differentiation-promoting transcription factors: STAT3, SOX3, SOHLH1, and SOHLH2 [88,90,100]. Also consistent with its having a crucial role, the Ngn3 gene is negatively regulated by the SSC maintenance factor, GDNF (Fig. 3B); presumably to prevent NGN3 from promoting the premature differentiation of SSCs still under the influence of GDNF [90]. In the future, it will be crucial to decipher the downstream targets of NGN3 in SSCs towards generating a complete map of the molecular circuitry revolving around NGN3 that drives SSC differentiation.
SOHLH1 and SOHLH2
The spermatogenesis- and oogenesis-specific bHLH transcription factor-1 (Sohlh1) gene was first identified as being of interest because it was found to exhibit a germ cell-specific expression pattern [101]. To determine its function, Ballow et al. (2006) generated Sohlh1-KO mice and found that they accumulated PLZF+ spermatogonia and contained few spermatocytes, suggesting they have a spermatogonial differentiation defect [102]. This was interpreted as resulting from a cell autonomous defect in spermatogonial differentiation given that SOHLH1 protein was found to be exclusively expressed in spermatogonia [102].
A related gene—Sohlh2—was identified that is also exclusively expressed in a subset of undifferentiated (GFRα1 negative) spermatognia and most differentiating spermatogonia [102–104]. This led to the hypothesis that Sohlh2 serves some or all of the same functions as Sohlh1. In support of this hypothesis, Sohlh2-KO mice were found to have essentially the same phenotype as Sohlh1-KO mice [102–104]. Furthermore, double knockout mice lacking both Sohlh1 and Sohlh2 had virtually same defects as individual gene knockout mice, providing evidence that they encode transcription factors that function cooperatively, perhaps in order to bind and regulate the same target genes. In support of this, these two SOHLH proteins were found to form not only homodimers, but also heterodimers [100,105].
As further evidence for an interrelationship between these two proteins, they were found to regulate each other. Sohlh1 expression depends on SOHLH2 (based on reduced Sohlh1 mRNA levels in Sohlh2-KO testes) and SOLHL2 has the potential to compensate for deficiencies in SOHLH1 (based on the increased Sohlh2 mRNA levels in Sohlh1-KO testes) [102,104]. This cross-regulation may be direct, based on ChIP analysis demonstrating that SOHLH1 and SOHLH2 bind each other’s promoters [100]. These two transcription factors also bind their own promoters, raising the possibility of autoregulatory control [100].
Since SSC differentiation and the subsequent steps of spermatogonial differentiation are interconnected process, it is difficult to distinguish between them. This is accentuated by the fact that there is no definite marker to distinguish between the spermatogonial cell types predicted to accumulate when one or the other of these differentiate events fail; i.e., SSCs and spermatogonial progenitors, respectively. To address this issue for SOHLH1 and SOHLH2, Suzuki et al. (2012) elected to assay the expression of differentiation- and self renewal-specific genes as “a readout” of differentiation events [100]. Their results led them to propose that these two transcription factors promote both SSC and spermatogonial differentiation. As one line of evidence for the former, they found that SOHLH1 and SOHLH2 negatively regulate many genes that promote SSC self-renewal, including c-Ret, Gfrα1, Nanos2, and Pou5f1. The regulation of the c-Ret and Gfrα1 genes is likely to be direct, based on ChIP analysis showing that both SOHLH1 and SOHLH2 occupy the promoters of these genes. This is likely to be functionally important since these two genes encode the two subunits of the receptor for GDNF, which is essential for SSC self-renewal (Fig. 2) [106]. In their KO testes (individual KO as well as double KO), GFRα1 was detected in 8 or 16 chains of Aal spermatogonia, which normally do not express GFRα1, suggesting that SOHLH1 and SOHLH2 suppress GDNF signaling to promote the differentiation of SSCs. Another line of evidence that these two transcription factors promote SSC differentiation is that they positively regulate other genes that promote SSC differentiation, such as Ngn3 and Sox3 [102,104]. This regulation appears to be direct given that both SOHLH1 and SOHLH2 bind to the Ngn3 and Sox3 promoter regions, as judged by ChIP analysis [100]. The evidence that the SOHLH proteins promote later steps of spermatogonial differentiation comes from the finding that they both positively regulate c-Kit, which is known to be specifically involved in A-spermatogonia differentiation [81,82]. c-Kit appears to be a direct target of the SOHLH proteins, based on (i) electrophoretic mobility shift and ChIP analyses showing that both can independently or cooperatively bind to a classic bHLH transcription factor binding site within the c-Kit promoter and (ii) overexpression of SOHLH proteins cooperatively activates expression of reporter containing the c-Kit promoter [105]. In summary, the available evidence suggests that SOHLH1 and SOHLH2 promote both SSC differentiation and spermatogonial differentiation by regulating different target genes.
