Abstract
The overall complexity of the microbial communities in the gastrointestinal (GI) tracts of mammals has hindered observations of dynamics and interactions of individual bacterial populations. However, such information is crucial for understanding the diverse disease-causing and protective roles that gut microbiota play in their hosts. Here, we determine the spatial distribution, interanimal variation, and persistence of bacteria in the most complex defined-flora (gnotobiotic) model system to date, viz., mice colonized with the eight strains of the altered Schaedler flora (ASF). Quantitative PCR protocols based on the 16S rRNA sequence of each ASF strain were developed and optimized to specifically detect as few as 10 copies of each target. Total numbers of the ASF strains were determined in the different regions of the GI tracts of three C.B-17 SCID mice. Individual strain abundance was dependent on oxygen sensitivity, with microaerotolerant Lactobacillus murinus ASF361 present at 105 to 107 cells/g of tissue in the upper GI tract and obligate anaerobic ASF strains being predominant in the cecal and colonic flora at 108 to 1010 cells/g of tissue. The variation between the three mice was small for most ASF strains, except for Clostridium sp. strain ASF502 and Bacteroides sp. strain ASF519 in the cecum. A comparison of the relative distribution of the ASF strains in feces and the colon indicated large differences, suggesting that fecal bacterial levels may provide a poor approximation of colonic bacterial levels. All ASF strains were detected by PCR in the feces of C57BL/6 restricted flora mice, which had been maintained in an isolator without sterile food, water, or bedding for several generations, providing evidence for the stability of these strains in the face of potential competition by bacteria introduced into the gut.
The mammalian body harbors large microbial populations on the skin, in the oral cavity, and in the genital and gastrointestinal (GI) tracts. The number of bacteria is thought to exceed the number of mammalian cells by as much as a factor of 10, and a majority of bacteria are present in the GI tract (26). Culture-dependent studies of the bacterial populations in the mouse gut have indicated about 109 bacteria/g of tissue in the stomach and a gradient of bacterial density along the intestine, with the numbers changing from 107 bacteria/g of tissue in the small intestine to 1011 to 1012 bacteria/g of tissue in the cecum and colon (5, 6, 26). No culture-independent estimates of bacterial density are available for the GI tract; however, comparison of direct counts and culturing indicates that between 40 and 80% of anaerobic bacteria in fecal matter have not been cultured (18). Although the culturability of bacteria in the GI tract exceeds that of many other environments studied (2), the lack of culture-independent cell counts indicates that the gut remains a poorly characterized ecosystem.
The nature of the GI tract changes along its length, with reduced flow rates and oxygen levels as ingested matter travels from the stomach to the large intestine. These factors, combined with the abundance of substrates, make the gut an ecosystem that could support a complex bacterial community with high spatial heterogeneity (5, 9, 26). Indeed, in culture-based studies, the bacterial populations mirror this gradient along the length of the gut. Aerotolerant bacteria, such as enterococci and lactobacilli, are predominant in the stomach and small intestine, and obligate anaerobic species, such as clostridia and bacteroides, are predominant in the large intestine (6). Overall, anaerobes are thought to exceed facultative aerobes by a ratio of 1,000:1 (5).
Both culture-based and molecular ecological studies have indicated that the gut microflora are very diverse. Studies of the human fecal flora using culture-based methods have suggested that over 400 species could be present (20). While the culture-based assessment of species diversity can be misleading due to subjectivity in defining species, recent molecular studies have indicated a similarly high level of diversity. Analysis of a 16S rRNA clone library constructed from human fecal DNA found 82 phylotypes, where phylotypes were defined as sequences with greater than 98% similarity (32). A similar study of the pig GI tract used amplification products from pooled ileal, cecal, and colonic DNA from a pig to generate a 16S rRNA clone library. A total of 375 phylotypes were detected when phylotypes were defined as sequences with greater than 97% similarity (19).
The GI microflora are thought to have a major effect on the host, but the extent and mechanisms of influence of the GI microbiota on the host are only beginning to be understood (11). Several studies have indicated a role for the GI microflora in providing essential nutrients such as vitamin K (27), differentiation of the host intestine (11), immune development of the host (7), and protection of the host from pathogenic infections (10, 15, 30, 33). Comparison of gnotobiotic mice colonized with a single bacterial species with germfree mice has shown a marked effect on gene expression in the tissues of the small intestine (12) and a significant increase in the survival rate following infection with a pathogen (13). On the other hand, the microflora are also postulated to play a role in the progression of various diseases. An example of the potentially harmful nature of the microbiota are their role in the development of inflammatory bowel disease (IBD) and cancer (3, 25). While the role of Helicobacter hepaticus in causing persistent hepatitis and liver cancer in mice has been known since the early 1990s, recent studies in immune dysregulated (C.B-17 SCID) gnotobiotic mice have shown that infection with H. hepaticus may result in IBD (3).
