Abstract
The field of synthetic biology seeks to engineer reliable and predictable behaviors in organisms from collections of standardized genetic parts. However, unlike other types of machines, genetically encoded biological systems are prone to changes in their designed sequences due to mutations in their DNA sequences after these devices are constructed and deployed. Thus, biological engineering efforts can be confounded by undesired evolution that rapidly breaks the functions of parts and systems, particularly when they are costly to the host cell to maintain. Here, we explain the fundamental properties that determine the evolvability of biological systems. Then, we use this framework to review current efforts to engineer the DNA sequences that encode synthetic biology devices and the genomes of their microbial hosts to reduce their ability to evolve and therefore increase their genetic reliability so that they maintain their intended functions over longer timescales.
Introduction
The field of synthetic biology seeks to construct complex biological devices with predictable behaviors from standardized genetic parts.1–4 It draws a conceptual framework and inspiration from disciplines such as computer science (e.g., refactoring the DNA code of biological systems to make them more modular and portable) and engineering (e.g. host organisms are chassis for genetic circuits). However, designing devices that can be deployed in living organisms presents new challenges when compared with engineering inanimate materials. One key difference is that biological systems are able to reproduce, and sequence errors are sometimes introduced in the genetic information of their offspring – simply put, these machines evolve.5–7 Copying errors will most often degrade the information in the DNA blueprint for a biological device and cause it to stop operating as intended. On rare occasions, however, these errors may improve the function of a complex biological device or even lead to unexpected, but useful new properties. Evolution is therefore an important process to account for in the design of genetically engineered machines.
For anticipating the effects of evolution on engineered biological devices, one must understand the fundamental properties that determine the evolutionary potential – or evolvability – of a system. The process of Darwinian evolution involves selection for the most-fit variants in a population of organisms. Genetic variation arises when mutations create new DNA sequence variant or when sexual recombination between genomes produces new combinations of DNA sequence variants in a genome. In addition to genetic changes that are directly beneficial or deleterious to the chances that an organism survives and reproduces, genetic changes may occur during evolution that impact the ability of an organism and its progeny to continue to further evolve or adapt.8–11 There are two general categories of explanations for differences in evolvability (Fig. 1). First, two organisms may have different mutation or recombination rates, such that there are likely to be more, fewer, or different genetic variants among their progeny.12 Second, there may be an effect of genetic background, such that the exact same set of mutations in the offspring of two organisms would impact their survival and reproduction in different ways due to genetic interactions with other evolved DNA sequence changes.13 Both types of evolvability differences have been found to arise spontaneously and determine long-term outcomes in experimental populations of adapting microorganisms.14
Fig. 1. What is evolutionary potential and how can it be engineered?
At its most basic level, an organism's evolutionary potential (or evolvability) is its capacity for producing viable offspring with genetic variation. Evolvability is often used to refer more specifically to the relative chances of accessing mutations or pathways consisting of multiple mutational steps that produce a beneficial change in the fitness of an organism, such that it can more rapidly reproduce or better survive to outcompete its progenitor. Different DNA sequences and host organism chassis can be used to engineer biological systems with similar or identical properties. Some of these choices may be more or less evolvable by two general mechanisms. First, there may be different rates of mutations in the genetic information used to encode two systems with the same function. Second, the same set of mutations may affect the properties of two systems with the same function in different ways. In the context of synthetic biology, it may be useful to reduce evolvability when mutations that inactivate an engineered biological device give the host organism a selective fitness advantage, such that malfunctioning systems rapidly take over a population of cells after they evolve.
What is a biological engineer to do when faced with the prospect of seemingly inevitable and unpredictable evolution of their carefully designed device? We posit that many synthetic biology efforts would benefit from taking one of two perspectives. First, evolution may be regarded as a nuisance parameter unique to biology that leads to unexpected variation in the function of genetically engineered machines and eventual malfunction over time.5, 6 Therefore, designed DNA sequences and host organisms should be made robust to this failure mode by minimizing error rates in genetic transmission and preventing certain evolutionary trajectories. Second, evolution may be regarded as a useful tool for optimizing the function of complex biological systems or for discovering variants of genetic parts with new properties.15 With these objectives in mind, living systems should be rationally engineered for increased evolvability by expanding the sequence space that can be explored or by creating specific types of genetic variation that are more likely to be beneficial for tuning or rewiring devices than random spontaneous mutations.
In this review, we discuss the first of these two perspectives on the relationship of evolution and synthetic biology: how the evolutionary potential of biological devices deployed in microorganisms can be reduced by engineering approaches. We do not discuss non-genetic sources of cell-to-cell phenotypic variability (e.g., stochastic gene expression) that introduce another source of variation that can confound the reliable function of biological systems.16, 17 Similarly, readers are directed elsewhere for discussions of how the dynamics of competition in populations of microorganisms affects the speed of evolution14, 18, 19. While great progress has already been made in some areas to tame and harness microbial evolution, many of the strategies for reducing evolvability that we will discuss have not yet been systematically tested or have not yet been applied on the scale of multi-gene biological devices or whole organisms.
Genetic reliability
Synthetic biology devices commonly impose a burden on their host organisms that is at odds with their long-term survival in a population competing for resources. This fitness cost may be due to the metabolic toll of constructing additional RNAs and proteins that are not required for cellular reproduction20, 21 or due to heterologous genetic parts that interfere with the efficient operation of native cellular processes.22 As a result of this cost, inactivating mutations that disable engineered DNA constructs may be strongly favored by selection. The genetic reliability of a biological device can been described in terms of an evolutionary half-life: the number of cell doublings (generations of growth) over which 50% of the cells in a population maintain a genetically intact copy of the original device (Fig. 2).1, 23 Typical engineered systems with a few genetic parts may be reliable for ~100 generations of Escherichia coli host growth1, 23. This limit may pose little problem for relatively simple devices intended to function over only one day of bacterial growth under laboratory conditions (6–25 generations), but greater design complexity in an engineered system and longer timescales of deployment make it progressively more difficult to maintain the desired function. Thus, mutations caused the evolutionary meltdown of circuits designed to limit population growth within 3–6 days.24 At the extreme, a system of engineered E. coli 'predator' and 'prey' cell types with quorum sensing circuits had to be studied in microchemostats because it became largely nonfunctional within the number of cell division required to achieve a normal bacterial culture size.25
Fig. 2. Measuring the genetic reliability of biological machines.