DMRT1
Doublesex and mab3-related transcription factor-1 (DMRT1) is best known for its essential roles in sex determination in many species [107–109]. DMRT1 achieves this through its expression in Sertoli cells [110]. In mice, DMRT1 is also expressed in all stages of spermatogonia [111]. To determine the role of DMRT1 in spermatogonia, Matson et al. (2010) used Ngn3-Cre mice to conditionally knockout the Dmrt1 gene in late undifferentiated spermatogonia. These Dmrt1-cKO mice had abnormal spermatogonia that exhibited premature initiation of meiosis and precocious expression of STRA8. As a result of premature initiation of meiosis, many of the undifferentiated spermatogonia in these cKO mutant mice skipped differentiating spermatogonial steps, leading to a reduced numbers of germ cells. While these data primarily suggest that DMRT1 inhibits meiosis in spermatogonia, it is also consistent with a model in which DMRT1 is essential for spermatogonial differentiation. As further evidence to support the latter, microarray analysis revealed that spermatogonial differentiation genes were overrepresented among the genes dysregulated in response to loss of Dmrt1 in the testes of a mouse model that accumulates undifferentiated spermatogonia [112].
To begin to understand the molecular mechanism of DMRT1 action, ChIP-chip analysis was performed on testes from early postnatal (P9) mice [113]. Among the genes identified as bound by DMRT1 in spermatogonia were Stra8 and Sohlh1. The former was of interest given that it is a key factor important for initiation of meiosis [114,115], while the latter is important since it drives spermatogonial differentiation, as described above. Sohlh1 gene is a positive direct target of DMRT1 based on several lines of evidence: (i) the Sohlh1 promoter contains a close match to the DMRT1 consensus binding site, (ii) SOHLH1 is coexpressed with DMRT1 in spermatogonia, (iii) Dmrt1-mutant mice spermatogonia lack detectable expression of SOHLH1 protein, and (iv) Dmrt1-mutant mice have reduced levels of Sohlh1 mRNA compared to control mice [112]. This data leads to a model in which DMRT1 promotes spermatogonial differentiation, in part, through SOHLH1 (Fig. 3B). Together, the available data suggest that DMRT1 is a transcription factor that helps guide the orderly developmental progression of spermatogonia by regulating key genes that drive their differentiation and prevent premature meiosis
5. Perspective
During the last decade, several transcription factors involved in the self-renewal and differentiation of spermatogonia have been uncovered. The field has been dramatically moved forward by two technical innovations: (i) the germ cell transplantation assay as a means to quantify SSCs and (ii) the development of in vitro SSC culture systems to manipulate and study SSCs. While these developments have benefited our understanding of SSCs and the factors important for their self-renewal, we remain largely in the dark about the factors important for the differentiation of SSCs and subsequent spermatogonial differentiation steps. In part, this has been because it has been challenging to distinguish between defects caused by SSC and post-SSC spermatogonial differentiation steps (see Fig. 1). Both defects cause an accumulation of undifferentiated spermatogonia with only subtle differences between the specific subsets affected in the two instances. In the future, it will be crucial to identify SSC- and spermatogonial progenitor-specific markers to address this issue. A candidate SSC-specific marker is ID4, which is only expressed in a subset of As spermatogonia [8]. As another approach, reporter mice need to be developed that distinguish between these spermatogonial subsets. Candidate reporter lines that may be useful in this regard are Id4-GFP mice [8] and Ngn3-GFP mice [98], which preferentially label SSCs and spermatogonial progenitors, respectively. However, whether they label such spermatogonial cell subsets exclusively remains to be determined.
