Abstract
Drought tolerance is an important trait being pursued by the agbiotech industry. Abscisic acid (ABA) is a stress hormone that mediates a multitude of processes in growth and development, water use efficiency (WUE), and gene expression during seed development and in response to environmental stresses. Arabidopsis B3-domain transcription factor Related to ABA-Insensitive3 (ABI3)/Viviparous1 (namely, AtRAV2) and basic leucine zipper (bZIPs) AtABI5 or AtABF3 transactivated ABA- inducible promoter: GUS reporter expression in a maize mesophyll protoplast transient assay and showed synergies in reporter transactivation when co-expressed. Transgenic cotton (Gossypium hirsutum) expressing AtRAV1/2 and/or AtABI5 showed resistance to imposed drought stress under field and greenhouse conditions and exhibited improved photosynthetic and WUEs associated with absorption through larger root system and greater leaf area. We observed synergy for root biomass accumulation in the greenhouse, intrinsic WUE in the field, and drought tolerance in stacked AtRAV and AtABI5 double-transgenic cotton. We assessed AtABI5 and AtRAV1/2 involvement in drought stress adaptations though reactive oxygen species scavenging and osmotic adjustment by marker gene expression in cotton. Deficit irrigation-grown AtRAV1/2 and AtABI5 transgenics had “less stressed” molecular and physiological phenotypes under drought, likely due to improved photoassimilation and root and shoot sink strengths and enhanced expression of endogenous GhRAV and genes for antioxidant and osmolyte biosynthesis. Over-expression of bZIP and RAV TFs could impact sustainable cotton agriculture and potentially other crops under limited irrigation conditions.
Keywords: abscisic acid, ABA-insensitive, RAV, maize protoplasts, transgenic cotton, drought stress resistance, reactive oxygen species, root biomass, less-stressed phenotype
Introduction
Engineering drought tolerance is an objective of the agbiotech industry and has the potential to create novel drought insurance models (e.g. purposeful imposition of drought stress during vegetative growth). Plant responses to the environment are not comprised of linear signaling pathways, but rather a complex network evolved to deal with ever-changing environments and plants’ immobility. Abscisic Acid (ABA) is a stress hormone that mediates a multitude of processes in growth and development, water use efficiency (WUE), and gene expression during seed development and in response to environmental stresses. Seed maturation and freezing/drought/salt tolerance likely share common protective mechanisms, as they all involve dehydration stress. Genes encoding cold-responsive, salt-inducible, and late embryogenesis-abundant protein homologues in wheat, maize, barley, carrot, and the resurrection plant Craterostigma are induced by ABA and dehydration stress (Dure 1993; Ingram and Bartels 1996) and have also been named RABs (Responsive to ABA), demonstrating the phenomenon of cross-tolerance to environmental stresses where exposure to one stress confers resistance to others. Although the exact roles of RAB genes in cross tolerance have not yet been drawn, there are strong evidences that support their adaptive functions in desiccation, freezing, and salt tolerance beyond the plant kingdom (Campos et al, 2013; Thomashow 1999). Altered expression of ABA signaling components can have utilitarian effects on stress adaptation of plants (Uno et al., 2000).
Transcription factors (TFs) control virtually all plant traits including yield, disease resistance, cold- and drought protection, and myriad value-added crop properties by coordinated regulation of multiple target genes of known or unknown functions. The Basic 3- DNA Binding Domain (B3-DBD) TFs, first identified as the viviparous1 (vp1) mutant from Zea mays (McCarty et al., 1991; Suzuki et al., 1997) and the orthologous Arabidopsis mutant ABA insensitive-3 (abi3) (Giraudat et al., 1992), play key roles in a hierarchical cascade of TF interactions that control seed maturation (McCarty et al., 1991; Finkelstein and Somerville 1990) via ABA signaling. B3-DBD TFs have been classified into five gene families: AUXIN RESPONSE FACTOR/ARF, ABI3, HIGH LEVEL EXPRESSION of SUGAR INDUCIBLE, RELATED to ABI3/VP1 (RAV), and REPRODUCTIVE MERISTEM (Peng and Weselake, 2013; Romanel et al., 2009). The DNA binding specificities of B3-DBD TFs has been studied in ABI3, RAVs, and ARFs, and the B3 domain of each of family binds to a different target DNA sequence. The RAV/TEMPRANILLO family of TFs contain an N-terminal APETELA2 (AP2)-like DBD that binds 5′-CAACA-3′ whereas the B3-DBD binds 5′- CACCTG-3′ (Kagaya and Hattori, 2009). Various RAVs from several species are induced in response to multiple hormone treatments or stresses (Xu et al., 2011) and over-expression of CaRAV1 in Arabidopsis or SlRAV2 in tomato results in the induction of pathogenesis-related genes, enhanced resistance against infection by bacterial pathogens, and tolerance to osmotic, salt, and cold stresses (Li et al., 2011; Sohn et al., 2006). Biotic and abiotic stresses generate Reactive Oxygen Species (ROS) that destroy membrane lipids and promote cell death. Virus induced- or RNA interference- gene silencing of CaRAV1 resulted in higher levels of lipid peroxidation, supporting that CaRAV1 is involved in ROS scavenging (Lee et al., 2010). Furthermore, CaRAV1 physically interacts with Oxidoreductase1/CaOX1 in yeast two-hybrid experiments (Lee et al., 2010).
There are 81 predicted Basic leucine zipper (bZIP) TFs in Arabidopsis, but only one bZIP subfamily (ABA INSENSITIVE-5/ABI5 and its close homologues ABA Responsive element Binding Factors/ABF1-4) has been genetically or functionally linked to ABA response in a pathway from Pyrabactin-Resistance-Like/Regulatory Control of ABA Receptors (PYR/RCAR) and downstream SnRK2-like protein kinases and type 2C protein phosphatases (Cutler et al., 2010; Lynch et al., 2012; Okamoto et al., 2013; Raghavendra et al., 2010; Soon et al., 2012). ABI5 is involved in seed-specific responses, whereas the ABFs play roles at the seedling and later stages. ABI5/ABFs are subject to proteolytic regulation by ubiquitylation mediated by 14-3-3 proteins and multiple E3 ligases (Chen et al., 2013). At the seedling stage, ectopic expression of ABI5 leads to higher expression of stress-induced genes (e.g. Cor6.6, Cor15a, Rab18) and over-expression is sufficient to confer hypersensitivity to exogenous ABA which inhibits root growth (Lopez-Molina et al., 2001; Brocard et al., 2002). ABI5 over-expression also results in high sensitivity to glucose and anthocyanin accumulation in response to sugar stress (Finkelstein et al., 2002). Analyses of transcript accumulation in abi5 mutants suggest that, similar to ABI3, ABI5 has both activator and repressor functions that may have either synergistic or antagonistic effects on gene expression, depending on the target gene. ABI5 protein accumulation is further enhanced by ABA-induced phosphorylation and resulting stabilization of the protein (Lopez-Molina et al., 2001; Wang et al., 2013).
In the present study, we show that B3-DBD effector RAVs, and ABA effector bZIPs transactivate ABA-inducible gene expression in a maize mesophyll protoplast transient assay and show synergy in their activities when co-expressed. Importantly, transgenic cotton expressing AtRAV1 or AtRAV2 and/or ABI5 TFs showed resistance to imposed drought stress under greenhouse and field conditions and had improved photosynthetic efficiency, likely due to improved absorption through a larger root system. Transgenic cotton expressing Arabidopsis RAVs and ABI5 has a “less stressed phenotype,” which may have broad utility for engineering abiotic stress tolerance in crops.
