Abstract
Minichromsome maintenance protein 10 (Mcm10) is an essential replication factor that is required for the activation of the Cdc45:Mcm2-7:GINS helicase. Mcm10's ability to bind both ds and ssDNA appears vital for this function. In addition, Mcm10 interacts with multiple players at the replication fork, including DNA polymerase-α and proliferating cell nuclear antigen with which it cooperates during DNA elongation. Mcm10 lacks enzymatic function, but instead provides the replication apparatus with an oligomeric scaffold that likely acts in the coordination of DNA unwinding and DNA synthesis. Not surprisingly, loss of Mcm10 engages checkpoint, DNA repair and SUMO-dependent rescue pathways that collectively counteract replication stress and chromosome breakage. Here, we review Mcm10's structure and function and explain how it contributes to the maintenance of genome integrity.
Keywords: DNA replication, cancer mutations, genome integrity, Mcm10, OB-fold, STUbL
1. Structure and function of Mcm10
Minichromosome maintenance protein 10 (Mcm10) is an essential replication factor first indentified by Lawrence Dumas and co-workers in budding yeast over 30 years ago. They screened 1100 temperature-sensitive mutants for S phase progression defects and initially cataloged the mcm10 strain as dna43 [1]. It took another decade until the mutant was further characterized and DNA43 was sequenced. In the late 1990s, the Tye laboratory discovered the gene independently as MCM10 [2], using a strategy formerly developed to uncover replication initiation mutants [3, 4]. Since then orthologs of this scaffold protein have been identified and studied in different model organisms. Mcm10 is unique to the eukaryotic replication machinery, neither bacteria nor archaea express proteins with homologous structural domains.
The core of Mcm10 harbors the evolutionarily conserved and essential internal domain (ID), which is required for DNA and protein binding [5–7]. The ID is flanked by an N-terminal domain (NTD), which displays a coiled coil (CC) motif of considerable sequence similarity among species [7, 8]. In contrast, the C-terminal domain (CTD) varies highly from uni- to multicellular organisms and is distinguished by a large metazoan-specific extension [9]. Mcm10 appears to lack any enzymatic activity and this is consistent with the overall absence of any known catalytic motifs [10]. In this review, we will focus on structural characteristics of Mcm10 that mediate well-documented functions of the protein (Figure 1).
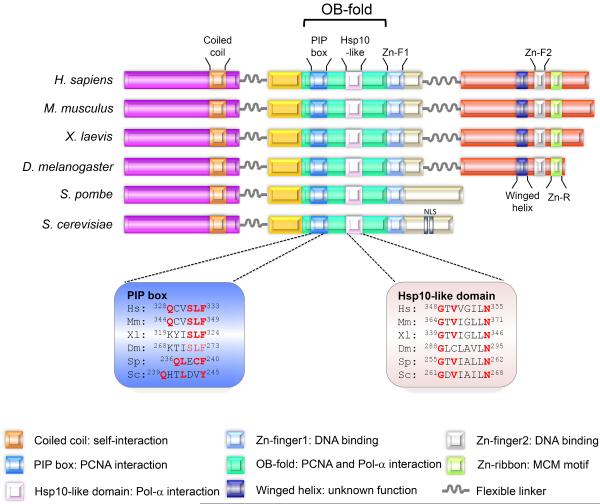
Figure 1. Structural architecture of Mcm10.
The functional domains of Mcm10 across different species are shown. The coiled-coil motif within the NTD mediates Mcm10 oligomerization. The ID includes a PIP box and Hsp10-like domain, both of which reside in the OB-fold. The PIP box and the Hsp10-like domain mediate Mcm10's interaction with PCNA and Pol-α, respectively, and sequence alignments of these regions are shown. (The positions of the indicated amino acids within these domains for HsMcm10 are in reference to isoform 1, consistent with the cancer mutations described in Figure 5a). The OB-fold and Zn-F1 provide a binding platform for DNA. Metazoan Mcm10 harbors additional DNA- and Pol-α-binding regions within the CTD. DNA binding within the CTD is primarily mediated by Zn-F2. How the Zn-R and winged helix motif contribute to protein function is currently unknown. Nuclear localization sequences (NLS) have only been identified in S. cerevisiae. Mcm10 orthologs are illustrated for Hs: Homo sapiens; Mm: Mus musculus, Xl: Xenopus laevis, Dm: Drosophila melanogaster, Sp: Schizosaccharomyces pombe, and Sc: Saccharomyces cerevisiae.
1.1. Mcm10 oligomerization
Several lines of evidence support the notion that Mcm10 acts as an oligomer and can assume dynamic multimeric formations. Independent studies of Schizosaccharomyces pombe (Sp) and Xenopus leavis (X) Mcm10 implicated the NTD in self-interaction [8, 10, 11]. Further support for oligomerization came from in vitro studies that characterized the binding of ScMcm10 to single-stranded (ss) DNA, which estimated that a Mcm10-trimer occupied up to 50 nucleotides [12]. In contrast, ~20 base pairs units of double-stranded (ds) DNA were bound by monomeric Mcm10, suggesting that DNA unwinding might trigger Mcm10 self-interaction during replication [12]. Several reports suggest that Mcm10 has a strong preference for ss over ds DNA [10, 12, 13], providing a rationale of how Mcm10 might contribute to the activation of the Cdc45:Mcm2-7:GINS (CMG) helicase [14]. Semi-quantitative chromatin binding studies are consistent with recent findings that revealed that the conserved CC domain in the NTD enables XMcm10 to dimer- and trimerize in a highly dynamic fashion [5, 8, 15]. Removal of the corresponding motif in ScMcm10 leads to severe hydroxyurea (HU) sensitivity when checkpoint function is compromised [8](Alver and Bielinsky, unpublished). The NTD has also been implicated in oligomerization of human (Hs) Mcm10 [16]. Gel-filtration of this domain identified a high-molecular weight complex that was interpreted to be either a trimer or hexamer [16]. HsMcm10 has been reported to form a hexameric ring based on electron microscopy (EM) reconstruction of full-length protein in which the ring-shaped archaeal Mcm helicase was utilized for molecular modeling [17]. However, the crystal structure of the ID of XMcm10 revealed an oligosaccharide/oligonucleotide binding fold (OB-fold) and Zn-F1 domain that assume a different configuration than in the archaeal Mcm helicase [6]. This might explain why the EM reconstruction of HsMcm10 is not fully compatible with the crystallographic data of XMcm10. Nevertheless, what appears to be consistent across species is that the NTD promotes oligomerization of the full-length protein. Multi-conformation scaffolding might happen during leading and lagging strand synthesis and could be crucial to maintain coordination between both activities during conditions of replication stress.
