Abstract
The genetic determinants for lactose utilization from Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 and galactose utilization from Lactococcus lactis subsp. cremoris MG 1363 were heterologously expressed in the lysine-overproducing strain Corynebacterium glutamicum ATCC 21253. The C. glutamicum strains expressing the lactose permease and β-galactosidase genes of L. delbrueckii subsp. bulgaricus exhibited β-galactosidase activity in excess of 1,000 Miller units/ml of cells and were able to grow in medium in which lactose was the sole carbon source. Similarly, C. glutamicum strains containing the lactococcal aldose-1-epimerase, galactokinase, UDP-glucose-1-P-uridylyltransferase, and UDP-galactose-4-epimerase genes in association with the lactose permease and β-galactosidase genes exhibited β-galactosidase levels in excess of 730 Miller units/ml of cells and were able to grow in medium in which galactose was the sole carbon source. When grown in whey-based medium, the engineered C. glutamicum strain produced lysine at concentrations of up to 2 mg/ml, which represented a 10-fold increase over the results obtained with the lactose- and galactose-negative control, C. glutamicum 21253. Despite their increased catabolic flexibility, however, the modified corynebacteria exhibited slower growth rates and plasmid instability.
The amino acid-producing bacterium Corynebacterium glutamicum is unable to metabolize lactose or galactose and is therefore unable to utilize dairy-based substrates such as whey (the liquid remaining following the removal of the curd formed by rennet enzymes or acid from milk during the manufacture of cheese and casein) to overproduce amino acids such as lysine. Initial experiments to achieve lysine production in whey-based medium with amino acid-producing bacteria (such as C. glutamicum) involved the hydrolysis of lactose by the addition of commercially prepared β-galactosidase or acid hydrolysis using HCl treatment (16). With this approach, lysine yields of 17 g/liter (16) were obtained. However, the addition of β-galactosidase to hydrolyze lactose is an expensive process and consequently hampers the use of whey and C. glutamicum for the industrial production of lysine. C. glutamicum has been used for the production of l-lysine for more than 40 years, with current production exceeding 600,000 tons per annum, all of which is produced through fermentation (22). Given its commercial significance, C. glutamicum has been the subject of intense research investigations over the last decade, with a number of genetic tools and systems being developed (12, 22).
A number of food-grade bacterial strains have been genetically engineered to facilitate metabolite overproduction and nutritional enhancement of dairy products (in particular, whey) (1). Furthermore, successful overexpression of the β-galactosidase and lactose permease genes of Escherichia coli in a non-lysine-producing C. glutamicum strain (with the result that cells were able to grow in minimal medium with lactose supplied as the sole carbon source) has been reported previously (3).
The aspartate-derived amino acids, including lysine, threonine, methionine, and isoleucine, are produced from oxaloacetate, a component of the tricarboxylic acid cycle which is essential for glucose metabolism (12). Commonly used substrates for industrial amino acid production by C. glutamicum include sucrose- or glucose-based medium from molasses, high-test molasses, and starch or sucrose hydrolysates (6); however, the use of a lactose-based medium is not possible due to the inability of C. glutamicum to metabolize this sugar. In addition, the use of molasses for lysine production can be problematic due to its seasonal availability, which can result in inconsistency in product quality during storage (6).
One possible alternative to the substrates currently used for lysine production is whey. In the past, many of the efforts to generate value-added whey products have been directed at the protein fraction for fortification of foods such as infant formula, leaving the lactose fraction largely unexploited. The purpose of this study was to introduce the lactose- and galactose-metabolizing genes from lactic acid bacteria into a lysine-overproducing C. glutamicum strain and to examine the ability of the novel transformant to produce lysine from lactose-containing whey-based medium.
MATERIALS AND METHODS
Bacterial strains, media, cultivation conditions and plasmids.
All bacterial strains used in this study are shown in Table 1. C. glutamicum ATCC 13032 and C. glutamicum ATCC 21253 were propagated at 37 and 30°C, respectively, in brain heart infusion (BHI) medium (Merck, Darmstadt, Germany). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added as previously described (23). TOP 10 chemically competent E. coli strains (Invitrogen BV, Groningen, The Netherlands), which were used as transformation hosts, were grown in cultures on Luria-Bertani medium (10 g of tryptone/liter, 5 g of NaCl/liter, 5 g of yeast extract/liter). Lactococcus lactis subsp. cremoris MG 1363 (9) was propagated at 30°C in M17 medium (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% (wt/vol) glucose (GM17) and was grown anaerobically. Anaerobic conditions were maintained in Anaerocult anaerobic jars (Merck) with Anaerocult A gas packs (Merck). Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 was grown at 37°C in MRS (Oxoid Ltd., Basingstroke, Hampshire, England).
