Abstract
Pseudo-phosphorylation of cardiac myosin regulatory light chain (RLC) has never been examined as a rescue method to alleviate a cardiomyopathy phenotype brought about by a disease causing mutation in the myosin RLC. This study focuses on the aspartic acid to valine substitution (D166V) in the myosin RLC shown to be associated with a malignant phenotype of familial hypertrophic cardiomyopathy (FHC). The mutation has also been demonstrated to cause severe functional abnormalities in transgenic mice expressing D166V in the heart. To explore this novel rescue strategy, pseudo-phosphorylation of D166V was used to determine whether the D166V-induced detrimental phenotype could be brought back to the level of wild-type (WT) RLC. The S15D substitution at the phosphorylation site of RLC was inserted into the recombinant WT and D166V mutant to mimic constitutively phosphorylated RLC proteins. Non-phosphorylatable (S15A) constructs were used as controls. A multi-faceted approach was taken to determine the effect of pseudo-phosphorylation on the ability of myosin to generate force and motion. Using mutant reconstituted porcine cardiac muscle preparations, we showed an S15D-induced rescue of both the enzymatic and binding properties of D166V-myosin to actin. A significant increase in force production capacity was noted in the in vitro motility assays for S15D-D166V vs. D166V reconstituted myosin. A similar pseudo-phosphorylation induced effect was observed on the D166V-elicited abnormal Ca2+ sensitivity of force in porcine papillary muscle strips reconstituted with phosphomimic recombinant RLCs. Results from this study demonstrate a novel in vitro rescue strategy that could be utilized in vivo to ameliorate a malignant cardiomyopathic phenotype. We show for the first time that pseudo-RLC phosphorylation can reverse the majority of the mutation-induced phenotypes highlighting the importance of RLC phosphorylation in combating cardiac disease.
Keywords: Cardiomyopathy, muscle contraction, mutation, myosin phosphorylation, in vitro motility, reconstituted cardiac system
Introduction
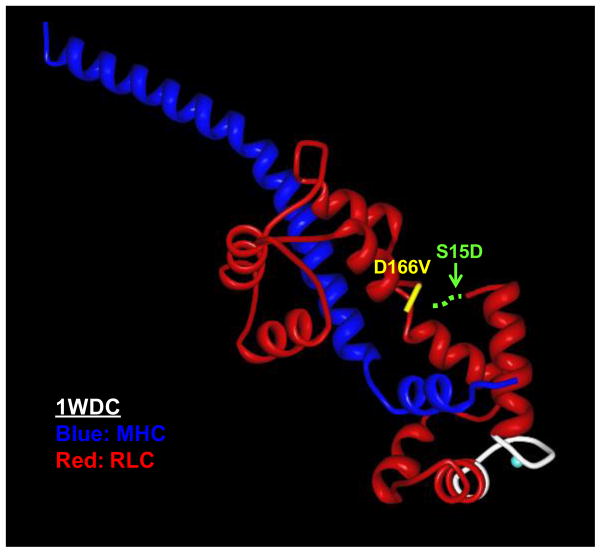
Familial hypertrophic cardiomyopathy1 (FHC) is a heritable form of cardiac hypertrophy caused by mutations in genes encoding for all major sarcomeric proteins [1–4]. The clinical manifestations of FHC range from asymptomatic to progressive heart failure and sudden cardiac death2 (SCD), and can vary from individual to individual even within the same family. Several mutations in the myosin regulatory light chain3 (RLC) have been implicated in the development of FHC and some of them are among the most prevalent mutations capable of affecting the thick filament structure and sarcomeric protein organization [5–7]. The RLC wraps around the α-helical neck region of the myosin head (lever arm) [8], and plays an important role in stabilizing its structure and function [9]. Considering the importance of the RLC in cardiac muscle contraction, it is understandable why subtle structural alterations in the RLC sequence would lead to cardiac disease. In particular, the D166V4 (aspartic acid to valine) mutation in the RLC was found to result in hypertrophic cardiomyopathy and SCD phenotypes [10, 11]. The mutation occurs at the last amino acid residue of the human ventricular RLC (Swiss-Prot: P10916) and substitutes valine for the normally occurring aspartic acid (Fig. 1). In the three dimensional structure of RLC, the site of mutation lies close to serine 15 (Ser15), a recognized site for the myosin light chain kinase5 (MLCK)-dependent RLC phosphorylation. As shown by many, myosin RLC phosphorylation plays important functional and structural roles in cardiac muscle contraction [12–16], and any changes in RLC phosphorylation are expected to affect heart performance and lead to cardiac disease [17–19]. Besides a highly conserved N-terminal phosphorylatable Ser15, the RLC also contains a Ca2+-Mg2+ binding site [20] which is thought to be occupied by Mg2+ when muscles are in the relaxed state [21] and partially saturated with Ca2+ during contraction [22]. The D166V mutation is located close to these two functionally important RLC domains and is also positioned in the elbow of the myosin heavy chain (MHC) region that links the cross bridge lever arm with the rod portion of myosin [8] (Fig. 1). Our previous studies on transgenic (Tg)-D166V mice revealed a mutation specific functional change in the Ca2+ dependent cardiac muscle contraction, which coincided with a significant decrease in the endogenous level of RLC phosphorylation [23]. This result implied a possible communication between all functional regions of RLC and in particular between the site of the D166V mutation and phosphorylatable Ser15 (Fig. 1). It also suggested an importance of RLC phosphorylation in the manifestation of the FHC-linked RLC phenotype. Our further study of this mutation showed that an MLCK-induced phosphorylation of Tg-D166V cardiac muscle preparations was able to partially reverse the negative functional consequences of the D166V mutation tested in myofibrils and skinned papillary muscle strips [24].
FIGURE 1.
The location of the D166V mutation in myosin RLC pictured in the regulatory domain of scallop myosin (1WDC) [49]: The MHC is labeled in dark blue and the RLC in red, the D166V mutation (in yellow) and the Ca2+-Mg2+ binding site (in white). The hypothetical serine phosphorylation site has been indicated (dashed green line) since the region of the RLC containing the Serine 15 site has not been resolved in any of the available myosin crystal structures.
In the current report we explore a novel rescue mechanism via constitutive RLC phosphorylation, using pseudo-phosphorylated RLC mimetic proteins. Porcine cardiac preparations were reconstituted with recombinant RLC constructs containing the following mutations: 1) S15D substitution to mimic a constitutively phosphorylated RLC; 2) S15A to mimic a non-phosphorylatable RLC; 3) S15D-D166V to mimic a constitutively phosphorylated RLC in D166V background; 4) S15A-D166V to mimic D166V-RLC protein that is incapable of phosphorylation; and 5) WT-RLC6 and D166V-RLC proteins as controls. The advantage of using porcine cardiac preparations reconstituted with phosphomimic RLCs was two-fold. First, in contrast to transgenic mouse cardiac samples expressing the α-MHC7, this study was performed on porcine preparations expressing the β-MHC, the same isoform of myosin found in the human heart. Hence, our experiments provide an essential control in studying human disease by using the proper MHC background. Second, the preparations of fixed levels of RLC phosphorylation allowed us to overcome any potential discrepancies between the samples due to potential varied levels of MLCK-induced phosphorylation of RLC.
In this study, we demonstrate the ability of the S15D phosphomimic RLC proteins to reverse or partially rescue the majority of the detrimental effects induced by the D166V mutation. We show an S15D-induced rescue of Vmax of the actin-activated myosin ATPase activity which was observed to be decreased due to the D166V mutation. A significant increase in force production capability was noted in the in vitro motility assays for S15D-D166V vs. D166V reconstituted myosin. In porcine papillary muscle strips reconstituted with the S15D-D166V construct, we monitored a similar pseudo-phosphorylation induced rescue of the D166V-elicited increase in Ca2+ sensitivity. Importantly, effects of pseudo-phosphorylation of D166V observed in this report are in accord with the results obtained in Tg-D166V preparations phosphorylated with Ca2+-calmodulin activated MLCK [24] suggesting that the mechanism by which RLC phosphorylation counteracts the effects of the D166V mutation is specific to the phosphorylation-induced RLC conformation rather than the enzymatic activity of the MLCK or its counteractive phosphatase.