(A) Genetic reliability has been defined as the evolutionary half-life (E½) of a synthetic biology device, measured as the number of cell doublings over which 50% of the engineered function (e.g., output of a reporter gene) is maintained in a microbial culture.1, 23 This value can be measured by monitoring device function while propagating cells through many cycles of serial transfer and regrowth or during continuous growth in a chemostat. Two different example decay curves are shown, for device 1 and for a more genetically reliable device 2. (B) Inactivation of many synthetic biology devices is dominated by a single mutational event that leads to a complete loss of the designed function. In this situation, all cells in a population have either 100% or 0% function at any given time. This distribution of activities present in the population at selected time points is shown on separate vertical axes for device 1 with each horizontal peak containing the cumulative number of cells with a given activity. This distribution would be obtained by measuring expression of a fluorescent protein reporter in each cell in the population by flow cytometry, for example. In this single-hit inactivation regime, competition within a microbial culture will lead to a relatively sharp decline in overall device function as one or more new mutant cells take over population when loss of the engineered function confers a fitness benefit. (C) The function of other synthetic biology designs like device 2 may degrade through accumulating multiple mutations that each partially reduce the activity (as shown) or as mutations that partially inactivate the engineered function activity compete with mutations that completely inactivate it. Competition within a microbial culture in this case may lead to a more gradual and complex decal curve for the level of function in the overall population over time.
In these situations, where mutations stochastically degrade the information in carefully planned synthetic biology devices and selection exponentially amplifies failures, one would like to stop or at least slow evolution. Conceptually, the simplest way to accomplish this aim is to lower the rates at which mutations that compromise devices arise in the first place. Genome-wide spontaneous mutation rates in microorganisms are thought to have been optimized by natural selection to balance: (i) the genetic load associated with generating offspring with deleterious or lethal mutations at some rate, (ii) the cost of encoding and expressing various DNA proofreading and repair activities, and (iii) the rare possibility that a new mutation is beneficial to a cell's fitness and enables its offspring to survive competition in the long term.26, 12
Synthetic biologists are not necessarily bound by these evolutionary trade-offs; they are free to manipulate mutation rates within the realm of biochemical possibility in ways that might never survive natural selection. Most obviously, one can choose DNA sequences to encode a device that are less prone to mutations or reduce the mutation rate globally within a chassis organism by manipulating its DNA copying and repair mechanisms. A second way to potentially prevent the evolutionary degradation of engineered biological systems is to functionally couple or overlap the DNA sequence encoding genetic parts with information for some activity that is required for cell survival, such that at least some mutations that result in loss of the engineered function are no longer favored by selection. Finally, it may also be possible to reduce the genetic and metabolic cost of maintaining any generic device in a host organism by eliminating unnecessary components from its genome or by utilizing an orthogonal gene expression or signaling system.
Choosing DNA sequences for devices
The most direct approach to decrease the evolvability of a synthetic biology device is to lower the rate at which new mutations appear in its DNA sequence as the host cell reproduces. DNA sequences may experience chemical damage and mistakes as they are copied from generation to generation, and microbial cells encode a variety of repair mechanisms to correct these errors before they are inherited as mutations.27, 28 The chance of a DNA base mutating in the E. coli genome is 2×10−10 per cell division, on average.29, 30 This roughly works out to just one new base-substitution mutation appearing in the entire E. coli genome every 1000 cell divisions, and similarly low rates have been found for yeast and other microbes.31, 32 The chance of a mutation causing an insertion or deletion of a base is approximately one-tenth this rate, and other types of errors are typically even rarer.30–32 Despite this high overall level of genetic stability, certain sites within a genome are more prone to mutations and experience greatly elevated mutation rates.27 The result can be a staggeringly uneven distribution of mutations within an engineered DNA construct; it is not unusual for one or several hotspots to dominate the mutations that inactivate a reporter gene (Fig. 3).33 Because different DNA sequences can be used to construct identical or equivalent biological devices in most cases, carefully choosing which sequences to use can be a straightforward way to increase the evolutionary reliability of a biological device.
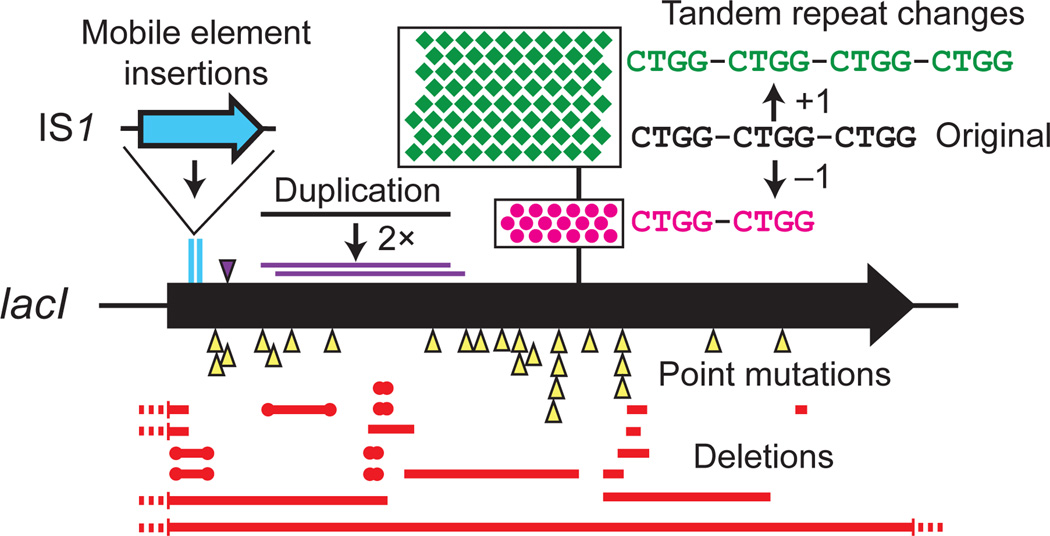
Fig. 3. Example of the molecular events causing evolutionary failure of a device.