TABLE 1.
Strains used in this study
| Strain | Relevant feature | Source or reference |
|---|---|---|
| C. glutamicum ATCC 13032 | Transformation host | ATCCb |
| C. glutamicum ATCC 13032 (pCLIK 5a MCS Pddh) | Transformation host bearing pCLIK 5a MCS Pddha | This study |
| C. glutamicum ATCC 13032 (pEB01) | Transformation host bearing pEB01a | This study |
| C. glutamicum ATCC 13032 (pEB02) | Transformation host bearing pEB02a | This study |
| C. glutamicum ATCC 21253 | Lysine producer | ATCC |
| C. glutamicum ATCC 21253 (pCLIK 5a MCS Pddh) | Lysine producer bearing pCLIK 5a MCS Pddha | This study |
| C. glutamicum ATCC 21253 (pEB01) | Lysine producer bearing pEB01a | This study |
| C. glutamicum ATCC 21253 (pEB02) | Lysine producer bearing pEB02a | This study |
| L. lactis subsp. cremoris MG 1363 | Source of galactose genes | 9 |
| E. coli TOP 10 one-shot chemically competent cells | Transformation host | Invitrogen BV, Groningen, The Netherlands |
| L. delbrueckii subsp. bulgaricus ATCC 11842 | Source of lactose genes | ATCC |
Kanamycin resistant.
ATCC, American Type Culture Collection, Manassas, Va.
The basal C. glutamicum minimal medium (designated BMC) used in this study was based on previously described BMC (18), with the addition of l-threonine, l-leucine, and l-methionine (25 mg/liter). The pH was first adjusted to pH 7.2 using 1 N NaOH and then sterilized by filtration through a 0.45-μm-pore-size bottle-top filter (Stericup; Millipore). A second minimal medium (designated MM) used throughout this study was based on a previously described defined medium (21) but without the addition of the base stock, which was not required for the growth of the C. glutamicum strains used in this study. In this case, the final pH was adjusted to pH 7.0 and the medium was filter sterilized through a 0.45-μm-pore-size bottle-top filter. Minimal medium contained either glucose, lactose, or galactose at the concentrations given below (see Results and Discussion).
The fermentation medium I1 was based on previously described I1 medium (10) with the following modifications: the incorporation of 6% (wt/vol) whey and 5% (wt/vol) CaCO3. Precultivation and growth curve determinations were performed with 25-ml sterile bottles containing 10 ml of medium at 30°C with shaking at 250 rpm. Fermentation studies of whey-based medium were performed with 150-ml bottles containing 30 ml of medium at 30°C and shaking at 250 rpm. All medium components were obtained from Sigma (St. Louis, Mo.), Difco Laboratories, Merck, or AnalaR (BDH Chemicals Ltd., Poole, England). All growth experiments were performed in triplicate.
BHIS (BHI with 0.5 M sorbitol) and SOC (Invitrogen) media were employed for the regeneration of transformed C. glutamicum and E. coli strains, respectively. The growth of C. glutamicum strains was measured by monitoring the optical density (OD) of cultures at 578 nm with a Genesys 5 thermospectronic spectrophotometer (Milton Roy, Rochester, N.Y.). Alternatively, growth was measured by performing serial dilutions of cells in maximum recovery diluent (Oxoid), spread plating 100 μl of diluent with cells onto BHI agar, and expressing numbers as CFU/ml.
The shuttle vector used for the construction of all plasmids in this study was the plasmid pCLIK 5a MCS Pddh (BASF, Ludwigshafen, Germany). This vector replicates in both E. coli and C. glutamicum and also contains a C. glutamicum promoter (ddh promoter) and a kanamycin gene which functions in both E. coli and C. glutamicum. Plasmids used in this study included plasmid pCLIK 5a MCS Pddh (BASF) and plasmids pEB01 and pEB02 (this study; see Results and Discussion). A detailed map and the strategies employed for the construction of plasmids are described in Results and Discussion.
DNA manipulations.
Genomic DNA was isolated from L. lactis subsp. cremoris and L. delbrueckii subsp. bulgaricus by a modification of the method of Hoffmann and Winston (11). Plasmid DNA was isolated from E. coli strains with a Qiagen plasmid purification kit (Qiagen Ltd., Crawley, West Sussex, England) and from transformed C. glutamicum 21253 strains with a modified Qiagen plasmid purification technique as described by Tauch et al. (24).
