Abstract
The yeast Saccharomyces cerevisiae can synthesize trehalose and also use this disaccharide as a carbon source for growth. However, the molecular mechanism by which extracellular trehalose can be transported to the vacuole and degraded by the acid trehalase Ath1p is not clear. By using an adaptation of the assay of invertase on whole cells with NaF, we showed that more than 90% of the activity of Ath1p is extracellular, splitting of the disaccharide into glucose. We also found that Agt1p-mediated trehalose transport and the hydrolysis of the disaccharide by the cytosolic neutral trehalase Nth1p are coupled and represent a second, independent pathway, although there are several constraints on this alternative route. First, the AGT1/MAL11 gene is controlled by the MAL system, and Agt1p was active in neither non-maltose-fermenting nor maltose-inducible strains. Second, Agt1p rapidly lost activity during growth on trehalose, by a mechanism similar to the sugar-induced inactivation of the maltose permease. Finally, both pathways are highly pH sensitive and effective growth on trehalose occurred only when the medium was buffered at around pH 5.0. The catabolism of trehalose was purely oxidative, and since levels of Ath1p limit the glucose flux in the cells, batch cultures on trehalose may provide a useful alternative to glucose-limited chemostat cultures for investigation of metabolic responses in yeast.
Trehalose [α-d-glucopyranosyl-(1-1)-α-d-glucopyranoside] is a nonreducing disaccharide of glucose discovered in 1832 by Wiggers in a mushroom crop and later in many other fungi, plants, and insects (13). In the yeast Saccharomyces cerevisiae, trehalose can accumulate up to 15% of the cell dry mass, depending on the growth conditions, the stage of the life cycle, or environmental stress (2, 27), and for this reason it has been considered a storage carbohydrate (15, 27). Moreover, the ability of this disaccharide to protect proteins and biological membranes against adverse conditions suggests that it also plays a stress-protectant role in this yeast (49, 50) and probably in other organisms that produce it (14).
Intracellular levels of trehalose result from a well-controlled balance between enzymatic synthesis and degradation. In the yeast S. cerevisiae, the synthesis of trehalose is catalyzed by a UDP-glucose-dependent trehalose synthase (TPS) protein complex encoded by four genes. TPS1 and TPS2 encode trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase, respectively, while TPS3 and TSL1 code for two regulatory subunits of the TPS complex (for a review, see reference 15). Hydrolysis of trehalose into glucose can be carried out by at least two enzymes: a cytosolic or neutral trehalase encoded by NTH1 (26) and a vacuolar or acid trehalase (25, 28) encoded by ATH1 (12). The yeast genome also harbors the NTH2 gene, whose product is 77% identical to Nth1p, but until now, no trehalase activity has been associated with this protein (37). The neutral trehalase Nth1p is responsible for the intracellular mobilization and/or recycling of trehalose (37, 38, 41). Lack of Ath1p does not alter intracellular levels of trehalose, probably because the vacuolar localization of this enzyme prevents its action on cytosolic trehalose stores (38).
Trehalose also is a potential carbon source for many yeast species (3). Nwaka et al. (39) hypothesized that acid trehalase is essential for trehalose assimilation since a mutant strain defective in this enzyme could not grow on this sugar. In addition, biochemical and genetic data were consistent with the existence of at least two trehalose transporters in S. cerevisiae: the high-affinity H+-trehalose symporter Agt1p and a low-affinity transporter system whose corresponding gene remains to be characterized (19, 42, 51-54). While Nwaka and coworkers (36) suggested that trehalose may reach the vacuole by simple endocytosis, Malluta et al. (30) proposed that the disaccharide could be taken up by Agt1p. However, to be effective, this model implies the protection of internalized trehalose against neutral trehalase and the existence of two additional transporter activities by which trehalose enters the vacuole to be degraded by Ath1p and the subsequent exit of glucose from this compartment.
The acid trehalase is extracellular in many other fungi (9, 10, 24, 29) and in prokaryotes (6). Ath1p may be localized in vacuoles in S. cerevisiae (1, 12, 25). We hypothesized that the S. cerevisiae acid trehalase also is extracellular, which would give a simple alternative explanation for the mechanism of trehalose assimilation. The occurrence of trehalose transporters (30, 42) prompted us to reconsider the cellular destination of trehalose molecules that are transported across the plasma membrane. In this report, we show that Ath1p is mainly localized in the periplasmic space and demonstrate that Ath1p and Agt1p belong to two independent systems for trehalose assimilation.
MATERIALS AND METHODS
Media and culture conditions.
Yeast cells were routinely cultured in shake flasks (1-liter Erlenmeyer flasks with a working volume of 200 ml) at 30°C in YP rich medium (yeast extract at 10 g liter−1, Bacto Peptone at 20 g liter−1) or YN synthetic medium (yeast nitrogen base without amino acids and ammonium at 1.7 g liter−1, supplemented with ammonium sulfate [Difco Laboratories, Detroit, Mich.] at 5 g liter−1). Since succinate is not used as a carbon source, the pH of the growth medium could be adjusted to 6.7 (neutral) or 4.8 (acid) by varying the NaOH/succinate ratio. Unless stated otherwise, YN medium was buffered at pH 4.8 by the addition of 14.3 g of succinic acid liter−1 and 6 g of NaOH liter−1 and contained the carbon source at a 2% (wt/vol) concentration. Growth was monitored by measuring the A600 with an Easyspec IV spectrophotometer (Safas, Monaco, France). Optical density at 600 nm (OD600) could be converted to cell dry mass by using a calibration curve established for the CEN.PK113-7D strain. Under this condition, 1 U of OD600 corresponds to 0.41 g of dry cells liter−1. The maximal specific growth rate (μmax) was calculated as previously described (40). Briefly, growth curves were fitted by exponential regression analysis (Excel software). Fitting curves (solid lines in Fig. 1 and 2) were drawn over the experimental points that were used for their calculation. These points were selected so that they led to a correlation coefficient (r2) of >0.998. The μ constant from the equation OD = b exp(μ t) is the maximal specific growth rate.
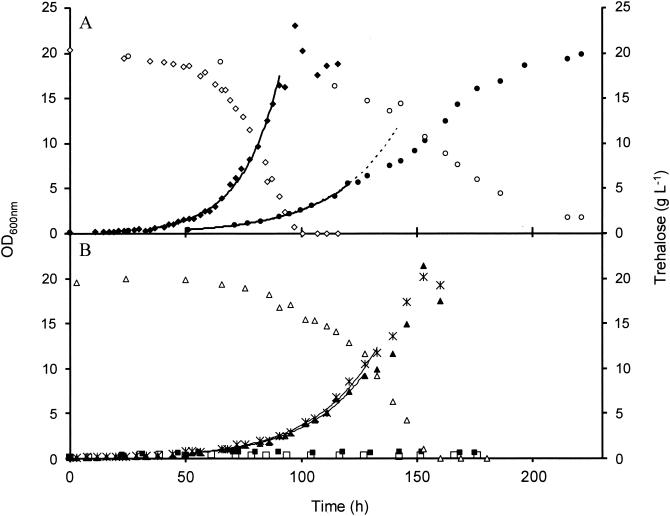
FIG. 1.