1.2. DNA binding and protein interaction
Unlike the NTD, the ID and CTD are essential and provide the main interaction surfaces for DNA binding. Mcm10 associates with DNA regardless of sequence context and topology [12, 18]. The ID displays high sequence similarity among species [6, 19]. It comprises a OB-fold, which forms a typical DNA binding cleft, but is also involved in protein:protein interactions with Pol-α, the Mcm2-7 complex and PCNA [6, 19–21]. The interaction with Pol-α is mediated through a conserved hydrophobic patch known as the Hsp10-like domain that resides in the concave of the cleft [6, 19], and PCNA binding requires a PCNA interacting peptide (PIP) box QxxM/I/LxxF/YF/Y [21]. In higher organisms, including humans, the PCNA interaction motif resembles the consensus of the prokaryotic β-clamp binding site QLsLF [13].
Site-directed mutagenesis of the ID in XMcm10 further suggested that the DNA binding region spans the hydrophobic cleft and positively charged residues on the adjacent Zn-F1 [6]. In vitro, the disruption of the XMcm10 Zn-F1 reduces overall protein stability and binding to dsDNA, but not ssDNA [10]. Mutation of residues that were identified by nuclear magnetic resonance chemical shift perturbation to make contact with DNA on the surface of Mcm10 significantly reduced ssDNA binding in vitro and increased sensitivity to replication stress in living cells [6]. These results strongly imply that the Zn-F1 domain binds DNA in vivo. Lastly, there is evidence that the ID of Mcm10 and Cdc45 bind DNA cooperatively [16].
1.3. Interplay of the ID and CTD in metazoa
The C-terminal extension found in metazoa provides an additional interface for DNA and protein interaction [9]. In conjunction with the ID, the CTD binds DNA and the catalytic subunit of Pol-α with higher affinity than either domain by itself [18]. Two Zn-coordinating structures reside within the CTD and form a globular domain, which is distinct from the Zn-F1 in the ID [9]. The first motif is the Zn-F2 that interacts with ssDNA, whereas the second bears homology to the Mcm2-7 helicase OB-fold Zn-ribbon, however, its function is not known [9]. The biological implication of having two distinct modules, namely the ID and CTD that can bind both ssDNA and Pol-α is compatible with a molecular hand-off mechanism that has been proposed by Eichman and colleagues [18]. According to this model, either the ID or CTD could grab a hold of ssDNA, while the other domain helps in the recruitment of Pol-α. This model predicts that both domains might be regulated individually in their ability to bind DNA [18]. Indeed, a recent report shows that acetylation of the HsMcm10 ID and CTD is reversible and affects chromatin binding of these domains in a non-uniform manner [22]. The modification is removed by the SIRT1 deacetylase, a homolog of budding yeast Sir2. In both humans and yeast, a direct interaction with the respective proteins is mediated by Mcm10's C-terminus [22, 23]. Whether ScMcm10 is acetylated remains an open question. Future studies are necessary to further substantiate the role of acetylation and deacetylation as a regulator of Mcm10 function.
1.4. Roles of Mcm10 in replication initiation
Multiple independent studies have implicated Mcm10 in both replication initiation and elongation. Mcm10's possible roles in these processes have been discussed in detail in a previous review [14], and thus we will focus here on common emerging themes (Figure 2). There is uniform consensus in the field that Mcm10 is loaded onto chromatin after “origin licensing” in the G1 phase of the cell cycle [14]. This process requires the assembly of the origin recognition complex (ORC), cell division cycle protein 6 (Cdc6) and Cdt1 all of which are needed to load double hexamers of the Mcm2-7 core helicase onto dsDNA [24]. During the G1-to-S-phase transition two kinases, the Dbf4-dependent kinase Cdc7 (DDK) and S-phase cyclin-dependent kinase (S-CDK) orchestrate the recruitment of two helicase co-activators, Cdc45 and GINS (go-ichi-nisan) [24]. DDK phosphorylates the Mcm2-7 complex, which allows for subsequent Cdc45 association. S-CDK targets specific adapter proteins (Sld2 and Sld3 for synthetically lethal with dpb11) that function to deliver GINS in conjunction with Pol-ε to nascent replication complexes [24]. GINS associates with Cdc45:Mcm2-7 to form the CMG complex – the functional replicative helicase [25]. What needs to follow after Cdc45 and GINS loading is a conformational rearrangement that allows the twin CMG complexes to separate and encircle the leading strand template rather than parental dsDNA [26]. These steps culminate in helicase activation and Mcm10 has been shown to be indispensable for this process, although the underlying mechanism of action has not been elucidated [27–29]. A requirement for Mcm10 in DNA unwinding was first recognized in a plasmid based replication system that employed Xenopus egg extract [15]. The Masukata laboratory has provided some evidence that Mcm10's ability to bind ssDNA is important in this context [27]. It is likely however, that additional properties of Mcm10 make it the ideal tool for this task, as it has been shown to interact with Cdc45 [16, 30, 31], distinct subunits of Mcm2-7 and GINS [2, 20, 30, 32–34], ORC [30, 33–35], the Sld2 ortholog Recql4 [36] and the ssDNA binding complex replication protein A (RPA) [5]. Although RPA has a 40-fold higher affinity for ssDNA than Mcm10 [13], the latter also binds dsDNA [10, 12, 13], uniquely enabling it to bind parental and unwound DNA. DNA unwinding subsequently allows the DNA synthesis machinery to strike. Although Pol-ε is the first DNA polymerase to arrive at replication origins, it is not equipped to initiate DNA synthesis. A specialized enzymatic complex, Pol-α/primase, produces small RNA/DNA primers that are extended into leading and lagging strands by Pol-ε and Pol-δrespectively [24]. Both polymerases are tethered to PCNA, which enhances processivity and ensures that DNA replication is completed in a timely manner [24].