A classic study of the spectrum of spontaneous mutations that inactivate the lacI gene in E. coli33 serves as a fairly typical example of the relative rates of different types of mutations that may inactivate a DNA design used in synthetic biology. Each symbol indicates one observation of a given type of mutation. Of 140 total mutations, 94 (67%) were copy number changes in a sequence consisting of three tandem repeats of a four-base sequence: 78 additions of one unit (green) and 18 deletions of one unit (magenta). Of the 19 deletions (red), 7 were flanked by a repeat of at least five bases (circles). Near the beginning of the gene, 2 insertions of an IS1 transposable element (cyan) and 1 duplication of 88 base pairs that resulted in a frameshift (purple) were observed. The remaining 24 point mutations were base substitutions or insertions or deletions of a few bases. Thus, mutations are not randomly distributed within a DNA sequence, and repeat sequences can be a significant source of local mutational hotspots.
Genetic parts in synthetic biology are ideally abstracted as modules that can be composed into different and more complex devices on the basis of their performance characteristics.34 However, when multiple copies of the same part are present within a DNA molecule, they create a hotspot for deletion mutations due to errors during DNA replication that skip from one copy to the next or due to homologous recombination processes that can excise the sequence between direct sequence repeats (Fig. 4a).35, 28 One of the first studies to characterize the genetic reliability of a biological device used a quorum sensing system that induced green fluorescent protein (GFP) expression in response to the addition of exogenous homoserine lactone.1 This device had an evolutionary half-life of <100 cell doublings and was always found to be inactivated by a deletion mediated by two copies of an exact 183-base sequence that was present because the same transcriptional terminator part was used twice in its construction. As expected if the cost of protein expression favored selection for malfunctioning variants, the device was less genetically reliable when GFP expression was induced to a higher level.1, 23
Fig. 4. Engineering DNA sequences for reduced mutation rates.
(A) Synthetic biology designs that rely on multiple copies of the same genetic part have inherent sequence homologies of tens to hundreds of bases that lead to instability due to the high rates of recombination events between these parts that will delete one copy of the repeat and any intervening sequences. This type of instability can be engineered against by designing systems to use alternative parts with fulfill the same function (e.g. different transcriptional terminators with the same strength) or by utilizing sequence re-coded variants of a part that function equivalently (e.g. by altering codon usage across an entire protein part). Eliminating the possibility of homologous recombination between copies of a part will lower the mutation rate in the designed DNA sequence, such that other types of mutations will now dominate the spectrum of inactivating mutations. (B) Simple sequence repeats experience slip-strand mutations that lead to locally elevated rates of base insertions and deletions that can cause frameshift mutations and inactivate a gene. Often these repeat sequences can be avoided by altering sequences locally to maintain the properties of the encoded protein (e.g. by substituting synonymous codons). (C) Certain DNA sequence sites result in DNA methylation by enzymes in common E. coli strains. In the case of Dcm methylation, the resultant 5-methyl cytosine can spontaneously deaminate directly to thymidine, leading to an increased mutation rate at these sites that cannot be repaired.92, 30 Avoiding sites recognized by these methylases or that are prone to other types of chemical damage can decrease the overall rates of inactivating base substitution mutations.
Further studies of this receiver-and-reporter device demonstrated that altering one copy of the terminator sequence could extend its evolutionary half-live when strongly induced from ~30 to ~120 generations.23 This improvement was coincident with a complete shift in the types of inactivating mutations observed, from deletions of the entire GFP reporter system to various smaller deletions, transposon insertions, and point mutations. In other inducible expression devices, repeated copies of transcription factor binding sites (operator sequences) consisting of 18–20 bases led to similar deletion hotspots that were not present when using constitutive promoters without repeats.23 Deletions mediated by sequence repeats also dominated the inactivating mutations observed in a three-color reporter plasmid engineered so that visible color changes could be used as read-out of the relative genetic stability of several inducible reporter genes at the same time.36 In summary, the genetic reliability of a device can often be increased several fold by avoiding the re-use of the exact same genetic part and any exact sequence repeats in a designed sequence of more than ~10 bases. Large combinatorial libraries of common parts, such as transcription and translation initiation sites37 and transcriptional termintors38 are increasingly being characterized. One should theoretically be able to pick distinct sequences with the same performance characteristics from these collections to avoid the use of identical sequences in device construction, thereby improving evolutionary reliability without compromising the predictability of engineered functions.
Other types of mutational hotspots depend primarily on the local composition of neighboring DNA bases. For example, expansion or contraction of simple sequence repeats, consisting of tandem copies of several bases (Fig. 3) or a single base (Fig. 4b), can occur with very high rates approaching once every 10 generations due to DNA polymerase slipping during replication.39 These mutations may commonly inactivate engineered devices by introducing frameshift mutations in protein open reading frames. In general, repeats of ≥4 of the same base experience detectably elevated rates, and instability increases in a predictable fashion for longer repeat sequence units and for more copies of a given repeat.30, 31, 40
Other local hotspots may arise due to host processes that chemically alter DNA for chromosomal maintenance (Fig. 4c). For example, the Dcm and Dam enzymes of E. coli methylate DNA bases in the context of specific 4–5 base sites. A side effect of Dcm function is that spontaneous chemical deamination of the product 5-methylcytosine produces the DNA base thymine. When this occurs, it cannot be recognized as damaged and leads to a mutation. Dam methylation also leads to an increased mutation rate, but by a different mechanism. Methylated bases in these two types of sites experience 3- to 5-fold elevated chances of mutations relative to other bases.30 Mutational hotspots caused by simple sequence repeats and methylation sites can typically be relatively easily designed against, by altering codon choice within protein open reading frame, for example.