Materials and Methods
Mutation, expression, and purification of wild-type (WT) human cardiac RLC and the FHC mutants
The cDNA for WT human cardiac RLC was cloned by reverse transcription-polymerase chain reaction using primers based on the published cDNA RLC sequence (GenBank™ accession no. AF020768) using standard methods as described previously [20]. The RLC mutants: S15D, S15A, D166V, S15D-D166V and S15A-D166V were generated using overlapping sequential polymerase chain reaction [25]. Briefly, WT and mutant cDNAs were constructed with an NcoI site at the N-terminal ATG and a BamHI site following the stop codon to facilitate ligation into the NcoI-BamHI cloning site of the pET-3d (Novagen) plasmid vector and transformation into DH5α cloning host bacteria for expression. The cDNAs were transformed into BL21 expression host cells and all proteins were expressed in large (16 liters) cultures. Expressed proteins were purified using a S-Sepharose column followed by a Q-Sepharose column, both equilibrated with 2 M urea, 25 mM Tris-HCl, 0.1 mM phenylmethylsulfonyl fluoride8 (PMSF), 1 mM dithiothreitol9 (DTT), 0.001% NaN3, pH 7.5. The proteins were eluted with a salt gradient of 0–450 mM NaCl. The final purity of the proteins was evaluated using 15% SDS-PAGE.
Preparation of porcine cardiac10 (PC) myosin
PC myosin was purified as described by Pant et al. [26]. Briefly, left ventricular muscle was chilled on ice, washed clear of blood with ice-cold dH2O, and minced. The muscle mince was rinsed in ice-cold dH20 until clear and extracted on ice with stirring for 1.5 h in 300 ml/100 g of muscle in Edsall-Weber solution (0.012 M Na2CO3, 0.04 M NaHCO3, and 0.6 M KCl, pH 9.0). The homogenate was then centrifuged at 13,000 g for 20 min, and the supernatant was precipitated with 13 vol of water containing 1 mM EDTA (ethylenediaminetetraacetic acid)11 and 1 mM DTT, followed by centrifugation at 13,000 g for 10 min. The pellet was resuspended in buffer containing 0.5 M KCl, 20 mM MOPS (3-(N-morpholino)propanesulfonic acid)12 (pH 7.0), 1 mM DTT, and 10 mM MgATP and centrifuged at 186,000 g for 1.5 h. Supernatant containing native PC myosin was precipitated with 13 vol of ice-cold water and centrifuged at 8,000 g for 10 min. The pellets were left on ice overnight. Then the pellet containing PC myosin was dissolved in the above buffer and clarified by centrifugation at 186,000 g for 1.5 h, mixed with glycerol (1:1), and stored at −20°C until used.
Preparation of muscle F-actin
Rabbit skeletal muscle actin was prepared as described previously [27] with modifications. Briefly, rabbit skeletal acetone powder was extracted with a G-actin buffer consisting of 2 mM Tris-HCl (pH 8.0), 0.2 mM Na2ATP, 0.5 mM β-mercaptoethanol, 0.2 mM CaCl2 and 0.0005% NaN3 at a ratio of 20 ml·g−1 for 30 min with stirring on ice. The extract was clarified by centrifugation at 7000 g at 4 °C for 1 h and the tissue pellet was discarded. The supernatant was adjusted to a final concentration of 40 mM KCl, 2 mM MgCl2 and 1 mM Na2ATP (pH 8.0) and the F-actin was allowed to polymerize for 2 h at 4 °C. The KCl concentration was then increased very slowly to a final concentration of 0.6 M and the solution was stirred slowly on ice for 30 min. This step was necessary to remove possible traces of tropomyosin–troponin13 (Tm-Tn). The F-actin pellet was then collected by ultracentrifugation at 160,000 g at 4 °C for 1.5 h. The supernatant was discarded and the F-actin pellet was re-dissolved in a buffer consisting of 10 mM MOPS (pH 7.0) and 40 mM KCl.
Labeling of F-actin with pyrene for actin-myosin binding assays
For fluorescence assays, rabbit skeletal actin was labeled with Pyrene Iodoacetamide14 (PIA) (Invitrogen) in accordance with the method described by Cooper et al. [28]. Briefly, 20–40 μM F-actin was incubated at room temperature, in the dark, for 16 h with a 10 molar excess of PIA in a buffer containing 10 mM MOPS (pH 7.0), 1 mM MgCl2 and 40 mm KCl. Then the reaction was quenched with 1 mM DTT and the preparation was centrifuged at 1,000 g for 1 h. Next, F-actin was dialyzed against G-actin buffer (2 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT) and polymerized overnight to form F-actin. The resulting molar ratio of pyrene/F-actin was 0.8 as determined using the molar extinction coefficient, ε344(pyrene) = 22,000 M−1·cm−1 [29].
Depletion of native RLC from porcine cardiac myosin and reconstitution with human cardiac WT or RLC mutants for myosin ATPase, in vitro motility assays and stopped flow measurements
Endogenous native RLC was depleted from PC myosin by treatment with 1% Triton X-100 and 5 mM CDTA ((cyclohexane-1,2-diaminetetraacetic acid)15, pH 8.5, as described previously [26]. Myosin depleted of endogenous RLC was then resuspended in a buffer composed of 0.4 M KCl, 50 mM MOPS, pH 7.0, 2 mM MgCl2, and 1 mM DTT, mixed in a 1:3 molar ratio with recombinant human cardiac RLC (WT, S15D, D166V or S15D-D166V), and dialyzed against the same buffer at 4°C for 2 h. RLC-reconstituted myosins were then dialyzed against 5 mM DTT overnight. The precipitated myosin-RLC complexes were then collected by centrifugation and resuspended in myosin buffer composed of 0.4 M KCl, 10 mM MOPS, pH 7.0, and 1 mM DTT. Samples were taken for SDS-PAGE analysis before they were mixed 1:1 with glycerol and stored at −20°C until needed for experiments. At least two isolated preparations of myosin reconstituted with the respective RLCs were used for all the assays involving reconstituted porcine myosin.
CDTA-extraction of endogenous RLC from skinned cardiac muscle strips
Freshly isolated porcine hearts were placed in oxygenated physiological salt solution of 140 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 1.8 mM NaH2PO4, 5.5 mM glucose, and 50 mM HEPES buffer, pH 7.4. The papillary muscles of the left ventricles were isolated, dissected into muscle bundles of about 20 × 3 mm, and chemically skinned in a 50% glycerol, 50% pCa 8 solution (10−8 M [Ca2+], 1 mM [Mg2+], 7 mM EGTA, 5 mM [Mg-ATP2+], 20 mM imidazole, pH 7.0, 15 mM creatine phosphate; ionic strength = 150 mM adjusted with potassium propionate) containing 1% Triton X-100 for 24 h at 4°C. Then the bundles were transferred to the same solution without Triton X-100 and stored at −20°C for about 2 months.
Endogenous RLC depletion from porcine cardiac muscle preparations was achieved in a buffer containing 5 mM CDTA, 40 mM Tris, 50 mM KCl, 1 μg/ml pepstatin A, 0.6 mM NaN3, 0.2 mM PMSF, and 1% Triton X-100, pH 8.4. This procedure was achieved in small muscle strips, ~100 μm wide and ~1.4 mm long, isolated from glycerinated papillary muscle bundles. The muscle strips were attached to the arms of a force transducer and incubated in this solution for 5 min at room temperature followed by incubation in the fresh solution of the same composition for another 30 min. The extent of RLC extraction was determined by analysis of the samples run on SDS-PAGE. Depletion of the endogenous RLC may result in partial extraction of the endogenous Troponin C16 (TnC), and therefore cardiac TnC along with the RLC WT or mutants were added back into the CDTA-treated strips.
Reconstitution of the CDTA-depleted muscle preparations with WT or mutants for steady state force and Ca2+ dependence of force measurements
Reconstitution of the RLC-depleted strips with porcine cardiac TnC and RLC-WT or S15D, S15A, D166V, S15D-D166V and S15A-D166V mutants was performed in muscle strips mounted to the force transducer in pCa 8 solution containing 40 μM RLC of interest and 15 μM TnC at room temperature. The solution of TnC was included in the reconstitution protein mixture during the first 30 min of incubation followed by 30-min incubation with fresh RLC solution. Reconstituted muscle strips were then washed in pCa 8 buffer and subjected to force measurements. The extent of RLC extraction and then reconstitution was tested by SDS-PAGE.
Binding of porcine myosin reconstituted with RLC to pyrene labeled F-actin
PC myosin exchanged with the RLC of interest was titrated at 0.1 μM increments against constant concentrations of actin (0.5 μM) until it reached a 2 fold molar excess over [actin]. Fluorescence measurements were done using a JASCO 6500 Spectrofluorometer. PIA was excited at 340 nm and fluorescence collected at 407 nm. The titration data were then fitted to the following quadratic equation (Eq. 1) to obtain the binding constant (Kd) and stoichiometry (n):
| Eq. 1 |
where m1=initial signal, m2=maximal amplitude (decrease in fluorescence intensity on myosin binding to pyrene-actin), n=stoichiometry of binding (mole of myosin per mole of actin), a=concentration of actin and x=total concentration of added myosin.