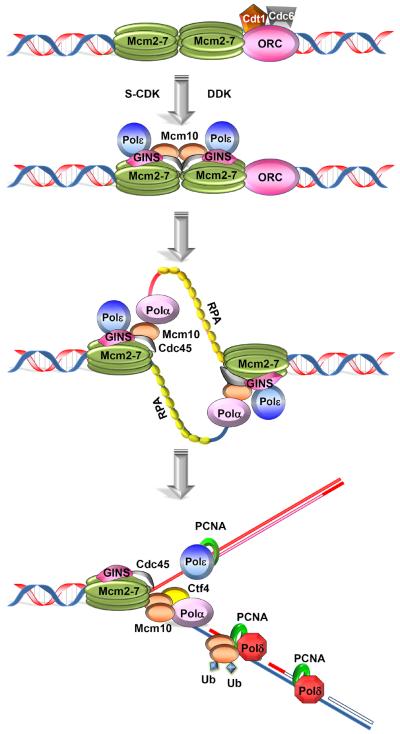
Figure 2. Roles of Mcm10 in replication initiation and elongation.
The assembly of replication complexes is shown. During the G1 phase of the cell cycle, ORC, Cdc6 and Cdt1 load double hexameric Mcm2-7 complexes onto origins, which completes origin licensing. To initiate replication, the functional Cdc45:Mcm2-7:GINS (CMG) helicase is formed and Pol-ε is delivered in conjunction with GINS. These processes are regulated by the subsequent actions of DDK and S-CDK. Mcm10 is recruited to chromatin during the G1 to S-phase transition. Mcm10 promotes origin unwinding and facilitates Pol-α binding to DNA together with Ctf4. This happens repeatedly during the initiation of each Okazaki fragment. Di-ubiquitination of Mcm10 releases Pol-α and promotes the interaction with PCNA, enabling processive DNA polymerization by Pol-ε and Pol-δ. The ssDNA binding protein RPA is shown in yellow coating the leading (red) and lagging strand (blue) templates. The steps described here are primarily based on experimental evidence in budding yeast.
1.5. Functions of Mcm10 during DNA elongation
Besides its essential role during replication initiation, Mcm10 is also required for DNA elongation and has been identified at replication forks by chromatin immunoprecipitation [5, 37]. Because Mcm10 interacts with the Mcm2-7 complex as well as Pol-α [2, 5, 18–20, 30, 32–34, 38], it was proposed to link DNA unwinding and DNA synthesis [5]. Moreover, Mcm10, Mcm2-7 and Pol-α are members of replisomes in budding yeast and Xenopus [39, 40]. These findings are consistent with the idea that Mcm10 helps in the recruitment of Pol-α to chromatin [5, 38, 41]. Mcm10 cooperates with another Pol-α binding protein, the cohesion factor Ctf4 [38]. Ctf4 is dispensable for DNA replication in budding yeast, but essential in higher eukaryotes. Co-immunoprecipitation studies in human cells and Xenopus egg extracts have demonstrated that Mcm10, Pol-α and Ctf4 are in a common complex and postulated that Mcm10 and Ctf4 connect the lagging strand polymerase, Pol-α, with the CMG helicase [38]. This notion is consistent with the finding that genetic disruption of Mcm10 and knockdown of Ctf4 result in very similar cellular phenotypes in Drosophila melanogaster [42, 43]. Moreover, depletion of either Ctf4 or Mcm10 has been implicated in regulating the turnover of Pol-α [5, 38, 44, 45]. Taken together, it appears that Mcm10, in conjunction with Ctf4, RPA and Pol-α, contributes to replication initiation and lagging strand synthesis. It is interesting to note that Pol-α synthesizes the 3' recessed dsDNA structure that serves as the substrate for PCNA loading. Mcm10 may have a role in facilitating the recruitment of PCNA via its central PIP box motif [21]. A single tyrosine substitution within the PIP box of Mcm10 renders budding yeast inviable [21]. The interaction between Mcm10 and PCNA in budding yeast is regulated by the mono-ubiquitination of Mcm10 at two distinct lysine residues, which occurs during G1 and S-phase of the cell cycle [21]. It is intriguing that ubiquitinated Mcm10 no longer interacts with Pol-α [21]. Therefore, ubiquitination of Mcm10 might regulate the release of Pol-α after the completion of RNA-DNA primer synthesis.
2. Understanding the genetic interaction network of Mcm10
Reviewing synthetic genetic interactions of mcm10 provides valuable insight into its role during DNA synthesis and suggests additional functions it may play in protecting genome integrity. These data also provide clues about the molecular players and DNA repair mechanisms involved in resolving replication stress induced by Mcm10 depletion. A compilation of genetic interactions reported for mcm10 by several different laboratories [32, 34, 35, 46, 47] and from a synthetic genetic array screen (SGA) (Data Repository of Yeast Genetic Interactions: http://drygin.ccbr.utoronto.ca/)[48] are illustrated in Figure 3. We propose that checkpoint-dependent and -independent functions including a novel pathway, mediated by the SUMO-targeted ubiquitin ligase (STUbL), Slx5/Slx8 (for synthetically lethal with sgs1), alleviate replication stress when Mcm10's function is compromised.