Another future need is to develop in vitro systems that recapitulate spermatogonial differentiation events. A movement in this direction was reported by Dann et al. (2008), who found that treatment of GS cells with RA increased the number of cells positive for the spermatogonial differentiation marker, c-KIT [63]. Given that RA is essential for spermatogonial differentiation in vivo [80], this provided a potentially physiological in vitro model system to study this differentiation step. However, Dann et al. (2008) noted that RA-treated GS cells also exhibited signs of premature meiosis, indicating a limitation to this approach [63]. To potentially improve the fidelity of this system, one could introduce gene products that are known to promote spermatogonial differentiation, such as KITL [82].
As detailed in this review, molecular analyses of mouse transcription factor knockouts/knockdowns have begun to enlighten us as to the transcription networks in spermatogonia (Fig. 3). A transcription factor that has emerged from these studies as potentially central for SSC self-renewal is ETV5. This transcription factor positively regulates several other SSC self-renewal transcription factors, raising the possibility that it is a master regulator of SSC self-renewal (Fig. 3A). This leads to the prediction that ETV5 and the transcription factors involved in SSC maintenance will share many gene targets. In contrast to this prediction, however, microarray analysis has demonstrated that the profile of genes regulated by ETV5 mostly differ from other transcription factors involved in SSC maintenance, including those downstream of ETV5 [6,22]. This may allow more versatility; e.g., each transcription factor can be regulated be different inputs that control self-renewal. ETV5 may control the levels of other SSC maintenance transcription factors for regulatory purposes, not because it is the master regulator in a hierarchical system. A non-mutually exclusive explanation for why SSC maintenance transcription factors regulate different genes is to protect against environmental and genetic perturbations. According to this hypothesis, each SSC self-renewal transcription factor drives a different pathway maintaining stemness; these pathways would be partially redundant so that if one pathway is rendered defective, the others would serve as backups. Consistent with this idea, loss of most SSC maintenance transcription factors does not completely eliminate SSCs, even in aged mice. Regardless of the underlying mechanisms responsible for why SSC maintenance transcription factors have different gene targets, it is also possible that the field is underestimating the number of common targets between them. This possibility derives from the fact that the microarray analysis used to identify the genes regulated by these various transcription factors does not distinguish between direct and indirect targets. If the indirectly regulated genes greatly outnumber the direct targets, this would likely obscure the identification of their common gene targets.
The gene networks downstream of the transcription factors that promote SSC and spermatogonial differentiation remain largely undefined. What is known is that many spermatogonial differentiation transcription factors regulate each other. For example, the Ngn3 gene is positively regulated by virtually all the other known SSC differentiation transcription factors: SOX3, STAT3, SOHLH1, and SOHLH2. The two SOHLH paralogs not only positively regulate the gene encoding NGN3, but also the genes encoding the pro-differentiation factors SOX3 and c-KIT (Fig. 3B), providing a mechanism by which these transcription factors promote spermatogonial differentiation. Potentially supplementing this, SOHLH1 and SOHLH2 cross-regulate each other and they negatively regulate the SSC self-renewal genes Gfra1 and c-Ret (Fig. 3B).
Many SSC-expressed transcription factors have been identified whose functions in SSCs remain unknown. It will be important in the future to determine whether these transcription factors promote SSC maintenance or differentiation. Candidates for the former category are the SSC marker proteins, UTF1 and SALL4, both of which exhibit conserved expression in human and rodent SSCs [116–121]. Both of them are also expressed in ES cells, where they promote ES self-renewal and/or pluripotency [122,123]. The role of Sall4 in SSCs has been a challenge to decipher because of the early lethality of Sall4-null mice [122]. In contrast, Utf1-null mice have surprisingly mild defects, raising the possibility of redundancy with other transcription factors or context-specific effects [124]. Another candidate to have a role in SSCs is the transcription factor RHOX10, which is encoded by a member of a large X-linked homeobox gene cluster selectively expressed in the reproductive tract [125]. The possibility that RHOX10 has roles in SSCs is consistent with its expression pattern, including expression very early during postnatal testes development [126], high expression in cultured GS cells in vitro [127], expression in spermatogonia in the adult testes [127], and enriched expression in purified undifferentiated (Oct4+) spermatogonia [128].