Major limiting factors in studying interactions of the GI microbiota with the host are both the complexity and potential variation of bacterial populations. Thus, defined-flora (gnotobiotic) animal models have become useful tools for the study of the effects of the microbiota on the host (7, 8), and germfree mice inoculated with a single bacterial species have been used as the simplest gnotobiotic model system for the study of bacterial effects. Another simplifying approach involves the use of fecal microflora as a substitute for colonic microflora (23, 26). The representative nature of the fecal flora has not been established (23), and a recent study comparing the qualitative molecular fingerprints of the colonic mucosal flora with those of fecal flora found significant differences (35). Thus, the development of a widely available defined-flora mouse model that is more complex, and thus more realistic, than monoassociated models would assist in the study of the consortium effects of the gut microflora.
Here, we introduce a novel application of mice colonized with the eight-member ASF as a model system for quantitatively studying the microecology of the murine gut. The original Schaedler flora, developed in 1965, was used to colonize germfree mice and consisted of eight aerobic and aerotolerant bacterial species isolated from mice. These flora were selected on the basis of their persistence from generation to generation in germfree mice and their ability to restore the cecal morphology of germfree mice to a morphology comparable to that of normal mice (29). The original Schaedler flora was later modified to include a spiral bacterium and more extremely oxygen-sensitive (EOS) strains to make it more representative of the GI flora (21). The defined flora substantially reduced the size of the cecum of germfree mice, indicating the functional similarity of the defined flora to the complex microbiota of healthy mice. This defined flora, ASF, has become the standard flora to colonize germfree rodents worldwide (21). Several members of this flora were also detected in a recent study that assessed the diversity of the mouse GI microbiota using clone libraries (24). Because gnotobiotic immunodeficient (SCID) mice colonized with the ASF strains have recently been used as a model system to study the development of IBD (3), a characterization of the ASF strains in this mouse model would greatly enhance its utility. The bacteria in the ASF have been phylogenetically characterized on the basis of their 16S rRNA gene sequences (4). The aerotolerant bacterial strains, ASF360 and ASF361, are lactobacilli. ASF360 clusters with Lactobacillus acidophilus, while the 16S gene sequence of ASF361 is identical to that of Lactobacillus murinus and Lactobacillus animalis. ASF519 is related to Bacteroides distasonis, while ASF356, ASF502, and ASF492 fall in Clostridium cluster XIV, with the 16S gene sequence of ASF492 identical to that of Eubacterium plexicaudatum. ASF457 is a spiral-shaped bacterium that clusters with the Flexistipes species, while ASF500 is not closely related to any of the sequences in the database and clusters with the low-G+C-content gram-positive bacteria (4).
The goals of this study were as follows: (i) develop protocols for quantification of ASF strains on the basis of their 16S rRNA genes, (ii) assess the variation of the ASF strains along the length of the GI tract, (iii) evaluate the differences between mice, and (iv) compare fecal levels to colonic levels.
MATERIALS AND METHODS
Strain culture.
The eight bacterial strains of ASF were obtained from Taconic (Germantown, N.Y.). Strains ASF360 and ASF361 were grown on MRS agar (Difco Laboratories, Detroit, Mich.) under microaerobic conditions generated using the Campypak system (Remel, Lenexa, Kans.). The rest of the ASF strains were cultured as recommended (22) on Schaedler agar supplemented with 5% defibrinated sheep blood (Remel) in an anaerobic glove box (Coy Laboratories, Grass Lakes, Mich.) containing a 10% CO2-10% H2-80% N2 atmosphere at 34 or 35°C. Schaedler agar plates were prereduced for 2 days prior to inoculation of the EOS strains, which require an anaerobic glove box for successful culture. However, even under these conditions, EOS strains ASF500 and ASF502 did not grow well. Whole-cell lysates of these two strains (kind gift of Bruce Paster, Forsyth Institute, Boston, Mass.) were used to obtain genomic DNA to test the specificity of the primers and generate quantitative PCR (QPCR) standards. This DNA was used as the template to amplify the 16S rRNA gene used to construct plasmids for use in QPCR (see below). Liquid cultures were grown at 37°C in MRS broth (ASF360 and ASF361) or in Schaedler broth supplemented with 5% heat-inactivated fetal calf serum (ASF356 and ASF519).
Mice.
Six-week-old female C.B-17 SCID mice inoculated with ASF strains were obtained from Taconic. This age was chosen because the GI microbiota become a climax community after 4 weeks of age (5, 6). The mice were transferred to autoclaved SCID mouse cages upon receipt and fed autoclaved food and water. To avoid contamination of the mice with additional species of bacteria, the mice were euthanized 1 day after arrival at the animal facility, as opposed to the standard 72-h period for recovery from the stress of transportation. There is a possibility that the mice might not have fully recovered from the stress of transportation in 1 day. The stability of the ASF strains in a more complex biota was also assessed using fecal pellets obtained from 9-month-old female C57BL/6 mice with a restricted flora (RF mice). The original founders of the RF breeding colony were mice with defined flora before they were bred and maintained in an isolator with nonsterile food, water, and bedding. Hence, they may have acquired a more complex flora than the original defined flora. There is evidence that these mice had a more complex microbiota from terminal restriction fragment length polymorphism analysis of their fecal bacterial DNA (unpublished results). The RF mice used in this experiment were maintained in a cube isolator (Charles River Laboratories, Wilmington, Mass.) with HEPA filtered air and were fed nonsterile feed and water. All mice were maintained and handled per the Institutional Animal Care and Use Committee guidelines.