Cleavage and ligations of DNA were performed as described by the supplier (New England Biolabs, Beverly, Mass. [NEB]). Restricted DNA involved in ligation reactions employing a single cleavage site was treated with the enzyme alkaline phosphatase (CIP; New England Biolabs) as described by the supplier.
Amplification of DNA with PCR.
PCR techniques were used for amplification of regions of the L. delbrueckii subsp. bulgaricus lactose operon, consisting of the β-galactosidase (lacZ) and lactose permease (lacY) genes (accession number M55068), and of the L. lactis subsp. cremoris MG1363 galactose operon, consisting of aldose-1-epimerase (galM), galactokinase (galK), UDP-glucose-1-P-uridylyltransferase (galT), and UDP-galactose-4-epimerase (galE) genes (accession number AJ011653).
Primers (Sigma-Genosys) with restriction sites (underlined) at their 5′ ends were designed for the lactose-utilizing genes as follows (sequences are written 5′ → 3′): forward primer, GAATTCCATATGTTATAAACAAGTTAACACACC; reverse primer, CGGGATCCTTATTTTAGTAAAAGGGGC. Primers (Sigma-Genosys) with restriction sites (underlined) at their 5′ ends were designed for the galactose-utilizing genes as follows (sequences are written 5′ → 3′): forward primer, CGGGATCCAAGTTGACCTCAGGTTAGC; reverse primer, CGGGATCCGATAATTAATCAGTAGCC.
DNA from L. lactis subsp. cremoris and L. delbrueckii subsp. bulgaricus was amplified in an Eppendorf Mastercycler Gradient (Eppendorf) with High Fidelity Expand as described by the supplier (Roche Diagnostics Ltd., Lewes, East Sussex, England). Annealing temperatures of 50°C for L. delbrueckii subsp. bulgaricus and 48°C for L. lactis subsp. cremoris were used for 30 s.
Transformation of bacteria.
C. glutamicum strains were transformed with all plasmids by electroporation (17). Transformations of chemically competent E. coli (Invitrogen) were performed as described by the manufacturer.
β-Galactosidase and lactose transport assays.
The β-galactosidase assay was a modification of the method described by Sambrook and Russell (23). The lactose transport assay was performed as described by Weisburg et al. (25). For both, enzyme activity levels are expressed as Miller units per milliliter of cells.
Analysis of free amino acids in whey-based medium.
The procedure for analyzing free amino acids in whey-based medium was based on the method previously described by Fenelon et al. (7). Samples for amino acid analysis were extracted from whey-based medium during or following growth of C. glutamicum strains, and assays were performed in duplicate. Free-amino-acid levels were expressed in micrograms per milliliter.
Analysis of sugars in MM.
The levels of sugars in supernatants were measured spectrophotometrically using Lactose/D-Galactose kits (Boehringer Mannheim, Biopharm GmbH, Darmstadt, Germany) as described by the manufacturer.
RESULTS AND DISCUSSION
Cloning and expression of β-galactosidase and lactose permease from L. delbrueckii subsp. bulgaricus in C. glutamicum.
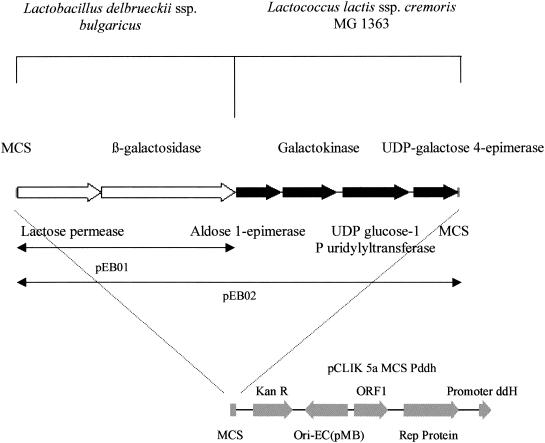
To improve the growth performance of C. glutamicum ATCC 13032 and ATCC 21253 in lactose-containing medium, the genes responsible for lactose catabolism (β-galactosidase and lactose permease) from the yogurt-producing bacterium L. delbrueckii subsp. bulgaricus were initially cloned. This involved amplification of the genes from the genome of L. delbrueckii subsp. bulgaricus ATCC 11842 to yield a PCR product of 4,914 bp. This was subsequently cloned downstream of a Corynebacterium promoter in the vector pCLIK 5a MCS Pddh to generate the plasmid pEB01. A schematic diagram for the construction of pEB01 is illustrated in Fig. 1.