Actin activated myosin ATPase assays
The kinetics of the actomyosin interaction was measured using the actin-activated myosin ATPase activity assays. Reconstituted porcine myosin preparations, previously stored in glycerol, were precipitated with 13 volumes of ice-cold 2 mM DTT and collected by centrifugation at 8,000 g for 10 min. Myosin pellets were resuspended in 0.4 M KCl, 10 mM MOPS (pH 7.0), and 1 mM DTT, and then dialyzed overnight at 4 °C against the same buffer. Concentrations of myosin preparations were determined using a Coomassie Plus protein assay (Pierce, Rockford, IL, USA). Myosin at a final concentration of 1.9 μM was titrated with increasing amounts of rabbit skeletal actin (in μM): 0.1, 5, 10, 15, 20, 25 and 30. The assays were performed in triplicate on 96-well microplates in a 120 μl reaction volume containing 25 mM imidazole (pH 7.0), 4 mM MgCl2, 1 mM EGTA and 1 mM DTT. The final KCl salt concentration was 77.7 mM. The reactions were initiated with the addition of 2.5 mM ATP with mixing in a Jitterbug incubator shaker and allowed to proceed for 20 min at 30°C and then terminated by the addition of ice-cold trichloroacetic acid17 (TCA) at a final concentration of 4%. Precipitated protein was cleared by centrifugation, and the inorganic phosphate was determined according to the Fiske and Subbarrow method [30]. Data were analyzed using the Michaelis–Menten equation, yielding Vmax and Km [31, 32].
Stopped-flow measurements
Reconstituted porcine myosins at a concentration of 0.25 μM were mixed with 0.25 μM pyrene labeled F-actin (stabilized by 0.25 μM phalloidin) in rigor buffer containing 0.4 M KCl, 1 mM DTT and 10 mM MOPS, pH 7.0. The complexes were then mixed in a 1:1 vol/vol ratio with increasing (25–300 μM) concentrations of MgATP dissolved in the same buffer in the stopped flow apparatus and the time course of the change in pyrene fluorescence on MgATP-dependent myosin dissociation was monitored. Measurements were performed using a BioLogic (Claix, France) model SFM-20 stopped-flow instrument outfitted with a Berger ball mixer and an FC-8 observation cuvette. The data were collected and digitized using a JASCO 6500 Fluorometer. The estimated dead time was 3.5 ms. The pyrene-F actin was excited at 347 nm and emission was monitored at 404 nm using monochromators set to 20-nm bandwidths. Typically, 6–23 stopped-flow records were averaged and fit to a single exponential equation to obtain the rate at a given MgATP concentration. A plot of the observed myosin dissociation rates as a function of [MgATP] was linear and the slope corresponded to the rate constant expressed in M−1s−1.
In vitro motility assays
To determine a mutant myosin force generation capability, we employed the frictional loading assay where a non motor and low affinity actin-binding protein18 (ABP) was used as a mechanical load in a standard in vitro motility assay. The assay was performed as previously described [33]. Briefly, a nitrocellulose cover slip was attached to a glass microscope slide with double stick tape. The resulting channel formed a flow chamber where experimental solutions could be added. Myosin at 100 μg/ml was mixed with several different amounts of α-actinin in high-salt buffer (300 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2 and 1 mM DTT, pH 7.6) and introduced into the flow chamber and incubated for 2 min. The chamber was then washed with 30 μl of 0.5 mg/ml BSA in high-salt buffer. After 1 min BSA incubation, the chamber was washed with 60 μl of low-salt buffer (25 mM KCl, 25 mM imidazole, 1 mM EGTA, 4 mM MgCl2, and 1 mM DTT, pH 7.6). To remove any inactive myosin, 1 μM unlabeled actin, prepared from chicken skeletal muscle acetone powder using the method of Straub [34] with the modification of Drabikowski and Gergely [35], was added in low-salt buffer and allowed to bind to the myosin for ~2 min. Actin was washed with low-salt buffer in the presence of 1 mM ATP twice and in the absence of ATP four times. Tetramethylrhodamine isothiocyanate19 (TRITC)-phalloidin labeled actin (prepared by incubating a 1:1 molar ratio of TRITC phalloidin and actin in actin buffer overnight at 4°C) were used for this assay. Labeled actin filaments (~ 10 nM) in low-salt buffer were added and allowed to bind to the myosin in the absence of ATP. The movement of actin was initiated by the addition of low-salt buffer with 1 mM ATP, oxygen scavengers (17 units/ml glucose oxidase and 125 units/ml catalase), and 0.5% methylcellulose. Filament movement was observed at 30°C with an intensified charge-coupled device (ICCD) camera (IC300; PTI, Birmingham, NJ, USA). Video images were captured in sequence using Scion Image and an AG-5 image grabber (Scion Corp., Frederick, MD, USA). The average velocity for a given filament was determined from the distance traveled by the filament between 5–10 consecutive video images taken at 5-0.5 s intervals using Retrac, the freeware written by Dr. Nick Carter (http://mc11.mcri.ac.uk/Retrac). Velocities of 20–30 filaments from each video segment were averaged, and ≥2 video segments/flow cell were analyzed.
The load induced by α-actinin opposes actin filament movement and thus, increasing amounts of added α-actinin were to slow actin filament movement. For each α-actinin concentration, >10 filaments were analyzed for at least 3 separate areas of the flow cell resulting in > 360 measurements for each force determination. A plot of actin filament velocity vs. α-actinin concentration was fit to Eq. 2 to derive maximum filament velocity and myosin force production [33].
| Eq. 2 |
where Vactin is the maximal sliding velocity, Fd is the force of the bed of myosins, κ is the system compliance associated with the ABP and the ABP linkages, L is the length of a typical actin filament, r is the reach of an ABP to bind to an actin filament, kA is the second order rate constant for ABP attachment to actin, kD is the ABP detachment rate and ζ and χ are constants that define the surface geometry of ABPs. Significance was determined from the errors of the fit.
Steady-state force measurements
The porcine papillary muscle strips, reconstituted with RLC, were tested for steady-state force development in a pCa 4 solution (composition is the same as pCa 8 buffer except the [Ca2+] = 10−4 M) and relaxed in pCa 8 solution. Maximal force (in Newtons) was calculated per cross-section of muscle strip and expressed in kN/m2. The cross-sectional area was calculated based on measurement of the strip width using an SZ6045 Olympus microscope (zoom ratio of 6.3:1, up to 189 maximum magnification) and the assumption that the muscle strip is circular in diameter. The measurement was taken at ~3 points along the strips and averaged. Maximal steady-state force (pCa 4) was determined for all RLC recombinant proteins: WT, S15D, S15A, D166V, S15D-D166V and S15A-D166V reconstituted in RLC-depleted skinned porcine cardiac muscle strips.
Ca2+ -dependence of force development
To determine the force–pCa dependence, the strips were exposed to solutions of increasing Ca2+ concentrations (from pCa 8 to pCa 4) and force development was monitored. The data were analyzed using the Hill equation yielding the pCa50 and nH (Hill coefficient) [36].
SDS-PAGE
Control, CDTA-depleted and WT, S15D, S15A, D166V, S15D-D166V and S15A-D166V reconstituted porcine cardiac myosin and skinned muscle strips were run on 15% SDS-PAGE according to Laemmli [37]. The respective RLC protein bands were quantified utilizing densitometry with the Odyssey Software. The percent of RLC depletion and/or reconstitution was calculated from the net intensity of the RLC bands compared with the control untreated native cardiac muscle strips (100%). Differences in gel loading were corrected by densitometry of the ELC bands, as myosin ELC protein was not affected by the RLC extraction/reconstitution procedure.
Statistical analysis
Data are expressed as the average of n experiments ± SE (standard error). Student’s t-test was used for simple comparisons of two groups (e.g. non-phosphorylated vs. phosphorylated). For multiple comparisons between groups, a One Way Analysis of Variance (Sigma Plot 11; Systat Software, Inc., San Jose, CA, USA) was used. Statistically significant differences were defined as P < 0.05.
