Abstract
The Down Syndrome Cell Adhesion Molecule (DSCAM) is required for regulation of cell number, soma spacing and cell type specific dendrite avoidance in many types of retinal ganglion and amacrine cells. In this study we assay the organization of cells making up the outer plexiform layer of the retina in the absence of Dscam. Some types of OFF bipolar cells, type 3b and type 4 bipolar cells, had defects in dendrite arborization in the Dscam mutant retina, while other cell types appeared similar to wild type. The cone synapses that these cells project their dendrites to were intact, as visualized by electron microscopy, and had a distribution and density that was not significantly different than wild type. The spacing of type 3b bipolar cell dendrites was further analyzed by Voronoi domain analysis, Density Recovery Profiling (DRP) analysis and Nearest Neighbor Analysis (NNA). Spacing was found to be significantly different when comparing wild type and mutant type 3b bipolar cell dendrites. Defects in arborization of these bipolar cells could not be attributed to the disorganization of inner plexiform layer cells that occurs in the Dscam mutant retina or an increase in cell number, as they arborized when Dscam was targeted in retinal ganglion cells only or in the bax null retina. Localization of DSCAM was assayed and the protein was localized near to cone synapses in mouse, macaque and ground squirrel retinas. DSCAM protein was detected in several types of bipolar cells, including type 3b and type 4 bipolar cells.
Keywords: Synapse, Development, Adhesion, Pedicle, Spherule, Rod, Cone, Horizontal Cell, Connectome, Mosaic, Differential Adhesion Hypothesis
Introduction
The first synapses in the photopic visual transduction pathway are those between cone photoreceptors and bipolar and horizontal cells. A single cone axon terminal, or pedicle, makes a series of complex synapses in which the dendrites of bipolar and horizontal cells are organized into multiple distinct signaling hubs. Each of these hubs at a given cone pedicle is composed of the invaginating dendrites of cone bipolar and horizontal cells, facing a presynaptic ribbon, while additional bipolar cell dendrite tips make flat contacts arrayed on the lateral edges of the invaginating ON bipolar and horizontal cell dendrites (Boycott and Hopkins, 1991; Haverkamp et al., 2000; Hopkins and Boycott, 1992). Mechanisms underpinning the developmental organization of this structure are largely unexplored. In this study we assay the role of the cell adhesion molecule DSCAM (Down Syndrome Cell Adhesion Molecule) in organization of the cells making the multiple synapses that compose the cone synapse.
Vertebrate Dscams differ from Drosophila Dscam1 in that they lack the extensive alternative splicing that occurs in the insect gene (Schmucker et al., 2000). Remarkably, despite this difference many of the protein’s reported functions are conserved between vertebrates and fly (Schmucker and Chen, 2009). For example Dscams in both fly and vertebrates have been implicated in axon guidance, self-avoidance and organization of synaptic pairing and targeting (Fuerst et al., 2008; Liu et al., 2009; Ly et al., 2008; Matthews et al., 2007; Millard et al., 2010; Neves et al., 2004; Schmucker et al., 2000; Wang et al., 2002; Yamagata and Sanes, 2008). In the retina, Dscams have been implicated in both a passive form of self-avoidance, early in development, and in synaptic lamination and coupling through adhesion (Fuerst et al., 2009; Fuerst et al., 2008; Yamagata and Sanes, 2008). Functional studies of DSCAM in the retinal outer plexiform layer have not been performed, while Dscaml1 is required for organization of the cells making up the mouse rod circuit, suggesting that Dscam may function in organization of cone circuits (Fuerst et al., 2009). Unlike the inner plexiform layer of the retina, in which the synapses are very small and difficult to individually image, the large cone synapses offer the opportunity to not only assay the development of the structure itself, but to also study the function of DSCAM in synapse formation and maintenance.
In this study we characterize the localization and function of DSCAM at the mammalian cone synapse. We find that DSCAM is localized on the postsynaptic face of the mouse, squirrel and macaque cone synapse. Defects in the arborization of some OFF populations of cone bipolar cells were observed in the absence of Dscam. These defects were not the result of increases in cell number or disorganization of cells contributing to the inner plexiform layer of the retina. This study therefore identifies DSCAM as an essential organizer of the dendritic fields of some populations of OFF bipolar cells.
Materials and Methods
Animals used in this study
The following mouse strains were used in this study.
Dscam2J mice
Dscam2J mice contain a four base pair insertion that disrupts the Dscam gene. A detectable DSCAM protein product is not made by these mice (Fuerst et al., 2010; Schramm et al., 2012). Dscam2J mice were maintained on a C3H/HeJ inbred background in which the Pde6b (rd1) gene is wild type. Wild type siblings were used as controls in these studies.
DscamFD and DscamF mice
The Dscam floxed allele was generated by flanking the exon encoding the Dscam transmembrane domain with loxP sites, allowing for tissue specific targeting of the Dscam gene (Fuerst et al., 2012). The DscamFD allele was generated by targeting the floxed exon in the germ line. No significant morphological differences have been detected when comparing the DscamFD allele to the Dscam2J allele or when comparing the DscamF allele to wild type. Both alleles were backcrossed to the rd1 corrected C3H/HeJ genetic background that the Dscam2J allele is carried on for ten generations after which they have been maintained by intercrossing siblings.
Brn3b-Cre mice
The Brn3b-Cre transgene is a knock in allele that expresses Cre recombinase in most retinal ganglion cells (Fuerst et al., 2012). It had been backcrossed to the rd1 corrected C3H/HeJ genetic background for four generations at the time of study.
Bax mutant mice
The bax null strain is maintained on a C57Bl/6J genetic background.
Mouse Care and Housing
All protocols were performed in accordance with the University of Idaho Institutional Animal Care and Use Committee. Mice were fed ad libitum under a 12-hour light/dark cycle.
Ground Squirrel
Ground squirrels were housed and eyes were obtained as previously described (Chen and Li, 2012).
Macaque
Eyes were obtained from a single 11 year old female macaque that was euthanized for other reasons at the Davis primate center.
Mouse genotyping
Mice were genotyped by PCR as previously described (Fuerst et al., 2012; Fuerst et al., 2009; Fuerst et al., 2010; Fuerst et al., 2008). Tail or toe tip biopsies were prepared for genotyping by boiling biopsies in 75 μl 25 μM sodium hydroxide and 0.2 μM EDTA for 15 minutes. Samples were neutralized with an equal volume of Tris Cl, pH 5.0. DNA was added to OneTaq Hot Start 2X Master Mix with standard buffer, along with primers and water to dilute the PCR mixture to 1X concentration (New England Biolabs, Ipswich, MA).
Tissue Preparation
Mice were anesthetized and perfused with phosphate buffered saline pH 7.4 (PBS) prior to enucleation. Eyes from all three species were hemisected in PBS and immediately fixed in PFA in the following conditions: ground squirrel and macaque eyes were fixed in 4% PFA for fifty minutes on ice. Mouse eyes were fixed in 4% PFA for fifty minutes on ice (DSCAM staining) or 4% PFA for 30 minutes at room temperature (all other staining). Tissue was sunk in 30% sucrose overnight prior to freezing. Tissue was then immersed in tissue-tek optimal cutting temperature media (Sakura Finetek, USA) and rapidly frozen by holding the tissue above the liquid phase of liquid nitrogen. Sections were cut at 10 μm on to charged slides.
Antibody Characterization
Please see Table 1 for a list of all antibodies used, the dilution they were used at and the source of these reagents.
Table 1.
Primary Antibodies
| Antigen | Immunogen | Manufacturer and Details | Dilution Used |
|---|---|---|---|
| NK3R | Synthetic peptide at the c-terminus of rat NK-3 (SFISSPYTSVDEYS) | Novus Biologicals, Littleton CO, USA. Catalog Number: 300-102. Rabbit Polyclonal. | 1:2,000 |
| Recoverin | Recombinant human recoverin | Millipore, Billerica, MA, USA. Catalog Number: AB5585. Rabbit Polyclonal. | 1:2,000 |
| HCN4 | Amino acids 119–155 of human HCN4 (HGHLHDSAEERRLIAEGDASPGEDRTPPGLAAEPERP) | Alomone, Jerusalem, Israel. Catalog Number: APC-052. Rabbit Polyclonal. | 1:500 |
| PKARIIβ | Amino acids 1–418 (full length) of human protein kinase A, regulatory subunit IIβ | BD Biosciences, Franklin Lakes, NJ, USA. Catalog Number: P54720. Mouse Monoclonal IgG1. | 1:1,000 |
| Calsenilin | Recombinant human Calsenilin | Upstate, Lake Placid, NY, USA. Catalog Number: clone 40A5. Mouse Monoclonal IgG1. | 1,1,000 |
| Syt2 | 1–5 day zebrafish embryo | Zebrafish International Resource Center, Eugene, OR, USA. Catalog Number: Znp-1. Mouse Monoclonal IgG2A. | 1,500 |
| Go alpha | Purified Goa from bovine brain | Millipore, Billerica, MA, USA. Catalog Number: MAB3073. Mouse Monoclonal IgG1. | 1:2,000 |
| PKCα | An epitope mapping between amino acids 645–672 at the C-terminus of PKCα of human origin (IANIDQSDFEGFSYVNPQFVHPILQSAV) | Santa Cruz Biotechnology, Santa Cruz CA, USA. Catalog Number: SC-8393. Mouse Monoclonal IgG1. | 1:2,000 |
| β-dystroglycan | C-terminal peptide β-dystroglycan (KNMTPYRSPPPYVPP) | Developmental Studies Hybridoma Bank, Iowa City, IA, USA. Catalog Number: Mandag 2. Mouse Monoclonal IgG1. | 1:1,000 |
| Cone arrestin | Synthetic peptide: EEFMQHNSQTQS | Millipore, Billerica, MA, USA. Catalog Number: AB15282. Rabbit Polyclonal. | 1:5,000 |
| GS | Sheep glutamine synthetase (amino acids 1–373) | BD Transduction Laboratories, Franklin Lakes, NJ, USA. Catalog Number: 610517. Mouse Monoclonal IgG2A. | 1:2,000 |
| Calbindin D28K | Purified rat calbindin D-28 | Swant, Bellinzona, Switzerland. Catalog Number: CB-38a. Rabbit Polyclonal. | 1:1,000 |
| DSCAM | Mouse myeloma cell line NS0-derived recombinant human DSCAM Long Isoform | R&D Systems, Minneapolis, MN, USA. Catalog Number: MAB36661. Mouse Monoclonal IgG2B. | 1:25 |
| DSCAM | Mouse myeloma cell line NS0-derived recombinant human DSCAM Long Isoform | R&D Systems, Minneapolis, MN, USA. Catalog Number: AF3666. Goat Polyclonal. | 1:100 |
| Bassoon | Fusion protein amino acids 738–1075 of ground squirrel bassoon | Custom made courtesy of Wei Li (Covance) Rabbit Polyclonal. | 1:100 |
| PSD95 | Fusion protein amino acids 77–299 (PDZ domains 1 and 2) of human PSD-95 | UC Davis NeuroMab Facility, Davis, CA, USA. Catalog Number: 75-028. | 1:1,000 |
| ChAT | Human placental choline acetyltransferase | Millipore, Billerica, MA, USA. Catalog Number: AB144P. Goat Polyclonal. | 1:500 |
| Brn3A | Amino acids 1–109 of Brn-3a of mouse origin | Santa Cruz Biotechnology, Santa Cruz, CA, USA. Catalog Number: SC-8429. Mouse Monoclonal IgG1. | 1:200 |
| Disabled (Dab1) | GST-mouse Dab1 fusion protein corresponding to residues 107–243 | Generous Gift of Brian Howell. Rabbit Polyclonal. | 1:500 |
| bNOS | Synthetic peptide corresponding to the C-terminal fragment of nNOS of rat brain origin (amino acids 1409–1429 with N-terminal added lysine: EDAGVFISRLRDDNRYHEDIF) | Sigma; St. Louis, MO, USA. Catalog Number: N7280. Rabbit Polyclonal. | 1:10,000 |
| Chx10 | a peptide encompassing the amino acids 44–61 of human CHX10 (PPSSHPRAALDGLAPGHL) | Santa Cruz Biotechnology, Santa Cruz, CA, USA. Catalog Number: sc-21690. Goat Polyclonal. | 1:200 |
NK3R
The NK3R antibody used in this study is specific to the NK-3 receptor. Specificity was verified by the localization of immunohistochemical staining as well as by pre-absorption studies with NK-3 peptide, which eliminates staining (manufacturer’s website) (Burke et al., 2006). NK3R has previously been characterized as a marker for type 1 and 2 cone bipolar cells and the staining pattern we observed in this study is consistent with what has been reported in previous studies (Haverkamp et al., 2003).