Growth curves of CEN.PK113-7D and mutant strains on trehalose medium at pH 4.8. Cultures were carried out in 2-liter bioreactors as described in Materials and Methods. The culture was started with 0.08 mg (equivalent dry mass) of cells from an overnight preculture in YN glucose medium. (A) Wild-type and ath1 mutant strains; (B) nth1, agt1, ath1 nth1, and ath1 agt1 mutant strains. Dry mass: wild type (♦), ath1 (•), nth1 (*), agt1(▴), ath1 nth1 (▪), and ath1 agt1(□). Trehalose: wild type (⋄), ath1 (○), and agt1 (▵). The μmax values calculated from fitting curves (solid lines) were 0.07 h−1 (r2 = 0.990) for the wild type, 0.035 h−1 (r2 = 0.994) for the ath1 mutant, 0.037 h−1 (r2 = 0.987) for the nth1 mutant, and 0.040 h−1 (r2 = 0.991) for the agt1 mutant. The dashed line is the theoretical projection of the exponential fitting curve from the ath1 mutant strain.
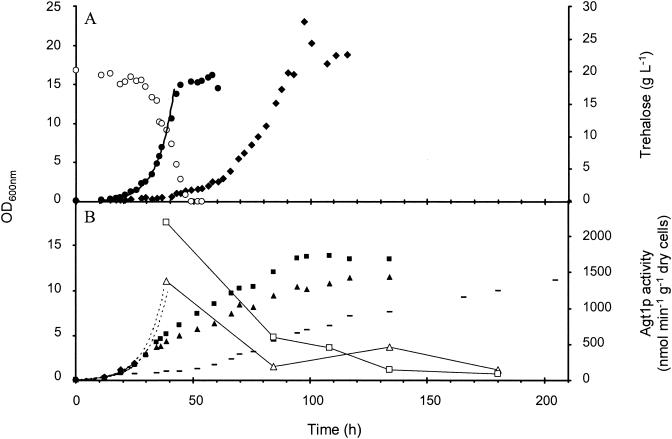
FIG. 2.
Growth curves of CEN.PK113-7D and mutant strains that overexpress ATH1, AGT1, and NTH1 on trehalose medium at pH 4.8. Procedures were as described in the legend to Fig. 1. (A) Wild type (dry mass, ♦) and CENPK113-5D transformed with pGRSd-ATH1 (dry mass, •; trehalose, ○); (B) ath1 (dry mass, -), ath1 ura mutant strain transformed with pGRSd-AGT1 (dry mass, ▴; transport activity, ▵) and ath1 ura his mutant strain transformed with both pGRSd-AGT1 and pGRSd-NTH1 (dry mass, ▪; transport activity, □). The μmax values calculated from exponential fitting curves were 0.07 h−1 (r2 = 0.990) for the wild type, 0.14 h−1 (r2 = 0.994) for the ura/pGRSd-ATH1 strain, 0.13 h−1 (r2 = 0.994) for the ath1 ura/pGRSd-AGT1 strain, and 0.13 h−1 (r2 = 0.998) for the ath1 ura his/pGRSd-AGT1/pGRSd-NTH1 strain. Dashed lines are theoretical projections of exponential fitting curves from ath1 mutant strains transformed with pGRSd-AGT1 and pGRSd-AGT1 plus pGRSd-NTH1 (B).
Batch cultures were performed in 2-liter bioreactors (Setric Genie Industriel, Toulouse, France) with an initial working volume of 1.5 liters of YN medium containing trehalose at 2% (wt/vol) and buffered at pH 4.8 with orthophosphoric acid. The temperature was kept constant at 30°C, and the pH of the medium was maintained at 4.8 by addition of 2 M NaOH. Growth was monitored independently by gas analysis, OD600, and cell dry mass. The dissolved-oxygen concentration was kept above 20% of air saturation in the liquid phase by using a dry-air flow of 10 liters h−1 and variable agitation. Online estimation of the O2, CO2, and N2 molar fractions of inlet and exhaust gases was performed by mass spectrometry (PRIMA 600S; VG gas, Manchester, United Kingdom) with a relative accuracy of 0.1%. Rates of gas consumption and production (rO2 and rCO2, respectively, in millimoles per hour) were used for calculation of the respiratory quotient (RQ [RQ = rCO2/rO2]).
Plasmid construction.
A 4.41-kb fragment bearing ATH1 was amplified by PCR with genomic DNA from wild-type strain CEN.PK113-7D as the template and primers ATH1−245 (5′-CGTATCACGACAAACCAACAGCC-3′) and ATH1+4162 (5′-CAAACCCTACTGACGAGAGAAG-3′). The PCR product was cloned into the pGEM-T Easy vector (Promega Corporation), and the 1.59-kb PvuII-EcoRV kanMX4 cassette from vector pUG6 (18) was inserted in place of the 2.44-kb EcoRV fragment of ATH1 to yield pATH1::KanMX4. Similarly, a 2.39-kb fragment of NTH2 was amplified by PCR from genomic DNA with the primers NTH2over-dir (5′-CGGGATCCATGTTCGTCAATATTATAGGTA-3′) and NTH2over-rev (5′-CGGGATCCAGACCTGCATATACTCAAAT-3′) (BamHI restriction sites underlined, start codon in bold). After cloning of the PCR product into the pGEM-T Easy vector (pNTH2 construct), the 1.65-kb BamHI LEU2 fragment from plasmid YDp-L (4) was inserted into the BglII site of pNTH2 to yield pNTH2::LEU2.