Results
Functional interactions of AtRAV2 with ABA effectors in transiently transformed maize mesophyll protoplasts
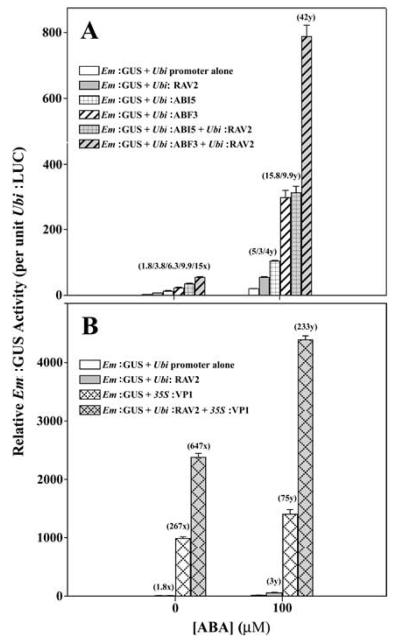
We previously showed by cotransformation of reporter and multiple effector plasmid constructs (all components in trans) that bZIP TFs ABI5 and ABF3 interact functionally and physically with maize B3-DBD TF VP1 (Finkelstein et al., 2005), and that a homozygous T-DNA insertion line (SALK_070847) in B3-DBD TF AtRAV2/At1g68840 show elevated transpiration rates (Luo, Gampala, and Rock. 2005. 16th Intl. Conf. Arabidopsis Res. http://www.arabidopsis.org), suggesting involvement of RAVs in ABA responses. Similarly, an AtRAV1/At1g13260 homozygous T-DNA insertion line (SALK_021865) showed modest ABA insensitivity to root growth inhibition, and conversely over-expression of AtRAV1 or AtRAV2 in transgenic Arabidopsis resulted in increased sensitivity to ABA inhibition of seed germination and root growth (data not shown; Mittal, 2012). In order to further test the involvement of AtRAV2 as a positive effector of ABA signaling, we over-expressed the AtRAV2 cDNA in transiently transformed maize mesophyll protoplasts (Sheen, 2001; Jia et al., 2009) and characterized its function on ABA-inducible reporter gene expression and interactions with known positive effectors (ABI5, ABF3, VP1) and the dominant negative ABA effector mutant abi1-1 encoding a protein phosphatase type 2C (PP2C) (Sheen, 1998). The results are shown in Fig. 1. The ABA effectors ABI5 and ABF3 (Finkelstein et al., 2005) resulted in transactivation of between four- to 16-fold above promoter of Early-methionine-rich:GUS reporter gene (Em:GUS) alone in the absence or presence of a saturating concentration (100 μM) ABA, respectively. ABA treatment resulted in five-fold induction of the Em:GUS reporter alone, whereas over-expression of AtRAV2 resulted in transactivation of two- to three-fold above the reporter gene alone in both the presence and absence of exogenous ABA (Fig. 1A), values somewhat less than over-expression of ABI5 or ABF3. Similar to previous results with B3-DBD TF VP1 and bZIPs (Finkelstein et al., 2005), we observed a synergy between AtRAV2 and ABI5 or ABF3 when co-transformed, where the observed transactivation was 10-fold and 42-fold for RAV2 plus ABI5 and RAV2 plus ABF3, respectively, above reporter gene alone in the presence of ABA. The interaction can be described as synergistic because the observed effects were about twice the sums of the two respective individual transgene effects in both the absence and presence of ABA (Fig. 1A). Unexpectedly, we also observed a synergistic interaction between over-expressed B3-DBDs AtRAV2 and VP1 (Fig. 1B).
Figure 1.
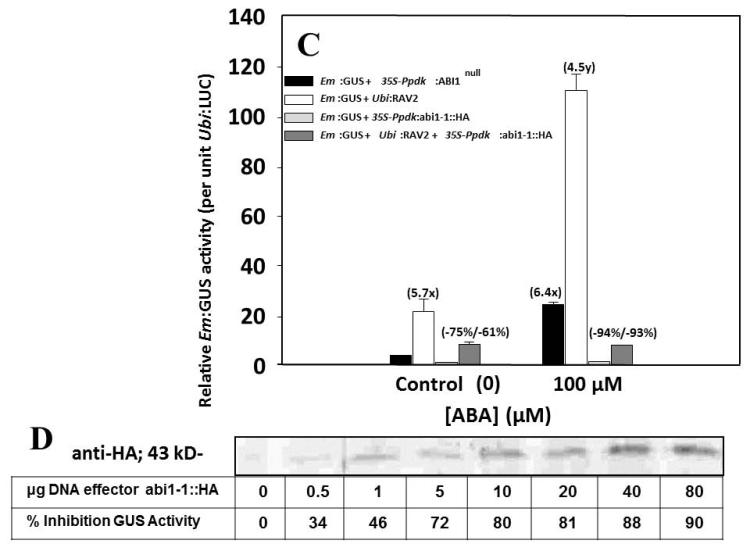
Synergistic interactions between known effectors of ABA signaling and AtRAV2 in transiently transformed maize mesophyll protoplasts. Numbers in parentheses (x) represent fold induction over ‘No ABA’ control and numbers in parentheses (y) represent fold transactivation by over-expressed AtRAV2 (and/or other effector[s]) compared to treatment with 100 μM ABA alone. (A) Synergy between B3 domain-containing AtRAV2 and bZIPs AtABI5 and AtABF3. (B) Synergy between B3-domain ZmVP1 and AtRAV2. (C) Antagonism of AtRAV2 transactivation by upstream ABA effector abi1-1 dominant negative (G180D) protein phosphatase 2C mutant. Negative numbers in parentheses indicate the percent inhibition of proEm:GUS expression relative to controls (without/with RAV2 cotransformation, respectively) by co-transformation of abi1-1 construct. (D) Dose-dependence of input effector DNA for abi1-1 antagonism of ABA (and AtRAV2) transactivation, shown by immunoblot of abi1-1::HA-tagged effector and corresponding percent decreases in relative GUS activities measured in the same protoplast extracts. Error bars are ± s.e.m. (n=4).
In order to further characterize the functional interaction between AtRAV2 and known ABA signaling components, we tested whether the upstream PP2C dominant negative mutant protein abi1-1, which abolishes phosphatase activity and physically interacts with Pyrabactin-like ABA receptors (Fujii et al., 2009), SnRK2, CPK11, and possibly bZIP TFs (Antoni et al., 2012; Lynch et al., 2012), could specifically antagonize the AtRAV2 transactivation of the ABA-inducible Em:GUS reporter. Fig. 1C shows the result that co-transformation of abi1-1 effector with AtRAV2 construct strongly antagonized the >fourfold specific RAV2 transactivation of ABA-inducible reporter gene expression in the presence and absence of ABA (>90% inhibition for ABA treatment). Fig. 1D shows an immunoblot of the dose-dependence of input DNA on protein expression of abi1-1::HA tagged effector, and excellent concordance of abi1-1 protein expression to inhibition of Em:GUS reporter gene expression. Taken together, these results suggest that AtRAV2 over-expression can positively impact ABA responses in monocot and dicot plants, so we generated transgenic cotton over-expressing AtABI5 and AtRAVs in order to test effects on drought stress tolerance.
Generation and molecular characterization of transgenic cotton lines expressing AtRAV1, AtRAV2, AtRAV2L, and AtABI5
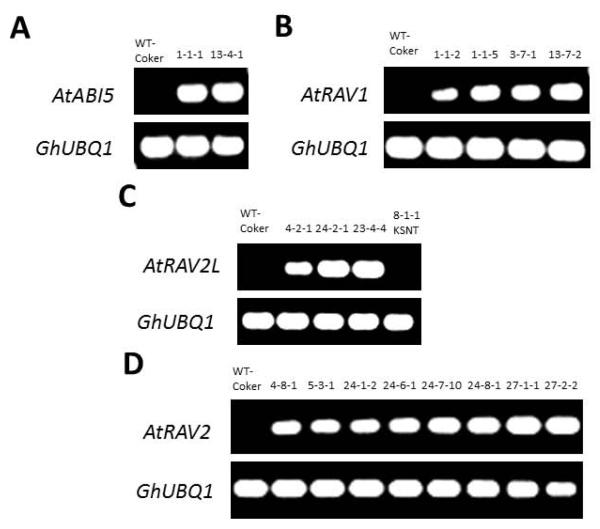
The pro35S-:RAV1, -:RAV2, -:RAV2L, and -:ABI5 constructs were introduced into cotton (Gossypium hirsutum) cultivar Coker312, which has a high capacity for regeneration from hypocotyl explants, via Agrobacterium-mediated transformation (Bayley et al., 1992). A total of four independent RAV1, eight RAV2, three RAV2L, and two ABI5 transgenic events were propagated in the greenhouse and subjected to RNA blot analysis to confirm stable expression (Supplemental Fig. S1). Several larger transcripts than the sizes of full-length coding sequences (RAV2=1,059 nt; RAV2L=1,086 nt; RAV1= 1,035 nt; ABI5= 1,329 nt) were consistently detected in independent transgenic lines, similar to our prior results in protoplasts (Jia et al., 2009) where we observed transcription termination at the bovine growth hormone genomic polyadenylation signal (Goodwin and Rottman 1992), but also predicted some read-through past this animal cis element based on bioinformatic analyses (Loke et al., 2005). Homozygous transgenic plants from these lines and one Kanamycin-Selected NonEffector Transgenic (KSNT, a regenerant line from RAV2L transformation experiments that either subsequently lost the effector DNA or was a false positive for Kanr), were subjected to semi-quantitative reverse transcriptase PCR (RTPCR) (Fig. 2) in the T4 generation to validate the effector transgene expression over several generations and to identify high expression lines for physiological assays.
Figure 2.
Transcript expression analysis for transgenes in cotton lines by Reverse Transcriptase-PCR. GhUBQ1 was used as an internal control. (A) AtABI5 overexpression in two independent lines compared to wild type. (B) AtRAV1 overexpression in four independent lines compared to wild type. (C) AtRAV2L overexpression in three independent lines compared to wild type. (D) AtRAV2 overexpression in eight independent lines compared to wild type.