FIG. 1.
Schematic diagram of the cloning of the lactose and galactose operons into pCLIK 5a MCS Pddh. pEB01 was constructed by cloning the lactose operon of L. delbrueckii subsp. bulgaricus into the plasmid pCLIK 5a MCS Pddh downstream of the promoter ddH, while pEB02 was constructed by cloning the galactose operon of L. lactis subsp. cremoris MG 1363 into the plasmid pEB01.
Both the wild-type C. glutamicum ATCC 13032 and the lysine-producing C. glutamicum ATCC 21253 strains were transformed with the recombinant plasmid pEB01 to examine whether the genes were expressed efficiently in both strains. Following incubation of the transformants on BHI plates with X-Gal and kanamycin (25 μg/ml), the resulting colonies emerged intensely blue in color (data not shown), which indicated high levels of β-galactosidase activity. However, it was noted that the blue colonies of C. glutamicum 21253 (pEB01) were smaller than the colonies of C. glutamicum 21253 (pCLIK 5a MCS Pddh) (data not shown) despite having been electroporated and incubated under identical conditions.
Measurements of the β-galactosidase activities in cell lysates of the recombinant C. glutamicum strains revealed that C. glutamicum 13032 (pCLIK 5a MCS Pddh) and 21253 (pCLIK 5a MCS Pddh) exhibited little or no activity and that C. glutamicum 13032 (pEB01) and 21253 (pEB01) both produced levels of enzyme activity in excess of 1,000 Miller units/ml of cells (Table 2). During the exponential-growth phase of C. glutamicum 21253 (pEB01) in 1% (wt/vol) lactose MM, the lactose concentration in the medium decreased over twofold whereas galactose levels increased to over 4 g/liter (data not shown).
TABLE 2.
Measured levels of β-galactosidase
| Strain | Units of β-galactosidase (Miller units/ml of cells) |
|---|---|
| C. glutamicum ATCC 13032 (pCLIK 5a MCS Pddh) | 3.8 ± 1.1 |
| C. glutamicum ATCC 13032 (pEB01) | 1,089.1 ± 124.7 |
| C. glutamicum ATCC 13032 (pEB02) | 884.2 ± 118.2 |
| C. glutamicum ATCC 21253 (pCLIK 5a MCS Pddh) | 6.2 ± 4.8-5 |
| C. glutamicum ATCC 21253 (pEB01) | 1,025.8 ± 117.9 |
| C. glutamicum ATCC 21253 (pEB02) | 734.9 ± 82.3 |
The active expression of the lacY (lactose permease) gene was verified by a lactose transport assay (25), in which C. glutamicum 21253 (pEB01) was examined for the ability to transport lactose into the cell by measuring ONPG (o-nitrophenyl-β-d-galactopyranoside) hydrolysis in nonpermeabilized cells. The level of enzyme activity in toluene-permeabilized cells was 825.0 ± 106.1 Miller units/ml of cells, that in non-toluene-permeabilized cells was 581.6 ± 18.8 Miller units/ml of cells, and that in non-toluene-permeabilized cells incubated in the presence of the lactose permease inhibitor 1-thio-β-d-digalactoside was 437.10 ± 41.18 Miller units/ml of cells. 1-Thio-β-d-digalactoside, which inhibits ONPG transport by binding to the E. coli M protein encoded by the lacY gene (14), was added at a final concentration (7.6 mM) in excess of the concentration required to completely inhibit E. coli lactose permease (2 mM) (20). The difference in measured Miller units between non-toluene-permeabilized cells and non-toluene-permeabilized cells treated with 1-thio-β-d-digalactoside was deemed to be due to lactose permease activity (20). A difference of 144.6 Miller units was detected between these two conditions, indicating that inhibition of the permease protein reduced ONPG hydrolysis, and suggests that this gene was also active in C. glutamicum 21253 (pEB01). It has been reported that ONPG hydrolysis by disrupted E. coli cells was 10 times greater than ONPG hydrolysis by undisrupted cells due to easier access to the substrate (20) and that the level of ONPG hydrolysis by C. glutamicum cells expressing only the E. coli β-galactosidase gene was approximately 15% of the level of cells expressing the entire lactose operon (2). However, a level of 437.1 ± 41.2 Miller units/ml of cells was detected in non-toluene-permeabilized cells treated with thiodigalactoside (data not shown), which is slightly more than 50% of the value obtained with permeabilized cells. This value appeared to be excessively high, suggesting membrane damage in these cells (which could have allowed entry of the large ONPG molecule [molecular mass, 301.3 Da] into the cells). A molecule of this size would not be expected to pass through an intact cell membrane (2, 19).