RESULTS
Binding of porcine myosin reconstituted with RLC proteins to pyrene labeled F-actin
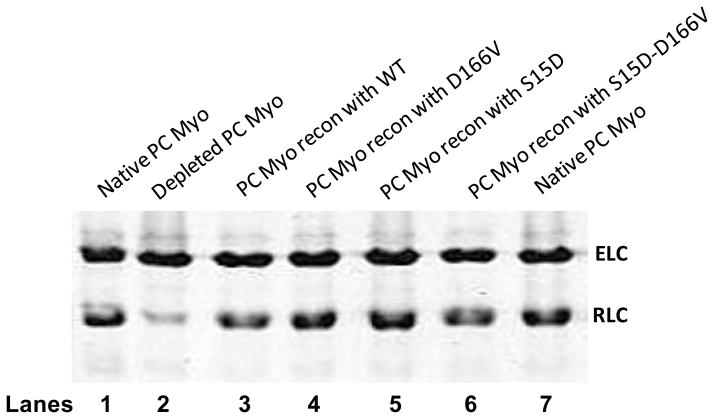
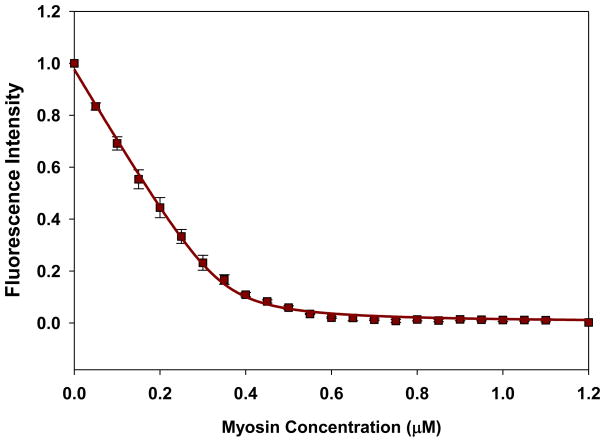
Porcine cardiac myosin was depleted of endogenous RLC in the presence of 1% Triton X-100 and 5 mM CDTA. Fig. 2 shows a representative gel image of native PC, PCdepl myosin and PCdepl myosin reconstituted with different RLC mutants. When compared with native PC myosin (lane 1), more than 80% of the RLC was removed from the CDTA treated myosin (lane 2). Incubation of PCdepl myosin with recombinant RLC proteins resulted in a reconstitution level of ~100% (lanes 3–6), thus showing that the depleted myosin was still able to bind the RLC after the extraction procedure. Mutant reconstituted myosin was then tested to measure its interaction with actin. First, we assessed the effect of RLC mutation or pseudo-phosphorylation on the binding of myosin to fluorescently labeled actin under rigor conditions. A decrease in the fluorescence intensity (quenching) induced by the binding of myosin to F-actin was measured as a function of increasing myosin concentrations and the titration data were fitted to Eq.1 (illustrated in Materials and Methods) to obtain the apparent dissociation constants (Kd) and stoichiometry of binding (n). Fig. 3 shows a representative curve and its fit. The binding of porcine myosin reconstituted with WT-RLC to actin was strong (in nM range) and the Kd value =19.3±2.4 nM, n=0.49 (n=10 experiments). We observed a slightly decreased binding [Kd =23.2±4.4 nM, n=0.51 (n=7)] for the RLC carrying the D166V mutation. Kd of S15D-myosin to F-actin was 10.3±2.7 nM, n=0.69 (n=7), suggesting a slight increase in binding compared with WT, while that of S15D-D166V was Kd=16.6±1.9 nM, n=0.67 (n=8) indicating a subtle rescue in binding affinity by pseudo-phosphorylated RLC. Thus the presence of S15D was able to increase binding (decrease Kd) in both WT and D166V. The differences in Kd values between D166V and S15D-D166V were statistically significant.
FIGURE 2.
Representative SDS-PAGE of porcine myosin reconstituted with recombinant RLC phosphomimetic proteins. The CDTA treatment of native porcine cardiac (PC) myosin (lanes 1 and 7) resulted in approximately 80% depletion of the endogenous RLC (lane 2). The RLC-depleted myosin was reconstituted with either WT (lane 3), D166V (lane 4), S15D (lane 5) or S15D-D166V (lane 6) RLC. The samples were run on 15% SDS PAGE gels and the percent of RLC reconstituted was calculated. ELC was used as a loading control. Abbreviations: Myo, myosin; ELC, myosin essential light chain; RLC, myosin regulatory light chain.
FIGURE 3.
A representative curve for fluorescence-based measurements of binding affinity of porcine cardiac myosin reconstituted with recombinant RLC-S15D protein to pyrene-labeled F-actin. F-actin labeled with pyrene iodoacetate was used at a concentration of 0.5 μM.
Actin activated myosin ATPase assays
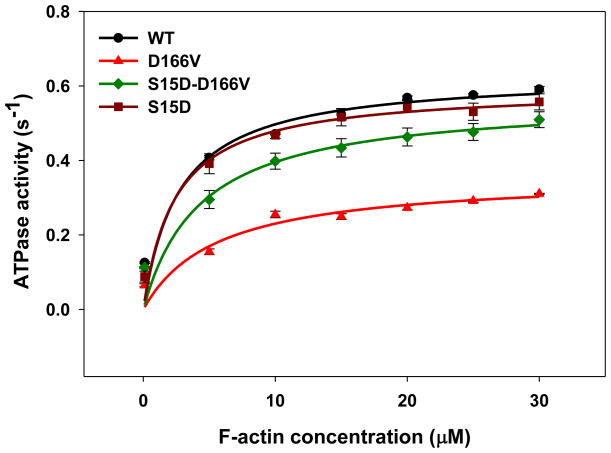
S15D and S15D-D166V reconstituted PC myosins were used to measure the effect of pseudo-phosphorylation of RLC on the ATPase activity compared to WT or D166V-reconstituted myosin preparations. Two different preps of myosins reconstituted with the respective RLC were used for this assay. The steady-state actin-activated myosin ATPase activity was determined as a function of increasing F-actin concentrations. As shown in Fig. 4, D166V largely decreased Vmax (maximal ATPase activity) compared to WT-myosin. Vmax for PC myosin reconstituted with D166V was 0.36±0.02 s−1 vs. 0.63±0.02 s−1 for WT, n=7 experiments. The difference between the groups was statistically significant (P<0.005). Therefore, similar to other effects of the disease causing mutation, D166V showed a 2-fold decrease in the maximal actin-activated myosin ATPase compared to WT. The presence of phosphomimic on the D166V background (S15D-D166V) rescued the maximal ATPase activity to 0.56±0.02 s−1 (n=8 experiments) (Fig. 4). Since Vmax represents the velocity of the cross bridge cycling, results from this experiment suggest that the D166V mutation slows down the cross-bridge turnover rate of the acto-myosin cycle. Decreased Vmax may also indicate a reduced number of myosin heads capable of interacting with actin. Remarkably, as above, the presence of pseudo-phosphorylated RLC (S15D-D166V) was able to recover the low level of ATPase activity to that observed for WT (Fig. 4). The Michaelis–Menten constant (Km in μM) for D166V-myosin: Km=5.6±0.9 (n=9 experiments) was significantly increased compared to WT-myosin: Km=2.7±0.5 (n=7), suggesting that a higher concentration of actin is needed to activate the D166V cross-bridges. As expected, myosin reconstituted with S15D-D166V RLC showed a small but significant increase in Km=4.3±0.9, n=8 experiments, compared with WT-myosin, P<0.001 (Fig. 4).
FIGURE 4.
Actin-activated myosin ATPase activity. Porcine myosins reconstituted with WT, D166V, S15D and S15D-D166V recombinant RLCs were used for this assay. Approximately, seven to eight independent experiments run in triplicate were performed for each group of myosins. Note that a significantly lower Vmax observed for D166V could be restored to the value close for WT myosin in the presence of the phosphomimic S15D-D166V.
Stopped flow measurements
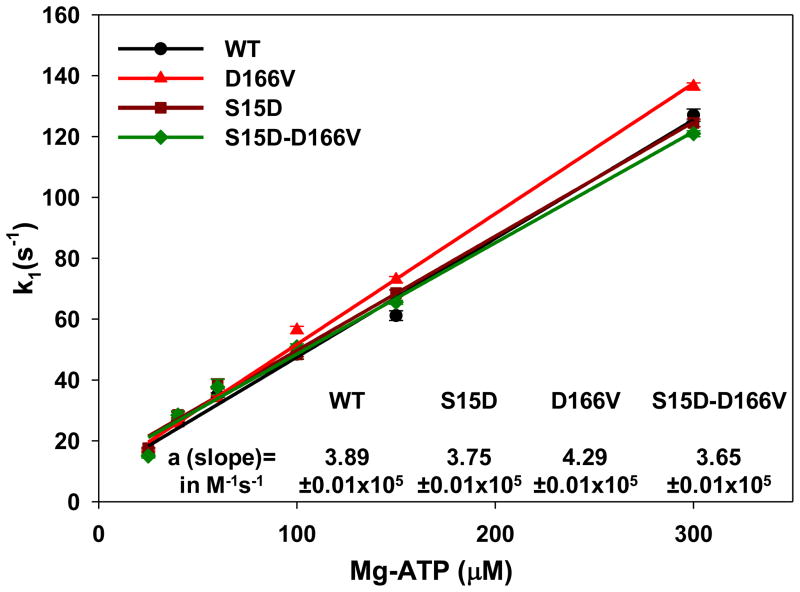
To further demonstrate the effect of the D166V mutation and the rescue by S15D-D166V on the interaction of myosin with actin, the MgATP-dependent transition of the strongly bound acto-myosin complex (M•A) to the weakly bound state (M•A•ATP) was measured using pyrene labeled F-actin. The time course of the recovery in the pyrene fluorescence was monitored as a function of ATP concentrations. Myosins were stoichiometrically mixed with pyrene-F-actin (two heads of myosin per actin monomer) and the complexes were mixed in a 1:1 volume ratio with the increasing concentrations of MgATP (25–300 μM) in a stopped flow apparatus. An increase in the fluorescence intensity on the addition of MgATP was monitored as a function of time. The observed rate constant (k1) for the M•A → M•A•ATP transition was derived from the averaged fluorescence traces and fitted with a single exponential dependence (Table 1). Compared to WT, slightly faster transition rates were monitored for S15D-reconstituted myosin at 25 and 40 μM MgATP while no differences were observed at higher MgATP concentrations. On the other hand, a small but significant effect of D166V on k1 was observed for 100, 150 and 300 μM MgATP indicating a faster k1 for D166V compared to WT (Table 1). No differences were seen for 25, 40 and 60 μM MgATP. As in previous assays, the presence of the S15D-D166V RLC reversed the subtle effect of D166V on k1 bringing it back to the values observed for WT-reconstituted myosin (Table 1). A plot of the observed transition rates (k1) as a function of [MgATP] is presented in Fig. 5 and shows a linear-type of dependence with the slope “a” values (in M−1 s−1) corresponding to the effective second-order MgATP binding rates. Significantly altered binding rates were observed for D166V compared to WT (Fig. 5). However, S15D-D166V once again recovered the second-order MgATP binding rates of D166V to those observed for WT.