Recoverin
The recoverin antibody labels a single band of 26 kDa by western blot analysis of protein extracted from human retinoblastoma Y79 cells (Wiechmann, 1996). Recoverin is expressed by bipolar cells in a number of species, including mouse, in which it labels type 2 cone bipolar cells and the staining observed in this study is consistent with what has been previously reported (Haverkamp et al., 2003; Milam et al., 1993; Wikler et al., 1998).
HCN4
Western blot analysis using the polyclonal HCN4 antibody detects a dark band of approximately 130 kDa in extracts of rat brain membrane, compared to a predicted protein size of 129 kDa. This band and several fainter bands were not detected after preincubation with the antigen (manufacturer’s website). HCN4 has been shown to be expressed by type 3 (later reclassified as type 3a) bipolar cells and the morphology and stratification of the cells visualized in this study matched what has been previously observed (Muller et al., 2003).
PKARIIβ
The PKARIIβ antibody recognizes a single band of 53 kDa by western blot analysis of protein extracted from human endothelial cells, close to the predicted size of the protein, 46 kDa (manufacturer’s website). This antibody has previously been shown to label type 3b bipolar cells and the staining we observed in this study is consistent with these results (Mataruga et al., 2007).
Calsenilin
The 40A5 antibody to calsenilin has previously been shown to label type 4 cone bipolar cells (Haverkamp et al., 2008). The specificity of the 40A5 monoclonal antibody used in Haverkamp et. al. was demonstrated to be specific by performing western blot analysis, in which a single band, the same size as recombinant human calsenilin, was detected in retina protein extracts and that this band was absent when the antibody was preabsorbed with calsenilin antigen (Haverkamp et al., 2008).
Syt2
The specificity of Syt2 immunoreactivity was demonstrated by western blot analysis, in which a single 60 kDa band was detected in lysates of zebrafish embryos or mouse cerebellum but not liver. Syt2 immunoreactivity has previously been described in the soma, axons and dendrites of type 2 bipolar cells and type 6 cone bipolar cell axons (Fox and Sanes, 2007; Wässle et al., 2009). Staining observed in this study matched what has previously been described in the mouse retina.
Go alpha
Specificity of the Go alpha antibody was verified by western blot analysis of Go alpha and related proteins expressed in bacteria, in which a single band between 39 and 42 kDa is detected, consistent with the protein’s predicted size of 40 kDA (manufacturer’s website). Localization of Go alpha to ON bipolar cells in the mouse retina has been previously described and the localization observed in this study was consistent with these previous findings (Haverkamp and Wässle, 2000).
PKCα
PKCα specificity was confirmed by western blot analysis, in which a dark band between the 91 and 42 kDa markers was detected in PMA treated Hela cell protein extracts, compared to the predicted molecular weight of the protein, 77 kDa (manufacturer’s website). PKCα is widely used as a marker for rod bipolar cells and the morphology of the stained cells that we observed was similar to what many others have reported.
β-dystroglycan
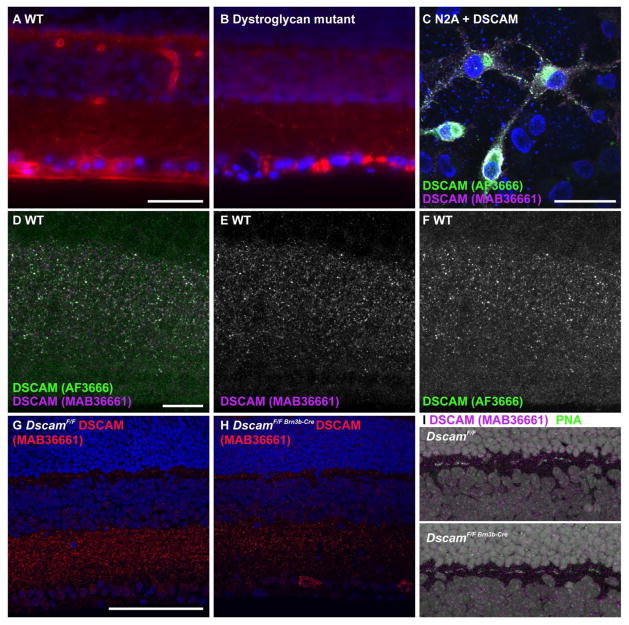
Specificity of the Mandag2 antibody was confirmed by staining retina tissue from a mouse lacking the region of the protein in which the epitope is located (courtesy of Dr. Kevin Campbell and Rolf Turk, University of Iowa). Staining of blood vessels and rod and cone synapses by this antibody was absent in mice lacking the epitope containing region, while staining in the inner plexiform layer was still present, indicating that the inner plexiform staining is non-specific (Figure 1A and B).
Figure 1. Antibody and strain characterization.
A and B, Sections of retina from wild type and dystroglycan mutant mice were stained with the Mandag2 antibody. Based on the differences between the two genotypes, blood vessel and photoreceptor synapse staining by the antibody is specific, while staining of ganglion cell axons, dendrites and cell bodies is not. C, Neuro2A cells transfected with a full length Dscam expression construct were immunostained with two different DSCAM antibodies. The two antibodies stained the same structures, with no staining present in untransfected cells. D–F, Section of wild type retina stained with AF3666 and MAB36661 antibodies. The two antibodies stain a largely overlapping set of puncta in the mouse inner nuclear layer. G–I, Sections of DscamF/F or DscamF/F Brn3b-Cre retina stained with antibodies to DSCAM. A reduction in DSCAM immunoreactivity is observed in the inner plexiform layer and cells in the retinal ganglion cell layer accumulate DSCAM protein in their soma in the presence of Brn-3b Cre. DSCAM protein is observed adjacent to the cone synapse in both genotypes. The scale bar in (A) is equivalent to 50 μm in A and B. The scale bar in (C) is equivalent to 20 μm in C. The scale bar in (D) is equivalent to 20 μm in D, E, F and I. The scale bar in (G) is equivalent to 100 μm in G and H.
Cone Arrestin
This antibody recognized a single band in protein lysates made from mouse retina, located between 37 and 50 kDa markers, consistent with the protein’s predicted size of 42 kDa, (see manufacturer’s website). Specificity was also confirmed by costaining with PNA and by the characteristic morphology of the cone pedicle (Nasonkin et al., 2011).
GS
Specificity of this antibody was demonstrated by western blot analysis in which a single band of 45 kDa was detected in protein extracted from rat brain, the predicted size of the protein (see manufacturer’s website). The antibody has also been used in retina and has a staining pattern consistent with specific labeling of Müller glia (Puller and Haverkamp, 2011).
Calbindin
This antibody stains a single band of 28 kDA in western blot analysis of brain extracts from a variety of species including mouse, the expected size of the protein. The D28K antibody is widely used in retinal research and the staining pattern observed in this study was very similar to what has been previously reported.
DSCAM antibodies
DSCAM antibodies were first assayed on neuro-2a cells transfected with a plasmid encoding the full length Dscam gene under control of the CAG promoter. Both mouse and goat antibodies co-labeled protein in the membrane and endomembrane systems of transfected cells (Figure 1C).
DSCAM (goat)
The polyclonal DSCAM antibody AF3666 stains a band of 220 kDa by western blot analysis of mouse retina, cortex and cerebellum. Staining is absent in protein extracts generated from Dscam2J mice or reduced in size when detecting protein extracts made from DscamFD mice, in which the DSCAM transmembrane domain is absent (Schramm et al., 2012). Staining in tissue sections showed overlap with the MAB36661 antibody, although some additional non-specific staining was present (Figure 1D–F).
DSCAM (mouse)
The staining pattern of the DSCAM MAB36661 antibody overlaps with the AF3666 antibody and its staining is eliminated in sections of Dscam2J retina, in which no protein product is detected by western blot analysis. Furthermore, DSCAM immunoreactivity was detected in retinal cell types in which Dscam mRNA has been detected by in situ hybridization and was absent in those in which mRNA was not detected. The epitope of this antibody was deletion mapped to the first four Ig repeats of the DSCAM protein (data not shown). The DSCAM MAB36661 antibody did not detect DSCAM by western blot analysis, suggesting the epitope is sensitive to denaturation.