Reducing host mutation rates
While employing design principles such as those outlined above in choosing the sequences of DNA devices can be useful for avoiding obvious sources of genetic unreliability, at some point mutation rates may be more effectively addressed at the level of the host machinery. Several efforts aiming to reduce, clean, and/or refactor microbial genomes have included systematic changes to tame their evolutionary potential. One example is that common cloning strains of E. coli (e.g., TOP10, DH5α) are recA−, and therefore lack a major cellular pathway for homologous recombination. This engineered change to the chassis is thought to allow more stable maintenance of plasmids by eliminating this potential type of inactivating mutation.41 More ambitious efforts to reduce the rates of inactivating mutations in a host strain have tackled genetic changes caused by selfish DNA elements and by point mutations.
Transposons are mobile DNA elements that have the ability to autonomously replicate and insert copies of themselves at new sites in a genome. New copies of insertion sequence (IS) elements, bacterial transposons which contain only the genes required for self-replication, have been found to accumulate at a rate comparable to that of other types of mutations during laboratory evolution experiments42, 19. The relatively high rate of these events makes them a common cause of mutations that inactivate synthetic biology devices when a new transposon copy inserts into and interrupts a gene (Fig. 3).23, 43 Eliminating this source of evolutionary instability is conceptually simple: delete all copies of self-replicating mobile elements from a genome (Fig. 5a). Indeed, one of the first systematic programs to improve a host organism chassis included the step-wise removal of the nearly 50 native IS elements from the E. coli K-12 strain MG1655 genome.44 The observation that five new IS copies appeared during this lengthy genome editing process that also had to be eliminated underscores their mutagenic potential.
Fig. 5. Engineering clean genome hosts to reduce evolvability.
(A) Deleting all copies of a self-replicating genomic element, such as a transposon, from a chassis genome can eliminate a major source of mutations that inactivate engineered biological systems. (B) E. coli encodes several error-prone DNA polymerases induced by stress or DNA damage. After a cell experiences DNA damage creating a genomic lesion, these error-prone mechanisms may compete with alternative pathways to repair the damage, or certain types of DNA damage may only be repairable by these mechanisms. By deleting the error-prone DNA polymerases, the rare cells that typically experience these types of damage under laboratory conditions either will have the damage repaired by high-fidelity mechanisms or will not survive. In either case, the overall mutation rate of the bacterial population is reduced.
The resulting clean-genome MDS42 strain was shown to be completely free of inactivating mutations caused by IS activity.44 Consequently, it was better able to stably propagate plasmids encoding intact copies of a toxic protein that was rapidly inactivated by IS insertions in other E. coli hosts.45 MDS42, with or without further knockout of recA, has also been shown to be superior to other host strains of E. coli for maintaining the genetic integrity of plasmids containing lentiviral DNA sequences flanked by 183-bp long terminal repeats that lead to the elimination of the intervening sequences.46 The native transposons of yeast (Ty elements) are similarly being eliminated during refactoring the yeast chromosomes,47 and the development of transposon-less versions of additional organisms in the future is likely to produce chassis with improved genetic reliability for a variety of biotechnology applications.
Point mutations affecting one or a few DNA bases are perhaps the most challenging mutations to prevent because they can result from so many different types of DNA damage and/or the action of different DNA repair pathways. Nevertheless, the overall mutation rate of strain MDS42 has been decreased a further ~50% by additional engineering.48 To understand how this improvement was achieved, one must first be aware that E. coli induces three auxiliary DNA polymerases that lack proofreading subunits in response to DNA damage and other stresses.49 Deleting these error-prone polymerases (PolB, DinB, UmuDC) to create strain MDS42pdu decreased the rate of spontaneous point mutations in a reporter gene under both stressful and non-stressful growth conditions48. This effect was attributed to the error-prone polymerases competing with processes able to repair the same types of DNA lesions with higher fidelity (Fig. 5b), under the hypothesis that stress-induced mutagenesis mediated by these enzymes has evolved to increase bacterial evolvability.50 However, the same result would also be observed if certain lethal DNA lesions require repair by this mutagenic pathway. If the pathway is not functional, this very rare but highly mutated sub-population is eliminated and does not contribute to genetic variation in the population in the MDS42pdu strain.
Is it possible to achieve even lower mutation rates in microbial chassis? It is not always obvious what source of DNA damage leads to the most mutations, and the dominant source of mutations may vary with environmental conditions27. Therefore, multiple DNA repair and proof-reading activities may need to be engineered simultaneously to observe any appreciable reduction in evolvability. Mutations in the main DNA polymerases used to replicate the E. coli genome that are able to compensate for the loss of DNA repair processes have been discovered.51 These antimutator polymerases can decreases the mutation rates in hypermutator strains that have a defect in a DNA repair process by as much as 100-fold. While this magnitude of improvement is not maintained when these mutations are transferred into a wild-type strain background with intact DNA repair mechanisms, at least one antimutator version of DNA polymerase III has been shown to reduce the mutability of wild-type E. coli by 30–50%.52
Overexpression of DNA repair proteins can also potentially reduce mutation rates if the types of damage they address make up a large proportion of the spontaneous mutations under relevant environmental conditions. The complexity of these interactions between environment and different mutagenic processes can be seen from a few representative studies. Overexpression of the MutL protein, one of the components of methyl-directed mismatch repair in E. coli reduced mutation rates, but only in stationary phase after cells stopped replicating.53 Overexpression of MutS and MutH, other components involved in the same repair pathway, did not have this effect. Interestingly, overexpression of NudG, a MutT-like enzyme that hydrolyzes free nucleotides with bases that have experienced oxidative damage so that they are not incorporated into DNA during genome replication, reduced E. coli mutation rates by roughly 50% under many conditions.54 The effectiveness of combining these types of antimutator phenotypes into a single host organism and how general the reduction in mutation rates will be with respect to different growth conditions remains to be determined by future work.