Sample collection.
Three C.B-17 SCID mice were euthanized by CO2 asphyxiation, and the entire GI tract of each mouse was collected aseptically. The GI tract was divided into five anatomically defined segments, the esophagus, stomach, small intestine, cecum, and colon. The stomach was incised, rinsed free of ingesta, and separated into the glandular and nonglandular portions, and 0.5-cm samples were taken from each portion. The cecum was divided into the basal (including the ileocecal valve) and apical halves, with the loss of some of the cecal ingesta. Samples (0.5 cm long) were then collected from the ileocecal valve and apical cecum. The small intestine was subdivided into six equal-length segments. Due to approximation errors and the accuracy of the measurement method, the last segment was consistently longer than the others. The colon was subdivided into three equal segments. Full-thickness colon samples, which include the tissue and lumenal contents, consisting of the first 0.5 cm of each intestinal segment, were collected. Prior to sectioning of the colon, formed fecal pellets were expelled from the colon and collected from each mouse for analysis. All samples were weighed immediately and stored at −80°C prior to DNA extraction.
DNA extraction. (i) Tissue samples.
DNA was extracted from tissue samples with the DNEasy tissue kit, using a modification of the DNEasy tissue extraction protocol (Qiagen, Valencia, Calif.) to ensure maximal lysis of both gram-positive and gram-negative bacteria. Following overnight digestion of tissue samples with proteinase K (20 μl, 12 milliAnson Units) and ATL buffer (Qiagen), the sample was centrifuged, and DNA was isolated from the supernatant. Briefly, the supernatant was treated with 400 μg of RNase A for 2 min at room temperature, incubated with a guanidium isothiocyanate-based buffer (AL buffer) for 10 min at 70°C, and applied to a spin column. Following washes with AW1 buffer and AW2 buffer, the DNA was eluted in Tris chloride elution buffer (AE buffer). The pellet formed from centrifugation of the lysed tissue was subjected to an extraction using the DNEasy protocol for gram-positive bacteria to ensure recovery of any possibly recalcitrant gram-positive bacteria. Briefly, the pellet was resuspended in a lysozyme buffer (20 mM Tris Cl [pH 8.0], 2 mM EDTA, 1.2% Triton X-100) and incubated at 85°C for 10 min to inactivate the proteinase K, and 3.6 mg of lysozyme (Sigma, St. Louis, Mo.) was added to the resuspended pellet. Following a 45-min incubation at 37°C, 400 μg of RNase A was added to the lysate and incubated at room temperature for 2 min. Proteinase K (25 μl, 15 milli-Anson units) and AL buffer were added to the lysate and incubated at 70°C for 30 min. Following the addition of 200 μl of ethanol, the lysate was applied to a spin column, washed with AW1 and AW2 buffers and eluted in elution buffer (AE buffer) as described above. DNA from both extractions was pooled and stored at −80°C prior to use for QPCR.
(ii) Fecal pellet samples.
Since bead-beating protocols have been shown to be more effective in extracting bacterial DNA from fecal samples (34), a bead-beating protocol adapted from the method of Stahl et al. (31) was used for the fecal pellet samples. Briefly, the fecal pellet samples were resuspended in phosphate-buffered saline (pH 7.4). Tris-saturated phenol (pH 7.4) (500 μl), 0.1-mm-diameter zirconium beads (0.5 g), and 20% sodium dodecyl sulfate (SDS) (50 μl) were added to the tube and shaken at 5,000 rpm for 2 min on a Bead beater (Biospec, Bartlesville, Okla.). This step was repeated after a 10-min incubation at 60°C, following which the supernatant was transferred to a new tube and an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added. The tube was mixed by inversion and centrifuged for 5 min at 12,000 × g. The supernatant was transferred to a new tube, and the same procedure was repeated twice. After this, an equal volume of chloroform (Sigma) was added to the aqueous phase, and the tube was mixed by inversion and centrifuged at 12,000 × g for 5 min. DNA from the supernatant was then precipitated using 0.1 volume of 3 M sodium acetate and a volume of isopropanol equal to that of the aqueous phase. The DNA was pelleted by centrifugation at 15,000 × g for 10 min, washed with 70% ethanol, dried under a vacuum using a SpeedVac concentrator (Thermo Electron Corporation, Milford, Mass.), and resuspended in Tris chloride buffer. Losses in extraction of different samples were kept constant by using constant volumes at all the steps.
The modified Qiagen tissue protocol was compared with the bead-beating protocol by extracting DNA from identical mixtures of Lactobacillus murinus ASF361 (gram positive) and Citrobacter rodentium (gram negative) using both protocols and quantifying the number of bacteria in the extracted DNA. The two protocols yielded comparable results for both gram-positive and gram-negative bacteria (data not shown).