Physiological characterization of cotton transgenic lines
All of the RAV2L and ABI5 over-expressing lines, and all but one of the RAV2 lines (24-7-10), generated higher seed cotton yield (fiber plus seed weight per plant) under WW greenhouse conditions (Supplemental Fig. S2A), whereas RAV1 and KSNT control regenerant line produced on par with wild-type Coker312. When tested in the field under the most extreme drought and heat conditions on record (2011) that resulted in a 43% overall yield reduction, deficit-irrigated (DI) transgenics, to a greater extent than WW transgenics (including RAV224-7-10), had yields on par with control genotypes (Supplemental Fig. S2B). Differences in yields for transgenics between greenhouse and field may have been due to effects of RAV and ABI5 transgenes on phenology (Castillejo and Pelaz, 2008; Wang et al., 2013); the phenotypes relating to flowering and fiber development will be described elsewhere. Leaf area was higher in transgenic lines under both WW and drought stress treatments in the field (Table 1). Both AtRAV1 cotton lines showed significant (p < 0.04) increases in leaf area per plant with concomitant significant increases in dry mass. Similar results were obtained with AtABI5 and AtRAV2/2L over-expressing lines (Table 1). These results showing increased leaf biomass and non-significant yield penalties during drought in the field suggested that RAV and ABI5 over-expressing lines may have higher WUE, possibly due to enhanced ABA response consistent with the protoplast transient expression results. Therefore, we analyzed photosynthetic assimilation rates (A) in transgenics in response to water deficit under greenhouse and field conditions.
Table 1.
Leaf area (cm2/plant) of transgenic lines grown 85 days in the field under well watered or deficit irrigation after day 42
| Well Watered |
Deficit Irrigation |
|||||
|---|---|---|---|---|---|---|
| Genotype | Average | %ΔWT‡ | ±SEM† | Average | %ΔWT‡ | ±SEM† |
| Coker312 | 2204 | 223 | 1611 | 115 | ||
| ABI51-1-1 | 3626 | 64.5 | 714 | 2596 § | 61.1 | 262 |
| ABI513-4-1 | 2494 | 13.2 | 87 | 1945 | 20.8 | 252 |
| RAV11-1-5 | 3559 | 61.5 | 507 | 3002 § | 86.3 | 319 |
| RAV113-7-2 | 4108 ¶ | 86.4 | 99 | 2805 § | 74.1 | 274 |
| RAV24-8-1 | 3092 | 40.3 | 412 | 2181 § | 35.4 | 172 |
| RAV25-3-1 | 3231 | 46.6 | 602 | 2051 | 27.3 | 468 |
| RAV224-8-1 | 5103 § | 131.5 | 519 | 2606 | 61.7 | 399 |
| RAV2L4-2-1 | 3103 § | 40.8 | 184 | 1778 | 10.4 | 282 |
| RAV2L24-2-1 | 3001 | 36.2 | 744 | 2211 § | 37.2 | 159 |
Average percent change of transgenic line from wild type Coker312
Standard error of the mean. n=3 except for Coker312: n=12.
Significantly different than Coker312 control, p < 0.0001 (Two-tailed Student’s t-test)
Significantly different than Coker312 control, p < 0.05
RAV1, RAV2, and ABI5 transgenic cotton maintains higher photosynthesis resulting in increased WUE, and RAVxABI5 stacked lines adapt better to drought treatments than individual parents
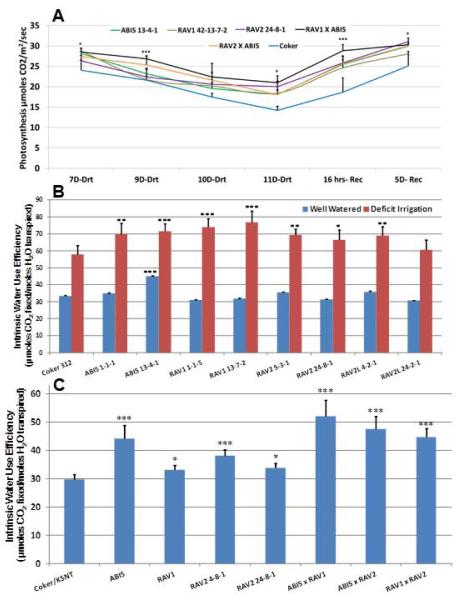
We analyzed drought resistance in the transgenics by conducting controlled DI experiments in both greenhouse and the field. A and other parameters (stomatal conductance Gs, transpiration) were measured on control and transgenic event lines RAV113-7-2, RAV224-8-1, ABI513-4-1, and F1 plants from RAVxABI5 crosses (RAV113-7-2xABI513-4-1 and RAV24-8-1xABI513-4-1) over a span of drought progression and recovery by re-watering in the greenhouse. The results for A are shown in Fig. 3A. Control Coker312 plants had a strong wilting phenotype in the afternoon on days 9, 10 and 11 of withholding water. The strong drought stress resulted in significant inhibitory effects on A compared to seven days drought, when no signs of afternoon wilting were observed either in wild type or transgenic plants. All the transgenic lines showed significantly better A under drought stress, especially at 11 days of drought (except ABI513-4-1 which also showed higher A, but at p = 0.14; Supplemental Datafile1) and did not show severe wilting symptoms upon withholding water. The stacked double transgenic RAV1×ABI5 cross showed the best A at 9, 10 and 11 days of drought and importantly, outperformed its parental lines. RAV2×ABI5 F1 resulted in better A at 9 and 10 days drought and outperformed its parental lines at these time points. Re-watering resulted in all the transgenics having significantly higher A recovery after 16 hr as compared to wild type. Importantly, RAV1×ABI5 showed better A recovery over its parental lines. All the transgenic lines and wild type recovered near full photosynthetic capacity five days after re-watering. Interestingly, wild-type plants never reached the same level of photosynthesis as transgenics (Fig. 3A) alluding to higher WUE in transgenics as predicted. Supplemental Fig. S3 shows the wilty phenotype seen in control Coker312 plants in the evening after 10 days drought treatment compared to individuals of RAV1×ABI5 stacked double transgenic line, which recovered by evening from afternoon wilting.
Figure 3.
Photosynthetic parameters in stacked and single transgene cotton under drought stress. (A) Elevated A of transgenics grown in greenhouse, relative to control Coker312 during several days imposed drought stress and recovery. (B) Significantly higher Intrinsic WUE in single gene transgenics in the field (2011) under WW (blue bars) and DI conditions (red bars). (C) Significantly higher Intrinsic WUE in single gene and stacked double gene transgenics in the field (2013) at peak bloom under conditions of marginal heat- and water-limiting stress. Error bars ± are s.e.m. (n=5 [A, C]; n=9 [B]). *Significantly different from control Coker312/KSNT at p ≤ 0.05; ** p ≤ 0.01; ***p ≤ 0.001 (two-tailed Student’s t-test, equal variance assumed).
In order to further substantiate the transgene effects on photosynthesis under drought stress, we measured parameters in two independent field trials (Supplemental Datafile1). Figure 3B and 3C shows reproducible, significantly higher intrinsic WUE (carbon fixed per unit water transpired, Gs) over two field trials for nearly all transgenics under drought stress (Fig. 3B), as well as synergistic effects of stacked RAVxABI5 double transgenics under water-sufficient but extreme heat conditions (Fig. 3C) and under DI (Supplemental Datafile1). To further characterize the physiological consequences of improved WUE, we quantified shoot and root biomass in transgenics under simulated repetitive DI conditions in the greenhouse.
AtABI5, AtRAVs, and ABI5×RAVs stacked double transgenic cotton plants have longer internodes, accumulate higher total dry biomass, and especially roots, compared to wild type under deficit irrigation treatments
Cotton plants subjected to drought stress in the greenhouse resulted in reduced internode lengths manifest as reduced height (Suppl. Fig. S4). All the transgenic lines subjected to 90 days of repeated DI grew taller than wild type due to increased internode length (Supplemental Table S1). Both ABI5 lines showed significant increases (6% and 18%) in internode length, whereas four out of seven RAV2 lines showed 12% to 21% significant increases (p < 0.03) and two others near significant increases (p = 0.08). All the RAV2 lines had correspondingly higher stem weight (Suppl. Table S2). Supplemental Fig. S5 shows a representative transgenic RAV2 line with 17% increased internode length compared to wild type Coker312. RAV1 lines had 5.8-7.9% increases in internode length and (Suppl. Table S1) significant increases in stem weight (9.7-33%; Suppl. Table S2).