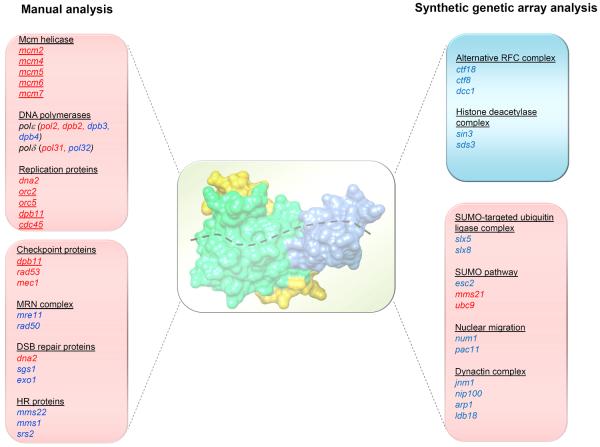
Figure 3. Genetic interaction network of MCM10.
Summary of genes that show synthetic loss of fitness or synthetic increase in survival with mcm10. The lists of genes on the left and right were generated from published reports [32, 34, 35, 46, 47] and a synthetic genetic array (SGA) screen ([48]; Thu, Nguyen and Bielinsky, unpublished; except mms21 and ubc9 [79]). Red letters indicate essential genes; blue letters mark non-essential genes. Red panels represent negative genetic interactions; the blue panel shows positive genetic interactions. Underlined genes encode proteins that physically interact with Mcm10. The space-filling model at the center shows the internal domain of XMcm10 with a trace of ssDNA [6]. The image was generated by importing the protein data bank (pdb) file 3EBE into the Chimera program (http://www.cgl.ucsf.edu/chimera) [80].
2.1. Mcm10 deficient cells require checkpoint signaling
The requirement for checkpoint activation in mcm10 mutants is indicated by the synthetic loss of fitness with mec1, rad53, dpb11 or dna2 [35, 46, 47] (Figure 3). These genetic interactions corroborate Mcm10's proposed functions during replication initiation and elongation. Due to its interaction with the Mcm2-7 complex and Pol-α, Mcm10 is thought to coordinate DNA unwinding and synthesis [5, 47]. Thus, Mcm10 deficient cells suffer from replication progression defects, accumulation of ssDNA and chronic checkpoint response, all of which have been supported by experimental evidence [2, 47, 49]. Checkpoint signaling protects stalled forks from collapse as mec1Δ and rad53Δ mutants cannot resume DNA synthesis after replication inhibition and this correlates with a loss of replisome components at stalled forks [50–54]. Therefore, the simplest explanation for the synthetic sickness with checkpoint mutants is that Mcm10 depletion creates a requirement for checkpoint mediators to prevent fork collapse.
The checkpoint kinases Mec1 and Rad53 phosphorylate multiple downstream effectors for different biological outcomes. The scaffold protein, Dpb11 and the Dna2 nuclease can converge on Mec1 activation through their ATR-activation domains and induce phosphorylation of the downstream kinase Rad53 during S-phase [55, 56] (Figure 4). Additional evidence underpinning the importance of checkpoint activation in the absence of Mcm10 comes from analyzing the mcm10–1 suppressor, mcm2-G400D. The G400D substitution partially inactivates the CMG helicase, thereby diminishing checkpoint activation [47]. These findings argue that elongation defects are the primary source of checkpoint activation in mcm10 mutants. The ability of the checkpoint kinases Mec1 and Rad53 to stabilize stalled forks logically explains their requirement in suppressing replication stress and the strong genetic interactions between mcm10 and mec1, rad53, dpb11 or dna2.
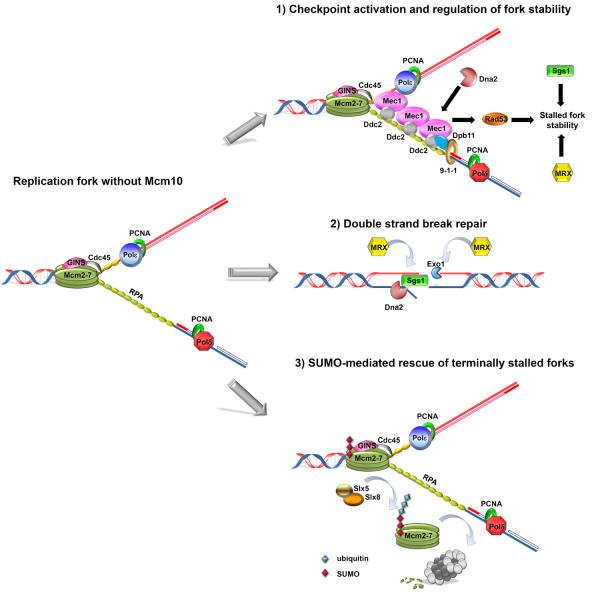
Figure 4. Proposed molecular mechanisms that alleviate replication stress following Mcm10 depletion.
On the left, a replication fork is illustrated in the absence of Mcm10. We hypothesize that Pol-α is recruited less efficiently to chromatin and that this defect is more pronounced on the lagging strand template. These events lead to the accumulation of RPA-coated ssDNA. Based on the genetic interaction, we propose that Mcm10 depletion triggers three possible scenarios: 1) Stretches of RPA-coated ssDNA are recognized by Mec1:Ddc2 complexes, initiating checkpoint activation. This response is enhanced by the 9-1-1 checkpoint clamp (Rad9-Hus1-Rad1), which binds to the 5' recessed ds RNA/DNA hybrid formed by an Okazaki fragment and the lagging strand template. The 9-1-1 clamp recruits Dpb11, which subsequently activates Mec1. In addition to Dpb11, Dna2 can also activate Mec1. Mec1 phosphorylates Rad53, leading to fork stabilization. Sgs1 and the MRX complex can also execute this function independently of the checkpoint. 2) Failure to stabilize the stalled fork in mcm10 mutants results in fork collapse and DSBs. DSB repair is initiated when the MRX complex and Sae2 mediate short-range resection of dsDNA ends. After this step, the MRX complex recruits Dna2, Sgs1/Top3/Rmi1 and Exo1, which perform additional resection to generate products compatible with homologous recombination (Sae2, Top3 and Rmi1 are not shown). 3) Possible mechanism to promote DSB formation in mcm10 mutants through Slx5/Slx8-mediated fork collapse. The Slx5/Slx8 complex ubiquitinates SUMOylated proteins. A hypothetical target shown here is the Mcm2-7 complex, which is degraded by the proteasome (shown in light and dark grey). Proteolysis results in disassembly of the replisome and collapse of terminally stalled forks.