Growth performance on lactose.
To investigate the efficiency of lactose utilization by C. glutamicum 21253 (pEB01), the strain was grown in MM broth containing either glucose (2% [wt/vol]) or lactose (2% [wt/vol]) and the growth performance was compared with the results obtained with a control medium without a carbon source. Even though MM broth is capable of sustaining the growth of these strains to some extent, inclusion of lactose (2% [wt/vol]) greatly improved the growth performance of the culture (data not shown). Indeed, the OD of C. glutamicum 21253 (pEB01) (ΔOD, ∼0.13/h; OD, ∼3.3 at 25 h) in lactose was considerably higher than that obtained without a carbon source (ΔOD, ∼0.07/h; OD, ∼1.9 at 25 h). However, the growth in lactose was not as efficient as growth in glucose (ΔOD, ∼0.15/h; OD, ∼4.1 at 25 h) or as the growth performance seen with C. glutamicum 21253 (pCLIK 5a MCS Pddh) (ΔOD, ∼0.17/h; OD, ∼4.2 at 25 h) in glucose MM.
The levels of growth of C. glutamicum 21253 (pEB01) in glucose (2% [wt/vol]) and lactose (2% [wt/vol]) MM with 25 μg of kanamycin/ml were similar in glucose and lactose, reaching final OD values of ∼4.41 and 4.21, respectively. However, greater growth performance of cells harboring pCLIK 5a MCS Pddh (OD, 5.14 at 29 h) suggested that expression of the lactose operon was associated with slower growth of the culture.
These experiments were also performed with both glucose BMC and lactose BMC, and similar results were obtained (data not shown). Long lag periods of ∼20 h were followed by a significant reduction in both kanamycin-resistant and lactose-positive cell numbers during the lag and exponential-growth phases for up to 40 h (data not shown). In the presence of lactose, after 25 h up to 70% of cells were lactose positive; in the presence of glucose, 25% of the cells were lactose positive.
Cloning and expression of the galactose operon from L. lactis subsp. cremoris MG1363 in C. glutamicum.
The genes responsible for galactose catabolism (aldose-1-epimerase, galactokinase, UDP-glucose-1-P-uridylyltransferase, and UDP-galactose-4-epimerase) were cloned from the bacterium L. lactis subsp. cremoris MG1363 and introduced into C. glutamicum 13032 (pEB01) and 21253 (pEB01) in an effort to improve their growth performance in lactose-based substrates such as whey. This initially involved amplification of the genes from the genome of L. lactis subsp. cremoris MG1363 to yield a PCR product of 4,996 bp. This was subsequently cloned downstream of the lactose operon in pEB01 to generate the plasmid pEB02 (Fig. 1). β-Galactosidase activities in cell lysates of the recombinant C. glutamicum 21253 (pEB02) strain were decreased compared with the results seen with C. glutamicum 21253 (pEB01) (Table 2). During the exponential-growth phase of C. glutamicum 21253 (pEB02) in 1% (wt/vol) lactose MM, the concentration of lactose in the medium decreased over twofold whereas the concentration of galactose increased to less than 1 g/liter.
Growth performance on galactose.