WW control Coker312 (WW Coker) generated 52% more biomass (p < 2 ×10−7) compared to DI Coker312, which clearly demonstrated the efficacy of the DI treatments (Suppl. Fig. S4). ABI513-4-1 and the two ABI513-4-1xRAV1 double transgenic lines generated ~17%, 11%, and 16% significantly higher total dry biomass, respectively (Suppl. Table S3). Importantly, the quasi-control KSNT line showed an 8% significant reduction in total dry mass compared to wild type, supporting that the transgenic effects observed are effector-mediated and not associated with somaclonal variation possible during regeneration of plants from hypocotyl explants. We next analyzed root biomass in order to investigate the basis of observed WUE increases.
Well watered (WW) Coker312 generated a mere 5% greater root mass compared to DI Coker312, and the increase was non-significant (p = 0.66). Compared to the above-described 52% significant elevation in total dry mass production for full water vs. deficit (Suppl. Fig. S4), the marginal effect of DI on root biomass shows that the sink strength of roots is very strong under drought stress, an adaptation advantageous for cotton (Pace et al., 1999). Both ABI5 lines, two of three RAV1 lines, six of seven RAV2 lines, two of three RAV2L lines, and all RAVxABI5 stacked double transgenic lines grew 21% to 96% more root mass under DI compared to wild type (Table 2). Supplemental Fig. S6 and Table 2 show several cases of higher root mass phenotypes for multiple independent RAV1 and RAV2 transgenic cotton lines and the compelling synergistic effects of ABI513-4-1xRAV24-8-1 stacked double transgenes compared to individual single transgene parents. As an independent validation of the results, the quasi-control KSNT line showed a 25% significant reduction (p < 0.05) in root biomass accumulation under DI (Table 2), further substantiating the transgene-specific effects on root growth.
Table 2.
Root# dry mass (g) in transgenic cotton lines subjected to 90 days of deficit irrigation in greenhouse conditions.
| Genotype | Average | %Δ WT | SEM (+/−) | p-value |
|---|---|---|---|---|
| ABI51-1-1 | 6.6 | 38.6 | 0.6 | 0.007 |
| ABI513-4-1 | 6.3 | 31.7 | 0.8 | 0.036 |
| RAV11-1-5 | 8.7 | 81.9 | 0.8 | 0.000006 |
| RAV13-7-1 | 6.2 | 30.5 | 0.4 | 0.039 |
| RAV113-7-2 | 5.3 | 11.6 | 0.5 | 0.372 |
| RAV24-8-1 | 5.8 | 21.4 | 0.4 | 0.107 |
| RAV25-3-1 | 4.7 | −1.0 | 0.6 | 0.9 |
| RAV224-1-2 | 5.8 | 22.5 | 0.4 | 0.061 |
| RAV224-6-1 | 6.3 | 31.7 | 0.4 | 0.011 |
| RAV224-7-10 | 9.3 | 96.1 | 0.4 | 0.0000002 |
| RAV224-8-1 | 8.4 | 77.1 | 1.3 | 0.00044 |
| RAV227-2-2 | 7.1 | 49.7 | 0.9 | 0.003 |
| RAV2L4-2-1 | 6.3 | 32.7 | 0.5 | 0.020 |
| RAV2L23-4-4 | 6.0 | 25.0 | 0.8 | 0.084 |
| RAV2L24-2-1 | 4.5 | −6.2 | 0.1 | 0.6 |
| RAV11-1-5xABI513-4-1 | 7.5 | 58.3 | 0.9 | 0.001 |
| RAV113-7-2xABI513-4-1 | 6.5 | 36.3 | 0.7 | 0.026 |
| RAV24-8-1xABI513-4-1 | 7.2 | 50.2 | 0.7 | 0.001 |
| RAV25-3-1xABI513-4-1 | 7.1 | 48.1 | 1.7 | 0.045 |
| Wild Type Coker312 | 4.8 | 0.5 | ||
| KSNT | 3.6 | −25.1 | 0.4 | 0.042 |
Root mass refers to lateral root mass excluding taproot.
Values in bold are shown for parental lines (RAV2 and ABI5)
Values in bold and larger font size are shown for crossed progeny (RAV2xABI5)
For S.E.M., n=6. For statistical significance analysis, a two-tailed Student’s t-test was applied and non-equal variance assumed.
Characterization of molecular marker expression associated with drought resistance in RAV and ABI5 transgenic lines
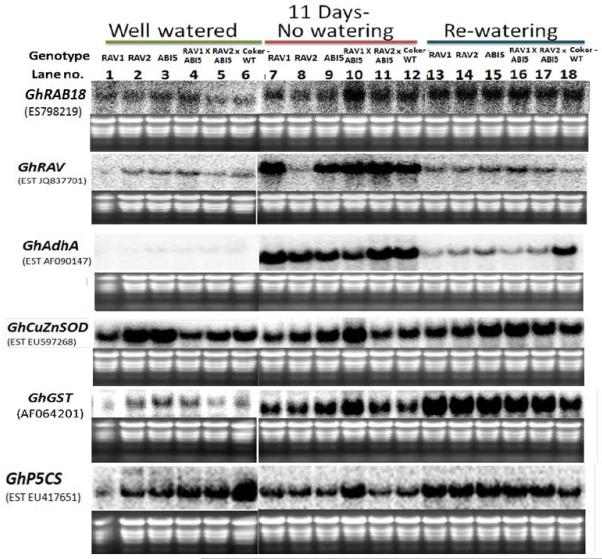
ABI5 over-expressing Arabidopsis plants, in response to ABA, accumulate transcripts of stress-inducible genes (e.g. Cor78, Cor6.6, Cor15a, and Rab18) (Brocard et al., 2002) and show high sensitivity to sugar stress resulting in anthocyanin accumulation (Finkelstein et al., 2002), suggesting ABI5 over-expression promotes vegetative stress adaptation. In order to further test the hypothesis that enhanced stress adaptation is due to elevated ABA response, we quantified the expression of cotton AtRAB18 homolog GhRAB18. In Arabidopsis, RAB18 accumulates in response to drought- and cold-stress in an ABA-dependent manner (Lång and Palva, 1992; Mantyla et al., 1995). A GhRAB18 transcript of low abundance was detected under WW conditions in cotton (Fig. 4). GhRAB18 transcript was induced ~two-fold in response to drought (Fig. 4; compare lanes 6 and 12). Re-watering did not alter the transcript level compared to drought (lanes 12 and 18), suggesting a role for GhRAB18 in stress adaptation and recovery in cotton. Remarkably, the expression of RAB18 was higher in RAV1 (~1.3 fold), in ABI5 (~1.5 fold) and RAV1×ABI5 (~1.5 fold) transgenics under WW conditions. Importantly, expression was ~1.6 fold higher in RAV1×ABI5 compared to Coker312 in response to drought, supporting that stress adaptation in RAV1×ABI5 is associated with an elevated ABA response.
Figure 4.
RNA blot assay for molecular marker gene expression associated with stress resistance/adaptation in RAV113-7-2-, RAV224-8-1-, and ABI513-4-1-over-expressing and stacked double transgenic lines. EtBr-stained gel of samples show equal loading. Lanes 1-6, 7-12, and 13-18 represent WW (24 DAS), 11 days of no watering (35 DAS), and overnight recovery (after re-watering) conditions, respectively.
In order to further explore the molecular basis of improved performance of transgenic cotton lines under DI and test the hypothesis that the transgenic lines have a “less stressed” or stress-adapted phenotype, we examined transcript abundances of several stress-related marker genes. The endogenous GhRAV transcript was detected at low abundance under WW conditions, and was highly induced by drought treatment. After 11 days of severe drought, GhRAV transcript expression was induced by ~six-fold in wild type Coker312 compared to WW conditions (Fig. 4). GhRAV transcript abundance was reduced ~90% in response to re-watering (compare lane 18 with lane 12). Interestingly, endogenous GhRAV transcript was ~1.9 to 2.7 times more highly expressed in all the single transgenics and stacked double transgenics in the recovery stage (Fig. 4, compare lanes 13-17 to lane 18). This result supports that endogenous GhRAV plays a role in drought response and recovery, possibly by functional interactions with AtRAV and AtABI5.
With the onset of anoxia and biotic or abiotic stresses, a rise in cytosolic Ca2+ acts as a signal for up-regulation of alcohol dehydrogenase (Adh) transcript in maize cells (Taiz and Zeiger, 2010). Fig. 4 shows that the GhAdhA transcript is of low abundance under WW conditions (Lanes 1-6), and was highly induced in response to drought (~40 fold; lane 12 compared to lane 6). Furthermore, expression was reduced to half in response to re-watering (lane 18 compared to lane 12). Remarkably, all the transgenic and double transgenic lines showed lower expression of GhAdhA in recovery stage (30-60% lower; lanes 13- 17 compared to lane 18). Lower GhAdhA levels in transgenics under different treatments corroborate the notion of a “less stressed phenotype” and correlate with higher photosynthesis and WUE under drought stress and recovery (Fig. 3, Suppl. Datafile1). In every case (transgenic lines and treatment) where endogenous transcript levels of GhRAV were high, the transcript levels of GhAdhA were lower (Fig. 4), supporting a role for GhRAV in reducing stress.