2.2. Mcm10 deficient cells require molecular mechanisms to prevent aberrant fork structures
In addition to checkpoint signaling, mcm10 mutants utilize alternative mechanisms to prevent replication fork catastrophe, as implied by the negative genetic interactions between mcm10 and dna2, sgs1, srs2, mre11 or rad50 [46, 47, 57] (Figure 3). These molecular players have been implicated in replication fork restart and the resolution of aberrant fork structures. The Dna2 nuclease, which is involved in the processing of stalled forks and mediating double-strand break (DSB) repair via DNA end resection, enhances survival of Mcm10 deficient cells. A study in S. pombe shows that Dna2 prevents extensive fork reversal by cleaving regressed nascent strands and thus stabilizing stalled forks [58]. It is conceivable that mcm10 mutants may utilize this specific function to suppress replication stress. In addition to Dna2, the RecQ helicase family member, Sgs1, is known to resolve aberrant structures at stalled forks. Since the helicase activity of Sgs1 is necessary in this process, Sgs1 is thought to perform this task by removing “chicken foot” structures at regressed forks [52, 59, 60]. However, Sgs1 also dissolves sister chromatid junctions (SCJs) during error-free post replicative repair [61]. Thus, mcm10 sgs1 mutant lethality is possibly due to an increased number of terminally stalled replication forks. Resolution of aberrant fork structures is not the only means to improve the viability of mcm10 mutants, as they also require the anti-recombinase, Srs2, for survival. A well-documented function of Srs2 is the suppression of illegitimate homologous recombination by dismantling Rad51 nucleofilaments [62]. Not surprisingly, deletion of Rad51 rescues the lethality of mcm10–1 srs2Δ double mutants [47].
Additional mechanisms are likely to be in place to ensure replication fork integrity upon loss of Mcm10. Survival of mcm10 depends on the Mre11-Rad50-Xrs2 (MRX) complex [57]. One possible explanation for this dependency is that the MRX complex stabilizes replisomes at stalled forks through maintaining sister chromatid cohesion [57]. In summary, proteins counteracting recombination, promoting the resolution of SCJs or facilitating specific steps in replication fork restart can support replication fork stability and genome integrity in mcm10 mutants. Further analyses of the relationships between these factors and Mcm10 can potentially provide better insights into molecular mechanisms by which Mcm10 deficient cells ameliorate replication stress.
2.3. Mcm10 deficient cells require double strand break repair
Synergistic loss of fitness between mcm10 and mutants of DSB repair genes (mre11, rad50, sgs1, exo1, and dna2) implicate Mcm10 as a suppressor of DSBs [46, 47] (Figure 3). These breaks possibly arise from fork collapse after checkpoint exhaustion. Indeed, DSBs have been observed in Mcm10 deficient mammalian cells [44, 63] as well as in yeasts (Becker and Bielinsky, unpublished). The MRX complex is one of the first responders at DSBs and signals downstream effectors to promote initial and bulk resections of DSB ends, facilitated by Mre11/Sae2 and Sgs1-Top3-Rmi1/Dna2/Exo1, respectively [64] (Figure 4). Therefore, it is conceivable that Mre11, Rad50, Sgs1, Dna2 and Exo1 are required for the repair of collapsed forks. Alternatively, the genetic interaction between mcm10 and mutants of DSB repair genes could imply a more direct role of Mcm10 in mediating DSB repair. This idea is supported by the observation that Mcm10 was found in a complex with Dna2 and Nbs1 in Xenopus egg extract [65]. However, there is no experimental evidence to date that Mcm10 is active in DSB repair.
2.4. Mcm10 deficient cells require the SUMO-targeted ubiquitin ligase Slx5/Slx8
In keeping with the notion that mcm10 mutants accumulate DSBs, SGA have revealed a dependency on Slx5/Slx8 [48] (Figure 3). These genetic interactions have been confirmed by tetrad dissections and generation of double mutants (Thu, Nguyen and Bielinsky, unpublished). The Slx5/Slx8 STUbL has been implicated in a nuclear pore-associated repair mechanism of collapsed forks [66]. Slx5/Slx8 and its mammalian homolog, really interesting gene (RING) finger protein 4 (RNF4) have multiple SUMO-interacting motifs (SIMs), which bind poly-SUMOylated proteins [67]. These enzymes conjugate K48-linked ubiquitin chains to substrates that are SUMOylated and/or to poly-SUMO chains, and promote the degradation of these target proteins [67]. To date, the targets of Slx5/Slx8 and RNF4 at collapsed replication forks are largely unknown. A recent proteomic study uncovered SUMOylated proteins in slx5Δ mutants, revealing potential substrates of the Slx5/Slx8 complex, which include replication factors [68]. Coincidentally, members at the replication fork, such as the Mcm2-7 complex and polymerase subunits are SUMOylated in response to methyl methanesulfonate treatment [69]. Importantly, this SUMO response occurs independently of checkpoint activation [69]. At this point, it is unclear whether any of these potential candidates constitutes a bona fide Slx5/Slx8 target. However, the idea of removing replication proteins from the stalled forks (Figure 4) is consistent with the role of RNF4 in ATR deficient mouse embryonic fibroblasts (MEFs). RNF4 suppresses replication restart in ATR depleted MEFs by regulating the turnover of chromatin-bound SUMOylated proteins [70]. Furthermore, RNF4 is required for the formation of DSBs in these cells [70]. These data suggest that RNF4-mediated degradation of SUMO conjugates induces fork collapse and DSBs when checkpoint function is compromised. In conclusion, STUbL-mediated regulation of SUMOylated replication proteins is a novel pathway to respond replication stress. The Slx5/Slx8 complex may participate in a tightly regulated process to remove terminally arrested forks in mcm10 mutants.