Primers. (i) Primer design.
The 16S rRNA gene sequences of the ASF strains (4) were aligned on the basis of secondary structure against the Escherichia coli 16S rRNA gene using Seqlab (Genetics Computing Group, Madison, Wis.). Candidate primers were designed on the basis of sequence stretches in the alignment that were unique for each ASF strain. These candidate primers were checked against the GenBank database using the BLAST algorithm (1), and hits that were perfect matches only for the ASF strain in question or other identical 16S sequences in the GenBank database were selected. If non-ASF sequences had perfect matches for the primers, forward and reverse primers that did not match the same non-ASF sequence were preferentially chosen. The other parameters considered in designing the primers, besides specificity, were primer length (preferably 18 to 22 bases), dissociation temperature (>58°C), and amplicon size (optimally <200 bp). Primer dissociation temperatures and amplicon sizes in this range are recommended to enhance QPCR performance. The melting temperatures of the selected sequences were determined using the PrimerExpress software (Applied Biosystems, Foster City, Calif.). Primers were synthesized and purified by polyacrylamide gel electrophoresis by IDT DNA Technologies (Coralville, Iowa).
(ii) Specificity testing.
The specificity of the primers was tested by amplifying DNA from the target organism in a background of fecal pellet DNA from RF mice and primers that yielded only a band of the expected size were selected for QPCR. Amplification was performed using the AmpliTaq kit (Applied Biosystems) on a Robocycler (Stratagene, La Jolla, Calif.) with the following conditions: (i) an initial step of 3 min at 94°C and (ii) 35 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C. After PCR, the amplification products were run on a 2% agarose gel (Invitrogen, Carlsbad, Calif.) in Tris-borate-EDTA buffer and visualized on the EagleEye gel documentation system (Stratagene).
QPCR standards.
Linearized plasmids containing the 16S rRNA gene of each ASF strain were used as standards. The 16S rRNA gene from each of the ASF strains was amplified using bacterium-specific 16S primers 27F and 1492R (17) and cloned into a pCR2.1 plasmid vector using the Topo-TA cloning kit (Invitrogen). The plasmids were sequenced using the BigDye terminator kit (Applied Biosystems) to ensure that the primer regions were not altered due to Taq polymerase error. Before the plasmids were used to generate standard curves, the plasmids were linearized with the restriction enzyme BamHI (New England BioLabs, Beverly, Mass.) that cuts the vector exactly once and does not have a recognition site in the 16S rRNA genes of the ASF strains. The concentrations of the linearized plasmid were determined on a Synergy HT microplate fluorescence reader (Bio-Tek, Winooski, Vt.) using the PicoGreen kit (Molecular Probes, Eugene, Oreg.) and dilutions of known concentrations of λ DNA as standards.
QPCR optimization. (i) MgCl2 optimization.
Increasing the concentration of MgCl2 in the QPCR mixture can make the reaction more efficient but can also lead to nonspecific amplification. To optimize the MgCl2 concentration, primers were tested in a range of MgCl2 concentrations (2 to 5 mM in increments of 0.5 mM) against all the ASF strains in a background of fecal pellet DNA from RF mice to ensure an absence of nonspecific amplification. PCR conditions were identical to those used for testing specificity. Amplification products were analyzed on a 2% agarose gel (Invitrogen) and visualized using the EagleEye gel documentation system (Stratagene). The highest MgCl2 concentration that did not yield nonspecific amplification products (4 mM) for any of the ASF strains was used as the optimal concentration for all reactions.
(ii) Primer concentration optimization.
Higher primer concentrations can increase the efficiency of the QPCR but can also lead to primer dimer formation. All combinations of 100, 200, and 300 nM forward and reverse primer concentrations were tested with approximately 106 copies of plasmid template. QPCR was performed in duplicate for each of the nine possible primer concentration pairs, along with corresponding no-template controls, using the SYBR green core reagent kit (Applied Biosystems) and the QPCR conditions specified below. The primer concentrations yielding the lowest threshold cycle (CT) value for the plasmid standard and highest threshold cycle value for the no-template control were chosen as the optimal concentrations for QPCR. Where no significant change between the different primer concentrations was seen, i.e., intra-assay variation was similar to interassay variation, the lowest concentration (100 nM) was considered optimal. None of the primers formed primer dimers at the optimal concentrations.
QPCR conditions.
All reactions were performed in triplicate on the DNA Engine Opticon continuous fluorescence detection system (MJ Research, Watertown, Mass.) in a 25-μl reaction mixture volume using the SYBR green core reagents kit (Applied Biosystems) and the following reaction conditions: (i) an initial step of 10 min at 95°C and (ii) 40 cycles, with 1 cycle consisting of 30 s at 95°C, 45 s at 60°C, and 75 s at 72°C. The MgCl2 concentration, optimized as specified above, was kept constant at 4.0 mM for all the reaction mixtures. Melting curves were generated after QPCR to test for purity by identification of the characteristic melting peak. This was done by measuring the fluorescence of the amplification product every 0.1°C while increasing the temperature from 50 to 95°C. Standard curves for individual ASF strains were generated using a 10-fold dilution series of the linearized plasmid standard ranging from 1 to 108 copies. The fluorescence level for determining the CT was set at 0.01 fluorescence unit in all reaction mixtures. It was observed that the small intestinal samples and some of the ileocecal samples had a very high background of eukaryotic DNA, which increased the fluorescence background in the QPCR mixtures. These samples were diluted 10-fold for QPCR.