Biotic and abiotic stresses generate excess ROS which oxidize membrane lipids. ROS scavenging in plants involves superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione peroxidase (GPX), and catalase (Apel and Hirt, 2004). There are numerous reports that show enhanced CuZnSOD and APX levels result in stress tolerance (Kim et al., 2010; Lee et al., 2010; Lin et al., 2011; Qin et al., 2012). There was no obvious change to CuZnSOD expression in response to drought or re-watering treatments, however CuZnSOD expression was elevated somewhat in ABI5 and RAVxABI5 stacked lines subjected to drought stress and recovery (Fig. 4; lanes 9 and 10 vs. lane 12 and lanes 15-17 compared to lane 18), supporting a synergistic effect of these TFs in stress control through ROS scavenging.
Glutathione S-transferases (GSTs) play roles in normal cellular metabolism, oxidative stress response, and detoxification of a wide variety of xenobiotic compounds (Apel and Hirt, 2004; Dixon and Edwards, 2010; Marrs, 1996, Sheeshan et al., 2001). The GhGST transcript was up-regulated ~two-fold in response to drought in wild type (Fig. 4). Furthermore, GST expression was elevated in response to re-watering compared to drought, suggesting a function in drought recovery in cotton. GhGST expression was higher in ABI5 and RAV1×ABI5 over-expressing lines under WW and drought conditions compared to respective treatments of wild type (Fig. 4). Remarkably, GhGST expression was higher in all the transgenic lines and stacked crosses during recovery from drought stress. These results support the hypothesis that faster recovery of photosynthetic capacity in transgenic cotton (Suppl. Datafile1) was due to higher ROS scavenging.
P5CS catalyzes the rate-limiting step of proline biosynthesis and is required for ROS reductions in response to drought, salinity and ABA in Arabidopsis (Gu et al., 2010; Qin et al., 2012; Székely et al., 2008). All the transgenic lines had reduced GhP5CS transcript abundances in comparison to wild type (Fig. 4). GhP5CS was reduced ~70% in wild type under imposed drought. Interestingly, RAV1×ABI5 stacked line accumulated more GhP5CS transcript compared to wild type under drought. All the transgenic lines including stacked crosses showed 1.5- two-fold higher transcript levels for GhRAV, GhGST, and GhP5CS and lower stress marker GhAdhA at the recovery stage (Fig. 4; lanes 13-18), supporting the notion of a “less stressed phenotype.”
Discussion
Over-expression of ABA-associated TFs such as Nuclear Factor-Y (NF-Y) can confer drought tolerance (Han et al., 2013; Li et al., 2008; Nelson et al., 2007) and is a focal point for development of the next generation of drought-tolerant crops. AtABI5 and AtRAV transgenic cotton exhibited higher or on par yields under WW greenhouse conditions and under drought stress in the field (Suppl. Fig. 2), reduced inhibition of photosynthesis and improved WUE in response to imposed drought (Fig. 3; Suppl. Datafile 1), and more carbon partitioned into leaves (Table 1), roots (Table 2), and stem (Suppl. Table S2, S3). Similar results have been reported recently for bZIPs, ABI3, NF-Ys, MYBs, AP2, WRKY, and Enhanced Drought Tolerance/HOMEODOMAIN GLABROUS11 (EDT1/HDG11) classes of TFs (Abdeen et al., 2010; Kumimoto et al., 2013; J. Li et al., 2013; Liu and Howell, 2010; Ni et al., 2013; Seo et al., 2009; Yotsui et al., 2013; Yu et al., 2008, 2013; Zhang et al., 2005), underscoring that there are many possible modes of TF engineering for vegetative drought stress tolerance. Remarkably, the stacked lines of RAVxABI5 exhibited synergistic effects on maintaining A in response to imposed drought (Fig. 3A) and concomitant adaptation to higher WUE by decreases in stomatal conductance Gs, which translated into generation of greater root mass (e.g. RAV2×ABI513-4-1; Table 2). Both RAV1×ABI5 crosses showed synergistic increases in stem weight (Suppl. Table S2) and intrinsic WUE (Fig. 3C), reminiscent of the drought resistance traits seen in drought-tolerant species such as Craterostigma, drought-tolerant wheat cultivars (Gupta et al., 2011), and transgenic wheat constitutively over-expressing TaNF-YB3 (Stephenson et al., 2011). A recent report also described over-expression phenotypes of a soybean RAV homologue in tobacco that largely resemble our cotton phenotypes of increased longevity and delayed flowering, increased lateral branching, however reduced root growth (Zhao et al., 2012).
Increased root mass at later stages of vegetative development under WW conditions might not benefit crops because assimilate deposition would generate root biomass unnecessarily. However, increased sink strength resulting in bigger root systems under water deficit would help plants maintain higher turgor pressure, resulting in higher photosynthesis. Deeper root growth into moist soil is second line of defense against drought (Pace et al., 1999; Taiz and Zeiger, 2010). The transgenic cotton lines generated bigger root systems (Table 2; Suppl. Fig. S6) under greenhouse drought conditions. Enhanced root growth generates a competition for assimilates between roots and fruits. RAV and ABI5 transgenic cotton lines showed higher yields in the greenhouse compared to wild type (Suppl. Fig. S2A) suggesting very high WUE and possibly increased sink strength in bolls, which is currently under investigation in the field.
Use of ethylene (ethephon) is common in production agriculture for enhancing root growth. We speculate increased root growth observed in RAV/ABI5 transgenic cotton might be a result of ABA and ethylene cross talk that alters sink strength. RAV TFs have also been described as Ethylene Response DNA binding Factors (EDFs) (RAV1= EDF4; RAV2= EDF2 and RAV2L= EDF1) and are ethylene-inducible (Alonso and Stepanova, 2004). Previous characterization of RAV functions in brassinosteroids (Hu et al., 2004), ROS scavenging (Lee et al., 2010), ethylene response (Alonso and Stepanova, 2004), suppression of RNA silencing by viruses (Endres et al., 2010), control of flowering time (Castillo and Pelaz, 2008; Mutasa-Göttgens et al., 2012; Osnato et al., 2012), cytokinin signaling (Zhao et al., 2012) and ABA signaling (present study) suggest that RAVs function as nodes in a crosstalk network, consistent with our unexpected observation (Fig. 1C) that RAV2 and VP1 synergize in ABA-inducible gene expression, whereas VP1 and bZIPs are known to interact physically to transactivate ABA-inducible promoters (Finkelstein et al., 2005). Recent reports on transcript profiling of the Lignon lintless-1 or fuzzless-lintless (fl) mutants of upland cotton show associations between fiber elongation and hormone pathways, especially ethylene biosynthesis and differential expression of AP2/ethylene and stress response TFs (Gilbert et al., 2013; Padmalatha et al., 2012), consistent with our results and known functions of AtABI5 and AtRAVs. Transcriptome profiling or CHIP-Seq of the cotton transgenics may shed some light on the issues of direct versus indirect/hierarchical interactions of RAVs with ABI5 and each other, and their target genes impacting agronomic traits such as assimilation under stress, yields, and fiber quality.
Altered carbon partitioning in the transgenic cotton lines resulted in bigger root systems and more leaf area, which if mechanistically conserved across species may facilitate engineering of crops to challenging environments and improve yields. Plants subjected to drought tend to reduce their internode length and become stunted. Remarkably, the cotton transgenics had longer internode lengths and a concomitant increases in stem, leaf, and total biomass (Table 1, Suppl. Tables S1-S3; Suppl. Fig. S5) under water deficit conditions in the greenhouse and field, and we interpret these traits as a “less stressed phenotype”.
Oxidoreductases play significant roles in response to biotic and abiotic stresses (Reddy et al., 2007; Jacquot et al., 2009). Lipid peroxidation in CaRAV1- and/or CaOXR1- silenced plants correlated with decreased tolerance to high salinity and drought (Lee et al., 2010). CaOXR1 positively controls CaRAV1-mediated plant defense during biotic and abiotic stresses (Lee et al., 2010). Importantly, RAV1×ABI5 stacked transgenic cotton lines had ~two-fold higher transcript levels of CuZnSOD (Fig. 4) under severe drought conditions and after re-watering. ABI5-over-expressing cotton also showed the same trend, emphasizing the synergistic effect of RAV1×ABI5 in combating drought stress, analogous to our observations for transient gene induction (Fig. 1). Higher GST transcript levels in RAV1×ABI5 under imposed drought, and a high level in all transgenics during stress recovery (Fig. 4) suggests the “less stressed” phenotype is due to combating stress through increased levels of ROS scavengers. Similar results were recently reported for Arabidopsis expressing a peanut ABI5 homologue (Li et al., 2013). Reduced AdhA transcript in transgenics (and synergistic reductions in RAV1×ABI5) under WW, drought- and recovery conditions (Fig. 4) supports the hypothesis of a less stressed phenotype for ABI5- and RAV over-expressing cotton. In drought experiments the transgenic plants were less wilty and leaves remained turgid due to increased water availability from bigger root systems (Table 2, Suppl. Fig. S6) and possibly osmotic adjustment (e.g. proline accumulation) mediated by two-fold higher GhP5CS during stress recovery (Fig. 4). Maintaining turgor enables the continuation of cell elongation and facilitates higher stomatal conductance at lower water potential (Taiz and Zeiger, 2010), observed in transgenic cotton as higher photosynthesis under drought and faster recovery (Fig. 3A).