TABLE 1.
Acto-myosin dissociation rates (k1 in s−1):
| System | MgATP concentration | |||||
|---|---|---|---|---|---|---|
| 25 μM | 40 μM | 60 μM | 100 μM | 150 μM | 300 μM | |
| WT | 15.70±0.69 | 26.25±0.38 | 35.04±0.73 | 48.45±1.62 | 61.13±1.59 | 127.03±1.99 |
| n=7 | n=7 | n=7 | n=7 | n=6 | n=7 | |
| S15D | 17.56±0.27 * | 28.52±0.93* | 38.72±1.69 | 48.38±0.78 | 68.55±1.07* | 124.48±1.24 |
| n=7 | n=7 | n=7 | n=7 | n=7 | n=7 | |
| D166V | 17.03±0.34 | 25.77±0.68 | 34.03±0.67 | 56.42±1.23* | 72.97±1.02* | 136.43±1.19* |
| n=23 | n=23 | n=23 | n=23 | n=23 | n=23 | |
| S15D-D166V | 15.00±0.66 | 28.46±1.37 | 37.69±2.53 | 50.88±0.99† | 65.51±0.66*† | 120.94±0.98*† |
| n=13 | n=13 | n=13 | n=13 | n=13 | n=13 | |
P value<0.05 compared to WT;
P value <0.01 compared to D166V
FIGURE 5.
The dissociation rate (k1) – [MgATP] dependence and the effective second-order MgATP binding rates (a=slope) for porcine myosin reconstituted with WT, S15D, D166V or S15D-D166V complexed with F-actin. The values of k1 ± SE for each MgATP concentration are presented in Table 1.
In vitro motility assays
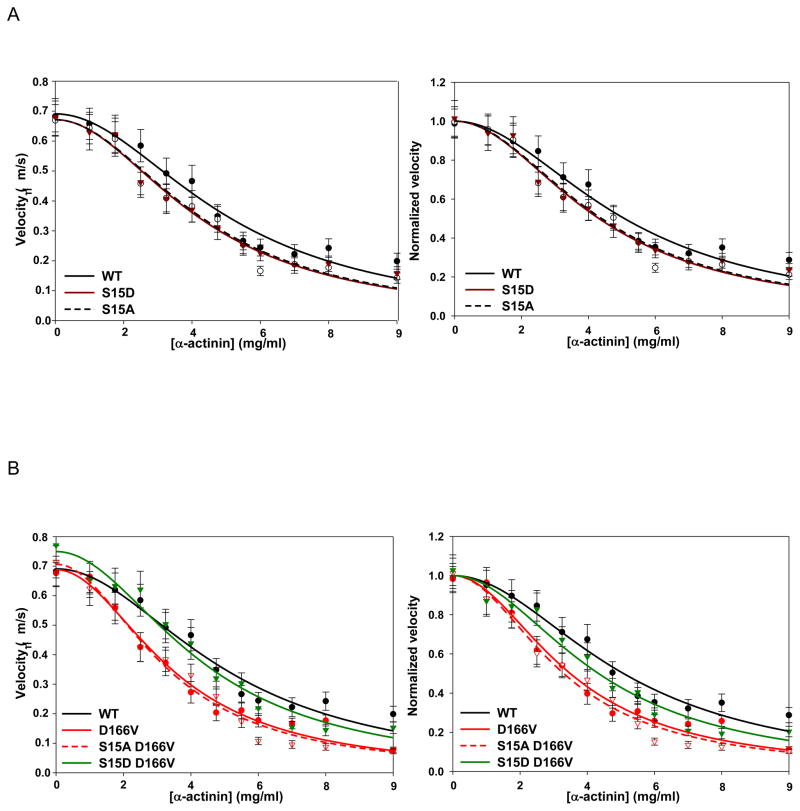
To determine whether the D166V mutation altered the force and motion generating capacity of myosin and the effect of pseudo-phosphorylation mutants, we used a frictional loading assay to measure loaded and unloaded velocity of actin filaments (Vactin) using in vitro motility assays (Fig. 6). In this experiment, α-actinin, a non-motor actin binding protein, was added to the conventional motility assay to apply a frictional load [33]. The α-actinin-induced slowing of actin filament motion provided a measure of the force-generating capacity of myosin [33]. Actin filament velocities driven by PC myosin reconstituted with WT, S15A, S15D, D166V, S15A-D166V and S15D-D166V were compared over a range of α-actinin concentrations. No differences in maximum velocities were observed between different RLC-reconstituted myosins under zero load (Table 2). On the other hand, WT, S15A and S15D were similarly sensitive to the frictional load imposed by α-actinin with S15A and S15D mutant myosins producing similar force values when compared to WT-reconstituted myosin (Fig. 6A; Table 2). Velocity of actin filaments propelled by the D166V mutant however, was more sensitive to the α-actinin load with a corresponding 2-fold decrease in driving force (2.04±0.20 μN) compared with WT (4.30±0.48 μN) (Fig. 6B; Table 2; P<0.0005). Importantly, S15D-D166V rescued the level of D166V-compromised force to 3.13±0.36 μN, showing statistically significant difference vs. D166V (Fig. 6B; Table 2; P<0.005). No statistically significant difference was observed between driving forces of WT and that of S15D-D166V rescue mutant (Table 2). Thus, a significant increase in force production ability was noted in the case of S15D-D166V reconstituted myosin.
FIGURE 6.
A. In vitro motility assay of porcine myosin reconstituted with WT and phosphomimic RLC proteins. Actin filament velocities over a range of α-actinin were determined and compared to porcine myosin reconstituted with WT, S15D and S15A recombinant RLC. Raw (left) and Vmax normalized (right) data are shown. The Fd values (in μN) for S15A and S15D were: 3.22±0.30 and 3.11± 0.33, and although slightly lower than WT value (4.30±0.48), the differences were not significant. B. Raw (left) and Vmax normalized (right) velocity data for mutant D166V; phosphorylation mimic, S15D-D166V; and control S15A-D166V. WT trace is shown for comparison. The Fd values (in μN) were 2.04±0.20 and 1.81±0.18 for D166V and S15A-D166V, while the Fd for S15D-D166V increased to 3.13±0.36. The Fd for S15D-D166V was significantly greater than D166V (P<0.005) and indistinguishable from WT. S15A-D166V Fd was not different from D166V, but significantly lower than WT (P<0.0001).
TABLE 2.