Construction of the expression vector (pGRSd) was carried out by inserting the 2.02-kb HindIII promoter-terminator cassette from expression vector pG3 (46) into the HindIII site of shuttle vector pRS316 (47), followed by further deletion of a 0.47-kb XbaI fragment that removed an extra sequence of this GPD promoter-PGK terminator cassette (the so-called GPD promoter is the promoter of the TDH3 gene, which encodes glyceraldehyde-3-phosphate dehydrogenase). Plasmids that overexpress ATH1, AGT1, NTH1, or NTH2 were constructed as follows. The ATH1 open reading frame (ORF) was amplified from CEN.PK113-7D genomic DNA with primers ATH1over-dir (5′-CGGGATCCATGAAAAGAATAAGATCGCTT-3′) and ATH1over-rev (5′-CGGGATCCTTAATCATTGAGAACAATTTC-3′). This 3.65-kb PCR product was cut with BamHI and cloned into the BamHI site of pGRSd (immediately downstream of the GPD promoter) to yield pGRSd-ATH1. Similarly, the AGT1 ORF (1.87-kb fragment), the NTH1 ORF (2.26-kb fragment), and the NTH2 ORF (2.38-kb fragment) were cloned into pGRSd to yield, respectively, pGRSd-AGT1 (primers AGT1over-dir [5′-CGGGATCCATGAAAAATATCATTTCATTGGT-3′] and AGT1over-rev [5′-CGGGATCCTTAACATTTATCAGCTGCATT-3′]), pGRSd-NTH1 (primers NTH1over-dir [5′-CGGGATCCATGAGTCAAGTTAATACAAGC-3′] and NTH1over-rev [5′-CGGGATCCTATAGTCCATAGAGGTTTC-3′]), and pGRSd-NTH2 (primers NTH2over-dir and NTH2over-rev [see above]). All PCR amplifications were carried out with the DyNAzyme EXT polymerase (Finnzymes, Espoo, Finland) in accordance with the standard procedure described in the DyNAzyme EXT instructions.
Strain construction.
The strains used in this study were CEN.PK113-7D (MATa MAL2-8c SUC2), a prototrophic MAL-constitutive strain from P. Kötter (Frankfurt, Germany) (56) and its auxotrophic derivatives (Table 1); BY4741 from the EUROSCARF collection (7), a maltose-inducible strain; and JF657, a prototrophic maltose-nonfermenting (mal0) strain from our collection. Construction of the agt1 mutant was carried out by homologous recombination with a PCR product that was obtained as follows. The 5′ ends of primers were complementary to 40 bp immediately downstream of the ATG and immediately upstream of the stop codon of AGT1, respectively. These primers amplified the lacZ reporter-kanMX4 marker cassette from plasmid pUG6loxlacZ. The ATH1, NTH1, and NTH2 genes also were disrupted by homologous recombination by using, respectively, the 3.57-kb SacII-SpeI fragment from pATH1::KanMX4 (this study), the 3.36-kb AflII-PvuII fragment from pTZ18RNTH1 (26), and the 3.98-kb BamHI fragment from pNTH2::LEU2 (this study). Correct replacement of the genes in the mutant strains was analyzed by PCR (58). ath1 agt1, ath1 nth1, and nth1 agt1 double-mutant cells were obtained by crosses and sporulation between the corresponding haploid strains, while construction of the nth1 nth2 mutant required successive disruption of NTH1 and NTH2 because of the centromeric linkage of the two genes.
TABLE 1.
Strains used in this work
| Background | Name | Genotype | Source |
|---|---|---|---|
| CEN.PK | CEN.PK113-7D | aMAL2-8c SUC2 | P. Kötter |
| CEN.PK | CEN.PK113-1A | α MAL2-8c SUC2 | P. Kötter |
| CEN.PK | CEN.PK113-5D | aMAL2-8c SUC2 ura3-52 | P. Kötter |
| CEN.PK | ath1 | aMAL2-8c SUC2 ath1Δ::kanMX4 | This study |
| CEN.PK | ath1 ura | aMAL2-8c SUC2 ura3-52 ath1Δ::kanMX4 | This study |
| CEN.PK | ath1 ura his | aMAL2-8c SUC2 ura3-52 his3Δ1 ath1Δ::kanMX4 | This study |
| CEN.PK | nth1 | aMAL2-8c SUC2 nth1Δ::kanMX4 | This study |
| CEN.PK | agt1 | aMAL2-8c SUC2 agt1Δ::lacZ loxP-kanMX4-loxP | This study |
| CEN.PK | nth2 | aMAL2-8c SUC2 leu2 nth2Δ::LEU2 | This study |
| CEN.PK | agt1 nth1 | aMAL2-8c SUC2 nth1Δ::kanMX4 agt1Δ::lacZ | This study |
| CEN.PK | ath1 nth1 | aMAL2-8c SUC2 ath1Δ::kanMX4 nth1Δ::kanMX4 | This study |
| CEN.PK | ath1 agt1 | aMAL2-8c SUC2 ath1Δ::kanMX4 agt1Δ::lacZ | This study |
| CEN.PK | ath1 nth2 | aMAL2-8c SUC2 leu2 ath1Δ::kanMX4 | This study |
| S288C | BY4741 | amet15Δ0 his3Δ1 leu2Δ0 ura3Δ0 | EUROSCARF |
| S288C | BY ath1 | amet15Δ0 his3Δ1 leu2Δ0 ura3Δ0 ath1Δ::kanMX4 | EUROSCARF |
| KT1112 | JF657 | aura3-52 URA3::GSY2-lacZ | J. L. Parrou |
| KT1112 | ath1 | aura3-52 URA3::GSY2-lacZ ath1Δ::kanMX4 | This study |
Preparation of extracts and enzyme assays.
Yeast cells (30-mg equivalent dry mass) were harvested by centrifugation (3,000 × g, 5 min, 4°C) and resuspended in 0.5 ml of extraction buffer (20 mM HEPES [pH 7.1], 1 mM EDTA, 100 mM KCl, completed just before use with 1 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluoride). The cell suspension was vigorously disrupted by vortexing in the presence of 1 g of glass beads for four 30-s periods at 4°C (41). After centrifugation (16,000 × g, 10 min, 4°C), the supernatant was used as a crude extract for enzyme assays.
Acid trehalase activity was measured at 30°C in a total volume of 0.5 ml containing 315 mM sodium citrate (pH 4.5), 1.4 mM EDTA, and 55 mM trehalose. The reaction was initiated by addition of 100 μl of crude extract. At different times, a 0.1-ml sample was withdrawn from the reaction mixture, mixed with 0.1 ml of hot (80°C) water, and inactivated by incubation for 5 min in a water bath at 80°C. The glucose liberated was measured by a glucose oxidase assay (kit 540; Sigma-Aldrich). Commercial trehalase from pig kidneys (T-8778; Sigma-Aldrich) was assayed as described above.