Assessment of persistence of ASF strains in RF mice.
To determine whether the ASF strains persisted in RF mice, DNA extracted from fecal pellets was amplified with the strain-specific primers using the conditions specified in Table 1 and the following amplification conditions: (i) an initial step of 3 min at 94°C and (ii) 35 cycles, with 1 cycle consisting of 1 min at 94°C, 45 s at 60°C, and 75 s at 72°C. After PCR, the amplification products were run on a 2% agarose gel (Invitrogen) in 0.5× Tris-borate-EDTA electrophoresis buffer and visualized using the EagleEye gel documentation system (Stratagene).
TABLE 1.
Primer sequences, specificity, rRNA copy number, and QPCR parametersa for the ASF strains
| Strain | Species or group | Primerb
|
Standard curve
|
Specificityc | No. of operons | ||
|---|---|---|---|---|---|---|---|
| Slope | Intercept | ||||||
| ASF356 | Clostridium sp. | 356-144F | CGGTGACTAATACCGCATACGG (100) | −3.50 | 35.83 | NHd | 5e |
| 356-538R | CCTTGCCGCCTACTCTCCC (100) | ||||||
| ASF360 | Lactobacillus sp. | 360-81F | CTTCGGTGATGACGCTGG (300) | −3.49 | 37.26 | Both primers hit L. intestinalis and 99-100% identical clones | 4e |
| 360-189R | GCAATAGCCATGCAGCTATTGTTG (200) | ||||||
| ASF361 | Lactobacillus murinus | 361-278F | GCAATGATGCGTAGCCGAAC (200) | −3.30 | 32.97 | Both primers hit L. animalis and 99-100% identical clones | 6e |
| 361-435R | GCACTTTCTTCTCTAACAACAGGG (300) | ||||||
| ASF457 | Flexistipes group | 457-130F | CCGAAAGGTGAGCTAATGCCGG (100) | −3.77 | 40.96 | NH | 14f |
| 457-219R | GGGACGCGAGTCCATCTTTC (100) | ||||||
| ASF492 | Eubacterium plexicaudatum | 492-57F | CTGCGGAATTCCTTCGGGG (100) | −3.72 | 38.15 | NH | 4f |
| 492-204R | CCCATACCACCGGAGTTTTC (100) | ||||||
| ASF500 | Low-G+C-content gram-positive group | 500-183F | GTCGCATGGCACTGGACATC (200) | −3.45 | 34.52 | Reverse primer hits environmental clones and human colonic clones | 11.2g |
| 500-445R | CCTCAGGTACCGTCACTTGCTTC (200) | ||||||
| ASF502 | Clostridium sp. | 502-195F | CGGTACCGCATGGTACAGAGG (200) | −4.65 | 48.25 | NH | 11.2g |
| 502-600R | CAATGCAATTCCGGGGTTGG (300) | ||||||
| ASF519 | Bacteroides sp. | 519-834F | CACAGTAAGCGGCACAGCG (200) | −3.78 | 38.57 | Forward primer hits | 5e |
| 519-1243R | CCGCTCACACGGTAGCTG (200) | uncultured pig GI and environmental clones, reverse primer hits identical mouse clone | |||||
MgCl2 concentration used in the QPCR mixtures was 4.0 mM.
The primer designation (F, forward; R, reverse), sequence, and nanomolar concentration of the primer used in QPCR (in parentheses) are shown.
Specificity determined by BLAST on 1 July 2003.
NH, no other hits.
Estimated by Southern blotting.
Estimated by QPCR see Materials and Methods.
Average for Clostridium spp. from Ribosomal RNA Operon Copy Number Database (16).
rRNA operon number determination.