Our results with transgenic cotton over-expressing ABI5 and RAV TFs show drought resistance in photosynthesis and traits of drought avoidance (bigger root and leaf systems) and tolerance (manifest as longer internode length and higher stem weight) that may lead to better establishment under limited water conditions due to synergy with endogenous GhRAV and enhanced antioxidant and osmolyte synthesis. Higher A in ABI5/RAV transgenic cotton begs the question whether the most economically important sink (the developing bolls) exhibit gains in strength that could contribute to observed improved yields (Suppl. Fig. 2) or enhance fiber quality and seed traits like oil and protein content. Better fiber quality under limited irrigation is a key trait for cotton producers, where staple prices are discounted because of immature or coarse fibers that result in poor yarn spinning performance. AtABI5 and AtRAV over-expressing transgenic cotton and potentially other crops could impact sustainable agriculture under limited irrigation and dryland farming, the ultimate consequence of continued depletion of the southern Ogallala Aquifer, the source of water for one-third of all U.S. cotton production.
Experimental Procedures
DNA constructs for transient gene expression
pBM207 contains the 650bp Triticum aestivum Early Methionine-labeled (Em) promoter driving uidA (E. coli β-glucuronidase, GUS) expression. pAHC18 contains the 2.0kbp Zea mays Ubiquitin (Ubi) promoter driving Photinus pyralis luciferase (LUC) (Bruce and Quail, 1990). The Viviparous-1 (VP1) cDNA effector driven by the 35S promoter is pCR349.13s (Hill et al., 1996). Plasmid p701-RAV2 is the Ubi promoter driving full length cDNA clone U09382/At1g68840/RAV2 (Yamada et al., 2003) and was constructed using the Cre-lox recombination system of bacteriophage P1 (Liu et al., 1998) in pCR701 as described (Jia et al., 2009). Plasmids Ubi:ABI5 and Ubi:ABF3 were as previously described (Finkelstein et al., 2005). Plasmid pG2 encodes the 35S-maize C4 pyruvate-orthophosphate dikinase basal promoter chimera (35S-Ppdk) driving the coding region of the Arabidopsis abi1-1 dominant-negative G180D mutant allele (Sheen, 1998). Plasmid pG1 is the same as pG2 except it encodes a G174D site-directed “null” mutation that abolishes phosphatase activity (Sheen, 1998). Plasmid pDirect2.6 contains the Ubi promoter alone in reverse orientation used as control to balance input DNAs. Plasmids were propagated in E. coli DH5α, TOP-10, or GC10 cells (Invitrogen, Carlsbad, CA) and prepared by CsCl density gradient ultracentrifugation (Ausubel et al., 1995).
Protoplasts
Zea mays seeds genotype FR37cms_X_FR49 (Illinois Foundation Seed, Champaign, IL) were imbibed in water overnight and sown in a 1:1 vermiculite:peat moss mix. Seeds were germinated in constant incandescent light for four days at 23°C and moved to a dark growth chamber when coleoptiles emerged. Protoplast isolation was according to Sheen (2001) with modifications (Jia et al., 2009). 50,000 protoplasts per electroporation sample in 300 μL were mixed with DNAs and transferred to pre-chilled 0.4 mm gap cuvettes (BioRad, Hercules, CA). After 10 min on ice, samples were electroporated (400 V, 200 μF; two pulses) with a BTX Electro Cell Manipulator 630 (Gentronics, San Diego, CA) and incubated on ice for 10 min. Protoplasts were then split into two aliquots in microfuge tubes and incubated with either wash solution only, or 100 μM ABA in wash solution. After 16 hrs incubation in the dark, cells were pelleted by centrifuging at 800 rpm for three min and 250 μL 1x Reporter Lysis Buffer (Promega, Madison, WI) was added to the pellet and thoroughly vortexed. The lysate was centrifuged at 10,000 rpm for three min and the supernatant removed to a fresh tube for reporter enzyme assays. Protein was quantified using Coomassie Protein Assay Reagent (Pierce, Rockford, IL).
Immunoblotting
SDS-PAGE was as described (Ausubel et al., 1995; Towbin et al., 1979), loading equal amounts (5 μg/lane) of protein along with pre-stained Low Range SDS-PAGE Standards (BioRad). The gel was electroblotted to Immobilon-P PVDF transfer membrane (Millipore) using the MiniBlot Module (Thermo EC). Immunoblotting was done according to the manufacturer’s instructions using ECL Advance Western Blotting Detection Kit (Amersham Pharmacia). The primary mouse anti-HA monoclonal antibody (clone HA-7, Sigma, St. Louis, MO) was used at 1:5000 dilution. The secondary was goat anti-mouse IgG1 conjugated horseradish peroxidase (sc-2060; 1:5000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA).
Reporter enzyme assays
Ten μL of sample extract was mixed with 50 μL of Luciferase substrate (Promega) and luciferase activity measured on a Zylux FB15 luminometer (Fisher Scientific, Pittsburgh, PA). GUS activities of 40 μL aliquots of samples (four timepoints) were determined (Jefferson, 1987) using 4-methylumbelliferone glucuronide (MUG; Rose Scientific, Edmonton, Canada) as substrate on a Biotek (Winooski, VT) Synergy HT microplate fluorimeter. The relative reporter gene activity was represented as the ratio of GUS to LUC activities, expressed in relative units (or μmoles) of 4-MU/40 μL extract/h and photons/10 μL extract/min, respectively.
Generation of recombinant vectors for transformation of cotton
We employed available pUNI51-derivative full length cDNA clones of Arabidopsis RAV family members RAV1 (U11954), RAV2 (U09382), RAV2L (U19336) and ABI5 (U85657; Arabidopsis Biological Resource Center, Ohio State University, http://abrc.osu.edu/) to recombine pKYLX-myc9-loxP binary acceptor vector (Guo and Ecker, 2003) in the presence of cre recombinase enzyme. For recombination, in a 20 μL reaction volume 500 ng each of acceptor and donor vector DNAs were mixed with 2 μL 10x recombination buffer (New England Biolabs; www.neb.com), 2 μL GST::CRE recombinase (Jia et al., 2009) and incubated at 37°C for 20 min. The DNAs were precipitated with EtOH, the pellet dissolved in 10 μL water and the DNA measured by a Nanodrop spectrophotometer (Willmington DE). About 200 ng of DNA products were electroporated into electrocompetent pir- E. coli GC10 cells (Invitrogen) and the bacterial colonies carrying recombinant fusion plasmid were selected on kanamycin-containing LB plates. The candidate transformation-ready constructs were restriction digested and were confirmed to be comprised of a dimer of one acceptor and one pUNI donor plasmid. Plasmids were electroporated into electrocompetent Agrobacterium tumifaciens strain GV3101.
Deficit irrigation treatments and gas exchange measurements
Field trials were conducted at the TTU New Deal Farm south plot with subsurface drip irrigation under USDA-APHIS permit #11-097-106n. Sowing was done on June 7th, 2011 for 17 transgenic lines and three Coker check groups in a randomized block design with a zone subjected to DI (1/4 acre-inches water/day until flowering stage [day 42 after sowing (DAS)] for DI) as well as a WW control treatment zone. All lines tested in the field condition were homozygous (confirmed by PCR sampling) and were planted in paired rows with other commercial genotypes included as needed to fill up the plot to minimize border effects. There were eight rows (40″ spacing) of ~140 feet for each watering treatment zone, giving an overall field plot of ~0.5 acre. Mechanical sowing was at the rate of four and one-half seeds per foot in eight feet-long subplots. For greenhouse experiments, potting mix, field soil, and sand were mixed in 3:1:1 volume proportions, respectively. Photosynthesis parameters were measured using a Licor-6400 XT (LI-COR Biosciences, Lincoln, NE) and taken in representative lines of all the transgenics starting one week after and until the sixth week of the DI treatment. For greenhouse experiments, the last watering was given on 24 DAS and measurements were commenced on 7 days after withholding water (7D-drt) and continued until five days after re-watering (5D-Rec) (10 days in total). Measurements were restricted to 10 am–1 pm when temperatures were not extreme, and plants from all lines were measured within half an hour using expanded source leaves (4th or 5th leaf from apical meristem). Greenhouse plants were not watered until more than 80% of wild type control plants did not show evening recovery from afternoon wilt. Several cycles of this treatment were repeated until 90 DAS. Tissue for stress marker gene analysis was collected from six individual greenhouse plants of each line for WW non-stressed condition (24 DAS), drought treatment (11 Days of no watering; 35 DAS), and recovery (overnight recovery from drought stress after re-watering).