Engineering genetic robustness
While decreasing mutation rates is clearly one effective approach to slow evolutionary failure, it may be difficult or impossible to completely prevent mutations within a biological device. Recall that changing the effects that mutations have on the function of a device or the fitness consequences of its inactivation on the host can also reduce evolvability (Fig. 1). One strategy of this type is to design the sequence or architecture of a device in such a way as to minimize the chance that any mutation that does occur will alter its function. This property of a system is known as mutational or genetic robustness.55, 56, 10 Genetic robustness can potentially be engineered at the level of individual proteins, where greater thermodynamic folding stability decreases the chance that a subsequent mutation will cause an enzyme to not achieve its active structural conformation and therefore not function.57, 58 Molecular chaperones such as Hsp90 can similarly prevent the misfolding of marginally stable proteins, which means that they can buffer the effects of some destabilizing mutations so that they are not deleterious to the function of a protein.59 Thus, overexpressing the GroEL/GroES molecular chaperonin during directed protein evolution experiments enabled more genetic variation and mutations to accumulate over time.60 There is also a report that overexpressing the GroEL chaperone in E. coli reduced the impact of random mutations that were deleterious to fitness genome-wide.61 It is currently unknown to what extent manipulating genetic robustness by stabilizing the folding of protein components would extend the evolutionary half-life of engineered biological devices.
Metabolic and regulatory networks may also exhibit genetic robustness due to their topological organization and the presence of redundant connections.62, 63 One straightforward way to introduce redundancy into an engineered device would be to include duplicate copies of the same gene or two copies of genes with equivalent function in its design. The clearest current demonstration of this approach is a more general method for producing up to ~40 stable copies of an entire engineered DNA construct in the E. coli chromosome.64 This method involves inserting the designed gene cassette flanked by exact repeat sequences into the genome and selecting for amplification of a weak antibiotic resistance gene included in this cassette to favor the formation of many tandem copies of the entire cassette in one genome via homologous recombination. Finally, one knocks out the recA gene in the host so that the array of DNA copies is stabilized (will not collapse due to homologous recombination causing deletions, as discussed above). Using this procedure greatly increased the genetic reliability of a polyhydroxybutyrate biosynthetic pathway from ~10 generations when encoded on a plasmid to >100 generations.64 Presumably, it would now take the accumulation of many separate point mutations to inactivate half the chromosomal copies of the pathway. It is important to note that although devices encoded on most plasmids used in synthetic biology are already present in multiple copies per cell, this setup does not confer the same type of genetic robustness; a highly beneficial mutation that occurs in one copy of the plasmid can rapidly assort to be present in all plasmid copies in a descendant cell due to the random segregation of plasmids into daughter cells.64
Constraining device evolution
Another way to engineer reduced evolvability by altering the effects of mutations is to link maintenance of a desired function to a trait required for survival of the host organism, effectively making it costly to lose rather than to maintain an engineered function. This situation can be engineered by designing architectures where a component required for expressing a gene important for device function is also required for expressing a gene that confers a property that is beneficial to host fitness such as antibiotic resistance (Fig. 6a). In this setup, mutations in the shared components have been converted from mutations that increase host fitness to ones that will be selected against (when antibiotic is present). In a study that tested designs where GFP and a kanamycin resistance gene shared a transcriptional promoter or were fused together into one reading frame, a subtle but not statistically significant increase in evolutionary lifetime of GFP expression was observed when kanamycin was present.23 The marginal utility of this approach in this case may have been due to the presence of mutations in the shared promoter that lowered GFP expression but maintained sufficient kanamycin resistance for cells to survive or due to the possibility of mutations in the host chromosome that affected antibiotic resistance. Therefore, further work is needed to validate this strategy for increasing evolutionary stability.
Fig. 6. Reducing the evolvability of DNA sequences through functional constraint.
(A) Overlapping the sequence information required for expressing a protein such as GFP (green) with a selectable marker such as antibiotic resistance (cyan) is one strategy to limit a device's evolvability. In principle, since stable expression of the selectable marker is required for growth, the more sequence information that the two genes share (yellow), the smaller the target size for possible mutations that inactivate the synthetic construct and maintain organismal viability. This overlap can include sharing promoters, ribosome binding sites, terminators, reading frame, and coding space. (B) Bidirectional promoter design which enables constitutive expression of genes on both the forward and reverse strands of DNA.43 Since the −35 and −10 elements of the forward and reverse promoters require conservation of some of the same bases, a mutation in their overlap would alter the expression of both genes. If one of the genes is a selectable marker, this change can be made lethal to the organism. In this case selecting for kanamycin resistance conferred by the forward gene (cyan) prevented a high-frequency deletion between sequence repeats that eliminated the −35 element of the forward promoter and led to the more stable maintenance of GFP expression in the reverse direction (green).
One can take this approach of decreasing the effective deleterious mutation target size one step further by designing multifunctional parts, such that a mutation that causes loss of a desired, yet costly activity also causes the loss of a function required for survival. This has been shown to work to a limited extent in an example of a bi-directional promoter (Fig. 6b) where the half-life of GFP expression from an engineered construct was increased from ~45 generations to ~80 generations by selecting for co-expression of an antibiotic resistance gene.43 The types of mutations that inactivated the construct also changed: from deletions between repeat sequences that removed a promoter element necessary for transcription in both directions to mutations that inserted transposable elements specifically disrupting expression of the GFP reporter gene. The development of multi-functionality more generally in a single protein would further constrain evolution, but may be technically challenging. As there will always be some mutations that can inactivate just one of the two overlapped functions, the usefulness of this strategy to reduce evolvability may be limited to specific circumstances.