Another important goal for the field is to define the transcription factors that function in human SSCs and spermatogonia. To our knowledge, no transcription factors have yet been identified that have roles in either SSC self-renewal or differentiation in humans. An obvious possibility is that the human orthologs of mouse transcription factors that control SSC/spermatogonial events in mice will have a similar activity in humans. Mitigating this possibility is the fact that there are significant differences between rodent and human spermatogonia. While rodent SSCs go through multiple rounds of proliferation to become spermatocytes (Fig. 1), human SSCs go through only two divisions to generate four spermatocytes per SSC. Therefore, the ratio of SSCs/total germ cells in humans is much higher than in mice. Human spermatogonia have also been traditionally classified in a different way than mouse spermatogonia: A-dark (Adark), A-pale (Apale) and B-spermatogonia. Adark are mostly quiescent, while Apale are actively dividing; it is not known whether one or both of these human spermatogonial subsets have SSC activity [15,129].
It will be important in the future to functionally define the human spermatogonial cell subsets and to determine their relationship with mouse spermatogonial subsets. In conjunction with this, it will be crucial to define the transcriptome of human spermatogonial subsets and to elucidate how these transcriptomes are influenced by transcription factors that dictate self-renewal vs. differentiation decisions. The possibility that mouse studies will be useful in this regard comes from Wu et al. (2009) who showed that there is a striking similarity in the gene expression profiles in prepubertal human spermatogonia and mouse gonocytes [130]. Furthermore, these investigators found that human orthologs of mouse SSC maintenance genes—including c-RET, GFRα1, ETV5 and BCL6B— were highly enriched in human spermatogonia relative to somatic cells. As further evidence for conservation, the human ortholog of the mouse SSC maintenance factor, PLZF, is expressed in human spermatogonia and its biallelic loss in humans is associated with genital hypoplasia [131]. Human and mouse spermatogonia also use the same extrinsic factor—GDNF—for their growth in vitro [130]. To screen for other factors important for SSC self-renewal and differentiation, investigators can take advantage of the recent development of in vitro SSC culturing methods and the xenotransplantation assay for human SSC activity [118,121,130,132]. By utilizing the new tools that are available to the field, the transcription networks functioning in spermatogonia that promote their self-renewal and differentiation can be rapidly mapped in the near future.
Highlights.
Several transcription factors have been identified that function in spermatogonial self renewal and differentiation
Many of these transcription factors form complex transcriptional networks in self-renewing and differentiating spermatogonia
Defining the transcriptional networks that drive spermatogonial maintenance and differentiation depend on recently developed in vivo and in vitro approaches
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- [1].Jan SZ, Hamer G, Repping S, de Rooij DG, van Pelt AMM, Vormer TL. Molecular control of rodent spermatogenesis. Biochim Biophys Acta. 2012;1822:1838–50. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- [2].Hogarth CA. Transcriptional/translational regulation of mammalian spermatogenic stem cells. Adv Exp Med Biol. 2013;786:105–28. doi: 10.1007/978-94-007-6621-1_7. [DOI] [PubMed] [Google Scholar]
- [3].Bettegowda A, Wilkinson M. Transcription and post-transcriptional regulation of spermatogenesis. … R …. 2010;365:1637–51. doi: 10.1098/rstb.2009.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell. 2010;7:214–24. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- [5].Nakagawa T, Nabeshima Y-I, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- [6].Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, et al. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–9. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- [7].Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–52. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- [8].Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod. 2011;85:347–56. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1663–78. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91:11303–7. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–6. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- [12].Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2004;101:16489–94. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- [14].Meng X, Lindahl M, Hyvönen ME, Parvinen M, de Rooij DG, Hess MW, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–93. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- [15].De Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776–98. [PubMed] [Google Scholar]