Biomass assays
Owing to the labor-intensive nature of hand harvesting, we measured fiber yields in one meter rows (in triplicate) from the interior regions of field plots (to discount border effects). For greenhouse studies, all the plant parts (root, stem, leaves, and fruits) were collected separately. Leaves were detached (leaving the petioles intact on the stem) and kept in ziplock bags at 4°C. Leaf area was measured using a LI-3100C Portable Leaf Area Meter. Fruits (flowers, immature and maturing bolls) and stems were detached and stored in paper bags in the greenhouse. After removing fruit, leaves, and stems, the pots were dipped in a wash-tub filled with water until saturated and manipulated to release the potting soil/earth/sand mixture from intact roots. After the roots were processed as a ball without any adhering soil, they were washed in several changes of fresh water and stored in ziplock bags until imaged. All the plant parts were dried in an oven at 74°C for 72 hrs and weighed. To obtain the weight of lateral roots the whole dried roots were weighed and then all the lateral roots were removed and the bare taproot was weighed. The difference gave the weight of lateral roots, shown in Results.
Semi-quantitative RT –PCR
Total RNA was extracted using Spectrum Plant RNA Mini Kit (Sigma-Aldrich, St. Louis, MO). Sigma On-column DNase1 digestion was used to remove DNA contamination in extracted RNA. Two μg RNA was reverse transcribed by M-MLV reverse transcriptase (Promega) with Anchored Oligo-dT (Thermo, Surrey, UK). 0.5 μL of cDNA template was used for a 25 μL PCR reaction. Gene-specific primers (Suppl. Table S4) for AtRAV1, AtRAV2, AtRAV2L and AtABI5 were used to amplify (35 cycles) the cDNA from transgenic lines. GhUBQ1 specific primers were used as an internal control.
RNA blot hybridization assay
10 μg of total RNA per sample was resolved on 1.2% denaturing agarose gel and blotted onto a Hybond-N+ membrane (GE healthcare, Piscataway, NJ). An RNA molecular weight marker lane was included to estimate mRNA transcript sizes (Ambion Millenium Marker, GE Healthcare). Primers (Suppl. Table S4) were designed based on BLAST results from NCBI plant EST database, and cDNA was amplified from reverse-transcribed Coker312 RNA. The PCR products were gel-purified and used as template for random-primed synthesis (Takara, Shiga, Japan) of radioactive probes with [α32P]-dCTP (PerkinElmer, Waltham, MA). PerfectHyb Plus hybridization buffer (Sigma) was used according to the manufacturer’s instructions. Autoradiography was with storage phosphor screen (GE Healthcare) scanned with Storm 860 PhosphorImager (GE Healthcare). Ethidium bromide-stained total RNA samples were quantified from gel images using ImageJ software (imagej.nih.gov/ij/download). The RNA blot band intensity was quantified using ImageQuant TL software (v2003, GE Healthcare). The ratio of ImageQuant to ImageJ values gives normalized transcript quantity for relative comparisons.
Supplementary Material
Acknowledgements
The authors thank DeeDee Laumbauch and Kay McCrary for transformations, Ying-wen Jiang, Fan Jia, Meenakshi Mittal, Sayani Mallick, Jacob Sanchez, Marie Syapin, Dustin Stidger, and Yuvisela Chavez (supported by NIH Plains Bridge to the Baccalaureate grant 5R25GM83730) for help with physiological assays, and Hong-Liang Zhu for help with molecular work. This work was supported by the TTU-USDA International Cotton Research Center, the Texas State Support Committee of Cotton Incorporated, and the USDA Ogallala Aquifer Program.
References
- Abdeen A, Schnell J, Miki B. Transcriptome analysis reveals absence of unintended effects in drought-tolerant transgenic plants overexpressing the transcription factor ABF3. BMC Genomics. 2010;11:69. doi: 10.1186/1471-2164-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN. The ethylene signaling pathway. Science. 2004;306:1513–1515. doi: 10.1126/science.1104812. [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PL. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 2012;158:970–980. doi: 10.1104/pp.111.188623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struh K. Current Protocols in Molecular Biology. Green Publishing Associates and Wiley-Interscience; New York: 1995. [Google Scholar]
- Bayley C, Trolinder N, Ray C, Morgan M, Quisenberry JE, Ow DW. Engineering 2,4-D resistance into cotton. Theor. Appl. Genet. 1992;83:645–649. doi: 10.1007/BF00226910. [DOI] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR. Regulation and role of the Arabidopsis ABA insensitive (ABI5) gene in ABA, sugar and stress response. Plant Physiol. 2002;129:1533–1543. doi: 10.1104/pp.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce WB, Quail PH. Cis-acting elements involved in photoregulation of an oat phytochrome promoter in rice. Plant Cell. 1990;2:1081–1089. doi: 10.1105/tpc.2.11.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos F, Cuevas-Velazquex C, Fares MA, Reyes JL, Covarrubias AA. Group 1 LEA proteins, an ancestral plant protein group, are also present in other eukaryotes, and in the archeae and bacteria domains. Molec. Genet. Genomics. 2013;288:503–517. doi: 10.1007/s00438-013-0768-2. [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Chen YT, Liu H, Stone S, Callis J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013;75:965–976. doi: 10.1111/tpj.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. The Arabidopsis Book. American Society of Plant Biology; Rockville, MD: 2010. Glutathione transferases; p. e0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L., III A repeating 11-mer amino acid motif and plant desiccation. Plant J. 1993;3:363–369. doi: 10.1046/j.1365-313x.1993.t01-19-00999.x. [DOI] [PubMed] [Google Scholar]
- Endres MW, Gregory BD, Gao Z, Foreman AW, Mlotshwa S, Ge X, Pruss GJ, Ecker JR, Bowman LH, Vance V. Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathogens. 2010;15:e1000729. doi: 10.1371/journal.ppat.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville CR. Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 1990;94:1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SS, Lynch TJ, Thomas TL, Rock CD. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MK, Turley RB, Kim HJ, Li P, Thyssen G, Tang YH, Delhom CD, Naoumkina M, Fang DD. Transcript profiling by microarray and marker analysis of the short cotton (Gossypium hirsutum L.) fiber mutant Ligon lintless-1 (Li-1) BMC Genomics. 2013;14:403. doi: 10.1186/1471-2164-14-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin EC, Rottman FM. The 3′-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J. Biol. Chem. 1992;267:16330–16334. [PubMed] [Google Scholar]
- Gu L, Liu Y, Zong X, Liu L, Li DP, Li DQ. Overexpression of maize mitogen activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol. Biol. Rep. 2010;37:4067–4073. doi: 10.1007/s11033-010-0066-6. [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Kaur K, Kaur N. Stem reserve mobilization and sink activity in wheat under drought conditions. Am. J. Plant Sci. 2011;2:70–77. [Google Scholar]
- Han X, Tang S, An Y, Zheng D-C, Xia X-L, Yin W-L. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 2013;64:4589–4601. doi: 10.1093/jxb/ert262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Nantel A, Rock CD, Quatrano RS. A conserved domain of the Viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J. Biol. Chem. 1996;271:3366–3374. doi: 10.1074/jbc.271.7.3366. [DOI] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Jacquot JP, Eklund H, Rouhier N, Schürmann P. Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 2009;14:336–343. doi: 10.1016/j.tplants.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 1987;5:387–405. [Google Scholar]
- Jia F, Gampala SSL, Mittal A, Luo QJ, Rock CD. Cre-lox univector acceptor vectors for functional screening in protoplasts: analysis of Arabidopsis donor cDNAs encoding ABSCISIC ACID INSENSITIVE1-like protein phosphatases. Plant Mol. Biol. 2009;70:693–708. doi: 10.1007/s11103-009-9502-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Hattori T. Arabidopsis transcription factors, RAV1 and RAV2, are regulated by touch-related stimuli in a dose-dependent and biphasic manner. Genes Genet. Sys. 2009;84:95–99. doi: 10.1266/ggs.84.95. [DOI] [PubMed] [Google Scholar]