Streamlining host genomes
The goal of reducing the evolvability of an organism is to increase the predictability of how engineered genetic constructs behave in a given cellular background in the face of mutations. Even with detailed knowledge of the topology of a regulatory network with well defined parts, it can be difficult to predict the performance of genetic circuits in cells,65 and these predictions may be confounded by the complexity of potential interactions with cellular components that are active in the host chassis. To reduce genetic complexity and subsequently increase the predictability of imported device performance — both immediately and after mutations accumulate — streamlining genomes by removing non-essential sequences to create ‘clean’ and ‘minimized’ genomes may be a promising solution. The genomes of a variety of model microorganisms have been subjected to genome reduction by directed66–69 and/or combinatorial70, 71 methods. The directed approach of removing specific regions of the genome utilizes previously acquired knowledge of genome architecture to identify areas for removal. In the creation of the reduced E. coli MDS42 strain, for instance, segments encompassing nonconserved genes were deleted from the genome to promote genomic stability, in addition to the IS removal discussed above44. Undirected methods of genome streamlining, such as the use of transposons carrying Cre/loxP sites71 to delete random regions of the genome, provide a useful alternative to the directed approach as they are not constrained by pre-existing notions of a genome region’s essentiality. Combining the two approaches by performing randomized deletions on a pre-streamlined genome may, in the future, allow for new levels of functional minimization beyond what either method could create alone.
In addition to reducing the chances that mutations to nonessential cellular processes might impact an engineered system, streamlining the genome can also potentially prevent the reactivation of latent gene products that could degrade the function of an engineered design. In E. coli K12, ~38% of genes are annotated as hypothetical without a known function.72 Many genes in this set may be mutationally inactivated pseudogenes73 or otherwise cryptic genes74 that are not normally expressed under lab conditions, but whose expression could have significant effects on cellular physiology and the function of engineered DNA devices. These genes can be reactivated for an adaptive benefit by mutations that disrupt their repression75, by the movement of transposons that can function as mobile promoters76, or by chromosomal rearrangements that place them under control of other promoters.77 As many hypothetical proteins can have toxic effects on the cell if expressed,78 removing non-essential and inactive regions of the genome could greatly decreases the availability of uncharacterized mutations that have significant functional consequences. Currently, these effects of streamlining host genomes on long-term synthetic biology device function have not been investigated in detail. As the evolutionary modes of failure of more engineered biological devices are characterized using methods such as whole-genome re-sequencing,19, 14 particularly in new model organisms with genomes that have not yet been domesticated by long periods of growth and manipulation in the lab, we expect many examples of genome streamlining improving the genetic reliability of devices to emerge.
Engineering orthogonality
The burden that an engineered device imposes on a host organism may arise because it perturbs native cellular processes or because it makes demands on shared cellular resources. One can potentially reduce the fitness cost of a device, and consequently increase its evolutionary reliability, by minimizing these types of unwanted crosstalk. Signal transduction and gene expression are notable examples of biological processes that have been engineered in this manner to create systems that are 'orthogonal' to host components.79 This goal is often accomplished in synthetic biology by some combination of transplanting genetic parts between disparate species, structure-guided re-design of protein components, and directed evolution. New variants of sensors and genetic circuit elements that can orthogonally transduce signals without interfering with normal cellular pathways have been created by these methods.80, 81
Devices that rely on orthogonal transcription and translation systems for gene expression will not sequester host machinery away from essential host processes or express genes without the direct control of the orthogonal components. For example, bacteriophage T7 RNA polymerase (RNAP) recognizes a heterologous promoter that may be used to express synthetic constructs while minimally affecting transcription of host genes.82 Directed evolution approaches have diversified83 and greatly improved the function of T7 RNAP as a tightly controlled orthogonal expression system.84 Orthogonal variants of E. coli ribosomes that will only translate distinct mRNA pools carrying altered translation initiation sequences have also been engineered.85 Since these systems require additional genetic parts to be added to a cell (e.g., T7 RNAP), they create new opportunities for a biological device under their control to be inactivated: by mutations in the gene expression system itself. Therefore, it remains to be seen under what circumstances their potential to reduce the fitness burden of an engineered device — by not sequestering native polymerases and ribosomes needed for proper expression of host genes or by more tightly repressing the costly expression of biological devices when they are not in use — outweighs this increased mutational hazard and leads to greater evolutionary stability.
Conclusions
Over billions of years, evolution has found efficient solutions to myriad biochemical challenges. This history provides synthetic biologists with a wealth of genetic parts to draw from, and examples of how to repurpose and improve these tools even further. However, when synthetic biologists take over from where evolution left off tinkering86 and begin designing their own systems and expecting predictable and reliable behaviors, evolution can become a nuisance. The studies discussed here have shown that it is possible to design the DNA sequences of devices in ways that extend their evolutionary lifetimes and to improve the host organism chassis in which these devices are deployed to slow the appearance of mutations that break engineered systems.
The unpredictability of biological evolution is one of the primary reasons that the risk of genetically engineered organisms is difficult to assess. Rational modifications to microorganisms that reduce evolvability can be used to extend the useful lifetimes of kill switches and reduce the frequencies of mutants that escape other types of laboratory biocontainment to reduce this risk.87 We anticipate that a synthetic biologist who is savvy to the effects of evolution on her or his device will soon routinely use a combination of improved chassis organisms and computer-aided design programs that perform not only functions such as optimizing codon usage for efficient protein expression, but systematically design against sequences that are genetically unstable.
In the future, we speculate that synthetic biologists may even be able to replace DNA with genetic molecules that are more chemically stable. This leap will likely first occur by importing orthogonal genetic systems into cells where information in a non-DNA plasmid is replicated and transcribed by dedicated polymerases created by directed evolution.88, 89 This area of synthetic genetics may be one place where the top-down genetic engineering discipline of synthetic biology discussed here intersects with the branch that extrapolates from synthetic organic chemistry to build complex life-like systems from the ground up.90 Although there are clearly ethical issues and technical challenges with editing the sequence and function of the human genome on a large scale, it may become possible in the not-so-distant future to achieve reduced mutation rates in humans with great medical benefits. Re-engineering known fragile sequences in the human genome, or perhaps administering anti-evolutionary drugs that 'overclock' native DNA repair processes, could reduce the incidence of inherited genetic diseases and neoplastic cancers,91 for example.