Acid trehalase in the medium was measured by adding 50 μl of a 20% (wt/vol) trehalose solution to 450 μl of broth medium that was filtered through a nylon filter (0.2-μm pore size; Sartorius AG) prior to use. The reaction was stopped as described above. On intact yeast cells, acid trehalase activity was determined by a procedure described for invertase activity measurement (48). The cell suspension (about 16-mg equivalent dry mass) was harvested by centrifugation (3,000 × g, 5 min), washed with cold water, and resuspended in 0.84 ml of 50 mM Na-citrate solution (pH 4.75) containing 50 mM NaF. After incubation at 30°C for 30 min, the reaction was initiated by addition of 160 μl of 580 mM trehalose, stopped at different times, and processed as described above. To estimate acid trehalase activity in the cell wall fraction, yeast cells (500-mg equivalent dry mass) were harvested by centrifugation (3,000 × g, 5 min, 4°C), resuspended in 12 ml of extraction buffer containing 10 g of glass beads, and vigorously shaken as described above for crude-extract preparation. Cell wall fractions were prepared as described by Dallies et al. (8). Acid trehalase activity was assayed by incubating 450 μl of cell wall preparation with 50 μl of a trehalose solution at 20% (wt/vol), and the reaction was stopped at different times as described above.
The neutral trehalase was assayed as previously described (35) in a total volume of 0.5 ml containing 50 mM HEPES (pH 7.1), 2.5 mM CaCl2 and 50 mM trehalose. The reaction was started with 20 μl of crude extract that was incubated for 15 min in the presence of 1 mM Mg-ATP and 0.1 mM cyclic AMP. The reaction was monitored as described for acid trehalase.
Invertase activity was assayed either in intact cells or in the cell wall fraction by using the same procedure and reaction buffer as those described for acid trehalase activity, except that 55 mM trehalose was replaced with 55 mM sucrose. One unit is defined as the amount of enzyme that catalyzes the hydrolysis of 1 μmol of trehalose or sucrose per min under the conditions of the assay.
Agt1-mediated transport of trehalose was determined by the amount of p-nitrophenol resulting from the uptake of p-nitrophenyl-α-d-glucopyranoside coupled to the hydrolysis of this substrate by the endogenous α-glucosidase (52). This assay is only functional in strains that constitutively express MAL2 genes encoding α-glucosidase, which is present in large excess over the uptake activity of the disaccharide. This assay could be applied to our CEN.PK background. The transport activity is expressed as nanomoles of p-nitrophenol produced per minute per gram of cell dry mass.
Blockage of glucose metabolism in yeast was carried out by incubating intact cells with 1.78 mM glucose in 40 mM citrate buffer (pH 4.75) in the presence of 50 mM NaF.
Analytical procedures.
Extracellular trehalose, glucose, acetic acid, ethanol, and other by-products were measured by high-performance liquid chromatography with an Aminex HPX-87H column (Bio-Rad Laboratories). The column was eluted at 48°C with 5 mM H2SO4 with a flow rate of 0.5 ml min−1. Concentrations of these compounds were determined by using a Waters 410 refractive-index detector.
RESULTS
Growth of S. cerevisiae on trehalose.
Since Ath1p is required for growth of S. cerevisiae on trehalose (39) and because the in vitro activity of this protein is optimal at pH 5.0 (5, 28, 34), we tested whether a medium buffered close to this pH may improve growth, as a first indication of the extracellular localization of the so-called vacuolar trehalase. The growth of the wild-type CEN.PK113-7D strain was assayed in synthetic or rich medium, buffered or not, with or without trehalose as the sole carbon source (Table 2 and Fig. 1A). In a synthetic medium buffered at pH 4.8, the culture of the wild-type strain exhibited a perfect exponential growth phase with a high biomass yield (YX/S = 0.45). Trehalose depletion coincided with growth arrest, and its consumption was accompanied by the production of neither ethanol nor other fermentative by-products. Moreover, the respiratory quotient was close to 1, indicating that the metabolism of trehalose was purely oxidative. This result also was consistent with complete growth inhibition on trehalose in the presence of antimycin (data not shown). In pH 6.7 medium, cell growth was severely reduced and cells only reached an OD600 of 1.8 with no significant trehalose consumption (Table 2). In nonbuffered synthetic medium, growth was initiated rapidly and arrested very early, probably because of a growth-dependent drop in pH from 5 to 3. On rich medium containing 2% (wt/vol) trehalose, whether buffered at pH 6.7 or not, the growth of the wild-type cells was extremely slow (μ = <0.03 h−1) and they only reached an OD600 of ∼5 after more than 200 h (Table 2). The OD600 under this condition was 10 times higher than in the same medium without trehalose, suggesting that the disaccharide may provide some carbon molecules that can be combined with consumption of the amino acids present in this medium to sustain this high growth rate. The cell growth rate was considerably improved when the medium was buffered at pH 4.8, with a μmax of 0.15 h−1and an OD600 of >20 (Fig. 1). Taken together, these results indicate that in contrast to yeast cultures on glucose, for which the growth is barely affected in the pH range of 3 to 7 (data not shown), the medium pH must be buffered to around 5.0 for efficient trehalose assimilation by S. cerevisiae.
TABLE 2.
Influence of the pH of trehalose medium on the growth of S. cerevisiaea
| Mediumc and trehalose concn (g liter−1) | Bufferb | pH
|
Lag phase (h) | Growth rate (h−1) | Final OD600 | Final trehalose concn (g liter−1) | |
|---|---|---|---|---|---|---|---|
| Initial | Final | ||||||
| YN | |||||||
| 0 | − | 5.1 | 5.1 | —d | —d | 0.5e | 0 |
| 20 | − | 5.1 | 3 | 40 | 0.04 | 6e | 16.8 |
| 20 | + | 6.7 | 6.7 | >50 | —d | 1.8e | 20 |
| 20 | + | 4.8 | 4.8 | 40 | 0.07 | 20f | 0 |
| YP | |||||||
| 0 | − | 6.9 | 6.9 | —d | —d | 0.6e | 0 |
| 20 | − | 6.9 | 6.9 | 20-30 | 0.03 | 5e | 17.5 |
| 20 | + | 6.7 | 6.7 | 20-30 | 0.03 | 5e | 17.5 |
| 20 | + | 4.8 | 4.8 | 20 | 0.14/0.16 | 16f | 0 |
Growth was monitored until the stationary phase or arbitrarily stopped after 200 h when it was severely impaired. The values are the mean of five independent experiments with a standard deviation of less than 5%.
Presence (+) or absence (−) of Na-succinate buffer setting the pH at 6.7 or 4.8.
Synthetic medium (YN) and rich medium (YP) as described in Materials and Methods.
—, Lag phase and growth rate could not be accurately estimated.
Value measured at 200 h.
Value measured at substrate depletion.
Extracellular activity of the acid trehalase encoded by ATH1.
We looked for acid trehalase in the growth medium from wild-type cells cultivated on trehalose and from stationary-phase cells on glucose. In both cases, the extracellular activity was <5% of the total activity measured in the crude extracts. This low activity was not due to cell lysis but was clearly attributed to the ATH1 gene product since no extracellular activity was detected in ath1 mutant cells.