Bassoon
The ground squirrel bassoon antiserum recognized two expected bands of around 400 kDa molecular weight on western blots of ground squirrel retina and stained a pattern of cellular morphology and distribution in the mouse retina that is identical with previous reports of mouse bassoon antiserum (Dick et al., 2003).
PSD95
Specificity of this antibody was determined by performing western blot analysis using brain tissue generated from wild type and knock out mice (manufacturer’s website). Tissue from wild type mice had bands between 95 and 110 kDA, consistent with the phosphorylation dependent size of the protein, while no bands were detected in tissue extracts made from mutant mice. Immunoreactivity observed in this study matched the localization pattern of PSD-95 in the retina.
ChAT
The antibody recognizes bands of 70–74 kDa by western blot analysis of protein extract of mouse brain, consistent with the protein’s predicted size of 68–70 kDa (manufacturer’s website). The retina has a population of cholinergic amacrine cells that stratify in the ON and OFF lamina of the inner plexiform layer. The antibody used in this study recognized this population of cells and is widely used to assay this cell population in the retina and to demarcate the inner plexiform layer, including in mouse, and the staining pattern we observed is similar to what many others have reported (Pérez De Sevilla Muller et al., 2007).
Brn3A
This antibody recognizes a single band of 47 kDa in ND7 cells transfected with Brn3A by western blot analysis, the predicted size of the Brn3A protein. Staining was observed in the appropriate layer of the retina and staining was observed in many Brn3b positive, and therefore retinal ganglion cells, but also recognized cells in the Brn3b null retinal ganglion cell layer, indicating that it is not cross reacting with this homologous transcription factor (data not shown).
Disabled
The Dab1 antibody has been used extensively to label type AII amacrine cells, which make a characteristic bistratified projection of their neurite arbors. Specificity of this antibody has been demonstrated by the absence of staining in knockout mice (Kuo et al., 2005).
bNOS
This antibody is used to label a mixed population of amacrine cells, which was observed in this study. Antibody specificity was assayed by western blot of brain lysate, in which the immunizing peptide blocked detection of a band between 150 and 160 kDa, consistent with the protein’s predicted size of 150–160 kDa (manufacturer’s website).
Chx10
Chx10 is expressed by bipolar cells and is used as a marker for this cell type in the retina (Burmeister et al., 1996). The antibody used in this study labels a single band slightly larger than the 43 kDa marker band in western blot analysis of eye extract, compared to the predicted protein size of 39.4 kDa (manufacturer’s website). The expected localization pattern was observed in this and previous studies (Martinez-Navarrete et al., 2008).
Cre pattern verification
Brn3b-Cre activity was previously observed in retinal ganglion cells but actual activity is dependent on the sensitivity of the target of recombination. For example activation of the highly sensitive Cre reporter ai14 can be observed in retinal ganglion cells as well as off target cells such as horizontal cells, while recombination and activation of less sensitive reporters such as Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J is not observed in any cells of the retina (data not shown). Brn3b-Cre activity was therefore verified by staining for DSCAM protein after targeting the Dscam transmembrane domain in the DscamF/F mouse, which results in accumulation of residual DSCAM protein in the endomembrane system of the cell. Induction of recombination targeting the Dscam conditional allele using Brn3b-Cre resulted in a reduction of DSCAM immunoreactivity in the inner plexiform layer and accumulation of DSCAM protein in cells in the retinal ganglion cell layer, while DSCAM protein was still observed in the outer plexiform layer, consistent with Cre activity being targeted to retinal ganglion cells and also consistent with the phenotypes reported here (Figure 1G–I).
Antibody Staining
Tissue was blocked in a blocking solution composed of PBS, 5% normal donkey serum and 0.1% triton (for staining sections) or 0.4% triton (for staining whole retinas) for 30 minutes (sections) or between two hours and overnight (whole retinas). Antibodies were diluted in blocking solution according to the concentrations listed in Table 1. Sections were incubated overnight at 4 °C, after which they were washed three times for ten minutes in PBS. Secondary antibodies (Jackson Immuno Research at 1:2,000), peanut lectin (Molecular Probes at 1:2,000 dilution of manufacturer’s recommended reconstitution volume) and the nuclear stain Draq5 (Cell Signaling Technology at 1:500) were diluted in blocking solution and incubated for two hours at room temperature. Following secondary incubation sections were washed three times for ten minutes in PBS. If DAPI was used to label nuclei the reagent was incorporated into the second wash at a dilution of 1:50,000 of a 1 mg/ml stock. Following washes, cover slips were applied to the slides using 80% glycerol as a mounting medium. Whole retinas were stained in a similar manner except that both primary and secondary incubations lasted two-four days and were performed at 4 °C. Washes were carried out for one hour each.
Fluorescent Microscopy
Sections and whole retina were imaged using either an Olympus spinning disk (DSU) confocal microscope or Olympus Fluoview confocal microscope. Images were cropped and rotated using Adobe photoshop software. Any changes to brightness or contrast were made across entire images.
Electron Microscopy
EM was performed as previously described (Fuerst et al., 2009). Briefly, retinas were fixed in buffered 2% glutaraldehyde and 2% formaldehyde overnight and then rinsed in PBS. Retinas were rinsed 3 times for at least 5 minutes in 0.1 M cacodylate buffer, followed by fixation using 4% osmium tetroxide overnight. After three further rinses in 0.1 M cacodylate buffer (5 minutes each), dehydration was performed by rinses in increasingly concentrated ethanol. Embedding was performed by first rinsing tissue twice in propylene oxide (10 minutes each), followed by a rinse in a 50/50 mix by volume of propylene oxide and Spurr’s resin, three long rinses in Spurr’s resin (at least 10 hours each), and curing overnight. Samples were sectioned using a Reichert ultramicrotome. Sections were stained in 4% uranyl acetate with potassium permanganate for 12 minutes, rinsed with distilled deionized water, dried (either overnight or 1 hr under a heat lamp). Sections were further stained with lead acetate for 8 minutes, briefly rinsed with 0.1 M NaOH, rinsed with distilled deionized water, and dried (overnight or 1 hr under heat lamp). Samples were imaged using either a Phillips CM200 200KV TEM equipped with a Gatan digital camera system and LaB6 cathode, or an FEI T20 200KV TEM.
Imaging for spatial analysis
Cone pedicles
Four wild type and four Dscam2J retinas were stained with cone arrestin and PNA and imaged. Four peripheral and four central 213 μm confocal stacks were collected from each retina. Peripheral stacks were collected within 213 μm of the edge of the retina (i.e., one field of view on this instrument) while central stacks were collected within 213 μm of the optic nerve. Stacks were projected and the center of each pedicle was marked for Voronoi domain analysis.
Type 3b bipolar cells
Four wild type and four Dscam2J retinas were imaged. Four 213 μm confocal stacks were collected from each retina midway between the optic nerve and the edge of the retina. Confocal stacks were opened in FIJI and the most distal portion of the dendrite (referred in this manuscript as the dendrite bulb) before projection of distal dendrites to the cone pedicles was marked.
Voronoi Domain Analysis
Voronoi analysis was performed with Ka-me Voronoi image analyzer software (Khiripet et al., 2012). Each cone pedicle or dendrite bulb was marked and an image of the Voronoi domains was exported along with the area of each domain. The coefficient of variation was calculated for each of the 16 images per genotype/location (cone pedicles: wild type central, wild type peripheral, mutant central and mutant peripheral or type 3b dendrite bulbs: wild type versus mutant) and used to perform a Students T-Test.
Density Recovery Profile Analysis
Density Recovery Profile Analysis was performed using WinDRP 1.6.4 (http://wvad.mpimf-heidelberg.mpg.de/abteilungen/biomedizinischeOptik/software/WinDRP/index.html). Images were imported as BMP files and the location of each dendrite bulb was marked. The cell diameter was set to 3 μm, the annulus width was set at 5 μm and cells (dendrite bulbs) within 15 annuli were counted. The soma correction was not used (the first annulus bin is sometimes discarded because two cells cannot occupy the same space). This was not performed because we would be able to measure dendrite bulbs in close proximity to each other. The number of cells in each bin was averaged for each genotype and to calculate an average bin density. The packing indexes of each genotype was used to compare wild type and mutant genotypes using a Student’s T-Test.
Nearest Neighbor Analysis
Nearest Neighbor Analysis was performed using the same software as that used for DRP analysis. The regularity index of each test is given and was used to compare the distribution with random simulations or across genotypes using a Student’s T-Test.
Tissue culture and transfection
Neuro-2a cells were obtained from American Type Culture Collection (ATCC) and maintained in DMEM medium supplemented with 10% fetal bovine serum, glutamine and pen/strep antibiotics. A plasmid carrying full length Dscam under control of the CAG promoter was transfected into cells grown on poly-l-lysine coated coverslips using fugene 6 reagent at a ratio of 1 μg/ DNA to 3 μl fugene reagent (Schramm et al., 2012). Cells were cultured for 24 hours and then collected and fixed in 4 % paraformaldahyde for five minutes. Cells were stained using the same procedure used for retina sections.
Results
The Dscam gene is required for normal regulation of cell number, dendrite arborization and spacing of multiple amacrine and ganglion cell types (Fuerst et al., 2009; Fuerst et al., 2008; Keeley et al., 2012). The Dscam homologue Dscaml1 is required for organization of cells making up the mouse rod circuit; rods, rod bipolar cells and AII amacrine cells (Fuerst et al., 2009). The organization of the outer plexiform layer was assayed to determine if Dscam influences the organization of the outer plexiform layer, particularly the cone synapse. To this end the organization of multiple bipolar cell types, horizontal cells, cones, cone synapses, Müller glia and rod synapses were imaged in retina sections of wild type and Dscam2J mice, the latter which do not make a detectable DSCAM protein. The markers used encompass five types of OFF bipolar cells in the mouse retina and two markers of ON bipolar cells, in addition to PNA, which labels the invaginating processes of ON bipolar cells (Koike et al., 2010). Most bipolar cell types had an organization that appeared similar in wild type and Dscam2J retinas (Figure 2 and Figure 3). Two types of OFF bipolar cells, type 3b (Figure 2M–T) and type 4 bipolar cells (Figure 3A–H) appeared to have clumped dendrites, reminiscent of the self avoidance defect observed in Dscam null amacrine and retinal ganglion cells. Markers to other cell types associated with the cone synapse were used to assay the organization of cones, horizontal cells, Müller glia and rod synapses (Figure 4). Organization of these cell types appeared similar in wild type and Dscam2J retinas.
Figure 2. Axon and dendrite arborization of bipolar cells in the wild type and Dscam loss of function retina.