The number of rRNA operons in the different ASF strains was determined to enable correlation of the number of copies of the gene with the number of bacteria. When the ASF strain could be grown in liquid culture and high-quality genomic DNA could be extracted (ASF356, ASF360, ASF361, and ASF519), a Southern hybridization-based protocol for the estimation of operon number was used. Genomic DNA (1 to 2 μg) extracted from the individual ASF strains was digested overnight with different restriction enzymes (six-cutters [BamHI, BglII, EcoRI, and HindIII] and four-cutters [HhaI and NciI]) that did not have a recognition site in the probe region. The digested genomic fragments were run for 4 to 5 h on a 0.7% agarose gel at 5 or 6 V/cm and then electrophoretically transferred onto a Zeta-probe GT membrane (Bio-Rad, Hercules, Calif.) using the Trans-Blot SD semidry transfer cell (Bio-Rad). The DNA was cross-linked to the membrane in a UV cross-linker (Stratagene). The probe consisted of a 500-bp fragment of the 16S rRNA from each ASF strain amplified using the 16S rRNA primers 27F and 519R (17) and gel purified. The probe was labeled by random priming (Amersham, Sunnyvale, Calif.) using [32P]dATP (Perkin-Elmer Life Sciences, Boston, Mass.). Unincorporated [32P]dATP was removed by purification with MicroBio-Spin 30 columns (Bio-Rad). The membrane was hybridized with the probe for 14 h in a hybridization buffer containing 50% formamide (Amersham), 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution and sheared salmon sperm DNA (100 μg/ml) (Brinkmann, Westbury, N.Y.). The membrane was then subjected to two low-stringency washes for 30 min each time (in 2× SSC-0.1% SDS at 37°C and in 1.5× SSC-0.1% SDS at 50°C) and one high-stringency wash for 1 h at 65°C (1× SSC-0.1% SDS). A phosphor-storage plate was exposed to the membrane for 5 to 30 min and scanned on a Cyclone phosphorimager (Perkin-Elmer Life Sciences). The amount of radioactive label in the bands was quantified using Optiquant software (Perkin-Elmer Life Sciences), and a comparison of the number and intensity of the bands for the different restriction enzymes was used to determine a consensus copy number.
Two of the strains (ASF457 and ASF492) could not be grown in sufficient quantities for Southern hybridization. Therefore, rRNA operon number was estimated by correlation of microscopic cell counts with target copy number estimated by QPCR. Cells were collected either from a liquid culture or a pure culture on Schaedler agar plates. Cells collected from plates were resuspended in Schaedler broth. Following vortexing to disrupt any cell clumps, two replicate 100-μl samples were centrifuged at 8,000 × g for 10 min, and DNA was extracted from the resultant cell pellet using the DNEasy tissue kit (Qiagen). Two replicate samples of the cell suspension diluted 1/100 and 1/50 were prepared in filter-sterilized phosphate-buffered saline (pH 7.4) (passed through a 0.2-μm-pore-size filter), fixed with 100 μl of formaldehyde, stained with 4′-6-diamidine-2′-phenylindole-dihydrochloride (DAPI) (Sigma), and collected on a Nucleopore membrane (Millipore, Bedford, Mass.). Cell numbers were estimated using an Axioskop epifluorescence microscope (Carl Zeiss, Thornwood, N.Y.). QPCR was performed on five replicates from each of the two cell pellet samples, using the conditions described above, and the number of copies in the template were estimated using the average CT numbers. The number of copies from QPCR was divided by the number of cell equivalents used as the template, which was calculated using the cell counts, to yield an estimate of the operon copy number. The average of the values obtained for the two replicates is reported as the estimated copy number. The method was tested using Lactobacillus murinus ASF361, for which the copy number had been estimated to be six by the Southern hybridization protocol. The QPCR-based protocol yielded an estimate of 5.79 copies, indicating its validity. While the protocol yields an estimate of the copy number, some error may be present due to the variability in bacterial counts and loss due to DNA extraction. ASF500 and ASF502 (both strains were most closely related to Clostridium spp.) did not grow sufficiently on plates to allow the use of even the alternate method. Hence, the average rRNA operon number of the Clostridium group (11.2) reported on the Ribosomal RNA Operon Copy Number Database (16) was used in all calculations of cell numbers.
RESULTS
Assay development.
All primers were sensitive and specific for the strain they were designed to target. Comparison against sequences in the GenBank database using BLAST (1) produced hits only against the ASF strains for four of the primer combinations, while the remaining four also hit some closely related sequences (≥99% or more sequence similarity) from intestinal clone libraries (Table 1). In all cases, amplification of each ASF strain in a background of fecal pellet DNA resulted in only one band of the same size as that of the strain-specific PCR product. Further, the shapes of the melting curves obtained for QPCR products amplified from DNA of pure strains, fecal pellet DNA from RF mice, and GI samples were identical for each primer pair. The products were also sequenced to confirm their identity. All these results indicate the specificity of the primers for the strain they amplify.
The sensitivity of the primers was determined by QPCR of a dilution series over 8 orders of magnitude of the plasmid standards for each strain. The slope of the standard curve, which is an indicator of amplification efficiency (a slope of −3.3 equals 100% efficiency), was in the range of −3.3 to −3.77 for most of the ASF strains, except ASF502 which had a slope of −4.65 (an efficiency of about 60%) (Table 1). The intercept, which is an estimate of the CT for the detection of one copy of the target gene, was less than 40 cycles for all strains except for strains ASF457 (40.96) and ASF502 (48.25). These considerations give confidence in the high sensitivity of the assay.
The rRNA operon number of the ASF strains was estimated to enable the correlation of QPCR results to cell numbers (Table 1). While the operon numbers for most of the ASF strains were estimated by either Southern hybridization or a QPCR-based method, two of the ASF strains (ASF500 and ASF502) that were closely related to the Clostridium group did not grow to adequate levels for either method to be used. The average operon number of the Clostridium group from the Ribosomal RNA Operon Copy Number Database (16) was used for all calculations for strains ASF500 and ASF502.