In order to examine whether Ath1p is secreted while being retained at the cell surface, we adapted the assay of invertase on whole yeast cells (48). This assay uses NaF, which is an inhibitor of enolase and indirectly prevents the uptake of glucose that is produced from sucrose by the extracellular invertase. As a control, we incubated intact cells with a glucose solution and found that the rapid consumption of this sugar (rate of about 8 nmol of glucose min−1 mg of dry mass−1) was totally prevented if the cells were preincubated with NaF (data not shown). Because neither acid trehalase from a yeast crude extract (Table 3) nor commercial acid trehalase from pig kidneys (data not shown) was sensitive to NaF, we applied this procedure to intact yeast cells incubated with trehalose and measured an extracellular activity of 1.53 mU/mg of dry mass (Table 3). This activity represented more than 90% of the total Ath1p activity measured in crude extracts but was undetectable without NaF (Table 3). This activity did not result from cell lysis since it was recovered in the pellet obtained after centrifugation of the NaF-treated cells, and no glycolytic enzymes such as neutral trehalase were detected in the supernatant of the treated cells (data not shown). Approximately 15% of extracellular Ath1p activity was measured in the cell wall fraction, while, as expected, more than two-thirds of extracellular invertase activity was found associated with the cell wall. Taken together, these results suggest that the acid trehalase could be secreted in the periplasmic space of the cells, as either a free or a membrane-bound protein.
TABLE 3.
Assay of acid trehalase, neutral trehalase, and invertase in different cellular fractionsa
| Fraction | Acid trehalase
|
Neutral trehalase
|
Invertaseb | ||
|---|---|---|---|---|---|
| +b | −c | +b | −c | ||
| Crude extract | 1.60 | 1.80 | 0.95 | 3.80 | 280 |
| Whole cells | 1.50 | 0 | 0 | 0 | 115 |
| Cell wall | 0.25 | 75 | |||
Enzyme activities are expressed in milliunits per milligram of dry cells. Each value is the mean of five independent experiments with a standard deviation of less than 5%.
With NaF at 50 mM.
No NaF.
Trehalose assimilation by the independent Ath1p and Agt1p-Nth1p systems.
The growth of ATH1-defective cells in a trehalose medium buffered at pH 4.8 started after a long lag phase similar to that observed in isogenic wild-type cells (Fig. 1A), with a μmax of 0.035 h−1. Unlike the wild type, the ath1 mutant could not maintain exponential cell growth and it entered an apparent linear phase even though less than 10% of the initial trehalose had been consumed (Fig. 1A). Moreover, growth of ath1 cells slowed significantly when the residual sugar concentration was <2 g liter−1 (approximately 6 mM; Fig. 1A) and complete consumption of the disaccharide took >300 h (data not shown).
Deletion of AGT1 reduced the growth rate from 0.070 to 0.040 h−1 (Fig. 1B). When AGT1 was disrupted in the ath1 mutant strain, the ath1 agt1 double-mutant strain could not grow on trehalose (Fig. 1B). Thus, the low-affinity transporter (53) plays no role in the growth of yeast on trehalose.
That an ath1 mutant still grew on trehalose suggested that the disaccharide transported by Agt1p must be hydrolyzed by neutral trehalase. The growth properties of the nth1 mutant were similar to those of the agt1 mutant (μmax of 0.037 and 0.040 h−1, respectively; Fig. 1B), but the ath1 nth1 mutant did not grow on trehalose as the sole carbon source (Fig. 1B), as did the agt1 ath1 mutant. This result also indicates that the second putative neutral trehalase encoded by NTH2 has no role in this process.
Trehalose metabolism when ATH1 and AGT1 are overexpressed.
The growth of yeast with any genetic background (CEN.PK, BY4741, or JF657) on trehalose was extremely slow, and the assimilation of this disaccharide was purely oxidative. Constitutive overexpression of ATH1 under the control of a strong TDH3 promoter increased acid trehalase activity 15- to 25-fold in crude extracts and 4-fold in intact cells. Thus, the proportion of periplasmic activity over total acid trehalase activity was reduced from 95% in wild-type cells to approximately 20% in cells overexpressing ATH1. The growth of ATH1-overexpressing cells was improved, as shown by a threefold reduction of time in lag phase and by an increase in μmax from 0.07 h−1 in the wild type to 0.14 h−1 (Fig. 2A). However, the metabolism of trehalose in the transformed cells remained purely oxidative (RQ ≈ 1).
Effects of increasing Agt1p-dependent trehalose assimilation were assessed in mutant cells defective in acid trehalase. ath1 mutant cells transformed with pGRSd-AGT1 (Fig. 2B) also had a reduced lag time and an increased growth rate (0.13 h−1), yet trehalose assimilation remained oxidative. However, these transformed cells rapidly lost their exponential growth (Fig. 2B). Finally, overexpression of NTH1 in the wild-type strain, the ath1 mutant strain, or the ath1 mutant strain transformed with pGRSd-AGT1 did not improve growth on trehalose, which in all cases remained purely oxidative.
Agt1p transporter activity during growth on trehalose.
Cells transformed with pGRSd-AGT1 contained 10 to 15 times higher transport activity than did wild-type cells, as measured by the p-nitrophenyl-α-d-glucopyranoside method (51) (Table 4). This very high activity was recorded at the beginning of growth and decreased steadily to levels found in wild-type cells at the end of growth on trehalose (Fig. 2B and Table 4). Constitutive overexpression of AGT1 in glucose-grown cells also led to a marked drop in permease activity, indicating that this event was not trehalose specific. In either wild-type or ath1 mutant cells cultivated on trehalose, a lower permease activity was also measured at the end of growth. This loss of Agt1p activity could explain the inability of ath1 null mutant cells to sustain exponential growth on trehalose; the rate of assimilation of the disaccharide is probably limited by the amount of Agt1p.
TABLE 4.
Agt1p transporter activity during growth on different carbon sourcesa
| Growth phaseb | Agt1p activity (nmol of p-nitrophenol min−1 g of dry biomass−1)
|
||||||
|---|---|---|---|---|---|---|---|
| YN trehalose
|
YN glucose
|
YN maltose (wild type) | |||||
| Wild type | ath1 | ath1 (pGRSd-AGT1) | Wild type | ath1 | pGRSd-AGT1 | ||
| Early exponential | 160 | 190 | 3,400 | 0 | 0 | 1,300 | 5,400 |
| Late exponential | 200 | 210 | 850 | 290 | 250 | 1,300 | 7,500 |
| Transition | 180 | 140 | 380 | 1400 | 1,700 | 310 | 120 |
| Stationary | 50 | 110 | 50 | 120 | 80 | 140 | |
Each condition, i.e., culture of one strain in a specific growth medium, was repeated at least three times. Values reported are from a representative culture.