- [16].Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci U S A. 2006;103:9524–9. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lee J, Kanatsu-Shinohara M, Inoue K, Ogonuki N, Miki H, Toyokuni S, et al. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development. 2007;134:1853–9. doi: 10.1242/dev.003004. [DOI] [PubMed] [Google Scholar]
- [18].Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem. 2007;282:25842–51. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–4. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tyagi G, Carnes K, Morrow C, Kostereva NV, Ekman GC, Meling DD, et al. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod. 2009;81:258–66. doi: 10.1095/biolreprod.108.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schmidt JA, Avarbock MR, Tobias JW, Brinster RL. Identification of glial cell line-derived neurotrophic factor-regulated genes important for spermatogonial stem cell self-renewal in the rat. Biol Reprod. 2009;81:56–66. doi: 10.1095/biolreprod.108.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu X, Goodyear SM, Tobias JW, Avarbock MR, Brinster RL. Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol Reprod. 2011;85:1114–23. doi: 10.1095/biolreprod.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2011;108:12740–5. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development. 2012;139:1734–43. doi: 10.1242/dev.076539. [DOI] [PubMed] [Google Scholar]
- [25].Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2:E75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choong ML, Yong YP, Tan ACL, Luo B, Lodish HF. LIX: a chemokine with a role in hematopoietic stem cells maintenance. Cytokine. 2004;25:239–45. doi: 10.1016/j.cyto.2003.11.002. [DOI] [PubMed] [Google Scholar]
- [27].Simon L, Ekman GC, Garcia T, Carnes K, Zhang Z, Murphy T, et al. ETV5 regulates sertoli cell chemokines involved in mouse stem/progenitor spermatogonia maintenance. Stem Cells. 2010;28:1882–92. doi: 10.1002/stem.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang Q-E, Kim D, Kaucher A, Oatley MJ, Oatley JM. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J Cell Sci. 2013;126:1009–20. doi: 10.1242/jcs.119826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Manders PM, Hunter PJ, Telaranta AI, Carr JM, Marshall JL, Carrasco M, et al. BCL6b mediates the enhanced magnitude of the secondary response of memory CD8+ T lymphocytes. Proc Natl Acad Sci U S A. 2005;102:7418–25. doi: 10.1073/pnas.0501585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hamra FK, Schultz N, Chapman KM, Grellhesl DM, Cronkhite JT, Hammer RE, et al. Defining the spermatogonial stem cell. Dev Biol. 2004;269:393–410. doi: 10.1016/j.ydbio.2004.01.027. [DOI] [PubMed] [Google Scholar]
- [31].Kobayashi A, Kwan K-M, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132:2809–23. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- [32].Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–30. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- [33].Ghazvini M, Mandemakers W, Jaegle M, Piirsoo M, Driegen S, Koutsourakis M, et al. A cell type-specific allele of the POU gene Oct-6 reveals Schwann cell autonomous function in nerve development and regeneration. EMBO J. 2002;21:4612–20. doi: 10.1093/emboj/cdf475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bermingham JR, Scherer SS, O’Connell S, Arroyo E, Kalla KA, Powell FL, et al. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–62. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- [35].Wu X, Oatley JM, Oatley MJ, Kaucher AV, Avarbock MR, Brinster RL. The POU domain transcription factor POU3F1 is an important intrinsic regulator of GDNF-induced survival and self-renewal of mouse spermatogonial stem cells. Biol Reprod. 2010;82:1103–11. doi: 10.1095/biolreprod.109.083097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–9. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- [37].Inman KE, Downs KM. Brachyury is required for elongation and vasculogenesis in the murine allantois. Development. 2006;133:2947–59. doi: 10.1242/dev.02454. [DOI] [PubMed] [Google Scholar]
- [38].Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell. 2013;27:516–29. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- [39].Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20:8317–25. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
- [40].Sablitzky F, Moore A, Bromley M, Deed RW, Newton JS, Norton JD. Stage- and subcellular-specific expression of Id proteins in male germ and Sertoli cells implicates distinctive regulatory roles for Id proteins during meiosis, spermatogenesis, and Sertoli cell function. Cell Growth Differ. 1998;9:1015–24. [PubMed] [Google Scholar]
- [41].De Rooij DG, van Beek ME a B. Computer simulation of the rodent spermatogonial stem cell niche. Biol Reprod. 2013;88:131, 1–11. doi: 10.1095/biolreprod.113.108639. [DOI] [PubMed] [Google Scholar]