Spatial distribution of ASF flora.
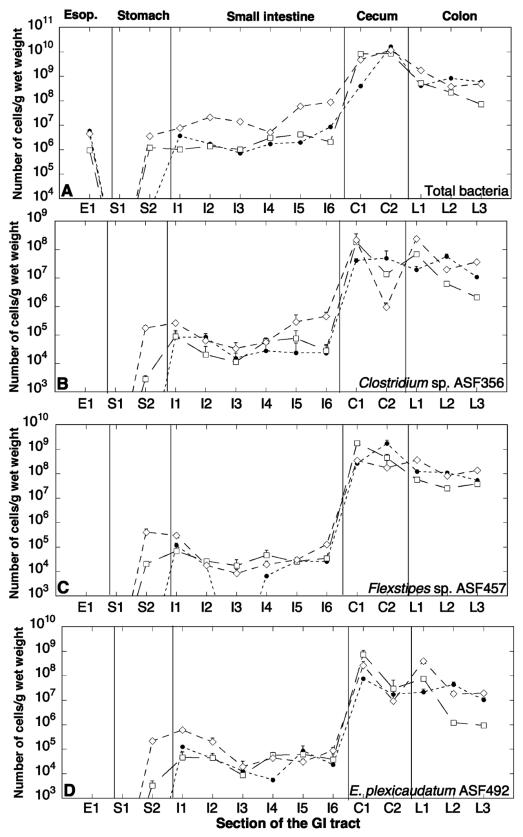
The total numbers (mean ± standard deviation) of the ASF strains varied over several orders of magnitude over the length of the GI tract as determined by averaging the sum of all ASF strains for the three mice (Fig. 1A). The esophagus harbored about 3.76 × 106 (±2.54 × 106) bacteria/g, while the number fell to about 1.59 × 106 (±1.04 × 106) bacteria/g in glandular stomach tissue. Surprisingly, no bacteria were detected in nonglandular stomach tissue. The number of bacteria increased to about 1.26 × 107 (±2.34 × 107) cells/g in the small intestine. A sharp increase in bacterial numbers was evident at the ileocecal valve, where the total number of bacteria climbed to 8.30 × 109 (±5.47 × 109) cells/g in the cecum and remained at about 5.78 × 108 (±4.76 × 108) and 1.13 × 109 (±3.09 × 108) cells/g in the colon and feces, respectively.
FIG. 1.
Distribution of ASF strains in different sections of the GI tracts of three defined-flora C.B-17 mice. The number of total bacterial cells of ASF strains (A), ASF356 (B), ASF457 (C), ASF492 (D), ASF500 (E), ASF361 (F), ASF502 (G), and ASF519 (H) in mouse 1 (solid circles), mouse 2 (open squares), and mouse 3 (open diamonds) are shown. The sections are taken from the esophagus (Esop.) (section E1), stomach (sections S1 and S2), small intestine (sections I1 to I6), ileocecal junction and apical cecum (sections C1 and C2, respectively), and colon (sections L1 to L3).
Overall, the diversity of strains and relative distribution changed substantially over the GI tract. The aerotolerant anaerobe Lactobacillus murinus ASF361 comprised close to 100% of the total bacteria in the esophagus, about 50% of the bacteria in the stomach and small intestine, and declined to about 0.01% of the bacteria in the cecum and colon. However, the spatial distribution of ASF361 in mouse 3 differed from that of the other two mice, with ASF361 accounting for about 0.2% of the total bacteria in the cecum and colon. The anaerobes were generally absent in the esophagus, and some were present at low numbers in the stomach, but they increased sharply over the length of the intestine from about 50% of the bacteria in the small intestine to about 99.99% of the bacteria in the cecum and colon.
Four of the ASF strains, Clostridium sp. strain ASF356 (Fig. 1B), Flexistipes sp. strain ASF457 (Fig. 1C), Eubacterium plexicaudatum ASF492 (Fig. 1D), and low-G+C-content, gram-positive bacterial strain ASF500 (Fig. 1E), showed a similar distribution over the length of the GI tract in all three mice. These four strains were present at low levels (104 to 106 cells/g) in the small intestine and increased sharply at the ileocecal valve (108 to 109 cells/g), sustaining this high level in the colon. Surprisingly, the EOS strain ASF500 was also seen at low levels (102 to 103 cells/g) in the esophagus of all three mice. In all three mice, aerotolerant Lactobacillus murinus ASF361 (Fig. 1F) maintained populations of 105 to 107 cells/g over the length of the GI tract, with about 106 cells/g in the esophagus, 104 to 106 cells/g in the stomach, 105 to 107 cells/g in the small intestine and cecum, and 103 to 105 cells/g in the colon. The apical cecum (C2) of mouse 3 had higher levels of Lactobacillus murinus ASF361 along with lower levels of the other strains compared to mouse 1 and mouse 2. The other lactobacillus, ASF360, was absent in most regions of the gut and had very low levels of 102 cells/g when present.