Early exponential, period of maximal growth rate; late exponential, period of decreasing growth rate; transition, time when the carbon source disappeared; stationary, phase after carbon source depletion.
Loss of Agt1p activity also occurred in wild-type cells cultivated on maltose. The high transport activity that is consistent with maltose being an inducer of AGT1 expression (59) dropped after sugar depletion (Table 4). In cells grown on glucose, a repressor of MAL genes, Agt1p transporter activity increased just before glucose depletion, reached a maximum activity at the diauxic transition, and then rapidly decreased (Table 4).
DISCUSSION
The most important conclusion from this study is that trehalose assimilation can occur by two distinct pathways. One pathway is associated with acid trehalase, in agreement with previous results of Nwaka and coworkers (39). These authors hypothesized that trehalose reaches the vacuole by an endocytotic process in which it is degraded by the vacuolar acid trehalase. Here, we propose an alternative and simpler explanation, which is based on the finding that more than 90% of the acid trehalase activity is extracellular and cleaves the disaccharide into glucose in the periplasmic space. Ath1p activity is partitioned similarly in stationary-phase cells on glucose, which indicates that the targeting of acid trehalase activity to the extracellular compartment is not specific to trehalose. Our results are consistent with results obtained for extracellular acid trehalase in fungi such as Candida albicans (43), Mucor rouxii (9), and Aspergillus nidulans (10). That the optimal pH for growth on trehalose is around 5.0, the in vitro optimum pH of acid trehalase (25, 34), also is consistent with an extracellular activity of acid trehalase.
The high glycosylation level of the ATH1 product (1) also is consistent with an extracellular location for this protein, similar to that of invertase and gp37, the product of YGP1 (11). It is not known whether these three proteins associate in vivo, although they can be copurified (5, 11, 34), but the physical association could suffice for the acid trehalase to be secreted by invertase and gp37 secretion pathways in the absence of known secretion signals for Ath1p (12). We found that acid trehalase is not localized in the cell wall, while secreted invertase was found primarily in the cell wall fraction of both yeast and Neurospora crassa (20). Thus, S. cerevisiae Ath1p could be released as a free protein in the periplasmic space or linked to the plasma membrane.
How acid trehalase can be partly localized in the vacuole and the physiological significance of this cellular localization remain to be clarified. One explanation is that unspecific protein sorting to the vacuoles occurs after saturation of the secretion pathway. This hypothesis relies on the fact that overproduction of sorted proteins often leads to their mislocalization, either to the vacuolar compartment (44) or to the cell cortex (45, 55). Our experiments are consistent with this hypothesis, since the ratio of intracellular to extracellular Ath1p activity increases when ATH1 is overexpressed. Alternatively, vacuolar localization of Ath1p could result from an endocytic process (36) or from direct targeting to the vacuole by a route that remains to be elucidated. To answer these questions and others regarding the secretory mechanisms of Ath1p, we currently are screening the EUROSCARF deletion strain collection (7) for mutant cells defective in trehalose assimilation. The presence of Ath1p in vacuoles may simply precede its proteolytic degradation, as is the case for other proteins. Alternatively, this localization may be required for the degradation of intracellular trehalose, which could be transported to the vacuole by autophagy as described for glycogen (60, 61).
We found a second route for trehalose assimilation in yeast that couples trehalose transport by Agt1p with trehalose hydrolysis by Nth1p, in contrast to previous work (39) that found Ath1p to be essential. We also found that deletion of ATH1 in some strains (BY4741 and JF657) resulted in the inability to grow on trehalose (data not shown). The apparent discrepancy between these results can be explained by the genetic background of the strains since the Agt1p-Nth1p pathway is under the control of the MAL system (19). The CEN.PK strains used in this study have the MAL2-8c allele of MAL3, which results in constitutive and non-glucose-repressible AGT1/MAL11 expression. In contrast, mal− mutant strains such as JF657 cannot use this pathway because they do not express AGT1. Similarly, MAL-inducible strains, such as those with the BY4741 background, do not express this pathway since trehalose cannot substitute for maltose in the induction of AGT1.
We also showed that the pH of the growth medium is a key parameter for effective growth on trehalose, in agreement with the in vitro optimal pH of both acid trehalase (28) and Agt1p permease (19). For this reason, Malluta and coworkers (30) concluded that AGT1 is essential for trehalose assimilation probably because agt1 mutant cells were cultured in nonbuffered trehalose synthetic medium, which prevents effective Ath1p-dependent hydrolysis of trehalose. Finally, the decrease in the trehalose consumption rate of ath1 mutant cells as the sugar concentration in the medium dropped below 6 mM is consistent with the affinity constant of Agt1p for trehalose (Km = 4 mM) (53).
The addition of a rapidly fermentable carbon source, such as glucose, to S. cerevisiae cells growing on maltose stimulates very rapid loss of maltose transport activity (22). We also found a rapid decline in the Agt1p-mediated trehalose transport activity during such growth. Interestingly, no other carbon source was needed to cause the loss of Agt1p, unlike the sugar-induced inactivation of maltose permease. Whether the loss of Agt1p activity during growth is similar to that described for the inactivation of maltose permease (21, 23, 31-33) remains to be tested. However, further study is needed to identify the triggering signal that occurs during growth on trehalose, since the extremely low glycolytic flux that is observed under these conditions contrasts with the high rate of sugar influx that is proposed as an important parameter for maltose permease inactivation (22). When the Agt1p-Nth1p pathway is the only means by which the yeast can grow on trehalose, loss of Agt1p activity, together with its moderate affinity for trehalose, leads to a linear pattern of growth probably because the permease activity becomes rate limiting for trehalose assimilation.
The catabolism of trehalose in S. cerevisiae is a nice illustration of the Kluyver effect (30), which is explicitly defined as the respiratory assimilation of sugars under aerobic conditions and the inability of these sugars to be assimilated when respiration is blocked (16). In practice, the Kluyver effect in K. lactis is linked to limiting sugar uptake, which results in a low glycolytic flux, since overexpression of S. cerevisiae sugar transporters enables K. lactis to ferment maltose (17). In the case of S. cerevisiae on trehalose, this effect is dependent not only on trehalose uptake by the Agt1p transporter (16, this study) but also on the extracellular Ath1p (this study). However, overexpression of ATH1 or AGT1 failed to shift trehalose metabolism from oxidative to fermentative mode despite a significant increase in in vitro trehalose transport and/or hydrolysis activities. This inconsistency between a moderate growth rate improvement and substantial increases in transport and/or hydrolysis abilities suggests that additional down-regulatory mechanisms for trehalose assimilation exist.