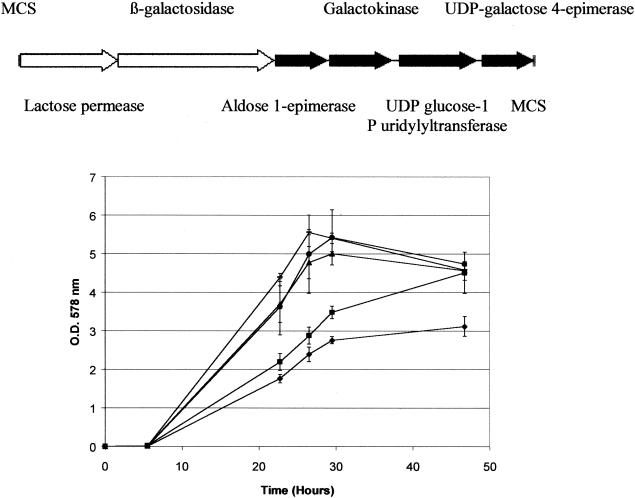
A direct comparison of the growth performance of C. glutamicum 21253 (pEB02) (in the presence of glucose, lactose, and galactose and in the absence of a carbon source as a control) was made with that of the parent strain (C. glutamicum 21253 [pCLIK 5a MCS Pddh]) in glucose to examine whether C. glutamicum 21253 (pEB02) utilized galactose and lactose as efficiently as it and the parent strain utilized glucose. Inclusion of lactose (2% [wt/vol]) and galactose (2% [wt/vol]) greatly improved the growth performance of the culture. Indeed, the OD of C. glutamicum 21253 (pEB02) in lactose (ΔOD, ∼0.12/h; OD, ∼3.5 at 29 h) was considerably higher than that obtained without a carbon source (ΔOD, ∼0.09/h; OD, ∼2.76 at 29 h) (Fig. 2). However, the growth performance of C. glutamicum 21253 (pEB02) in lactose was poorer than in glucose (ΔOD, ∼0.17/h; OD, ∼5 at 29 h) and was also poorer than the growth performance of C. glutamicum 21253 (pCLIK 5a MCS Pddh) in glucose MM (ΔOD, ∼0.18/h; ∼OD, 5.4 at 29 h). In the presence of galactose (2% [wt/vol]), in addition, C. glutamicum 21253 (pEB02) grew to the same extent (OD, ∼5.4 at 29 h) as C. glutamicum 21253 (pCLIK 5a MCS Pddh) in the presence of glucose MM (OD, ∼5.4 at 29 h), indicating that strains harboring pEB02 grew as well on galactose as C. glutamicum 21253 (pCLIK 5a MCS Pddh) grew on glucose MM. This contrasted with the inability of strains harboring pEB02 to grow on lactose (OD, ∼3.5 at 29 h) to the same extent as on glucose (OD, ∼5 at 29 h) (Fig. 2).
FIG. 2.
Comparison of the levels of growth of C. glutamicum 21253 (pEB02) in 2% (wt/vol) glucose (▴), lactose (▪), and galactose (•) MM and without a carbon source (⧫) with the growth of C. glutamicum 21253 (pCLIK 5a MCS Pddh) (—) in 2% (wt/vol) glucose.
When kanamycin was included in the medium, the level of growth performance of C. glutamicum 21253 (pEB02) was higher in glucose than in lactose, reaching OD values after 25 h of ∼5 and 3.42, respectively (data not shown). The similarity of the level of growth performance of cells harboring pCLIK 5a MCS Pddh to that of cells harboring pEB02 in glucose suggested that expression of the lactose operon was not maintained in C. glutamicum 21253 (pEB02) in the presence of kanamycin. The growth performance of C. glutamicum 21253 (pEB02) in galactose MM was similar to that of C. glutamicum 21253 (pCLIK 5a MCS Pddh) in glucose, with growth reaching an OD of ∼4.2 (data not shown).
These experiments were also performed with both glucose BMC and lactose BMC, and similar results were obtained (data not shown). The vast majority of cells eventually lost the ability to metabolize lactose after 43 h of growth (when less than 1% of colonies remained blue on an X-Gal plate) (data not shown). Furthermore, not all of the kanamycin-resistant cells were lactose positive on X-Gal plates. This may explain why the growth performance of cells harboring pEB02 was better in glucose MM than in lactose MM in the presence of kanamycin and why the level of β-galactosidase activity of cells was lower following insertion of the galactose operon. During the incubation of cells harboring pEB01/pEB02, it was evident that larger and healthier yellow colonies on X-Gal medium emerged and became the dominant phenotype in the culture, with the concomitant loss of the ability to metabolize lactose. Some of these cells contained a plasmid with a restriction pattern similar to that of pCLIK 5a MCS Pddh, indicating that the inserted DNA had been deleted from the plasmid vector. The growth of C. glutamicum 21253 pEB02 in galactose BMC was similar to its growth in BMC without a carbon source; therefore, galactose utilization was only observed in MM.
Lysine production and growth of C. glutamicum 21253 (pEB02) in whey-based medium.