In Vitro motility assays:
| Maximum velocities | |||
|---|---|---|---|
| Myosin | Vactin (μm/s) | WT Comparison | D166V Comparison |
| WT | 0.69 ± 0.02 | ----- | No difference |
| S15A | 0.67 ± 0.02 | No difference | No difference |
| S15D | 0.67 ± 0.02 | No difference | No difference |
| D166V | 0.69 ± 0.02 | No difference | ----- |
| S15A-D166V | 0.71 ± 0.02 | No difference | No difference |
| S15D-D166V | 0.75 ± 0.03 | No difference | No difference |
| Myosin driving forces | |||
|---|---|---|---|
| Myosin | Force (μN) | WT Comparison | D166V Comparison |
| WT | 4.30 ± 0.48 | ----- | ↑ (P < 0.0005) |
| S15A | 3.22 ± 0.30 | No difference | ↑ (P < 0.005) |
| S15D | 3.11 ± 0.33 | No difference | ↑ (P < 0.05) |
| D166V | 2.04 ± 0.20 | ↓ (P < 0.0005) | ----- |
| S15A-D166V | 1.81 ± 0.18 | ↓ (P < 0.0001) | No difference |
| S15D-D166V | 3.13 ± 0.36 | No difference | ↑ (P < 0.005) |
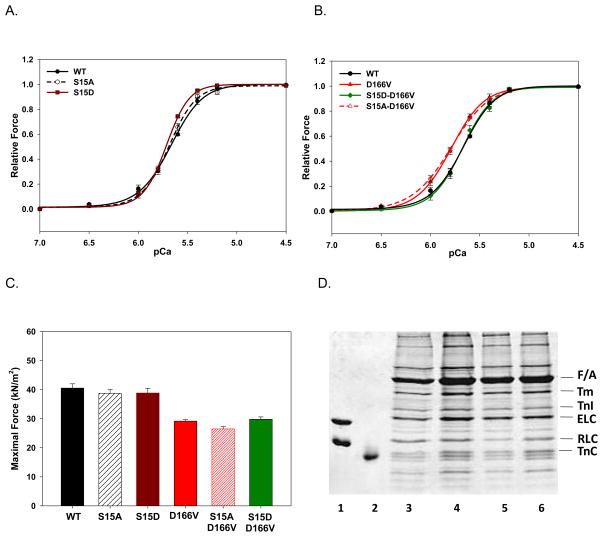
Ca2+ sensitivity and force development in reconstituted papillary muscle strips
Fig. 7 demonstrates the effect of D166V and pseudo-phosphorylation on the maximal force and force-pCa dependence measured in reconstituted skinned porcine papillary muscle strips. There was a small but significant increase in the Ca2+ sensitivity of force between WT and S15D reconstituted preparations (Fig. 7A, P<0.05). As we showed before [23], the presence of D166V caused a leftward shift toward lower [Ca2+] (ΔpCa50 ~0.12) of the force-pCa relationship compared to WT-reconstituted strips (Fig. 7B). Importantly, the presence of phosphomimetic mutation in the background of D166V (S15D-D166V) brought the Ca2+ sensitivity back to that observed for the WT reconstituted strips (Fig. 7B). Therefore, compromised myofilament calcium sensitivity of D166V was able to be rescued by pseudo-phosphorylated S15D-D166V (Fig. 7B). When the strips were reconstituted with non-phosphorylatable S15A-D166V protein, there was no change in the Ca2+ sensitivity of force compared with the D166V-reconstituted muscle preparations (Fig. 7B).
FIGURE 7.
The Ca2+ sensitivity of force development (A, B) and maximal tension (C) in porcine papillary muscle strips reconstituted with WT and phosphomimetic RLCs (D). Reconstitution of the RLC-depleted preparations with porcine TnC and RLC- WT, S15D, S15A, D166V, S15D-D166V and S15A-D166V was performed in pCa 8 solution containing 40 μM RLC and 15 μM TnC. The solution of TnC was included in the reconstitution protein mixture during the first 30 min of strip incubation followed by a 30-min incubation with fresh RLC solution at room temperature. The addition of TnC was to assure that the strips were not deficient in TnC as its partial extraction could affect the Ca2+ sensitivity of force development and the maximal level of force. After the strips were washed with the pCa 8 solution, they were subjected to steady-state force measurements. The respective midpoint values (pCa50) of force-pCa curves were: A. WT: 5.67±0.01 (n =18); S15A: 5.69±0.01 (n =10); S15D: 5.72±0.01 (n=12). B. D166V: 5.79±0.01 (n=11); S15A-D166V: 5.80±0.026 (n=12) and S15D-D166V: 5.68±0.01 (n=9). The Hill coefficients (nH) for the force-pCa dependences are depicted. Data represent mean values ± SE of n experiments. The WT curve is shown for comparison. C. Maximal force measured in pCa 4 after RLC and TnC reconstitution. Note that the D166V mutation decreased maximal force to 29.2±0.5 kN/m2, n=12 compared to WT (40.5±1.5 kN/m2, n=12), S15A (38.7±1.3 kN/m2, n=9) or S15D (38.8±1.6 kN/m2, n=8)- reconstituted strips. The phosphomimic S15D-D166V protein did not cause any significant changes in force (29.8±0.8 kN/m2, n=12) compared to D166V-reconstituted strips. But there was a significant difference in force between S15A-D166V (26.5±0.8 kN/m2, n=9) and S15D-D166V strips. Data represent mean values ± SE of n experiments. D. Representative SDS–PAGE of CDTA depleted and RLC/TnC reconstituted porcine papillary muscle strips. Lane 1: Human WT ELC and RLC standards (ELC was used as a loading control). Lane 2: TnC standard. The amount of RLC present in the strips was as follows: Native muscle strips, 100% (lane 3); RLC-depleted strips collected after initial depletion for 5 min, ~50% (lane 4); RLC-depleted strips collected after additional 30 min of depletion, <20% (lane 5); RLC and TnC-reconstituted strips, ~90% (lane 6). Abbreviations: Tm, tropomyosin; TnI, Troponin I; TnC, Troponin C.
To further understand the effect of pseudo-phosphorylation on contractile force developed at saturating calcium concentrations (pCa 4), the level of tension per cross-section of muscle strip in the preparations reconstituted with all phosphomimic proteins was measured (Fig. 7C). As before [23], a large decrease in maximal force was observed for strips reconstituted with D166V-RLC (~29 kN/m2) compared with WT (~41 kN/m2) (Fig. 7C). However, neither S15A-D166V (~39 kN/m2) nor S15D-D66V (~30 kN/m2) changed the low level of D166V force (Fig. 7C). Noteworthy, there was a small but significant difference in force between S15A-D166V (~26 kN/m2) and S15D-D166V strips (Fig. 7C, P<0.05), but no recovery in maximal force to the level of WT was achieved with S15D-D166V (Fig. 7C). This result was in accord with the data reported previously [24], where MLCK-treated D166V strips showed the same level of force as non-phosphorylated D166V strips. Therefore, the force data obtained in the in vitro motility assay using monomeric myosin reconstituted with S15D-D166V were not supported by the results from S15D-D166V reconstituted muscle preparations. Perhaps, ~20% difference in the efficiency of S15D-D166V reconstitution between myosin (100%) vs. muscle strips (~80%) (Fig. 7D) accounted for this discrepancy. Other possibilities are discussed below.
DISCUSSION
This study was designed to explore the effectiveness of pseudo-RLC phosphorylation as a rescue technique for treating the D166V-induced detrimental phenotype shown in humans and also manifested in transgenic mice [10, 11, 23]. We used porcine cardiac preparations that were previously depleted of the endogenous RLC protein and subsequently reconstituted with D166V and S15D-D166V mutants. The results were compared with those obtained for WT, S15A and S15D - reconstituted preparations. Our findings suggest a novel rescue strategy to ameliorate a malignant cardiomyopathic phenotype associated with the D166V mutation in myosin RLC.
The D166V mutation occurs at the last amino acid residue of the human cardiac RLC with the negatively charged and polar aspartic acid being replaced by the hydrophobic bulky valine residue (Fig. 1). The mutation lies at a critical region of the RLC that makes contact with the bend structure of the myosin lever arm [8]. It is well accepted that the role of the myosin lever arm is to amplify the small conformational changes that occur at the nucleotide and/or actin-binding sites of the myosin head into the large changes that ultimately result in directed movement, sarcomeric shortening and force generation in muscle [9]. Being hydrophobic and C-β branched, the valine residue introduces more bulkiness near the protein backbone and it is possible that the mutation may cause some steric inhibition of the lever arm swing. This could result in somewhat increased restrictions in the conformation that the myosin lever arm can adopt during the cross bridge cycle. On the other hand, it has been suggested that phosphorylation of the RLC may increase the mobility of myosin cross-bridges and perturb the relaxed orientation of the myosin heads [38–40]. In this study we hypothesized that pseudo-phosphorylation of D166V, by introducing a negative charge at Ser15, could ameliorate the steric constraints introduced by the valine residue in this conformation-sensitive region of the myosin lever arm and consequently rescue the functional effects of D166V. The study was performed using increasing levels of system complexity (myosin, acto-myosin and muscle strips).