A–D, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to NK3R to label type 1 and type 2 cone bipolar cells, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. E–F, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to recoverin to label type 2 cone bipolar cells and rods, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. I–L, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with HCN4 to label type 3a cone bipolar cells and PNA (N=4 retinas). Lamination of HCN4 positive neurites in the ON half of the inner plexiform layer are longer in the Dscam2J/2J retina compared to wild type controls (I versus J; arrow). No differences were detected in the outer plexiform layer. M–T, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to PKARIIβ to label type 3b cone bipolar cells and PNA (N>4 retinas). PKARIIβ immunopositive bipolar cells have clumped dendrite arbors proximal to cone pedicles, compared to wild type (P, S and T; mutant vs. O, Q and R; wild type). The scale bar in (A) is equivalent to 100 μm in A, B, E, F, I, J, M and N. The scale bar in (C) is equivalent to 20 μm in C, D, G, H, K, L, O and P. The scale bar in (Q) is equivalent to 10 μm in Q–T.
Figure 3. Axon and dendrite arborization of bipolar cells in the wild type and Dscam loss of function retina.
A–H, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to calsenilintolabel type 3b cone bipolar cells, and PNA (N>4 retinas). Calsenilin immunopositive bipolar cells have clumped dendritic arbors proximal to cone pedicles, compared to wild type (D, G and H; mutant versus C, E and F; wild type). I–L, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to Syt2 to label type 2 and type 6 cone bipolar cells, and PNA (N=4 retinas). The terminals of some Syt2 positive bipolar cells terminated in the inner nuclear layer (arrows). Differences when comparing the outer plexiform layer of wild type and Dscam2J retina were not detected. M–P, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to Go-alpha to label ON bipolar cells, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. Q–R, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to PKC-α to label rod bipolar cells, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. The scale bar in (A) is equivalent to 100 μm in A, B, I, J, M, N, Q and R. The scale bar in (C) is equivalent to 20 μm in C, D, K, L, O, P, S and T. The scale bar in (E) is equivalent to 10 μm in E–H.
Figure 4. Organization of rod synapses, cone pedicles, Müller glia and horizontal cells in the wild type and Dscam loss of function retina.
A–D, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to dystroglycan to label rod and cone synapses, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. E–H, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to cone arrestin to label cone cells, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. I–L, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to GS to label Müller glia, and PNA (N=4 retinas). Differences when comparing wild type and Dscam loss of function retina were not detected. M–P, Sections of adult (>P42) wild type and Dscam2J/2J retina were stained with antibodies to calbindin to label horizontal cells, and PNA (N=4 retinas). Disorganization of calbindin positive amacrine cells was observed in the Dscam mutant retina. Differences when comparing horizontal cells in wild type and Dscam loss of function retina were not detected. The scale bar in (A) is equivalent to 100 μm in A, B, E, F, I, J, M and N. The scale bar in (C) is equivalent to 20 μm in C, D, G, H, K, L, O and P.
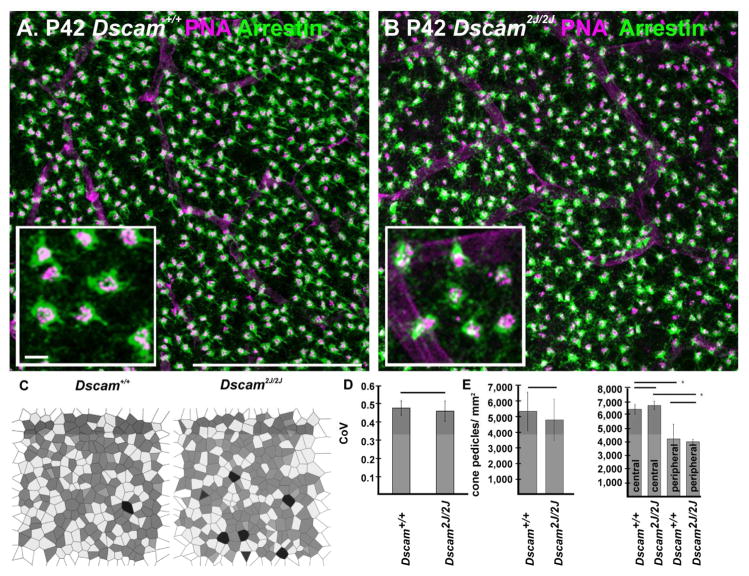
Arborization defects in cell types contributing to the cone synapse suggested that Dscam might be required for normal development of the cone synapse. Electron microscopy was performed to determine if the ultrastructure of the cone pedicle was intact in the Dscam2J retina. Ribbon synapses were observed in both the wild type and Dscam2J retina, organized around the processes of invaginating bipolar and horizontal cells (Figure 5). Cone pedicles were labeled to determine if Dscam is required for their spatial organization. The organization of cone pedicles appeared similar in wild type and Dscam2J retinas and telodendria were observed to project from the pedicles of both wild type and Dscam2J pedicles (Figure 6A and B). Voronoi domain analysis was performed to test if there was a significant difference in the space occupied by wild type and Dscam2J pedicles. Voronoi domain analysis is a measurement of the area around a given object, in this case the terminal of the cone axon, that is closer to that object than any other selected object, in this case other cone pedicles (Figure 6C). The area around wild type and Dscam2J pedicles was assayed and the coefficient of variation, a measure of the variability of the area of Voronoi domains, of each genotype were compared by a Student T-Test. No significant difference in the variability of the domain size was detected (Figure 6D). The density of cone terminals was also assayed in the wild type and Dscam2J retina. No significant difference in the density of pedicles was detected when comparing wild type and Dscam2J retinas (Figure 6E). This was repeated by assaying the central and peripheral retinas, which are more dense and sparse, respectively, in cones. A significant difference in the density of cones was detected when comparing central and peripheral regions of retina within or between genotypes, but no significant difference was detected when comparing like regions of retina between genotypes (Figure 6D and E). Therefore while Dscam may contribute to the organization of some cells that make synapses at the cone pedicle, we did not detect defects in the number or spacing of cone synapses.
Figure 5. Cone synapse is present in the absence of Dscam.
A and B, Electron microscopy of the adult (>P42) wild type and Dscam2J/2J cone pedicle and associated synapses, with the cone axon terminal labeled in pink and adjacent neurites labeled in green (N=2 retinas each). Ribbons and associated structures appeared equivalent comparing wild type and Dscam2J/2J retinas. Examples of high-resolution images of synaptic ribbons in which invaginating horizontal and bipolar cell processes can be observed opposed a ribbon are shown underneath each genotype (arrow pointing at ribbon, HC; horizontal cell, BC; invaginating bipolar cell). The scale bar in the upper left panel is equivalent to 1 μm in the upper panels and the scale bar in the lower left panel is equivalent to 200 nm in the lower panels.
Figure 6. Spacing and number of cone pedicles is maintained in the absence of Dscam.
A and B, Wild type and Dscam2J/2J retinas were stained with antibodies to cone arrestin, to visualize cone pedicles, and PNA (higher magnification in inset). C, The spatial arrangement of cone pedicles was assayed by Voronoi domain analysis, which identifies the area around a pedicle that is closer to that pedicle than any other pedicle. D, No significant difference was detected in the coefficient of variance, the degree to which spatial domains vary in size, when comparing wild type and mutant. E and F, No significant difference in the number of pedicles was detected comparing wild and mutant retina, although the density of pedicles was significantly higher in the central portion of the retina of both genotypes compared to the peripheral retina. The scale bar in (A) is equivalent to 100 μm in A and B. The inset scale bar is equivalent to 5 μm.
We next focused on the two OFF bipolar cell types, type 3b and type 4, that appeared to have self-avoidance defects in the absence of Dscam. The organization of type 3b and type 4 dendrites were assayed by confocal microscopy in whole retinas. Clumping of type 4 bipolar cell dendrites (Figure 7A–D) can be clearly observed in the Dscam mutant retina, while the axons and dendrites of other cell types appeared to be normal (Figure 7E and F and data not shown).
Figure 7. Clumping of calsenilin positive bipolar cell dendrites in the Dscam mutant retina.
A–D, Wild type and Dscam2J/2J retinas were stained with antibodies to calsenilin, to label type 4 cone bipolar cells, and PNA (N>4). Clumping of type 4 cone bipolar cell dendrites was observed in the Dscam2J/2J retina compared to wild type. C, and D, At high magnification the projection of type 4 cone bipolar cell dendrites to cone pedicles can be observed in both the wild type and Dscam2J/2J retina. E and F, Wild type and Dscam2J/2J retinas were stained with antibodies to calbindin, to label horizontal cells, and PNA (N>4). A change in arborization of horizontal cell neurites was not observed in the Dscam2J/2J retina compared to wild type. The scale bar in (A) is equivalent to 100 μm in A and B. The scale bar in (C) is equivalent to 10 μm in C and D. The scale bar in (E) is equivalent to 10 μm in E and F.
Type 3b bipolar cells also appeared to have clumped dendrites in the Dscam2J retina (Figure 8 A and B). Bipolar cell bodies are not organized in mosaics in the mouse retina (Reese, 2011; Wässle and Riemann, 1978). The microanatomy of type 3b bipolar cells includes the projection of a dendrite stalk that forms a bulb-like structure (heretofore referred to as the dendrite bulb) immediately before distal dendrites project out towards cone pedicles (Figure 8A and B; insets) (Mataruga et al., 2007). The dendrite bulbs appeared to be spaced in the wild type retina and clustered in the Dscam2J retina, and provided a fixed point that could be used for spacing analysis, which typically uses the cell soma as a reference point. Spacing analysis was performed to test if the dendrite bulbs of type 3b bipolar cells were clumped in the Dscam2J retinas compared to wild type. Voronoi domain analysis was performed, using the most distal portion of the dendrite bulb (Figure 8C). An apparent increase in dendrite bulbs with small or large Voronoi domains was noted, consistent with these structures being clumped in the Dscam2J retina. A significant difference in the coefficient of variation was detected comparing wild type and Dscam2J type 3b bipolar cell dendrite bulbs (Figure 8D). Next, density recovery profiling (DRP) was performed to determine if there is a change in the spacing of Dscam2J type 3b dendrite bulbs compared to wild type. DRP analysis is a measurement of the number of like structures within a given distance from a reference structure, measured for all such structures in a field (Rodieck, 1991). These data are then plotted out with the number of like events within a given distances from all of the objects collectively referred to as the reference soma (in this case the reference dendrite bulb). When objects are organized such that they avoid each other an exclusion zone will exist wherein the number of counted objects falls below the average cell density. Because the size of such an exclusion zone would be expected to vary with cell density the size of the exclusion zone is translated into a value that is independent of cell density, the packing index. The packing index can then be used in statistical tests comparing different populations. A significant increase in the density of type 3b bipolar cells was detected in the Dscam2J retina compared to wild type (WT average cell density 3912 +/− 214 vs. mutant average cell density 4570 +/− 221; P=0.02). A significant difference (P=0.03) was also detected when comparing the packing index of wild type and Dscam2J type 3b bipolar cell dendrite bulbs (Figure 8E and F). The exclusion zone of wild type 3b bipolar cells extended out to 10 microns from the reference soma, as indicated by a decrease in cell density compared to average cell density, whereas the exclusion zone reached average cell density in the Dscam2J retina by 10 microns (Figure 8E and F). Finally nearest neighbor analysis (NNA) was performed. Nearest neighbor analysis is a measurement of the distance to the nearest neighbor, plotted for all objects in a field, and here is compared to a random distribution of a like number of objects in a like sized field. The nearest neighbor distances of both wild type and Dscam2J type 3b bipolar cell dendrite bulbs were significantly different than a random distribution, but the Dscam2J type 3b bipolar cell dendrite bulbs were shifted towards random compared to wild type (P value comparing wild type and random = 0.00000009 versus Dscam2J versus random = 0.007; Figure 8G and H).