Acknowledgements
We thank Daniel Deatherage, Gabriel Suarez, Jamie Bacher, Alvaro Rodriguez, and members of our lab for helpful discussions. We acknowledge funding from the U.S. National Institutes of Health (R00-GM087550), the U.S. Army Research Office (W911NF-12-1-0390), the U.S. National Science Foundation BEACON Center for the Study of Evolution in Action (DBI-0939454), and the Welch Foundation (F-1780).
Notes and references
- 1.Canton B, Labno A, Endy D. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y-H, Wei KY, Smolke CD. Annu. Rev. Chem. Biomol. Eng. 2013;4:69–102. doi: 10.1146/annurev-chembioeng-061312-103351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng AA, Lu TK. Annu. Rev. Biomed. Eng. 2012;14:155–178. doi: 10.1146/annurev-bioeng-071811-150118. [DOI] [PubMed] [Google Scholar]
- 4.Khalil AS, Collins JJ. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endy D. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 6.Arkin AP, Fletcher DA. Genome Biol. 2006;7:114. doi: 10.1186/gb-2006-7-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danchin A. FEBS Lett. 2012;586:2129–2137. doi: 10.1016/j.febslet.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Pigliucci M. Nat. Rev. Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- 9.Arenas CD, Cooper TF. FEMS Microbiol. Rev. 2012;37:572–582. doi: 10.1111/1574-6976.12008. [DOI] [PubMed] [Google Scholar]
- 10.Lenski RE, Barrick JE, Ofria C. PLoS Biol. 2006;4:e428. doi: 10.1371/journal.pbio.0040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altenberg L, Wagner G. Evolution (N. Y) 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 12.Tenaillon O, Taddei F, Radmian M, Matic I. Res. Microbiol. 2001;152:11–16. doi: 10.1016/s0923-2508(00)01163-3. [DOI] [PubMed] [Google Scholar]
- 13.Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. Science. 2011;331:1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrick JE, Lenski RE. Nat. Rev. Genet. 2013;14:827–839. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voigt CA, Kauffman S, Wang ZG. Adv. Prot. Chem. 2000;55:79–160. doi: 10.1016/s0065-3233(01)55003-2. [DOI] [PubMed] [Google Scholar]
- 16.Kaern M, Elston TC, Blake WJ, Collins JJ. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 17.Ryall B, Eydallin G, Ferenci T. Microbiol. Mol. Biol. Rev. 2012;76:597–625. doi: 10.1128/MMBR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragosits M, Mattanovich D. Microb. Cell Fact. 2013;12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conrad TM, Lewis NE, Palsson BO. Mol. Syst. Biol. 2011;7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shachrai I, Zaslaver A, Alon U, Dekel E. Mol. Cell. 2010;38:758–767. doi: 10.1016/j.molcel.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Glick B. Biotechnol. Adv. 1995;13:247–261. doi: 10.1016/0734-9750(95)00004-a. [DOI] [PubMed] [Google Scholar]
- 22.Neubauer P, Lin HY, Mathiszik B. Biotechnol. Bioeng. 2003;83:53–64. doi: 10.1002/bit.10645. [DOI] [PubMed] [Google Scholar]
- 23.Sleight SC, Bartley BA, Lieviant JA, Sauro HM. J. Biol. Eng. 2010;4:12. doi: 10.1186/1754-1611-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You L, Sidney R, Cox R, Weiss R, Arnold FH. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 25.Balagaddé FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. Mol. Syst. Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Maki H. Annu. Rev. Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- 28.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair Mutagenesis. 2nd edn. Washington, DC: ASM Press; 2006. [Google Scholar]
- 29.Wielgoss S, Barrick JE, Tenaillon O, Cruveiller S, Chane-Woon-Ming B, Medigue C, Lenski RE, Schneider D, Andrews BJ. G3 Genes|Genomes|Genetics. 2011;1:183–186. doi: 10.1534/g3.111.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Popodi E, Tang H, Foster PL. Proc. Natl. Acad. Sci. U. S. A. 2012;109:e2774–e2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, Thomas WK. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M. Proc. Natl. Acad. Sci. U. S. A. 2012;109:18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farabaugh PJ, Schmeissner U, Hofer M, Miller JH. J. Mol. Biol. 1978;126:847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- 34.Shetty RP, Endy D, Knight TF. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DasGupta U, Weston-Hafer K, Berg DE. Genetics. 1987;115:41–49. doi: 10.1093/genetics/115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sleight SC, Sauro HM. Artificial Life 13: Proceedings of the Thirteenth International Conference on the Synthesis Simulation of Living Systems; 2012. pp. 481–488. [Google Scholar]
- 37.Mutalik VK, Guimaraes JC, Cambray G, Lam C, Christoffersen MJ, Mai Q-A, Tran AB, Paull M, Keasling JD, Arkin AP, Endy D. Nat. Methods. 2013;10:354–360. doi: 10.1038/nmeth.2404. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y-J, Liu P, Nielsen AAK, Brophy JAN, Clancy K, Peterson T, Voigt CA. Nat. Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 39.Moxon R, Bayliss C, Hood D. Annu. Rev. Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 40.Egbert RG, Klavins E. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16817–16822. doi: 10.1073/pnas.1205693109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vapnek D, Alton NK, Bassett CL, Kushner SR. Proc. Natl. Acad. Sci. U. S. A. 1976;73:3492–3496. doi: 10.1073/pnas.73.10.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 43.Yang S, Sleight SC, Sauro HM. Nucleic Acids Res. 2013;41:e33. doi: 10.1093/nar/gks972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pósfai G, Plunkett G, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 45.Umenhoffer K, Fehér T, Balikó G, Ayaydin F, Pósfai J, Blattner FR, Pósfai G. Microb. Cell Fact. 2010;9:38. doi: 10.1186/1475-2859-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakiath C, Esposito D. Biotechniques. 2007;43:466–470. doi: 10.2144/000112585. [DOI] [PubMed] [Google Scholar]