- [42].David G, Alland L, Hong SH, Wong CW, DePinho RA, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–56. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- [43].Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–7. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–72. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- [45].Yeyati PL, Shaknovich R, Boterashvili S, Li J, Ball HJ, Waxman S, et al. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925–34. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- [46].McConnell MJ, Chevallier N, Berkofsky-Fessler W, Giltnane JM, Malani RB, Staudt LM, et al. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol Cell Biol. 2003;23:9375–88. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology. 1999;140:5894–900. doi: 10.1210/endo.140.12.7172. [DOI] [PubMed] [Google Scholar]
- [48].Filipponi D, Hobbs RM, Ottolenghi S, Rossi P, Jannini EA, Pandolfi PP, et al. Repression of kit expression by Plzf in germ cells. Mol Cell Biol. 2007;27:6770–81. doi: 10.1128/MCB.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142:468–79. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- [51].Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–89. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, et al. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci U S A. 2008;105:19384–9. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Puszyk W, Down T, Grimwade D, Chomienne C, Oakey RJ, Solomon E, et al. The epigenetic regulator PLZF represses L1 retrotransposition in germ and progenitor cells. EMBO J. 2013;32:1941–52. doi: 10.1038/emboj.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–8. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- [56].Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- [57].Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- [58].Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- [59].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- [60].Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, et al. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–92. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- [61].Schöler HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–95. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Pesce M, Wang X, Wolgemuth DJ, Schöler H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- [63].Dann CT, Alvarado AL, Molyneux L a, Denard BS, Garbers DL, Porteus MH. Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells. 2008;26:2928–37. doi: 10.1634/stemcells.2008-0134. [DOI] [PubMed] [Google Scholar]
- [64].Kanatsu-Shinohara M, Ogonuki N, Miki H, Inoue K, Morimoto H, Takashima S, et al. Genetic influences in mouse spermatogonial stem cell self-renewal. J Reprod Dev. 2010;56:145–53. doi: 10.1262/jrd.09-153n. [DOI] [PubMed] [Google Scholar]
- [65].He Z, Jiang J, Kokkinaki M, Dym M. Nodal signaling via an autocrine pathway promotes proliferation of mouse spermatogonial stem/progenitor cells through Smad2/3 and Oct-4 activation. Stem Cells. 2009;27:2580–90. doi: 10.1002/stem.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Farmer SR. The forkhead transcription factor Foxo1: a possible link between obesity and insulin resistance. Mol Cell. 2003;11:6–8. doi: 10.1016/s1097-2765(03)00003-0. [DOI] [PubMed] [Google Scholar]
- [67].Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–52. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- [68].Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–7. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest. 2011;121:3456–66. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Biggs WH, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci U S A. 1999;96:7421–6. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- [72].Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, et al. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- [73].White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 2010;139:11–21. doi: 10.1530/REP-09-0083. [DOI] [PubMed] [Google Scholar]
- [74].Kimmins S, Kotaja N, Davidson I, Sassone-Corsi P. Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction. 2004;128:5–12. doi: 10.1530/rep.1.00170. [DOI] [PubMed] [Google Scholar]
- [75].Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, et al. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–9. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–7. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- [77].Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, et al. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 2005;19:794–803. doi: 10.1101/gad.1290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–7. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Yoshida S. Elucidating the identity and behavior of spermatogenic stem cells in the mouse testis. Reproduction. 2012;144:293–302. doi: 10.1530/REP-11-0320. [DOI] [PubMed] [Google Scholar]