Figure 8. Disruption of type 3b cone bipolar cell dendrite arborization in the Dscam2J/2J retina.
A and B, Whole adult (>P42) wild type and Dscam2J/2J retinas were labeled with antibodies to PKARIIβ and PNA (N>8 retinas). The dendrites of PKARIIβ positive bipolar cells are clumped in the Dscam2J/2J retina compared to wild type. C and D, The location of the dendrite bulb, the portion of the dendrite stalk from which distal dendritic branches project, of each PKARIIβ bipolar cell was plotted and used to determine the Voronoi domains of these cells’ dendrite bulbs in wild type and Dscam2J/2J retinas (arrows point at sample dendrite bulbs; insets A and B). D, Voronoi domain analysis revealed a statistically significant increase in the coefficient of variation (CoV) when comparing the Voronoi domains of wild type and Dscam2J/2J type 3b bipolar cell dendrite bulbs. E and F, DRP analysis of PKARIIβ dendritic bulb spacing. A reduced exclusion zone was observed in close proximity to the reference soma in the Dscam2J/2J retina compared to wild type controls (for example at 10 μm from the reference soma) and a significant difference in the packing factor was detected when comparing genotypes (P=0.03). G and H, Nearest neighbor analysis (NNA) of type 3b bipolar cells in wild type and Dscam2J/2J retina. Distribution of dendrite bulbs in both genotypes was significantly different than random simulations and a decrease in the average distance to the nearest neighbor was detected in the Dscam2J/2J retina compared to wild type. The scale bar in (A) is equivalent to 100 μm in A and B. The inset scale bar is equivalent to 5 μm.
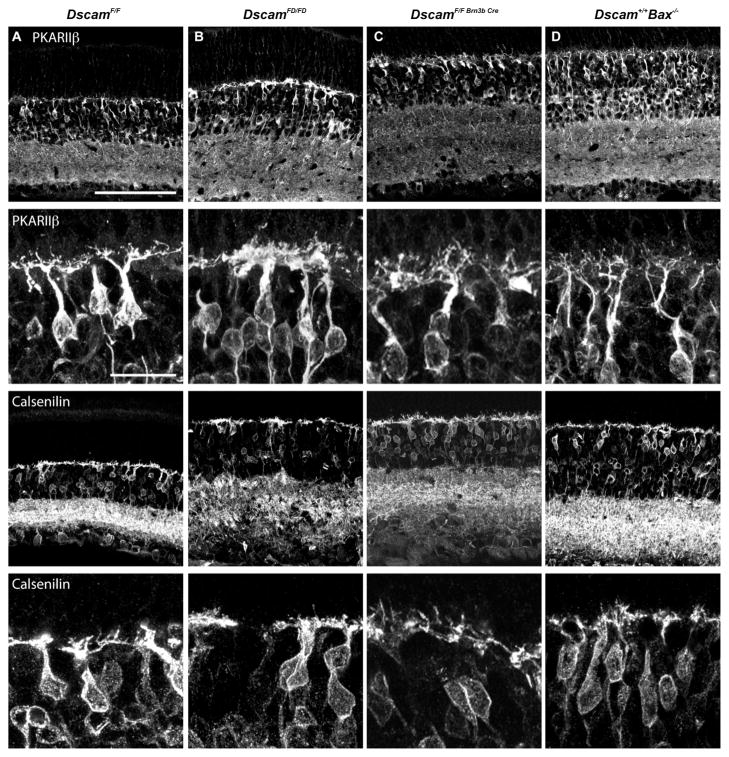
To understand the mechanism underlying the clumped type 3b and type 4 cone bipolar cell dendrites observed in the Dscam2J retina we tested if this clumping was a secondary effect caused by changes in cell number and or disorganization of cells contributing to the inner plexiform layer. Dscam was targeted for deletion specifically in retinal ganglion cells using Brn3b-Cre to test the possibility that type 3b and type 4 cone bipolar cell dendrites are clumped secondarily to other cell types. Defects in type 3b and 4 cone bipolar cell dendrite arborization was observed in the DscamFD retina compared to the DscamF retina, indicating that deletion of the Dscam transmembrane domain, targeted in the conditional allele, is sufficient to cause this dendrite clumping and that arborization resembles wild type in the floxed DscamF allele in the absence of Cre recombinase (Figure 9A and B). Type 3b and type 4 cone bipolar cell dendrites do not clump if Dscam is targeted for deletion in retinal ganglion cells, despite the widespread disorganization of retinal ganglion cells and some secondary disorganization of amacrine cells when the floxed allele of Dscam is targeted with Brn3b-Cre (Figure 9A–C) (Fuerst et al., 2012). A large increase in cell number is also observed in the Dscam mutant retina and we sought to test if increased cell number was sufficient to cause clumping of bipolar cell dendrites. To do so we assayed the organization of dendrites in the bax null retina, in which normal developmental cell death is blocked and a large increase in cell number is observed (Knudson et al., 1995; Mosinger Ogilvie et al., 1998). While deletion of bax is sufficient to cause clumping of cell soma and dendrites of some retinal cell types, similar to what is observed in the Dscam heterozygous retina, defects in type 3b and type 4 cone bipolar cell dendrites were not observed in the bax null retina (Figure 9D) (Chen et al., 2013; Keeley et al., 2012).
Figure 9. Clumping of type 3b and type 4 bipolar cell dendrites is not observed when Dscam was specifically targeted in RGCs or in the bax null retina.
A–D, Sections of retina from adult (P42 or older) unrecombined conditional allele of Dscam (DscamF/F; A) or the conditional allele of Dscam targeted for deletion in the germ line (DscamFD/FD; B) or the conditional allele of Dscam targeted in retinal ganglion cells (DscamF/F Brn3b-Cre; C) or bax null (Dscam+/+ Bax−/−754 ; D) mice were stained with antibodies to PKARIIβ or calsenilin, to label type 3b or type 4 cone bipolar cells, respectively (N=3 for each genotype). A, The dendrites of type 3b and type 4 cone bipolar cells were not clumped in the DscamF/F retina. B, The dendrites of type 3b and type 4 cone bipolar cells were clumped in the DscamFD/FD retina. C and D, The dendrites of type 3b and type 4 cone bipolar cells were not clumped in the DscamF/F Brn3b Cre or bax−/− retinas. The scale bar in the first panel in the first row is equivalent to 100 μm in the top and third rows. The scale bar in the first panel of the second row is equivalent 20 μm in the second and bottom rows.
The localization and cell type specific expression of Dscam was assayed to understand why some populations of cone bipolar cells had clumped dendrites in the absence of DSCAM. Previous studies of DSCAM localization found limited staining in the retina and did not detect protein in many cell types in which Dscam mRNA was detected, likely as a result of antigen inactivation during the prolonged fixation used in this study (Fuerst et al., 2008). We therefore screened DSCAM antibodies using transfected cells and wild type and Dscam null retinas and optimized staining procedures for DSCAM. Multiple antibodies were identified that stained DSCAM with a high level of specificity in cell culture (Figure 1C). One of these, a monoclonal antibody, was identified that had a high degree of specificity in immunohistochemical staining of the retina with low background. DSCAM protein was localized throughout the synaptic inner plexiform layer of the adult mouse retina and in the synaptic outer plexiform layer (Figure 10A). Specificity was verified by the lack of staining in Dscam2J mice, which have previously been shown to lack DSCAM by western blot analysis (Figure 10B) (Schramm et al., 2012). Sections of mouse retina were labeled with PNA, which labels the invaginating tips of ON bipolar cells, and DSCAM. DSCAM immunoreactivity was concentrated adjacent to PNA labeling (Figure 10C). The localization of DSCAM in two cone dominant species, macaque and ground squirrel, was also assayed. DSCAM staining was observed on the inner nuclear layer facing side of PNA staining in the macaque retina (Figure 11A–C). A similar pattern of post-synaptic localization was observed in ground squirrel retina (Figure 11D).
Figure 10. DSCAM localization in the mouse retina.
A–C, Sections of wild type and Dscam2J/2J retina were stained with antibodies to DSCAM (N>8 for both genotypes). A, DSCAM immunoreactivity was observed throughout the inner plexiform layer of the wild type adult retina. B, This immunoreactivity was not observed in the Dscam2J/2J retina. C, DSCAM immunoreactivity was observed in close proximity to the cone synapse in the outer plexiform layer. The scale bar in (A) is equivalent to 100 μm in A and B. The scale bar in (C) is equivalent to 10 μm in C.
Figure 11. DSCAM localization in macaque and ground squirrel retina.