In summary, we found two distinct pathways for trehalose assimilation in the yeast S. cerevisiae. One of them, the Agt1p-Nth1p-dependent route, is dependent on the genetic background of the strains and can be almost completely inactivated. The acid trehalase-dependent pathway thus appears to be the most important system for trehalose assimilation. As yeast cells have very efficient hexose transporters, the rate of extracellular trehalose hydrolysis into extracellular glucose, which can be easily monitored by the amount of Ath1p at the level of gene expression, limits glucose flux into yeast cells. This situation mimics glucose-limited continuous cultures in which the rate of glucose feeding fixes the specific growth rate (i.e., dilution rate). Consequently, batch growth of yeast on trehalose can be an alternative to labor-intensive glucose-limited chemostat cultures for studying rapid metabolic responses of cells challenged by pulse addition of various carbon sources or physicochemical perturbations (57). In particular, this cultivation process should be useful in searching for alterations in these responses by screening large collections of mutant cells.
Acknowledgments
This work was supported in part by the Microbiology and Pathogenicity program of the French Ministry of Education. M.J. was supported by a doctoral grant from the French Ministry of Education and Research.
We thank E. Boles (University of Frankfurt, Frankfurt, Germany) for providing primers and plasmids for the construction of agt1 mutant cells.
REFERENCES
- 1.Alizadeh, P., and D. J. Klionsky. 1996. Purification and biochemical characterization of the ATH1 gene product, vacuolar acid trehalase, from Saccharomyces cerevisiae. FEBS Lett. 391:273-278. [DOI] [PubMed] [Google Scholar]
- 2.Attfield, P. V. 1997. Stress tolerance: the key to effective strains of industrial baker's yeast. Nat. Biotechnol. 15:1351-1357. [DOI] [PubMed] [Google Scholar]
- 3.Barnett, J. A. 1997. Sugar utilization by Saccharomyces cerevisiae, p. 35-43. In F. K. Zimmermann and K. D. Entian (ed.), Yeast sugar metabolism. Technomic Publishing Co., Inc., Lancaster, Pa.
- 4.Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, N., and A. K. Ghosh. 1998. Regulation of acid trehalase activity by association-dissociation in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1379:245-256. [DOI] [PubMed] [Google Scholar]
- 6.Boos, W., U. Ehmann, E. Bremer, A. Middendorf, and P. Postma. 1987. Trehalase of Escherichia coli: mapping and cloning of its structural gene and identification of the enzyme as a periplasmic protein induced under high osmolarity growth conditions. J. Biol. Chem. 262:13212-13218. [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Dallies, N., J. Francois, and V. Paquet. 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall: application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14:1297-1306. [DOI] [PubMed] [Google Scholar]
- 9.de Almeida, F. M., A. K. Lucio, M. L. Polizeli, J. A. Jorge, and H. F. Terenzi. 1997. Function and regulation of the acid and neutral trehalases of Mucor rouxii. FEMS Microbiol. Lett. 155:73-77. [DOI] [PubMed] [Google Scholar]
- 10.d'Enfert, C., and T. Fontaine. 1997. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol. Microbiol. 24:203-216. [DOI] [PubMed] [Google Scholar]
- 11.Destruelle, M., H. Holzer, and D. J. Klionsky. 1994. Identification and characterization of a novel yeast gene: the YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol. Cell. Biol. 14:2740-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Destruelle, M., H. Holzer, and D. J. Klionsky. 1995. Isolation and characterization of a novel yeast gene, ATH1, that is required for vacuolar acid trehalase activity. Yeast 11:1015-1025. [DOI] [PubMed] [Google Scholar]
- 13.Elbein, A. D. 1974. The metabolism of α,α-trehalose. Adv. Carbohydr. Chem. Biochem. 30:227-256. [DOI] [PubMed] [Google Scholar]
- 14.Elbein, A. D., Y. T. Pan, I. Pastuszak, and D. Carroll. 2003. New insights on trehalose: a multifunctional molecule. Glycobiology. 13:17-27. [DOI] [PubMed] [Google Scholar]
- 15.François, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125-145. [DOI] [PubMed] [Google Scholar]
- 16.Fukuhara, H. 2003. The Kluyver effect revisited. FEMS Yeast Res. 3:327-331. [DOI] [PubMed] [Google Scholar]
- 17.Goffrini, P., I. Ferrero, and C. Donnini. 2002. Respiration-dependent utilization of sugars in yeasts: a determinant role for sugar transporters. J. Bacteriol. 184:427-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, E. K., F. Cotty, C. Sottas, H. Jiang, and C. A. Michels. 1995. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol. Microbiol. 17:1093-1107. [DOI] [PubMed] [Google Scholar]
- 20.Hecker, L. I., and A. S. Sussman. 1973. Localization of trehalase in the ascospores of Neurospora: relation to ascospore dormancy and germination. J. Bacteriol. 115:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, H., I. Medintz, and C. A. Michels. 1997. Two glucose sensing/signaling pathways stimulate glucose-induced inactivation of maltose permease in Saccharomyces. Mol. Biol. Cell. 8:1293-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, H., I. Medintz, B. Zhang, and C. A. Michels. 2000. Metabolic signals trigger glucose-induced inactivation of maltose permease in Saccharomyces. J. Bacteriol. 182:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, H., K. Tatchell, S. Liu, and C. A. Michels. 2000. Protein phosphatase type-1 regulatory subunits Reg1p and Reg2p act as signal transducers in the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:411-422. [DOI] [PubMed] [Google Scholar]
- 24.Jorge, J. A., M. L. Polizeli, J. M. Thevelein, and H. F. Terenzi. 1997. Trehalases and trehalose hydrolysis in fungi. FEMS Microbiol. Lett. 154:165-171. [DOI] [PubMed] [Google Scholar]
- 25.Keller, F., M. Schellenberg, and A. Wiemken. 1982. Localization of trehalase in vacuoles and of trehalose in the cytosol of yeast (Saccharomyces cerevisiae). Arch. Microbiol. 131:298-301. [DOI] [PubMed] [Google Scholar]
- 26.Kopp, M., H. Muller, and H. Holzer. 1993. Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J. Biol. Chem. 268:4766-4774. [PubMed] [Google Scholar]
- 27.Lillie, S. H., and J. R. Pringle. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Londesborough, J., and K. Varimo. 1984. Characterization of two trehalases in baker's yeast. Biochem. J. 219:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucio, A. K., M. L. Polizeli, J. A. Jorge, and H. F. Terenzi. 2000. Stimulation of hyphal growth in anaerobic cultures of Mucor rouxii by extracellular trehalose. Relevance of cell wall-bound activity of acid trehalase for trehalose utilization. FEMS Microbiol Lett. 182:9-13. [DOI] [PubMed] [Google Scholar]