Although the role of RLC phosphorylation on cardiac contractility is still elusive, clinical studies have linked the changes in myosin RLC phosphorylation with the pathology of the human heart. For example, patients with heart failure demonstrated complete dephosphorylation of RLC [12, 17, 19, 41]. Therefore, increasing or decreasing levels of RLC phosphorylation have been associated with a positive or negative inotropic state of the heart [16]. The Stull laboratory has reported that ablation of RLC phosphorylation in cardiac MLCK knockout mice resulted in cardiomyocyte hypertrophy with histological evidence of necrosis and fibrosis [42]. Recent investigations also demonstrated that pressure overload could cause severe heart failure in cardiac MLCK knockout mice but not in mice with MLCK over-expression [13]. In addition, another group reported on a direct correlation between the lack of RLC phosphorylation and abnormal cardiac chamber enlargement and wall thinning in genetically altered mice; thus showing a predisposition of the hearts to cardiomyopathy [43]. Very recent studies on permeabilized rat trabeculae preparations demonstrated the importance of increased RLC phosphorylation in the up-regulation of myocardial performance [12].
The focus of this report was to determine whether pseudo-phosphorylation of myosin RLC could have a positive impact on cardiac muscle function in a system containing the FHC-linked D166V - RLC mutation. We used β-MHC containing porcine cardiac myosin preparations reconstituted with various RLC phosphomimic proteins. Noteworthy, the β-MHC is the same MHC isoform found in the human heart. In addition, it has been postulated that expression levels of α- and β-MHC isoforms can be altered in disease states [44]. Studies on failing mouse hearts revealed a shift from the normally predominant α-MHC toward β-MHC [45]. Therefore, our study on porcine reconstituted preparations was performed in the physiologically relevant background of β-MHC. Porcine myosin/muscle strips were depleted of endogenous RLC and reconstituted with the RLC of interest. Importantly, using recombinant pseudo-phosphorylation proteins prevented any discrepancies that could be introduced by disproportionate phosphorylation of RLC WT vs. mutant by MLCK [24]. We also aimed to determine whether phosphorylation-mediated rescue could be achieved independently of the endogenous activity of the MLCK-phosphatase enzymes.
Since the RLC plays an important role in stabilizing the structure and the position of the myosin heads relative to the thin filaments [8, 16], the effect of the mutation on the acto-myosin interaction was examined. First, fluorescence binding measurements were applied to test the effect of the mutation on the ability of myosin to strongly bind to actin (under rigor conditions). The results showed a small decrease in the affinity in the presence of the D166V mutation compared with WT, which was subsequently increased on addition of S15D-D166V-reconstituted myosin. This result suggested that pseudo-phosphorylation of RLC could indeed play a role in the recovery process to regain normal RLC function. Results from the stopped-flow measurements assayed for the effective second-order ATP binding rates to the acto-myosin complex(slope of the k1-[MgATP] dependence) revealed significant differences between WT and D166V reconstituted myosins and a recovery of a D166V-induced increase in binding rates by S15D-D166V (Fig. 5). The data indicated a predisposition of porcine myosins reconstituted with the D166V mutant to undergo the faster M•A → M•A•ATP transition.. These results suggested that the cross bridges carrying the mutation might be predisposed to spend more time in the weakly bound state compared to WT. This could result in a lower force exerted by the D166V cross-bridges as observed here (Fig. 7C) and shown previously [23, 24]. Noteworthy, pseudo-phosphorylation of D166V decreased the effective second-order ATP binding rates to the level of WT demonstrating its capacity to retune the cross bridge kinetics to the optimal level and reverse any subtle conformational distortion in the myosin head structure that might be induced by the mutation.
We then evaluated the effect of mutations on myosin enzymatic properties and its interaction with actin. Actin-activated myosin ATPase activity assays demonstrated an approximate 50% decrease in the maximal ATPase activity in D166V myosin compared with the WT myosin. The D166V mutation also increased the Km constant compared with WT (Fig. 4). Based on the above studies, it is likely that the mutation causes a lower affinity for actin compared to WT and decreases the cross-bridge turnover rate. Remarkably, pseudo-phosphorylated S15D-D166V mutant increased the binding of myosin to actin (decreased Km) and increased the cross-bridge turnover rate (increased Vmax) to the level observed for the WT myosin. One can speculate that the S15D-D166V cross-bridges may cycle with a higher turnover rate compared to D166V or that the S15D mutation restores the activity of D166V cross bridges to participate in the ATPase cycle resulting in higher Vmax. These data were supported by our in vitro motility assays depicting the force generating capacity of myosin. Porcine myosin reconstituted with the D166V mutant significantly decreased Fd compared to WT (Table 2) while the phosphomimetic S15D-D166V mutant rescued the D166V-induced phenotype (Fig. 6, Table 2). Interestingly, we have observed a similar effect with the MLCK-induced phosphorylation of myosin purified from Tg-D166V muscle (data not shown). It is worth mentioning that in both assays the rescuing effect of S15D was only manifested through mitigating the D166V-induced phenotype while not changing the properties of WT myosin.
Measurements of force in porcine papillary muscle strips reconstituted with different RLC proteins showed a D166V-dependent increase in myofilament Ca2+ sensitivity (Fig. 7B), the phenomenon observed previously in transgenic D166V mouse preparations [23, 24, 46]. Noteworthy, increased Ca2+ sensitivity of contraction was observed in many FHC mutant proteins and has been considered a hallmark trait of FHC [47]. Ca2+ sensitization due to FHC causing mutations has been shown to cause increases in cytosolic Ca2+ buffering and ultimately resulting in arrhythmogenic changes in Ca2+ homeostasis in the intact mouse heart [48]. Consistently with other assays, pseudo-phosphorylation of D166V cross-bridges (with S15D) reversed this abnormal shift in calcium sensitivity bringing the pCa50 value to the level observed for WT (Fig. 7B). As before [23], we observed a decrease in maximal pCa 4 force in D166V compared with the WT reconstituted strips (Fig. 7C). Impaired myofilament contractile function manifested by increased Ca2+ sensitivity and decreased contractile force has been suggested to be the most common feature of FHC accounting for compensatory hypertrophy and diastolic dysfunction in FHC patients [47]. At the molecular level, one can hypothesize that the D166V mutation leads to a decreased number of strongly bound cross bridges resulting in decreased contractile force measured per cross-section of muscle at maximal Ca2+ activation. However, unlike in the in vitro motility assays where S15D-D166V rescued the low level of the D166V force (Fig. 6, Table 2), no effect was observed in S15D-D166V muscle preparations (Fig. 7C). The latter result is in line with the lack of force recovery observed in MLCK-phosphorylated Tg-D166V strips [24]. Based on these results, the S15D-D166V rescue mutation may mitigate the disease phenotype by desensitizing the heart to calcium, thus preventing an early activation of contraction observed in the D166V myocardium.
In conclusion, several important findings were noted in this report. First, we demonstrated that the RLC reconstituted porcine cardiac model system is well suited to replicate the profound functional phenotype associated with the D166V mutation observed in humans [10, 11] and in transgenic mice [23]. Secondly, similar to our reports from the α-MHC containing transgenic mice, we showed that D166V mutation affects the kinetics and function of myosin and its interaction with actin. Finally, we show that a constitutive S15D-D166V phosphomimic can reverse the majority of D166V-induced phenotypes in vitro. Follow up studies in vivo, using rescue animal models, are necessary to further investigate the beneficial role of pseudo-phosphorylation in restoring the compromised function of D166V mutated myocardium and to conclude on the effectiveness of pseudo-phosphorylation in combating malignant FHC-RLC phenotypes.
Highlights.
We examined the functional effects of pseudo-phosphorylation of myosin RLC mutant.
The D166V-RLC mutant was shown to cause FHC and abnormal cardiac function in mice.
Pseudo-phosphorylation of D166V reversed many of its detrimental phenotypes.
Phosphorylated RLC can serve as a potential therapeutic target for cardiomyopathy.
Acknowledgments
We thank Dr. Katarzyna Kazmierczak and Ms. Michelle Jones for their help in cloning, protein purification and stopped-flow experiments. This work was supported by National Institutes of Health Grants HL071778 and HL108343 (to D.S-C.), HL077280 (to J.R.M.) and American Heart Association Grant 10POST3420009 (to P.M.).