3. Mcm10 in cancer
Accumulating evidence suggest that Mcm10 contributes to genetic diseases associated with aberrant proliferation and genome instability, such as cancer. Consistent with this notion, MCM10 has been identified as a suppressor of chromosome breakage in two independent genome-wide screens [63, 71]. There is strong evidence for a causal link between replication stress and chromosomal instability [72], which can fuel transformation and tumor heterogeneity. Moreover, recent advances have allowed us to better understand the contribution of replication stress to tumorigenesis. For example, the model of oncogene-induced replication stress is gaining increasing importance [73]. Since Mcm10 deficiency can lead to replication stress and Mcm10 plays a pivotal role during genome duplication, it is conceivable that its misregulation facilitates cancer development. In this last section, we will briefly review mutations and the alterations of MCM10 that have been identified in cancer (Figures 5a and b).
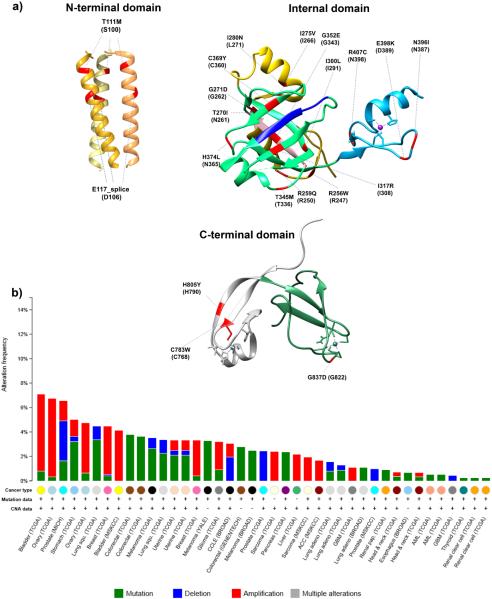
Figure 5. Genetic alterations in MCMC10 associated with cancer.
a) Residues corresponding to reported cancer mutations (shown in red) are projected on the available crystal structures of the NTD, ID, and CTD of XMcm10 (Xenopus residues are shown in parantheses). The CC motif within the NTD is shown as a homotrimer with each monomer depicted in a different shade of yellow. In the ID, the α-helical/random coil is shown in gold, the OB-fold in green, and Zn-F1 in blue. Pink marks the PIP box and dark blue the Hsp10-like domain. In the CTD, the Zn-F2 is labeled in white and the Zn-ribbon in green. A complete list of mutations is available at the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal/) [81, 82]. In the cBioPortal, mutations in MCM10 are assigned in reference to isoform 1. The images were generated by importing the following pdb data files 4JBZ (NTD), 3EBE (ID) and 2KWQ (CTD) into the Chimera program (http://www.cgl.ucsf.edu/chimera) [80]. The coordinates of HsMcm10 corresponding to the Xenopus NTD, ID and CTD crystal structures are 106–133, 243–422 and 770–857, respectively. (b) Frequencies of copy number alterations and mutations of MCM10 discovered in different types of cancer [80]. The figure was downloaded from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal/).
3.1. Expression changes of Mcm10 in cancer
The prioritization of cancer-associated genes is currently an area of intense research. Gene expression arrays can be analyzed to delineate recurring patterns that are common between independent data sets. One such study has recently uncovered the polo-like kinase 1 (PLK1)-MCM complex-S-phase kinase-associated protein 2 (SKP2) network in both breast and non-small cell lung cancers. Within this network, which included components of the Mcm2-7 complex, Plk1 and the E3 ligase Skp2, Mcm10 emerged as a “top-ten” hit [74]. It remains to be seen whether this type of integrated expression analysis will be valuable as a diagnostic or predictive tool. However, similar trends for Mcm10 up-regulation have also been documented for cervical cancer [75]. The expression level of Mcm10 correlated with stages of cancer progression, arguing that it might contribute to tumor aggressiveness [75]. This observation also raises the possibility that Mcm10 might be a target of certain oncogenes. Indeed, MCM10 expression is regulated by N-MYC and Ewing's sarcoma (EWS)-derived oncogenes in neuroblastoma and Ewing's tumors, respectively [76, 77]. In addition to transcriptional up-regulation, high expression levels of Mcm10 may result from gene amplification, as observed in bladder, ovarian and breast cancers (Figure 5b). Together, these observations suggest two possibilities: 1) Mcm10 may simply be one of the replication proteins upregulated as tumor cells increase their rate of proliferation or 2) MCM10 overexpression might act as an augmenting force during transformation. It is intriguing that MCM10 overexpression in yeast can drive genome instability [45]. Therefore, whether Mcm10 plays a causal role in tumor cell proliferation is an interesting question, which awaits further exploration. In addition to amplifications, other copy number variations, e.g., homo- or heterozygous deletions have been reported in prostate and lung cancers (Figure 5b). The significance of these deletions is currently unknown.