- 30.Malluta, E. F., P. Decker, and B. U. Stambuk. 2000. The Kluyver effect for trehalose in Saccharomyces cerevisiae. J. Basic Microbiol. 40:199-205. [DOI] [PubMed] [Google Scholar]
- 31.Medintz, I., H. Jiang, E. K. Han, W. Cui, and C. A. Michels. 1996. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J. Bacteriol. 178:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medintz, I., H. Jiang, and C. A. Michels. 1998. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J. Biol. Chem. 273:34454-34462. [DOI] [PubMed] [Google Scholar]
- 33.Medintz, I., X. Wang, T. Hradek, and C. A. Michels. 2000. A PEST-like sequence in the N-terminal cytoplasmic domain of Saccharomyces maltose permease is required for glucose-induced proteolysis and rapid inactivation of transport activity. Biochemistry 39:4518-4526. [DOI] [PubMed] [Google Scholar]
- 34.Mittenbuhler, K., and H. Holzer. 1988. Purification and characterization of acid trehalase from the yeast suc2 mutant. J. Biol. Chem. 263:8537-8543. [PubMed] [Google Scholar]
- 35.Neves, M. J., and J. François. 1992. On the mechanism by which a heat shock induces trehalose accumulation in Saccharomyces cerevisiae. Biochem. J. 288:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nwaka, S., and H. Holzer. 1998. Molecular biology of trehalose and the trehalases in the yeast Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 58:197-237. [DOI] [PubMed] [Google Scholar]
- 37.Nwaka, S., M. Kopp, and H. Holzer. 1995. Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J. Biol. Chem. 270:10193-10198. [DOI] [PubMed] [Google Scholar]
- 38.Nwaka, S., B. Mechler, M. Destruelle, and H. Holzer. 1995. Phenotypic features of trehalase mutants in Saccharomyces cerevisiae. FEBS Lett. 360:286-290. [DOI] [PubMed] [Google Scholar]
- 39.Nwaka, S., B. Mechler, and H. Holzer. 1996. Deletion of the ATH1 gene in Saccharomyces cerevisiae prevents growth on trehalose. FEBS Lett. 386:235-238. [DOI] [PubMed] [Google Scholar]
- 40.Parrou, J. L., B. Enjalbert, L. Plourde, A. Bauche, B. Gonzalez, and J. François. 1999. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast 15:191-203. [DOI] [PubMed] [Google Scholar]
- 41.Parrou, J. L., M. A. Teste, and J. François. 1997. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology 143:1891-1900. [DOI] [PubMed] [Google Scholar]
- 42.Plourde-Owobi, L., S. Durner, J. L. Parrou, R. Wieczorke, G. Goma, and J. François. 1999. AGT1, encoding an alpha-glucoside transporter involved in uptake and intracellular accumulation of trehalose in Saccharomyces cerevisiae. J. Bacteriol. 181:3830-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ram, S. P., L. K. Romana, M. G. Shepherd, and P. A. Sullivan. 1984. Exo-(1-3)-β-glucanase, autolysin and trehalase activities during yeast growth and germ-tube formation in Candida albicans. J. Gen. Microbiol. 130:1227-1236. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, C. J., S. F. Nothwehr, and T. H. Stevens. 1992. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J. Cell Biol. 119:69-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rothman, J. H., C. P. Hunter, L. A. Valls, and T. H. Stevens. 1986. Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc. Natl. Acad. Sci. USA. 83:3248-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schena, M., D. Picard, and K. R. Yamamoto. 1991. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 194:389-398. [DOI] [PubMed] [Google Scholar]
- 47.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silveira, M. C., E. Carvajal, and E. P. Bon. 1996. Assay for in vivo yeast invertase activity using NaF. Anal. Biochem. 238:26-28. [DOI] [PubMed] [Google Scholar]
- 49.Singer, M. A., and S. Lindquist. 1998. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell 1:639-648. [DOI] [PubMed] [Google Scholar]
- 50.Singer, M. A., and S. Lindquist. 1998. Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol. 16:460-468. [DOI] [PubMed] [Google Scholar]
- 51.Stambuk, B. U., M. A. da Silva, A. D. Panek, and P. S. de Araujo. 1999. Active α-glucoside transport in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 170:105-110. [DOI] [PubMed] [Google Scholar]
- 52.Stambuk, B. U., and P. S. de Araujo. 2001. Kinetics of active α-glucoside transport in Saccharomyces cerevisiae. FEMS Yeast Res. 1:73-78. [DOI] [PubMed] [Google Scholar]
- 53.Stambuk, B. U., P. S. de Araujo, A. D. Panek, and R. Serrano. 1996. Kinetics and energetics of trehalose transport in Saccharomyces cerevisiae. Eur. J. Biochem. 237:876-881. [DOI] [PubMed] [Google Scholar]
- 54.Stambuk, B. U., A. D. Panek, J. H. Crowe, L. M. Crowe, and P. S. de Araujo. 1998. Expression of high-affinity trehalose-H+ symport in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1379:118-128. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, T. H., J. H. Rothman, G. S. Payne, and R. Schekman. 1986. Gene dosage-dependent secretion of yeast vacuolar carboxypeptidase Y. J. Cell Biol. 102:1551-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Dijken, J. P., J. Bauer, L. Brambilla, P. Duboc, J. M. Francois, C. Gancedo, M. L. Giuseppin, J. J. Heijnen, M. Hoare, H. C. Lange, E. A. Madden, P. Niederberger, J. Nielsen, J. L. Parrou, T. Petit, D. Porro, M. Reuss, N. van Riel, M. Rizzi, H. Y. Steensma, C. T. Verrips, J. Vindelov, and J. T. Pronk. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb. Technol. 26:706-714. [DOI] [PubMed] [Google Scholar]
- 57.Visser, D., G. A. van Zuylen, J. C. van Dam, A. Oudshoorn, M. R. Eman, C. Ras, W. M. van Gulik, J. Frank, G. W. van Dedem, and J. J. Heijnen. 2002. Rapid sampling for analysis of in vivo kinetics using the BioScope: a system for continuous-pulse experiments. Biotechnol. Bioeng. 79:674-681. [DOI] [PubMed] [Google Scholar]
- 58.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 59.Wang, X., M. Bali, I. Medintz, and C. A. Michels. 2002. Intracellular maltose is sufficient to induce MAL gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 1:696-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, Z., W. A. Wilson, M. A. Fujino, and P. J. Roach. 2001. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 21:5742-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson, W. A., Z. Wang, and P. J. Roach. 2002. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol. Cell. Proteomics 1:232-242. [DOI] [PubMed] [Google Scholar]