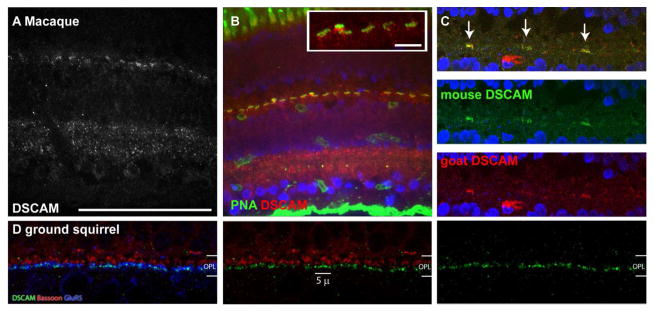
Sections of macaque and ground squirrel were stained with antibodies to DSCAM and PNA (macaque) or DSCAM, GluR5 and Bassoon (ground squirrel) (N=2 retinas). A, DSCAM immunoreactivity was observed in the macaque inner and outer plexiform layers. In the inner plexiform layer DSCAM was concentrated in the lamina proximal to the two cellular layers and less concentrated in the central inner plexiform layer. B, DSCAM immunoreactivity was observed proximal to the inner nuclear layer with respect to PNA staining in the macaque retina. C, Specificity of DSCAM immunoreactivity in the macaque outer plexiform layer was confirmed by using DSCAM antibodies generated in two different species. D, DSCAM immunoreactivity in the ground squirrel outer plexiform layer was observed running along GluR5, a receptor expressed by OFF bipolar cells, but not running alongside Bassoon, a presynaptic marker. The scale bar in (A) is equivalent to 100 μm in A–C. The scale bar in the inset in B is equivalent to 5 μm. The scale bar in (D) is equivalent to 5 μm in D.
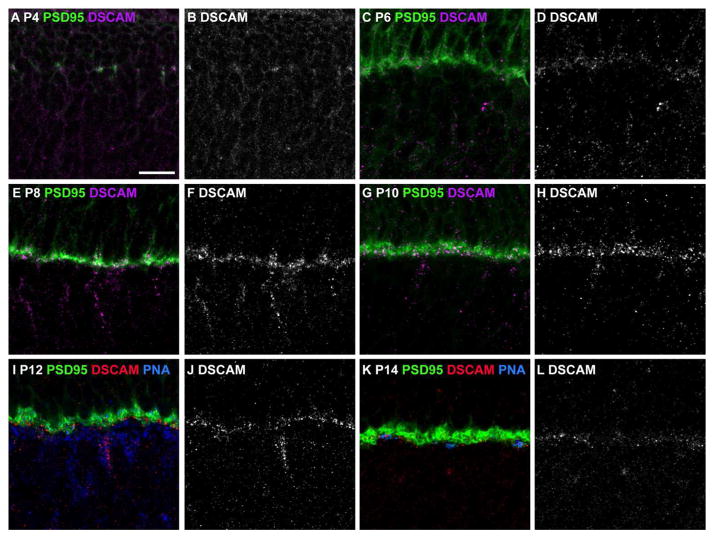
Next DSCAM was localized during development of the outer plexiform layer, identified with PSD95 staining. DSCAM protein was observed to roughly overlap with PSD95 staining until approximately postnatal day 12, after which DSCAM is observed proximal to the inner nuclear layer with respect to PNA and PSD95 staining (Figure 12). Some DSCAM staining was also observed in outer plexiform layer proximal cells in the inner nuclear layer, especially between postnatal days 8 and 12 (Figure 12).
Figure 12. Developmental localization of DSCAM in the outer plexiform layer.
A–H, Sections of mouse retina stained with antibodies to DSCAM and PSD95 (N=4 retinas at each age). A and B, At postnatal day 4 (P4) DSCAM and PSD95 immunoreactivity overlap. C and D, At P6 DSCAM immunoreactivity has taken on a punctate pattern nested within PSD95 immunoreactivity. E and F, At P8 DSCAM immunoreactivity is observed as puncta within areas immunoreactive for PSD95 and within the soma of cells in the developing inner nuclear layer. G and H, At P10 DSCAM immunoreactivity is observed in puncta overlapping with areas stained by PSD95 and in cell soma in the developing inner nuclear layer. Puncta of DSCAM and PSD95 immunoreactivity that do not overlap with each other become more apparent. I and J, By P12 DSCAM immunoreactivity begins to become confined to the inner face of PSD95 immunoreactivity. K and L, By P14 DSCAM immunoreactivity is closely associated with PNA staining and is largely distinct from PSD95 immunoreactivity. The scale bar in (A) is equivalent to 10 Tm.
The expression of Dscam in bipolar and horizontal cells was assayed to determine if cell type specific expression could explain the clumping of some OFF bipolar cells in the absence of DSCAM. We took advantage of the retention of DSCAM protein produced by the DscamFD allele, which lacks a transmembrane domain. DSCAM accumulates around the soma in the DscamFD retina (Figure 13A and B). Staining with DSCAM and one or more cell type specific antigens was performed to identify cell types that express Dscam, for example some calbindin-positive cells express Dscam, but not the cholinergic subset (Figure 13C). Next, cell types in which Dscam expression was previously assayed by in situ hybridization were assayed to verify this technique (Fuerst et al., 2009; Fuerst et al., 2008). Protein was observed in cell types previously shown to express Dscam (bNOS-positive amacrine cells and retinal ganglion cells) but was not observed in those in which Dscam transcript was not previously observed (cholinergic and AII amacrine cells) (Figure 14A–D). Next the presence of DSCAM was assayed in bipolar and horizontal cells. DSCAM protein was observed in HCN4, calsenilin, PKARIIβ and some Syt-2 positive bipolar cells, but was not observed in PKCα, NK3R or recoverin positive bipolar cells or in horizontal cells (Table 2 and Figure 14E–P). The presence of DSCAM in type 3b and type 4 bipolar cells is consistent with the dendrite arborization defects observed in these cell types. The lack of a dendrite arborization phenotype in type 3a and Syt2 immunopositive phenotype could be due to an absence of DSCAM protein at the dendrites (i.e., if it is localized to the bipolar cell axon of these cells). The localization of DSCAM to dendrite tips was assayed in type 3a, type 3b, type 4 and Syt2-immunopositive bipolar cells. The number of puncta localizing at the tips of each of these bipolar cell types was counted compared to the total number of puncta. Type 3b and type 4 bipolar cells were found to have more DSCAM localized to their tips (30 and 54% of puncta, respectively), compared to type 3a and Syt2 positive cells (11 and 9%, respectively) (Figure 15). The number of puncta overlapping with type 3a and type 4 dendrites was reduced when the channel containing the bipolar cell dendrites was flipped along the horizontal axis (type 3b: 30% to 18% and type 4: 54% to 16%). The number of puncta overlapping with the dendrites of type 3a and Syt2 immunopositive remained similar when the channel containing the dendrite staining was flipped along the horizontal axis (type 3a: 11% to 14% and Syt2 immunopositive 9% to 14%), suggesting that the amount of DSCAM protein observed at the dendritic tips of these cells may reflect coincident staining.
Figure 13. Use of a cytoplasmic isoform of DSCAM to identify cell types that produce the protein.
A–D, Sections of DscamFD/FD retina were stained with antibodies to DSCAM and ChAT and stained with Draq5, to label nuclei. The DSCAM protein produced by the DscamFD/FD allele lacks a transmembrane domain and accumulates in the cell cytosol. A and B, DSCAM immunoreactivity is observed in most cells in the retinal ganglion cell layer, in many cells in the RGL proximal portion of the inner nuclear layer and in some cells in the outer portion of the inner nuclear layer. DSCAM immunoreactivity was not observed in ChAT immunoreactive cells. C and D, Higher magnification of section from A and B. E and F, Sections of DscamFD/FD retina were stained with antibodies to DSCAM, calbindin and ChAT. E, ChAT is expressed by some but not all calbindin-positive cells. DSCAM immunoreactivity was not observed in ChAT positive cells but was observed in a subset of calbindin-positive ChAT-negative cells. The scale bar in (A) is equivalent to 10 μm in A and B. The scale bar in (C) is equivalent to 5 μm in C–F.
Figure 14. Dscam is expressed by some OFF bipolar cell populations.
A, Section of DscamFD/FD retina stained with antibodies to DSCAM and Brn3A (N=2). DSCAM immunoreactivity was observed in the soma of Brn3A positive cells in the retinal ganglion cell layer. B, Section of DscamFD/FD retina stained with antibodies to DSCAM and disabled (Dab) to stain AII amacrine cells (N=2). AII amacrine cells did not contain DSCAM in their soma. C, Section of DscamFD/FD retina stained with antibodies to DSCAM and ChAT, to label cholinergic amacrine cells (N=2). Cholinergic amacrine cell soma in the inner nuclear layer (top) or retinal ganglion layer (bottom) did not contain DSCAM protein. D, Section of DscamFD/FD retina stained with antibodies to DSCAM and bNOS (N=2). bNOS positive amacrine cells in both the inner nuclear layer (top) and retinal ganglion cell layer (bottom) contained DSCAM protein in their soma. E, Section of DscamFD/FD retina stained with antibodies to DSCAM and Chx10, to label bipolar cells (N=4). DSCAM immunoreactivity was observed in the soma surrounding some Chx10 positive cells. F and G, Section of DscamFD/FD retina stained with antibodies to HCN4 to label a subset of amacrine cells (G) and type 3a cone bipolar cells (F) (N=2 retinas). F, HCN4-positive bipolar cells, distinguished from amacrine cells by projection of dendrites to the outer plexiform layer and location, contained DSCAM protein in their soma. G, HCN4-positive amacrine cells, identified by their location proximal to the inner plexiform layer, did not contain DSCAM protein. H, Sections of DscamFD/FD retina were stained with antibodies to DSCAM and PKARIIβ to label type 3b bipolar cells. DSCAM immunoreactivity was observed in PKARIIβ positive cells. I, Sections of DscamFD/FD retina were stained with antibodies to DSCAM and Syt2, to label type 2 and type 6 bipolar cells (N=2 retinas). Some Syt2 immunopositive cells contained DSCAM immunoreactivity while others did not. J, Sections of DscamFD/FD retina were stained with antibodies to DSCAM and PKCα, to label rod bipolar cells. PKCαimmunopositive cells did not contain DSCAM. K and L, Sections of DscamFD/FD retina were stained with antibodies to Calbindin to label horizontal cells (K) and a subset of amacrine cells (L) (N=4 retinas). K, Horizontal cells, identified by their location close to the outer plexiform layer, did not contain DSCAM. L, Dimly staining calbindin-positive (ChAT-negative) amacrine cells located within the inner nuclear layer contained DSCAM protein. M and N, Sections of DscamFD/FD retina were stained with antibodies to calsenilin to label type 4 cone bipolar cells (M) and a subset of amacrine cells (N) (N=4 retinas). M, DSCAM immunoreactivity was observed in calsenilin positive bipolar cells. N, DSCAM protein was observed in some calsenilin positive amacrine cells, identified by their location in the inner nuclear layer. O, Sections of DscamFD/FD retina were stained with antibodies to DSCAM and NK3R, to label type 1 and 2 cone bipolar cells. NK3Rimmunopositive cells did not contain DSCAM. P, Sections of DscamFD/FD retina were stained with antibodies to DSCAM and recoverin, to label type 2 cone bipolar cells. Recoverinimmunopositive cells did not contain DSCAM. The scale bar in (A) is equivalent to 5 μm in A, F, H and I. The scale bar in (B) is equivalent to 5 μm in B and C. The scale bar in (D) is equivalent to 5 μm in D. The scale bar in (E) is equivalent to 5 μm in E. The scale bar in (G) is equivalent to 5 μm in G, J and L. The scale bar in (M) is equivalent to 5 μm in M–P.