3.2. Cancer-associated mutations in MCM10
Other genetic alternations that have been linked to pathological conditions include mutations that may influence the stability and function of Mcm10. Recent exome sequencing of early gastric carcinoma tissues identified mutations within Mcm10 that mapped to T217 and P570 [78]. Additional coding changes have been discovered in various cancer genomes and this list will probably grow with increased availability of sequencing information. Several of these mutations lie within the conserved regions of Mcm10 and are likely to affect their crucial functions during replication (Figure 5a). For example, the G352E mutation observed in bladder cancer (Figure 5a) resides within the Hsp10-like domain of Mcm10, a structural motif which mediates the interaction with Pol-α (Figures 1 and 5a) [19]. Importantly, the glycine residue in this position is conserved in higher eukaryotes (Figure 1). Other examples include the C783W and H805Y substitutions observed in esophageal cancer and lung adenocarcinoma, respectively (Figure 5a). Based on the structural data available for XMcm10, the corresponding residues in HsMcm10 are positioned within the Zn-F2 motif, responsible for DNA binding in higher eukaryotes (Figures 1 and 5a) [9]. In the same vein, mutations that map within the OB-fold of Mcm10 may affect protein stability and/or its interaction with DNA, Pol-α or PCNA (Figure 5a). If and how these mutations alter Mcm10 function and cell proliferation will be the subject of future studies.
Acknowledgements
The authors wish to acknowledge funding from the NIH (GM074917) to A-K.B. and thank Drs. Brandt Eichman and Nadine Shaban for advice regarding the crystal structures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Dumas LB, et al. New temperature-sensitive mutants of Saccharomyces cerevisiae affecting DNA replication. Mol Gen Genet. 1982;187:42–6. doi: 10.1007/BF00384381. [DOI] [PubMed] [Google Scholar]
- [2].Merchant AM, et al. A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3261–71. doi: 10.1128/mcb.17.6.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maine GT, et al. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–85. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tye BK. Minichromosome maintenance as a genetic assay for defects in DNA replication. Methods. 1999;18:329–34. doi: 10.1006/meth.1999.0793. [DOI] [PubMed] [Google Scholar]
- [5].Ricke RM, et al. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell. 2004;16:173–85. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- [6].Warren EM, et al. Structural basis for DNA binding by replication initiator Mcm10. Structure. 2008;16:1892–901. doi: 10.1016/j.str.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Du W, et al. Structural biology of replication initiation factor Mcm10. Subcell Biochem. 2012;62:197–216. doi: 10.1007/978-94-007-4572-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Du W, et al. Mcm10 self-association is mediated by an N-terminal coiled-coil domain. PLoS One. 2013;8:e70518. doi: 10.1371/journal.pone.0070518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Robertson PD, et al. Solution NMR structure of the C-terminal DNA binding domain of Mcm10 reveals a conserved MCM motif. J Biol Chem. 2010;285:22942–9. doi: 10.1074/jbc.M110.131276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Robertson PD, et al. Domain architecture and biochemical characterization of vertebrate Mcm10. J Biol Chem. 2008;283:3338–48. doi: 10.1074/jbc.M706267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fien K, et al. Fission yeast Mcm10p contains primase activity. J Biol Chem. 2006;281:22248–60. doi: 10.1074/jbc.M512997200. [DOI] [PubMed] [Google Scholar]
- [12].Eisenberg S, et al. Novel DNA binding properties of the Mcm10 protein from Saccharomyces cerevisiae. J Biol Chem. 2009;284:25412–20. doi: 10.1074/jbc.M109.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fien K, et al. Primer utilization by DNA polymerase alpha-primase is influenced by its interaction with Mcm10p. J Biol Chem. 2004;279:16144–53. doi: 10.1074/jbc.M400142200. [DOI] [PubMed] [Google Scholar]
- [14].Thu YM, et al. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci. 2013;38:184–94. doi: 10.1016/j.tibs.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wohlschlegel JA, et al. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell. 2002;9:233–40. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- [16].Di Perna R, et al. The physical interaction of Mcm10 with Cdc45 modulates their DNA-binding properties. Biochem J. 2013;454:333–43. doi: 10.1042/BJ20130059. [DOI] [PubMed] [Google Scholar]
- [17].Okorokov AL, et al. Hexameric ring structure of human MCM10 DNA replication factor. EMBO Rep. 2007;8:925–30. doi: 10.1038/sj.embor.7401064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Warren EM, et al. Physical interactions between Mcm10, DNA, and DNA polymerase alpha. J Biol Chem. 2009;284:24662–72. doi: 10.1074/jbc.M109.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ricke RM, et al. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-alpha in budding yeast. J Biol Chem. 2006;281:18414–25. doi: 10.1074/jbc.M513551200. [DOI] [PubMed] [Google Scholar]
- [20].Lee JK, et al. The Cdc23 (Mcm10) protein is required for the phosphorylation of minichromosome maintenance complex by the Dfp1-Hsk1 kinase. Proc Natl Acad Sci U S A. 2003;100:2334–9. doi: 10.1073/pnas.0237384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Das-Bradoo S, et al. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol Cell Biol. 2006;26:4806–17. doi: 10.1128/MCB.02062-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fatoba ST, et al. Human SIRT1 regulates DNA binding and stability of the Mcm10 DNA replication factor via deacetylation. Nucleic Acids Res. 2013;41:4065–79. doi: 10.1093/nar/gkt131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liachko I, et al. Mcm10 mediates the interaction between DNA replication and silencing machineries. Genetics. 2009;181:379–91. doi: 10.1534/genetics.108.099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Masai H, et al. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- [25].Moyer SE, et al. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–41. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–41. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanke M, et al. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. Embo J. 2012;31:2182–94. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van Deursen F, et al. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. Embo J. 2012;31:2195–206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Watase G, et al. Mcm10 Plays a Role in Functioning of the Eukaryotic Replicative DNA Helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–9. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- [30].Christensen TW, et al. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol Biol Cell. 2003;14:2206–15. doi: 10.1091/mbc.E02-11-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sawyer SL, et al. Mcm10 and Cdc45 cooperate in origin activation in Saccharomyces cerevisiae. J Mol Biol. 2004;340:195–202. doi: 10.1016/j.jmb.2004.04.066. [DOI] [PubMed] [Google Scholar]
- [32].Homesley L, et al. Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins. Genes Dev. 2000;14:913–26. [PMC free article] [PubMed] [Google Scholar]