Footnotes
FHC, familial hypertrophic cardiomyopathy
SCD, sudden cardiac death
RLC, regulatory light chain of myosin
D166V, aspartic acid to valine mutation
MLCK, myosin light chain kinase
WT-RLC, human ventricular wild-type RLC, Swiss-Prot: P10916
MHC, myosin heavy chain
PMSF, phenylmethylsulfonyl fluoride
DTT, dithiothreitol
PC, porcine cardiac
EDTA, ethylenediaminetetraacetic acid
MOPS, 3-(N-morpholino)propanesulfonic acid
Tm-Tn, tropomyosin-troponin complex
PIA, pyrene iodoacetamide
CDTA, cyclohexane-1,2-diaminetetraacetic acid
TnC, troponin C
TCA, trichloroacetic acid
ABP, actin-binding protein
TRITC, tetramethylrhodamine isothiocyanate
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Teekakirikul P, Padera RF, Seidman JG, Seidman CE. J Cell Biol. 2012;199:417–421. doi: 10.1083/jcb.201207033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olivotto I, Girolami F, Nistri S, Rossi A, Rega L, Garbini F, Grifoni C, Cecchi F, Yacoub MH. J Cardiovasc Transl Res. 2009;2:349–367. doi: 10.1007/s12265-009-9137-2. [DOI] [PubMed] [Google Scholar]
- 3.Alcalai R, Seidman JG, Seidman CE. J Cardiovasc Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 4.Maron BJ. JAMA: the journal of the American Medical Association. 2002;287:1308–1320. [Google Scholar]
- 5.Harris SP, Lyons RG, Bezold KL. Circulation research. 2011;108:751–764. doi: 10.1161/CIRCRESAHA.110.231670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szczesna D. Curr Drug Targets Cardiovasc Haematol Disord. 2003;3:187–197. doi: 10.2174/1568006033481474. [DOI] [PubMed] [Google Scholar]
- 7.Muthu P, Huang W, Kazmierczak K, Szczesna-Cordary D. In: Cardiomyopathies – From Basic Research to Clinical Management. Veselka J, editor. Vol. 17. InTech; Croatia: 2012. pp. 383–408. [Google Scholar]
- 8.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 9.Geeves MA. Nature. 2002;415:129–131. doi: 10.1038/415129a. [DOI] [PubMed] [Google Scholar]
- 10.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet J-P, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M for the EUROGENE Heart Failure Project, . Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 11.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet J-P, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M for the EUROGENE Heart Failure Project, . Circulation. 2004;109:3258. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 12.Toepfer C, Caorsi V, Kampourakis T, Sikkel MB, West TG, Leung MC, Al-Saud SA, MacLeod KT, Lyon AR, Marston SB, Sellers JR, Ferenczi MA. J Biol Chem. 2013;288:13446–13454. doi: 10.1074/jbc.M113.455444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren SA, Briggs LE, Zeng H, Chuang J, Chang EI, Terada R, Li M, Swanson MS, Lecker SH, Willis MS, Spinale FG, Maupin-Furlowe J, McMullen JR, Moss RL, Kasahara H. Circulation. 2012;126:2575–2588. doi: 10.1161/CIRCULATIONAHA.112.116202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamm KE, Stull JT. J Biol Chem. 2011;286:9941–9947. doi: 10.1074/jbc.R110.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colson BA, Locher MR, Bekyarova T, Patel JR, Fitzsimons DP, Irving TC, Moss RL. J Physiol. 2010;588:981–993. doi: 10.1113/jphysiol.2009.183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morano I. J Mol Med. 1999;77:544–555. doi: 10.1007/s001099900031. [DOI] [PubMed] [Google Scholar]
- 17.van der Velden J, Papp Z, Boontje NM, Zaremba R, de Jong JW, Janssen PM, Hasenfuss G, Stienen GJ. Adv Exp Med Biol. 2003;538:3–15. doi: 10.1007/978-1-4419-9029-7_1. [DOI] [PubMed] [Google Scholar]
- 18.van der Velden J, Papp Z, Boontje NM, Zaremba R, de Jong JW, Janssen PML, Hasenfuss G, Stienen GJM. Cardiovascular Research. 2003;57:505–514. doi: 10.1016/s0008-6363(02)00662-4. [DOI] [PubMed] [Google Scholar]
- 19.van der Velden J, Papp Z, Zaremba R, Boontje NM, de Jong JW, Owen VJ, Burton PBJ, Goldmann P, Jaquet K, Stienen GJM. Cardiovasc Res. 2003;57:37–47. doi: 10.1016/s0008-6363(02)00606-5. [DOI] [PubMed] [Google Scholar]
- 20.Szczesna D, Ghosh D, Li Q, Gomes AV, Guzman G, Arana C, Zhi G, Stull JT, Potter JD. J Biol Chem. 2001;276:7086–7092. doi: 10.1074/jbc.M009823200. [DOI] [PubMed] [Google Scholar]
- 21.Holroyde MJ, Robertson SP, Johnson JD, Solaro RJ, Potter JD. J Biol Chem. 1980;255:11688–11693. [PubMed] [Google Scholar]
- 22.Robertson SP, Johnson JD, Potter JD. Biophys J. 1981;34:559–569. doi: 10.1016/S0006-3495(81)84868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerrick WGL, Kazmierczak K, Xu Y, Wang Y, Szczesna-Cordary D. FASEB J. 2009;23:855–865. doi: 10.1096/fj.08-118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muthu P, Kazmierczak K, Jones M, Szczesna-Cordary D. J Cell Mol Med. 2012;16:911–919. doi: 10.1111/j.1582-4934.2011.01371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. 1995 [Google Scholar]
- 26.Pant K, Watt J, Greenberg M, Jones M, Szczesna-Cordary D, Moore JR. FASEB J. 2009;23:3571–3580. doi: 10.1096/fj.08-126672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardee JD, Spudich JA. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 28.Cooper A, Walker SB, Pollard TD. J Muscle Res Cell Mot. 1983;4:253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 29.Kazmierczak K, Muthu P, Huang W, Jones M, Wang Y, Szczesna-Cordary D. Biochem J. 2012;442:95–103. doi: 10.1042/BJ20111145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiske CH, Subbarow Y. J Biol Chem. 1925;66:375–400. [Google Scholar]
- 31.Hanson KR, Ling R, Havir E. Biochem Biophys Res Commun. 1967;29:194–197. doi: 10.1016/0006-291x(67)90586-4. [DOI] [PubMed] [Google Scholar]
- 32.Trybus KM. Methods. 2000;22:327–335. doi: 10.1006/meth.2000.1085. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg MJ, Kazmierczak K, Szczesna-Cordary D, Moore JR. Proc Natl Acad Sci USA. 2010;107:17403–17408. doi: 10.1073/pnas.1009619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Straub FB. In: Stud Inst Med Chem Univ Szeged. 1942;2:3–16. [Google Scholar]
- 35.Drabikowski W, Gergely J. In: Biochemistry of muscle contraction. Gergely J, editor. Boston: Little, Brown, & Co; 1964. pp. 125–31. [Google Scholar]
- 36.Hill TL, Einsenberg E, Greene LE. Proc Natl Acad Sci. 1980;77:3186–3190. doi: 10.1073/pnas.77.6.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Cooke R. Biophys Rev. 2011;3:33–45. doi: 10.1007/s12551-011-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craig R, Alamo L, Padron R. J Mol Biol. 1992;228:474–487. doi: 10.1016/0022-2836(92)90836-9. [DOI] [PubMed] [Google Scholar]
- 40.Midde K, Rich R, Marandos P, Fudala R, Li A, Gryczynski I, Borejdo J. J Biol Chem. 2013 doi: 10.1074/jbc.M112.434209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morano I. Basic Res Cardiol. 1992;87:129–141. doi: 10.1007/978-3-642-72474-9_11. [DOI] [PubMed] [Google Scholar]
- 42.Ding P, Huang J, Battiprolu PK, Hill JA, Kamm KE, Stull JT. J Biol Chem. 2010;285:40819–40829. doi: 10.1074/jbc.M110.160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheikh F, Ouyang K, Campbell SG, Lyon RC, Chuang J, Fitzsimons D, Tangney J, Hidalgo CG, Chung CS, Cheng H, Dalton ND, Gu Y, Kasahara H, Ghassemian M, Omens JH, Peterson KL, Granzier HL, Moss RL, McCulloch AD, Chen J. J Clin Invest. 2012;122:1209–1221. doi: 10.1172/JCI61134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadal-Ginard B, Mahdavi V. J Clin Invest. 1989;84:1693–1700. doi: 10.1172/JCI114351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harada K, Sugaya T, Murakami K, Yazaki Y, Komuro I. Circulation. 1999;100:2093–2099. doi: 10.1161/01.cir.100.20.2093. [DOI] [PubMed] [Google Scholar]
- 46.Borejdo J, Szczesna-Cordary D, Muthu P, Metticolla P, Luchowski R, Gryczynski Z, Gryczynski I. Methods Mol Biol. 2012;875:311–334. doi: 10.1007/978-1-61779-806-1_17. [DOI] [PubMed] [Google Scholar]
- 47.Marston SB. J Cardiovasc Transl Res. 2011;4:245–255. doi: 10.1007/s12265-011-9266-2. [DOI] [PubMed] [Google Scholar]
- 48.Schober T, Huke S, Venkataraman R, Gryshchenko O, Kryshtal D, Hwang HS, Baudenbacher FJ, Knollmann BC. Circulation research. 2012;111:170–179. doi: 10.1161/CIRCRESAHA.112.270041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houdusse A, Cohen C. Structure. 1996;4:21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]