Table 2.
Dscam expressing cell types in the inner nuclear layer
| Cell type | Marker | Dscam expression |
|---|---|---|
| Type 1 and 2 bipolar cells | NK3R | Negative |
| Type 2 bipolar cells | Recoverin | Negative |
| Type 3a bipolar cells | HCN4 | Positive |
| Type 3b bipolar cells | PKARIIβ | Positive |
| Type 4 bipolar cells | Calsenilin | Positive |
| Type 2 or Type 6 bipolar cell | Syt2 | Mixed |
| Rod bipolar cells | PKCα | Negative |
| Horizontal cells | Calbindin | Negative |
| HCN4+ amacrine cells | HCN4 | Negative |
| Calsenilin positive amacrine cells | Calsenilin | Mixed |
| Calbindin+ Chat -amacrine cells | Calbindin/Chat | Mixed |
Figure 15. DSCAM localization on bipolar cell dendrites.
A–C, Sections of wild type adult (>P42) retina stained with antibodies to DSCAM and PKARIIβ, calsenilin, HCN4 or Syt2 and PNA (N=4 retinas). DSCAM immunoreactivity was observed at the ends of some cone bipolar cell dendrite tips that terminated at the cone pedicle. The percent of DSCAM puncta that were observed within the dendrites of the respective bipolar cell types was counted for twenty pedicles and is reported in the upper corner of the left most panel. The result of this count when the channel containing bipolar cell staining was flipped along the horizontal axis is reported in the lower left hand corner. The scale bar in (A) is equivalent to 5 μm.
Discussion
Understanding how individual cells are integrated into the functional circuitry of the nervous system is a central goal of neuroscience. In recent years loss and gain of function analysis has begun to identify genes required for the integration of cells into neural circuits, but much work remains in order to understand how the proteins these genes encode mediate their functions.
DSCAMs were first functionally studied in Drosophila, where the incredible splice diversity of Dscam1 mediates specificity of neural development (Schmucker et al., 2000). Other Drosophila Dscam proteins that do not undergo extensive alternative splicing also play important neuro-developmental roles. For example Drosophila Dscam2 mediates synaptic specificity in conjunction with Dscam1 at the fly tetrad synapse by providing a repulsive cue (Millard et al., 2010). Indeed, it will be interesting to test if mouse DSCAM mediates innervation of the multi-contact cone synapse so as to limit the number of connections made at each pedicle, analogous to the function of Dscams in organization of the tetrad synapse in Drosophila. Dscams in the mouse retina are required for a number of different developmental events, including regulation of cell number, normal spacing of soma, arborization of dendrites and refinement of retinal ganglion cell axons in the lateral geniculate nucleus (Blank et al., 2011; Fuerst et al., 2009; Fuerst et al., 2008). While Dscams appear to mediate repulsion in both fly and mouse the neurites of many cell types in the mouse retina that clump in the absence of Dscam do not actively avoid each other, unlike Drosophila type IV DA neurons, (Keeley and Reese, 2010). This suggests the adhesion observed in the Dscam mutant retina is not because DSCAM directly mediates repulsion and has been interpreted to suggest that DSCAM prevents adhesion of cell type specific identifiers (Garrett et al., 2012). This is consistent with the observation that many spatially overlapping cell types require Dscam to prevent adhesion but when they do adhere they do so according to cell type. Indeed a simple explanation to explain why some cell types that express Dscam do not adhere in its absence is that they may not produce a protein that causes such adhesion in the absence of Dscam.
In further considering the function of Dscam in development of the cone synapses one might first consider the various requirements necessary for organization of such a structure. Each cone axon terminal is contacted by multiple bipolar and horizontal cells, which in turn send dendrites to distinct cone terminals. Therefore cells must not only make the correct synaptic contacts but also must avoid making multiple contacts at the same pedicle, so as to sample multiple pedicles. At a minimum this would require cells to have a mechanism to recognize their synaptic targets (heteroneuronal recognition between cell types), like cells (heteroneuronal recognition within cell types) and processes originating from the same cell (isoneuronal recognition). Mechanisms to mediate this include but are not limited to synaptic activity, glial stimulation and neural recognition through cell adhesion molecules. While glia are important facilitators of neural development and connectivity in other parts of the nervous system their role in development of the cone synapse appears to be limited in that normal development occurs in their absence (Williams et al., 2010). Likewise, while activity is required for maintenance of photoreceptor synapses they appear to develop normally in its absence (Claes et al., 2004). Based on these and other studies better candidates for mediating organization of the cone synapse appear to be cell adhesion molecules.
Current research in understanding the role of adhesion molecules in developmental organization of neurons is focused on identifying factors such as Dscams, sidekicks, semaphorins and plexins and the gamma-protocadherins that provide adhesive and repulsive cues required to properly wire the nervous system. Evidence for the role of cell adhesion molecules in organization of the cone synapse can be inferred by defects in development that occur in their absence. For example, horizontal cells utilize MEGF proteins, semaphorins and plexins to mediate self-avoidance (Kay et al., 2012; Matsuoka et al., 2011) and NGL-2 to properly innervate the rod spherule (Soto et al., 2013). The identification of factors regulating organization of the dendrites and axons of other cells composing the photoreceptor synapses is not currently as advanced, although homotypic regulation clearly regulates organization of the dendrite field of type 7 cone bipolar cells (Lee et al., 2011). Previously we have shown that Dscaml1 is required for arborization of rod bipolar cell dendrites, but a role for DSCAM in organization of outer plexiform layer cells had not been assayed.
In this study we identify DSCAM as a regulator of cell type specific avoidance in two populations of OFF bipolar cells, analogous to the role of MEGF proteins in horizontal cells. The role of DSCAM in heteroneuronal interactions between cell types at the cone pedicle remains an open question, as does the resulting physiology at disorganized cone synapses. Recently the role of Dscam1 over expression in the Drosophila nervous system has been demonstrated and linked to fragile-X syndrome (Cvetkovska et al., 2013; Kim et al., 2013). Preliminary results indicate that DSCAM over and ectopic expression in the cells contributing to the mouse cone synapse disrupt this structure, and that this line of approach together with targeted loss of function will be an extremely powerful tool in understanding the function of DSCAM in heteroneuronal interactions between like and non-like cell types, its role in human diseases and the developmental organization of the cone synapse and other synapses.
Acknowledgments
This research was supported by the National Eye Institute Grant EY020857. Imaging support was provided by NIH Grant Nos. P20 RR016454, P30 GM103324-01 and P20 GM103408. Additional thanks to Robert Burgess and Deborah Stenkamp for helpful suggestions and to Jingxing Ou for assistance imaging ground squirrel retina and Kevin Campbell and Rolf Turk for providing dystroglycan mutant retinas. We would also like to thank Christine Davitt and the Franceschi Microscopy and Imaging core (Washington State University) for assistance with electron microscopy.
Footnotes
Conflict of Interest The authors have no conflict of interest with respect to this work.
Author Contributions All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: PGF. Acquisition of data: GBA, SSL, HF, WL and PGF. Analysis and interpretation of data: PGF and WL. Drafting of the manuscript: PGF. Critical revision of the manuscript for important intellectual content: PGF. Statistical analysis: PGF. Obtained funding: PGF. Administrative, technical, and material support: PGF and WL. Study supervision: PGF.