- [80].Van Pelt AM, de Rooij DG. Synchronization of the seminiferous epithelium after vitamin A replacement in vitamin A-deficient mice. Biol Reprod. 1990;43:363–7. doi: 10.1095/biolreprod43.3.363. [DOI] [PubMed] [Google Scholar]
- [81].Koshimizu U, Sawada K, Tajima Y, Watanabe D, Nishimune Y. White-spotting mutations affect the regenerative differentiation of testicular germ cells: demonstration by experimental cryptorchidism and its surgical reversal. Biol Reprod. 1991;45:642–8. doi: 10.1095/biolreprod45.4.642. [DOI] [PubMed] [Google Scholar]
- [82].De Rooij DG, Okabe M, Nishimune Y. Arrest of spermatogonial differentiation in jsd/jsd, Sl17H/Sl17H, and cryptorchid mice. Biol Reprod. 1999;61:842–7. doi: 10.1095/biolreprod61.3.842. [DOI] [PubMed] [Google Scholar]
- [83].Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–44. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- [84].Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–55. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- [85].Weiss J, Meeks JJ, Hurley L, Raverot G, Frassetto A, Jameson JL. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol. 2003;23:8084–91. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Woods KS, Cundall M, Turton J, Rizotti K, Mehta A, Palmer R, et al. Over-and underdosage of SOX3 is associated with infundibular hypoplasia and hypopituitarism. Am J Hum Genet. 2005;76:833–49. doi: 10.1086/430134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Albrecht KH, Young M, Washburn LL, Eicher EM. Sry expression level and protein isoform differences play a role in abnormal testis development in C57BL/6J mice carrying certain Sry alleles. Genetics. 2003;164:277–88. doi: 10.1093/genetics/164.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Raverot G, Weiss J, Park SY, Hurley L, Jameson JL. Sox3 expression in undifferentiated spermatogonia is required for the progression of spermatogenesis. Dev Biol. 2005;283:215–25. doi: 10.1016/j.ydbio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- [89].Laronda MM, Jameson JL. Sox3 functions in a cell-autonomous manner to regulate spermatogonial differentiation in mice. Endocrinology. 2011;152:1606–15. doi: 10.1210/en.2010-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kaucher AV, Oatley MJ, Oatley JM. NEUROG3 is a critical downstream effector for STAT3-regulated differentiation of mammalian stem and progenitor spermatogonia. Biol Reprod. 2012;86:164, 1–11. doi: 10.1095/biolreprod.111.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Raz R, Lee CK, Cannizzaro LA, d’Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96:2846–51. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Murphy K, Carvajal L, Medico L, Pepling M. Expression of Stat3 in germ cells of developing and adult mouse ovaries and testes. Gene Expr Patterns. 2005;5:475–82. doi: 10.1016/j.modgep.2004.12.007. [DOI] [PubMed] [Google Scholar]
- [93].Oatley JM, Kaucher AV, Avarbock MR, Brinster RL. Regulation of mouse spermatogonial stem cell differentiation by STAT3 signaling. Biol Reprod. 2010;83:427–33. doi: 10.1095/biolreprod.109.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294:2546–9. doi: 10.1126/science.1066700. [DOI] [PubMed] [Google Scholar]
- [95].Martello G, Bertone P, Smith A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 2013;32:2561–74. doi: 10.1038/emboj.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–97. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–57. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- [98].Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, et al. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol. 2004;269:447–58. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
- [99].Gradwohl G, Dierich a, LeMeur M, Guillemot F. Neurogenin3 Is Required for the Development of the Four Endocrine Cell Lineages of the Pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–11. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361:301–12. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Rajkovic A, Yan MSC, Klysik M, Matzuk M. Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril. 2001;76:550–4. doi: 10.1016/s0015-0282(01)01966-5. [DOI] [PubMed] [Google Scholar]
- [102].Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–7. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- [103].Hao J, Yamamoto M, Richardson TE, Chapman KM, Denard BS, Hammer RE, et al. Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells. 2008;26:1587–97. doi: 10.1634/stemcells.2007-0502. [DOI] [PubMed] [Google Scholar]
- [104].Toyoda S, Miyazaki T, Miyazaki S, Yoshimura T, Yamamoto M, Tashiro F, et al. Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Dev Biol. 2009;325:238–48. doi: 10.1016/j.ydbio.2008.10.019. [DOI] [PubMed] [Google Scholar]
- [105].Barrios F, Filipponi D, Campolo F. SOHLH1 and SOHLH2 control Kit expression during postnatal male germ cell development. J Cell Sci. 2012;125:1455–64. doi: 10.1242/jcs.092593. [DOI] [PubMed] [Google Scholar]