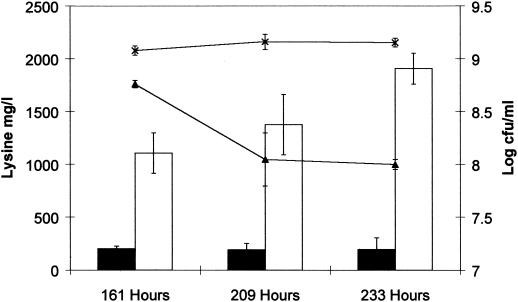
The purpose of this study was to generate a lactose-utilizing C. glutamicum strain that could exploit the lactose component of whey as a carbon source for the production of lysine during fermentation. Consequently, C. glutamicum 21253 and C. glutamicum 21253 (pEB02) were grown in whey-based medium to examine whether expression of the lactose-metabolizing genes improved the growth performance of the strain. Although there were no apparent differences in growth performance of the two strains on whey-based medium (data not shown), extended incubation of the cultures for more than 150 h showed a dramatic difference in viability of the cultures (Fig. 3). Following 225 h, the viability of the parent strain had decreased from 109 CFU/ml to 108 CFU/ml whereas the clone expressing the lactose-metabolizing genes retained viability greater than 109 CFU/ml. This increase in growth performance was associated with an accumulation of lysine in the medium to 1.9 mg/ml at 233 h of incubation, representing a 10-fold increase over the results seen with the control. However, growth and lysine production by C. glutamicum 21253 (pEB02) was extremely slow and inefficient in whey-based medium compared with yields obtained from lactose-hydrolyzed whey (10, 15) or with the results seen with current alternatives involving substrates such as molasses (22).
FIG. 3.
Comparison of the levels of growth of C. glutamicum 21253 (pCLIK MCS 5a Pddh) (▴) and C. glutamicum 21253 (pEB02) (×) in whey-based medium (I1). Levels of lysine production by C. glutamicum 21253 (pCLIK MCS 5a Pddh) (▪) and C. glutamicum 21253 (pEB02) (□) in whey-based media (I1) are shown.
Overexpression of heterologous lacY genes may have a toxic effect on cells (4, 5) which might be due to severe membrane damage accompanied by a decline in the growth of transformants in rich or minimal medium (22). This phenomenon may have occurred during our study. Indeed, the lactose permease protein has previously been expressed in C. glutamicum (without membrane damage) (2); therefore, the rate of expression may be crucial in determining whether membrane damage occurs in cells. Since it was suggested that lactose permease accounts for no more than 1% of membrane proteins (13), overexpression from the Pddh promoter could conceivably have resulted in an excess of lactose permease in the membrane, thereby damaging it. The problem of plasmid instability may be overcome by reducing the level of expression of the heterologous genes. Furthermore, integrating vectors have been used during transconjugation transformation procedures to avoid problems such as plasmid instability (8) and may be used as an alternative approach to the genetic engineering of C. glutamicum for growth in whey-based medium.
Conclusions.
This study reports the expression of the lactose permease and β-galactosidase genes in a lysine-overproducing C. glutamicum strain. Furthermore, a strain was developed which was able to efficiently utilize galactose as a sole carbon source. With this engineered strain, C. glutamicum 21253 (pEB02), lysine yields of up to ∼2 g/liter were achieved in a whey-based medium, representing 10-fold-higher yields than those seen with the control. However, growth and lysine production by C. glutamicum 21253 (pEB02) in whey-based media was extremely slow and inefficient; this result is most likely attributable to plasmid instability, which coincided with overexpression of the lactose operon.
Acknowledgments
E. Barrett is in receipt of a Teagasc Walsh Fellowship. This work was funded by the Irish Government under National Development Plan 2000-2006.
We are grateful to BASF, Ludwigshafen, Germany, for the C. glutamicum strains and the plasmid pCLIK 5a MCS Pddh. We are grateful to Paula O'Connor for amino acid analyses and to Richard Fitzpatrick for useful discussions.
REFERENCES
- 1.Barrett, E., C. Stanton, G. Fitzgerald, and R. P. Ross. 2003. Bio/technology of food cultures for the nutritional enhancement of milk and dairy products. In C. Shortt and J. O' Brien (ed.), Handbook of functional dairy products. CRC Press, Boca Raton, Fla.
- 2.Brabetz, W., W. Liebl, and K. H. Schleifer. 1993. Lactose permease of Escherichia coli catalyzes active β-galactoside transport in a gram-positive bacterium. J. Bacteriol. 175:7488-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brabetz, W., W. Liebl, and K. H. Schleifer. 1991. Studies on the utilization of lactose by Corynebacterium glutamicum, bearing the lactose operon of Escherichia coli. Arch. Microbiol. 155:607-612. [DOI] [PubMed] [Google Scholar]
- 4.De Vos, W. M., and G. Simons. 1988. Molecular cloning of lactose genes in dairy lactic streptococci: the phospho-beta-galactosidase and beta-galactosidase genes and their expression products. Biochimie 70:461-473. [DOI] [PubMed] [Google Scholar]
- 5.Donnelly, C. E., and A. C. Sonnenshein. 1982. Genetic fusion of E. coli lac genes to a B. subtilis promoter, p. 63-72. In A. T. Ganesan, Shing Chang, and James A. Hoch (ed.), Molecular cloning and gene regulation in bacilli. Academic Press Inc., New York, N.Y.