References
- Blank M, Fuerst PG, Stevens B, Nouri N, Kirkby L, Warrier D, Barres BA, Feller MB, Huberman AD, Burgess RW, Garner CC. The Down syndrome critical region regulates retinogeniculate refinement. J Neurosci. 2011;31(15):5764–5776. doi: 10.1523/JNEUROSCI.6015-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycott BB, Hopkins JM. Cone bipolar cells and cone synapses in the primate retina. Vis Neurosci. 1991;7(1–2):49–60. doi: 10.1017/s0952523800010932. [DOI] [PubMed] [Google Scholar]
- Burke MC, Letts PA, Krajewski SJ, Rance NE. Coexpression of dynorphin and neurokinin B immunoreactivity in the rat hypothalamus: Morphologic evidence of interrelated function within the arcuate nucleus. J Comp Neurol. 2006;498(5):712–726. doi: 10.1002/cne.21086. [DOI] [PubMed] [Google Scholar]
- Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, Roderick TH, Taylor BA, Hankin MH, McInnes RR. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12(4):376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Chen S, Li W. A color-coding amacrine cell may provide a blue-off signal in a mammalian retina. Nat Neurosci. 2012;15(7):954–956. doi: 10.1038/nn.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Chew KS, McNeill DS, Keeley PW, Ecker JL, Mao BQ, Pahlberg J, Kim B, Lee SC, Fox MA, Guido W, Wong KY, Sampath AP, Reese BE, Kuruvilla R, Hattar S. Apoptosis regulates ipRGC spacing necessary for rods and cones to drive circadian photoentrainment. Neuron. 2013;77(3):503–515. doi: 10.1016/j.neuron.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes E, Seeliger M, Michalakis S, Biel M, Humphries P, Haverkamp S. Morphological characterization of the retina of the CNGA3(−/−)Rho(−/−) mutant mouse lacking functional cones and rods. Invest Ophthalmol Vis Sci. 2004;45(6):2039–2048. doi: 10.1167/iovs.03-0741. [DOI] [PubMed] [Google Scholar]
- Cvetkovska V, Hibbert AD, Emran F, Chen BE. Overexpression of Down syndrome cell adhesion molecule impairs precise synaptic targeting. Nat Neurosci. 2013;16(6):677–682. doi: 10.1038/nn.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick O, tom Dieck S, Altrock WD, Ammermuller J, Weiler R, Garner CC, Gundelfinger ED, Brandstätter JH. The presynaptic active zone protein bassoon is essential for photoreceptor ribbon synapse formation in the retina. Neuron. 2003;37(5):775–786. doi: 10.1016/s0896-6273(03)00086-2. [DOI] [PubMed] [Google Scholar]
- Fox MA, Sanes JR. Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol. 2007;503(2):280–296. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Rounds RP, Erskine L, Burgess RW. Cell autonomy of DSCAM function in retinal development. Dev Biol. 2012;361(2):326–337. doi: 10.1016/j.ydbio.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron. 2009;64(4):484–497. doi: 10.1016/j.neuron.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Harris BS, Johnson KR, Burgess RW. A novel null allele of mouse DSCAM survives to adulthood on an inbred C3H background with reduced phenotypic variability. Genesis. 2010;48(10):578–584. doi: 10.1002/dvg.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW. Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature. 2008;451(7177):470–474. doi: 10.1038/nature06514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett AM, Tadenev AL, Burgess RW. DSCAMs: restoring balance to developmental forces. Front Mol Neurosci. 2012;5:86. doi: 10.3389/fnmol.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack I, Peichl L, Brandstätter JH. An alternative pathway for rod signals in the rodent retina: rod photoreceptors, cone bipolar cells, and the localization of glutamate receptors. Proc Natl Acad Sci U S A. 1999;96(24):14130–14135. doi: 10.1073/pnas.96.24.14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Ghosh KK, Hirano AA, Wässle H. Immunocytochemical description of five bipolar cell types of the mouse retina. J Comp Neurol. 2003;455(4):463–476. doi: 10.1002/cne.10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Grünert U, Wässle H. The cone pedicle, a complex synapse in the retina. Neuron. 2000;27(1):85–95. doi: 10.1016/s0896-6273(00)00011-8. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Specht D, Majumdar S, Zaidi NF, Brandstätter JH, Wasco W, Wässle H, Tom Dieck S. Type 4 OFF cone bipolar cells of the mouse retina express calsenilin and contact cones as well as rods. J Comp Neurol. 2008;507(1):1087–1101. doi: 10.1002/cne.21612. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wässle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424(1):1–23. [PubMed] [Google Scholar]
- Hopkins JM, Boycott BB. Synaptic contacts of a two-cone flat bipolar cell in a primate retina. Vis Neurosci. 1992;8(4):379–384. doi: 10.1017/s0952523800005125. [DOI] [PubMed] [Google Scholar]
- Hughes TE. Are there ionotropic glutamate receptors on the rod bipolar cell of the mouse retina? Vis Neurosci. 1997;14(1):103–109. doi: 10.1017/s0952523800008804. [DOI] [PubMed] [Google Scholar]
- Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483(7390):465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Reese BE. Morphology of dopaminergic amacrine cells in the mouse retina: independence from homotypic interactions. J Comp Neurol. 2010;518(8):1220–1231. doi: 10.1002/cne.22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Sliff BJ, Lee SC, Fuerst PG, Burgess RW, Eglen SJ, Reese BE. Neuronal clustering and fasciculation phenotype in Dscam- and Bax-deficient mouse retinas. J Comp Neurol. 2012;520(7):1349–1364. doi: 10.1002/cne.23033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khiripet N, Khantuwan W, Jungck JR. Ka-me: a Voronoi image analyzer. Bioinformatics. 2012;28(13):1802–1804. doi: 10.1093/bioinformatics/bts253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Wang X, Coolon R, Ye B. Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron. 2013;78(5):827–838. doi: 10.1016/j.neuron.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C, Obara T, Uriu Y, Numata T, Sanuki R, Miyata K, Koyasu T, Ueno S, Funabiki K, Tani A, Ueda H, Kondo M, Mori Y, Tachibana M, Furukawa T. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc Natl Acad Sci U S A. 2010;107(1):332–337. doi: 10.1073/pnas.0912730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270(5233):96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- Kuo G, Arnaud L, Kronstad-O’Brien P, Cooper JA. Absence of Fyn and Src causes a reeler-like phenotype. J Neurosci. 2005;25(37):8578–8586. doi: 10.1523/JNEUROSCI.1656-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Cowgill EJ, Al-Nabulsi A, Quinn EJ, Evans SM, Reese BE. Homotypic regulation of neuronal morphology and connectivity in the mouse retina. J Neurosci. 2011;31(40):14126–14133. doi: 10.1523/JNEUROSCI.2844-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, Wu JY. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc Natl Acad Sci U S A. 2009;106(8):2951–2956. doi: 10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier-Lavigne M, Stein E. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133(7):1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Navarrete GC, Angulo A, Martín-Nieto J, Cuenca N. Gradual morphogenesis of retinal neurons in the peripheral retinal margin of adult monkeys and humans. J Comp Neurol. 2008;511(4):557–580. doi: 10.1002/cne.21860. [DOI] [PubMed] [Google Scholar]
- Mataruga A, Kremmer E, Müller F. Type 3a and type 3b OFF cone bipolar cells provide for the alternative rod pathway in the mouse retina. J Comp Neurol. 2007;502(6):1123–1137. doi: 10.1002/cne.21367. [DOI] [PubMed] [Google Scholar]
- Matsuoka RL, Nguyen-Ba-Charvet KT, Parray A, Badea TC, Chedotal A, Kolodkin AL. Transmembrane semaphorin signalling controls laminar stratification in the mammalian retina. Nature. 2011;470(7333):259–263. doi: 10.1038/nature09675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell. 2007;129(3):593–604. doi: 10.1016/j.cell.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Milam AH, Dacey DM, Dizhoor AM. Recoverin immunoreactivity in mammalian cone bipolar cells. Vis Neurosci. 1993;10(1):1–12. doi: 10.1017/s0952523800003175. [DOI] [PubMed] [Google Scholar]
- Millard SS, Lu Z, Zipursky SL, Meinertzhagen IA. Drosophila dscam proteins regulate postsynaptic specificity at multiple-contact synapses. Neuron. 2010;67(5):761–768. doi: 10.1016/j.neuron.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosinger Ogilvie J, Deckwerth TL, Knudson CM, Korsmeyer SJ. Suppression of developmental retinal cell death but not of photoreceptor degeneration in Bax-deficient mice. Invest Ophthalmol Vis Sci. 1998;39(9):1713–1720. [PubMed] [Google Scholar]
- Müller F, Scholten A, Ivanova E, Haverkamp S, Kremmer E, Kaupp UB. HCN channels are expressed differentially in retinal bipolar cells and concentrated at synaptic terminals. Eur J Neurosci. 2003;17(10):2084–2096. doi: 10.1046/j.1460-9568.2003.02634.x. [DOI] [PubMed] [Google Scholar]
- Nasonkin IO, Lazo K, Hambright D, Brooks M, Fariss R, Swaroop A. Distinct nuclear localization patterns of DNA methyltransferases in developing and mature mammalian retina. J Comp Neurol. 2011;519(10):1914–1930. doi: 10.1002/cne.22613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. 2004;36(3):240–246. doi: 10.1038/ng1299. [DOI] [PubMed] [Google Scholar]
- Pérez De Sevilla Muller L, Shelley J, Weiler R. Displaced amacrine cells of the mouse retina. J Comp Neurol. 2007;505(2):177–189. doi: 10.1002/cne.21487. [DOI] [PubMed] [Google Scholar]
- Puller C, Haverkamp S. Cell-type-specific localization of protocadherin beta16 at AMPA and AMPA/Kainate receptor-containing synapses in the primate retina. J Comp Neurol. 2011;519(3):467–479. doi: 10.1002/cne.22528. [DOI] [PubMed] [Google Scholar]
- Puller C, Haverkamp S, Grünert U. OFF midget bipolar cells in the retina of the marmoset, Callithrix jacchus, express AMPA receptors. J Comp Neurol. 2007;502(3):442–454. doi: 10.1002/cne.21315. [DOI] [PubMed] [Google Scholar]
- Puller C, Ivanova E, Euler T, Haverkamp S, Schubert T. OFF bipolar cells express distinct types of dendritic glutamate receptors in the mouse retina. Neuroscience. 2013;243:136–148. doi: 10.1016/j.neuroscience.2013.03.054. [DOI] [PubMed] [Google Scholar]
- Reese BE. Development of the retina and optic pathway. Vision Res. 2011;51(7):613–632. doi: 10.1016/j.visres.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW. The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis Neurosci. 1991;6(2):95–111. doi: 10.1017/s095252380001049x. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. 2009;23(2):147–156. doi: 10.1101/gad.1752909. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101(6):671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Schramm RD, Li S, Harris BS, Rounds RP, Burgess RW, Ytreberg FM, Fuerst PG. A Novel Mouse Dscam Mutation Inhibits Localization and Shedding of DSCAM. PLoS One. 2012;7(12):e52652. doi: 10.1371/journal.pone.0052652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto F, Watkins KL, Johnson RE, Schottler F, Kerschensteiner D. NGL-2 Regulates Pathway-Specific Neurite Growth and Lamination, Synapse Formation, and Signal Transmission in the Retina. J Neurosci. 2013;33(29):11949–11959. doi: 10.1523/JNEUROSCI.1521-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zugates CT, Liang IH, Lee CH, Lee T. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron. 2002;33(4):559–571. doi: 10.1016/s0896-6273(02)00570-6. [DOI] [PubMed] [Google Scholar]
- Wässle H, Puller C, Müller F, Haverkamp S. Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J Neurosci. 2009;29(1):106–117. doi: 10.1523/JNEUROSCI.4442-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wässle H, Riemann HJ. The mosaic of nerve cells in the mammalian retina. Proc R Soc Lond B Biol Sci. 1978;200(1141):441–461. doi: 10.1098/rspb.1978.0026. [DOI] [PubMed] [Google Scholar]
- Wiechmann AF. Recoverin in cultured human retinoblastoma cells: enhanced expression during morphological differentiation. J Neurochem. 1996;67(1):105–110. doi: 10.1046/j.1471-4159.1996.67010105.x. [DOI] [PubMed] [Google Scholar]
- Wikler KC, Stull DL, Reese BE, Johnson PT, Bogenmann E. Localization of protein kinase C to UV-sensitive photoreceptors in the mouse retina. Vis Neurosci. 1998;15(1):87–95. doi: 10.1017/s0952523898151155. [DOI] [PubMed] [Google Scholar]
- Williams PR, Suzuki SC, Yoshimatsu T, Lawrence OT, Waldron SJ, Parsons MJ, Nonet ML, Wong RO. In vivo development of outer retinal synapses in the absence of glial contact. J Neurosci. 2010;30(36):11951–11961. doi: 10.1523/JNEUROSCI.3391-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451(7177):465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]