- [33].Izumi M, et al. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 2000;28:4769–77. doi: 10.1093/nar/28.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hart EA, et al. Fission yeast Cdc23 interactions with DNA replication initiation proteins. Curr Genet. 2002;41:342–8. doi: 10.1007/s00294-002-0316-9. [DOI] [PubMed] [Google Scholar]
- [35].Kawasaki Y, et al. Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae. Genes Cells. 2000;5:975–89. doi: 10.1046/j.1365-2443.2000.00387.x. [DOI] [PubMed] [Google Scholar]
- [36].Xu X, et al. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. Embo J. 2009;28:3005–14. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Taylor M, et al. Mcm10 interacts with Rad4/Cut5(TopBP1) and its association with origins of DNA replication is dependent on Rad4/Cut5(TopBP1) DNA Repair (Amst) 2011;10:1154–63. doi: 10.1016/j.dnarep.2011.09.001. [DOI] [PubMed] [Google Scholar]
- [38].Zhu W, et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–99. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–66. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- [40].Pacek M, et al. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–7. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- [41].Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Apger J, et al. Multiple functions for Drosophila Mcm10 suggested through analysis of two Mcm10 mutant alleles. Genetics. 2010;185:1151–65. doi: 10.1534/genetics.110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gosnell JA, et al. Drosophila Ctf4 is essential for efficient DNA replication and normal cell cycle progression. BMC Mol Biol. 2011;12:13. doi: 10.1186/1471-2199-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chattopadhyay S, et al. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell. 2007;18:4085–95. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Haworth J, et al. Ubc4 and Not4 regulate steady-state levels of DNA polymerase-alpha to promote efficient and accurate DNA replication. Mol Biol Cell. 2010;21:3205–19. doi: 10.1091/mbc.E09-06-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Araki Y, et al. Budding yeast mcm10/dna43 mutant requires a novel repair pathway for viability. Genes Cells. 2003;8:465–80. doi: 10.1046/j.1365-2443.2003.00648.x. [DOI] [PubMed] [Google Scholar]
- [47].Lee C, et al. Alternative mechanisms for coordinating polymerase alpha and MCM helicase. Mol Cell Biol. 2010;30:423–35. doi: 10.1128/MCB.01240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Koh JL, et al. DRYGIN: a database of quantitative genetic interaction networks in yeast. Nucleic Acids Res. 2010;38:D502–7. doi: 10.1093/nar/gkp820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Park JH, et al. Knockdown of human MCM10 activates G2 checkpoint pathway. Biochem Biophys Res Commun. 2008;365:490–5. doi: 10.1016/j.bbrc.2007.11.004. [DOI] [PubMed] [Google Scholar]
- [50].Lopes M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- [51].Tercero JA, et al. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–7. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- [52].Cobb JA, et al. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. Embo J. 2003;22:4325–36. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cobb JA, et al. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–69. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Raveendranathan M, et al. Genome-wide replication profiles of S-phase checkpoint mutants reveal fragile sites in yeast. Embo J. 2006;25:3627–39. doi: 10.1038/sj.emboj.7601251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pfander B, et al. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. Embo J. 2011;30:4897–907. doi: 10.1038/emboj.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kumar S, et al. Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev. 2013;27:313–21. doi: 10.1101/gad.204750.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tittel-Elmer M, et al. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. Embo J. 2009;28:1142–56. doi: 10.1038/emboj.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hu J, et al. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell. 2012;149:1221–32. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- [59].Bjergbaek L, et al. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. Embo J. 2005;24:405–17. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hegnauer AM, et al. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. Embo J. 2012;31:3768–83. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Karras GI, et al. Noncanonical role of the 9-1-1 clamp in the error-free DNA damage tolerance pathway. Mol Cell. 2013;49:536–46. doi: 10.1016/j.molcel.2012.11.016. [DOI] [PubMed] [Google Scholar]
- [62].Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–9. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- [63].Paulsen RD, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mimitou EP, et al. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011;10:344–8. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wawrousek KE, et al. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle. 2010;9:1156–66. doi: 10.4161/cc.9.6.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nagai S, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sriramachandran AM, et al. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta. 2014;1843:75–85. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- [68].Albuquerque CP, et al. Distinct SUMO ligases cooperate with Esc2 and Sl×5 to suppress duplication-mediated genome rearrangements. PLoS Genet. 2013;9:e1003670. doi: 10.1371/journal.pgen.1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cremona CA, et al. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol Cell. 2012;45:422–32. doi: 10.1016/j.molcel.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ragland RL, et al. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–73. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–53. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- [72].Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–6. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Halazonetis TD, et al. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- [74].Wu C, et al. Integrating gene expression and protein-protein interaction network to prioritize cancer-associated genes. BMC Bioinformatics. 2012;13:182. doi: 10.1186/1471-2105-13-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Das M, et al. Over expression of minichromosome maintenance genes is clinically correlated to cervical carcinogenesis. PLoS One. 2013;8:e69607. doi: 10.1371/journal.pone.0069607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Koppen A, et al. Direct regulation of the minichromosome maintenance complex by MYCN in neuroblastoma. Eur J Cancer. 2007;43:2413–22. doi: 10.1016/j.ejca.2007.07.024. [DOI] [PubMed] [Google Scholar]
- [77].Garcia-Aragoncillo E, et al. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing's tumor cells. Oncogene. 2008;27:6034–43. doi: 10.1038/onc.2008.203. [DOI] [PubMed] [Google Scholar]
- [78].Kang G, et al. Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PLoS One. 2013;8:e82770. doi: 10.1371/journal.pone.0082770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Makhnevych T, et al. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol Cell. 2009;33:124–35. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- [80].Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- [81].Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]