- [106].De Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J Androl. 2012;33:1085–95. doi: 10.2164/jandrol.112.016832. [DOI] [PubMed] [Google Scholar]
- [107].Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–63. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- [108].Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–95. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zarkower D. Establishing sexual dimorphism: conservation amidst diversity? Nat Rev Genet. 2001;2:175–85. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- [110].Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–4. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–27. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell. 2010;19:612–24. doi: 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, et al. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci U S A. 2010;107:13360–5. doi: 10.1073/pnas.1006243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Baltus AE, Menke DB, Hu Y-C, Goodheart ML, Carpenter AE, de Rooij DG, et al. In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet. 2006;38:1430–4. doi: 10.1038/ng1919. [DOI] [PubMed] [Google Scholar]
- [115].Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, van Pelt AMM, et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 2008;105:14976–80. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Gassei K, Orwig KE. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One. 2013;8:e53976. doi: 10.1371/journal.pone.0053976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Eildermann K, Aeckerle N, Debowski K, Godmann M, Christiansen H, Heistermann M, et al. Developmental expression of the pluripotency factor sal-like protein 4 in the monkey, human and mouse testis: restriction to premeiotic germ cells. Cells Tissues Organs. 2012;196:206–20. doi: 10.1159/000335031. [DOI] [PubMed] [Google Scholar]
- [118].Dovey SL, Valli H, Hermann BP, Sukhwani M, Donohue J, Castro CA, et al. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest. 2013;123:1833–43. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Van Bragt MP a, Roepers-Gajadien HL, Korver CM, Bogerd J, Okuda A, Eggen BJL, et al. Expression of the pluripotency marker UTF1 is restricted to a subpopulation of early A spermatogonia in rat testis. Reproduction. 2008;136:33–40. doi: 10.1530/REP-07-0536. [DOI] [PubMed] [Google Scholar]
- [120].Von Kopylow K, Kirchhoff C, Jezek D, Schulze W, Feig C, Primig M, et al. Screening for biomarkers of spermatogonia within the human testis: a whole genome approach. Hum Reprod. 2010;25:1104–12. doi: 10.1093/humrep/deq053. [DOI] [PubMed] [Google Scholar]
- [121].Kossack N, Terwort N, Wistuba J, Ehmcke J, Schlatt S, Schöler H, et al. A combined approach facilitates the reliable detection of human spermatogonia in vitro. Hum Reprod. 2013;28:3012–25. doi: 10.1093/humrep/det336. [DOI] [PubMed] [Google Scholar]
- [122].Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133:3005–13. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- [123].Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 1998;17:2019–32. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Nishimoto M, Katano M, Yamagishi T, Hishida T, Kamon M, Suzuki A, et al. In vivo function and evolution of the eutherian-specific pluripotency marker UTF1. PLoS One. 2013;8:e68119. doi: 10.1371/journal.pone.0068119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].MacLean JA, Wilkinson MF. The Rhox genes. Reproduction. 2010;140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Maclean J a, Chen M a, Wayne CM, Bruce SR, Rao M, Meistrich ML, et al. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–82. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- [127].Song H-W, Dann CT, McCarrey JR, Meistrich ML, Cornwall GA, Wilkinson MF. Dynamic expression pattern and subcellular localization of the Rhox10 homeobox transcription factor during early germ cell development. Reproduction. 2012;143:611–24. doi: 10.1530/REP-11-0479. [DOI] [PubMed] [Google Scholar]
- [128].Yang L, Wu W, Qi H. Gene expression profiling revealed specific spermatogonial stem cell genes in mouse. Genesis. 2013;51:83–96. doi: 10.1002/dvg.22358. [DOI] [PubMed] [Google Scholar]
- [129].Ehmcke J, Schlatt S. A revised model for spermatogonial expansion in man: lessons from non-human primates. Reproduction. 2006;132:673–80. doi: 10.1530/rep.1.01081. [DOI] [PubMed] [Google Scholar]
- [130].Wu X, Schmidt J a, Avarbock MR, Tobias JW, Carlson C a, Kolon TF, et al. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Natl Acad Sci U S A. 2009;106:21672–7. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Fischer S, Kohlhase J, Böhm D, Schweiger B, Hoffmann D, Heitmann M, et al. Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet. 2008;45:731–7. doi: 10.1136/jmg.2008.059451. [DOI] [PubMed] [Google Scholar]
- [132].Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AMM. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA. 2011;305:2416–8. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]