- Kim MD, Kim YH, Kwon SY, Yun DJ, Kwak SS, Lee HS. Enhanced tolerance to methyl viologen-induced oxidative stress and high temperature in transgenic potato plants overexpressing the CuZnSOD, APX and NDPK2 genes. Physiol. Plant. 2010;140:153–162. doi: 10.1111/j.1399-3054.2010.01392.x. [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Siriwardana CL, Gayler KK, Risinger JR, Siefers N, Holt BF., III NUCLEAR FACTOR Y transcription factors have both opposing and additive roles in ABA-mediated seed germination. PLoS ONE. 2013;8:e59481. doi: 10.1371/journal.pone.0059481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lång V, Palva ET. The expression of a RAB-related gene, RAB18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 1992;20:581–582. doi: 10.1007/BF00027165. [DOI] [PubMed] [Google Scholar]
- Lee SC, Choi DS, Hwang IS, Hwang BK. The pepper oxidoreductase CaOXR1 interacts with the transcription factor CaRAV1 and is required for salt and osmotic stress tolerance. Plant Mol. Biol. 2010;73:409–424. doi: 10.1007/s11103-010-9629-0. [DOI] [PubMed] [Google Scholar]
- Li C-W, Su R-C, Cheng C-P, Sanjaya, You S-J, Hsieh T-H, Chao T-C, Chan M-T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011;156:213–227. doi: 10.1104/pp.111.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Besseau S, Toronen P, Sipari N, Kollist H, Holm L, Palva ET. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013;200:455–472. doi: 10.1111/nph.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-X, Oono Y, Zhu J, He X-J, Wu J-M, Iida K, Lu X-Y, Cui X, Jin H, Zhu J-K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20:2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Liu X, Yao Y, Li YH, Liu S, He CY, Li JM, Lin YY, Li L. Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int. J. Mol. Sci. 2013;14:12827–12842. doi: 10.3390/ijms140612827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Jih PJ, Lin HH, Lin JS, Chang LL, Shen YH, Jeng ST. Nitric oxide activates superoxide dismutase and ascorbate peroxidase to repress the cell death induced by wounding. Plant Mol. Biol. 2011;77:235–249. doi: 10.1007/s11103-011-9805-x. [DOI] [PubMed] [Google Scholar]
- Liu J-X, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796. doi: 10.1105/tpc.109.072173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge S. The Univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138:1457–1468. doi: 10.1104/pp.105.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T, Erickson BJ, Finkelstein RR. Direct interactions of ABA-insensitive (ABI)-clade protein phosphatase (PP)2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol. Biol. 2012;80:647–658. doi: 10.1007/s11103-012-9973-3. [DOI] [PubMed] [Google Scholar]
- Mantyla E, Lång V, Palva ET. Role of abscisic acid in drought-induced freezing tolerance, cold acclimation, and accumulation of LT178 and RAB18 proteins in Arabidopsis thaliana. Plant Physiol. 1995;107:141–148. doi: 10.1104/pp.107.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione S transferases in plants. Annu. Rev. Plant. Physiol. Plant Mol. Biol. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. The Viviparous- 1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- Mittal A. Ph.D. dissertation. Texas Tech University; 2012. Overexpression and interactions of Arabidopsis thaliana RAV1 (Related to Abscisic Acid Insensitive 3/ Viviparous 1), RAV2, RAV2-Like and ABI5 in transgenic cotton (Gossypium hirsutum): Effects on drought avoidance and fiber quality; p. 161. [Google Scholar]
- Mutasa-Göttgens ES, Joshi A, Holmes HF, Hedden P, Göttgens B. A new RNASeq-based reference transcriptome for sugar beet and its application in transcriptome-scale analysis of vernalization and gibberellin responses. BMC Genomics. 2012;13:99. doi: 10.1186/1471-2164-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Repetti PP, Adams TR, Creelman RA, Wu J, Warner DC, Anstrom DC, Bensen RJ, Castiglioni PP, Donnarummo MG, Hinchey BS, Kumimoto RW, Maszle DR, Canales RD, Krolikowski KA, Dotson SB, Gutterson N, Ratcliffe OJ, Heard JE. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Hu Z, Jiang Q, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013;82:113–129. doi: 10.1007/s11103-013-0040-5. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl. Acad. Sci. U.S.A. 2013;110:12132–12137. doi: 10.1073/pnas.1305919110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Comm. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- Pace PF, Cralle HT, El-Halawany SHM, Cothren JT, Senseman SA. Drought-induced changes in shoot and root growth of young cotton plants. J. Cotton Sci. 1999;3:183–187. [Google Scholar]
- Padmalatha KR, Patil DP, Kumar K, Dhandapani G, Kanakachari M, Phanindra ML, Kumar S, Mohan TC, Jain N, Prakash AH, Vamadevaiah H, Katageri IS, Leelavathi S, Reddy MK, Kumar PA, Reddy VS. Functional genomics of fuzzless-lintless mutant of Gossypium hirsutum L. cv. MCU5 reveal key genes and pathways involved in cotton fibre initiation and elongation. BMC Genomics. 2012;13:624. doi: 10.1186/1471-2164-13-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor. Appl. Genet. 2013;126:1305–1319. doi: 10.1007/s00122-013-2054-4. [DOI] [PubMed] [Google Scholar]
- Qin Y, Wang M, Tian Y, He W, Han L, Xia G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012;39:7183–7192. doi: 10.1007/s11033-012-1550-y. [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signaling. Trends Plant Sci. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Reddy AM, Reddy VS, Scheffler BE, Wienand U, Reddy AR. Novel transgenic rice overexpressing anthocyanidin synthase accumulates a mixture of flavonoids leading to an increased antioxidant potential. Metab. Eng. 2007;9:95–111. doi: 10.1016/j.ymben.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE. 2009;4:e5791. doi: 10.1371/journal.pone.0005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. U.S.A. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001;127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- Soon F-F, Ng L-M, Zhou XE, West GM, Kovach A, Tan MHE, Suino-Powell KM, He Y, Xu Y, Chalmers MJ, Brunzelle JS, Zhang H, Yang H, Jiang H, Li J, Yong E-L, Cutler S, Zhu J-K, Griffin PR, Melcher K, Xu HE. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2012;335:85–88. doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson TJ, McIntyre CL, Collet C, Xue G-P. TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct. Integr. Genom. 2011;11:327–340. doi: 10.1007/s10142-011-0212-9. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, Koncz C, Szabados L. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008;53:11–28. doi: 10.1111/j.1365-313X.2007.03318.x. [DOI] [PubMed] [Google Scholar]
- Taiz L, Zeiger E. Plant Physiology. 5th edn Sinauer Associates Inc.; Sunderland: 2010. [Google Scholar]
- Thomashow MF. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–99. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Lu Y, Chen X, Wu Y. The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 2013;64:675–684. doi: 10.1093/jxb/ers361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integ. Plant Biol. 2011;53:570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, Pham P, Cheuk R, Karlin-Newmann G, Liu SX, Lam B, Sakano H, Wu T, Yu G, Miranda M, Quach HL, Tripp M, Chang CH, Lee JM, Toriumi M, Chan MMH, Tang CC, Onodera CS, Deng JM, Akiyama K, Ansari Y, Arakawa T, Banh J, Banno F, Bowser L, Brooks S, Carninci P, Chao Q, Choy N, Enju A, Goldsmith AD, Gurjal M, Hansen NF, Hayashizaki Y, Johnson-Hopson C, Hsuan VW, Iida K, Karnes M, Khan S, Koesema E, Ishida J, Jiang PX, Jones T, Kawai J, Kamiya A, Meyers C, Nakajima M, Narusaka M, Seki M, Sakurai T, Satou M, Tamse R, Vaysberg M, Wallender EK, Wong C, Yamamura Y, Yuan S, Shinozaki K, Davis RW, Theologis A, Ecker JR. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yotsui I, Saruhashi M, Kawato T, Taji T, Hayashi T, Quatrano RS, Sakata Y. ABSCISIC ACID INSENSITIVE3 regulates abscisic acid-responsive gene expression with the nuclear factor Y complex through the ACTT-core element in Physcomitrella patens. New Phytol. 2013;199:101–109. doi: 10.1111/nph.12251. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong Y-Y, Wang Y, Xu P, Ke S-D, Liu H-Y, Zhu J-K, Oliver DJ, Xiang C-B. Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell. 2008;20:1134–1151. doi: 10.1105/tpc.108.058263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen X, Wang Z, Wang S, Wang Y, Zhu Q, Li S, Xiang C. Arabidopsis Enhanced Drought Tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY. Overexpression of WXP1, a putative Medicago truncatula AP2 domain containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa) Plant J. 2005;5:689–707. doi: 10.1111/j.1365-313X.2005.02405.x. [DOI] [PubMed] [Google Scholar]
- Zhao L, Hao D, Chen L, Lu Q, Zhang Y, Li Y, Duan Y, Li W. Roles for a soybean RAV-like orthologue in shoot regeneration and photoperiodicity inferred from transgenic plants. J. Exp. Bot. 2012;63:3257–3270. doi: 10.1093/jxb/ers056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.