- 47.Dymond JS, Richardson SM, Coombes CE, Babatz T, Muller H, Annaluru N, Blake WJ, Schwerzmann JW, Dai J, Lindstrom DL, Boeke AC, Gottschling DE, Chandrasegaran S, Bader JS, Boeke JD. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Csörgo B, Fehér T, Tímár E, Blattner FR, Pósfai G. Microb. Cell Fact. 2012;11:11. doi: 10.1186/1475-2859-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. Proc. Natl. Acad. Sci. U. S. A. 2011;108:13659–13664. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galhardo RS, Hastings PJ, Rosenberg SM. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herr AJ, Williams LN, Preston BD. Crit. Rev. Biochem. Mol. Biol. 2011;46:548–570. doi: 10.3109/10409238.2011.620941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oller AR, Schaaper RM. Genetics. 1994;138:263–270. doi: 10.1093/genetics/138.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harris RS, Feng G, Ross KJ, Sidhu R, Thulin C, Longerich S, Szigety SK, Winkler ME, Rosenberg SM. Genes Dev. 1997;11:2426–2437. doi: 10.1101/gad.11.18.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamiya H, Iida E, Murata-Kamiya N, Yamamoto Y, Miki T, Harashima H. Genes to Cells. 2003;8:941–950. doi: 10.1046/j.1365-2443.2003.00688.x. [DOI] [PubMed] [Google Scholar]
- 55.Visser J, Hermisson J, Wagner GP, Meyers LA, Bagheri Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 56.Wagner A. Proc. R. Soc. B Biol. Sci. 2008;275:91–100. doi: 10.1098/rspb.2007.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tokuriki N, Tawfik DS. Curr. Opin. Struct. Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Queitsch C, Sangster TA, Lindquist S. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 60.Tokuriki N, Tawfik DS. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 61.Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- 62.Albert R, Jeong H, Barabasi A. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 63.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 64.Tyo KEJ, Ajikumar PK, Stephanopoulos G. Nat. Biotechnol. 2009;27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 65.Guet CC, Elowitz MB, Hsing W, Leibler S. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 66.Connor TJO, Adepoju Y, Boyd D, Isberg RR. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kolisnychenko V, Plunkett G, Herring CD, Fehér T, Pósfai J, Blattner FR, Pósfai G. Genome Res. 2002;12:640–647. doi: 10.1101/gr.217202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki N, Okayama S, Nonaka H. Appl. Environ. Microbiol. 2005;71:3396–3372. doi: 10.1128/AEM.71.6.3369-3372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westers H, Dorenbos R, van Dijl JM, Kabel J, Flanagan T, Devine KM, Jude F, Seror SJ, Beekman AC, Darmon E, Eschevins C, de Jong A, Bron S, Kuipers OP, Albertini AM, Antelmann H, Hecker M, Zamboni N, Sauer U, Bruand C, Ehrlich DS, Alonso JC, Salas M, Quax WJ. Mol. Biol. Evol. 2003;20:2076–2090. doi: 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- 70.Leprince A, de Lorenzo V, Völler P, van Passel MWJ, Martins dos Santos VAP. Environ. Microbiol. 2012;14:1444–1453. doi: 10.1111/j.1462-2920.2012.02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu BJ, Kim C. Methods Mol. Biol. 2008;416:261–277. doi: 10.1007/978-1-59745-321-9_17. [DOI] [PubMed] [Google Scholar]
- 72.Blattner FR. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 73.Ochman H, Davalos LM. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 74.Hall BG, Yokoyama S, Calhoun DH. Mol. Biol. Evol. 1983;1:109–124. doi: 10.1093/oxfordjournals.molbev.a040300. [DOI] [PubMed] [Google Scholar]
- 75.Hall BG. Mol. Biol. Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- 76.Zafarullah M, Charlier D, Glansdorff N. J. Bacteriol. 1981;146:415–417. doi: 10.1128/jb.146.1.415-417.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kimelman A, Levy A, Sberro H, Kidron S, Leavitt A, Amitai G, Yoder-Himes DR, Wurtzel O, Zhu Y, Rubin EM, Sorek R. Genome Res. 2012;22:802–809. doi: 10.1101/gr.133850.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filipovska A. ACS Chem. Biol. 2008;3:51–63. doi: 10.1021/cb700185e. [DOI] [PubMed] [Google Scholar]
- 80.Haseltine EL, Arnold FH. Annu. Rev. Biophys. Biomol. Struct. 2007;36:1–19. doi: 10.1146/annurev.biophys.36.040306.132600. [DOI] [PubMed] [Google Scholar]
- 81.Morey KJ, Antunes MS, Barrow MJ, Solorzano FA, Havens KL, Smith JJ, Medford J. Biotechnol. J. 2012;7:846–855. doi: 10.1002/biot.201100487. [DOI] [PubMed] [Google Scholar]
- 82.Studier FW, Moffatt BA. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 83.Temme K, Hill R, Segall-Shapiro TH, Moser F, Voigt CA. Nucleic Acids Res. 2012;40:8773–8781. doi: 10.1093/nar/gks597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ellefson JW, Meyer AJ, Hughes RA, Cannon JR, Brodbelt JS, Ellington AD. Nat. Biotechnol. 2013 doi: 10.1038/nbt.2714. [DOI] [PubMed] [Google Scholar]
- 85.Rackham O, Chin JW. Nat. Chem. Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- 86.Jacob F. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 87.Moe-Behrens GHG, Davis R, Haynes KA. Front. Microbiol. 2013;4:5. doi: 10.3389/fmicb.2013.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Z, Chen F, Alvarado JB, Benner SA. J. Am. Chem. Soc. 2011;133:15105–15112. doi: 10.1021/ja204910n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chaput JC, Yu H, Zhang S. Chem. Biol. 2012;19:1360–1371. doi: 10.1016/j.chembiol.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 90.Malinova V, Nallani M, Meier WP, Sinner EK. FEBS Lett. 2012;586:2146–2156. doi: 10.1016/j.febslet.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 91.Greaves M, Maley CC. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Nature. 1978;274:775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]