- 6.Eggeling, L., and H. Sahm. 1999. l-Glutamate and l-lysine: traditional products with impetuous developments. Appl. Microbiol. Biotechnol. 52:146-153. [Google Scholar]
- 7.Fenelon, M. A., P. O'Connor, and T. P. Guinee. 2000. The effect of fat content on the microbiology and proteolysis in cheddar cheese during ripening dairy foods. J. Dairy Sci. 83:2173-2183. [DOI] [PubMed] [Google Scholar]
- 8.Fitzpatrick, R. 1994. Metabolic manipulation to improve tryptophan productivity in industrially important strains of Corynebacterium glutamicum. Ph.D. thesis. University College Galway, Galway, Ireland.
- 9.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadj Sassi, A., N. Coello, A. M. Deschamps, and J. M. Lebeault. 1990. Effect of medium composition on l-lysine production by a variant strain of Corynebacterium glutamicum ATCC 21513. Biotechnol. Lett. 12:295-298. [Google Scholar]
- 11.Hoffmann, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 12.Jetten, M. S., and A. J. Sinskey. 1995. Recent advances in the physiology and genetics of amino acid-producing bacteria. Crit. Rev. Biotechnol. 15:73-103. [DOI] [PubMed] [Google Scholar]
- 13.Kaczorowski, G. J., G. LeBlanc, and H. R. Kaback. 1980. Specific labeling of the lac carrier protein in membrane vesicles of Escherichia coli by a photoaffinity reagent. Proc. Natl. Acad. Sci. USA 77:6319-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy, E. P. 1970. The lactose operon. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, N.Y.
- 15.Ko, Y., and J. Chipley. 1983. Microbial production of lysine and threonine from whey permeate. Appl. Environ. Microbiol. 45:610-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lebeault, J. M., A. M. Deschamps, and J. C. Patte. November1983. Lysine from lactose by fermentation. France patent GB2120240.
- 17.Liebl, W., A. Bayerl, B. Schein, U. Stillner, and K. H. Schleifer. 1989. High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol. Lett. 53:299-303. [DOI] [PubMed] [Google Scholar]
- 18.Liebl, W., R. Klamer, and K. H. Schleifer. 1989. Requirement of chelating compounds for the growth of Corynebacterium glutamicum in synthetic media. Appl. Microbiol. Biotechnol. 32:205-210. [Google Scholar]
- 19.Nottebrock, D., U. Meyer, R. Kramer, and S. Morbach. 2003. Molecular and biochemical characterization of mechanosensitive channels in Corynebacterium glutamicum. FEMS Microbiol. Lett. 218:305-309. [DOI] [PubMed] [Google Scholar]
- 20.Nunn, W. D., and J. E. Cronan. 1974. Unsaturated fatty acid synthesis is not required for induction of lactose transport in Escherichia coli. J. Biol. Chem. 249:724-731. [PubMed] [Google Scholar]
- 21.Otto, R., B. Ten Brink, H. Veldkamp, and W. N. Konings. 1983. The relationship between the growth rate and electrochemical proton gradient of Streptococcus thermophilus. FEMS Microbiol. Lett. 16:69-74. [Google Scholar]
- 22.Pfefferle, W., B. Mockel, B. Bathe, and A. Marx. 2003. Biotechnological manufacture of lysine. Adv. Biochem. Eng. Biotechnol. 79:59-112. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Tauch, A., O. Kirchner, L. Wehmeier, J. Kalinowski, and A. Puhler. 1994. Corynebacterium glutamicum DNA is subjected to methylation-restriction in Escherichia coli. FEMS Microbiol. Lett. 123:343-347. [DOI] [PubMed] [Google Scholar]
- 25.Weisburg, L. J., J. E. Cronan, and W. D. Nunn. 1975. Induction of lactose transport in Escherichia coli during the absence of phospholipid synthesis. J. Bacteriol. 123:492-496. [DOI] [PMC free article] [PubMed] [Google Scholar]