ASF502 (Fig. 1G) and ASF519 (Fig. 1H) showed greater variation in the three mice. The distribution of Bacteroides sp. strain ASF519 was similar for all three mice in the stomach (104 to 105 cells/g), small intestine (105 to 106 cells/g), and colon (107 to 109 cells/g). In the cecum, however, the number of ASF502 was much higher in mouse 2 than in mouse 1 and mouse 3 (108 versus 104 to 106 cells/g). The distribution of ASF502 in the small intestine varied in the three mice. Mouse 3 had higher numbers of ASF502 in the stomach (106 versus 104 cells/g) and small intestine (107 versus 106 cells/g) than mouse 1 and mouse 2. However, for all three mice, the numbers of ASF502 were comparable in the cecum (109 to 1010 cells/g) and colon (107 to 108 cells/g), with mouse 1 having a lower number (106 cells/g) at the ileocecal junction.
Comparison of fecal and intestinal bacterial distribution.
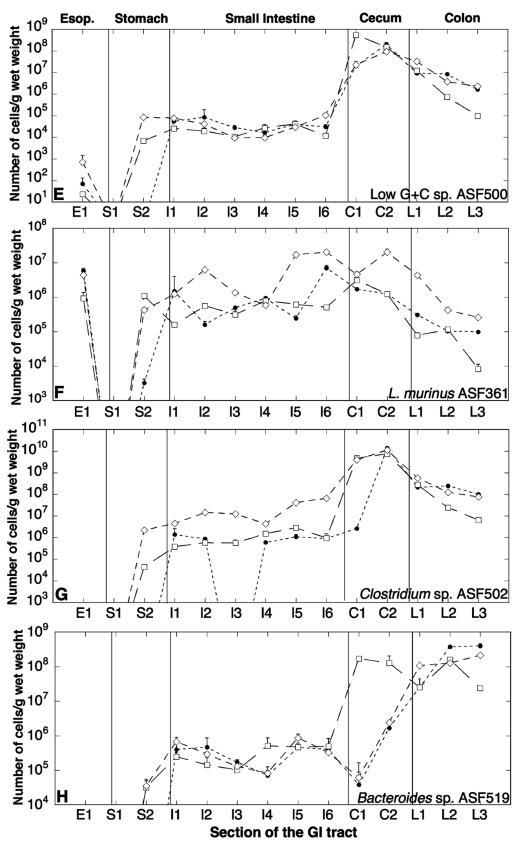
The numbers of individual ASF strains in fecal matter varied over 3 orders of magnitude for the different ASF species but were similar for all three mice. The nonparametric Kruskal-Wallis test indicated no significant differences between the fecal distributions for the three mice (P = 0.75; Kruskal-Wallis statistic = 0.564); therefore, the ASF strains had the same population distribution in the feces of all three mice. ASF502 and ASF519 were present at about 109 cells/g of feces, while ASF361 and ASF457 were present at about 108 cells/g of feces. ASF492 and ASF500 were present at about 107 cells/g of feces, and ASF356 was present at about 106 to 107 cells/g of feces (Fig. 2).
FIG. 2.
Fecal levels of the ASF strains in three defined-flora C.B-17 mice were determined by QPCR. The numbers of ASF bacteria/g (wet weight) in mouse 1 (white columns), mouse 2 (shaded columns), and mouse 3 (black columns) are shown.
Since fecal bacteria are often studied as a substitute for colonic microbiota, we compared the levels of bacteria in feces to the levels in the colon and found a considerable difference in the distribution of the ASF species in the colon and feces. The data were normalized to make the samples more comparable, since a gram of tissue is not comparable to a gram of feces. The numbers of the different ASF strains in each section of the colon and in feces were divided by the numbers of Lactobacillus murinus ASF361 in the same section. The ratios in the different sections were then compared for similarity. The fecal bacterial numbers of the ASF strains were within an order of magnitude of the number of ASF361. However, the colonic bacterial ratios varied greatly, with Flextipes sp. strain ASF457, Clostridium sp. strain ASF502, and Bacteroides sp. strain ASF519 present at 2 to 3 orders of magnitude higher than ASF361. The other strains were 1 to 2 orders of magnitude higher in abundance (Table 2). Thus, the correspondence between the levels of the ASF strains in feces and in the colon was poor.
TABLE 2.
Comparison of the ratios of the ASF strains in different sections of the colon and in fecal matter
| Strain | ASF strain ratioa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1
|
L2
|
L3
|
F1
|
|||||||||
| m1 | m2 | m3 | m1 | m2 | m3 | m1 | m2 | m3 | m1 | m2 | m3 | |
| ASF356 | 62.4 | 875.7 | 54.1 | 547.6 | 53.9 | 46.5 | 110.6 | 253.7 | 142.4 | 0.1 | 0.0 | 0.1 |
| ASF361 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| ASF457 | 392.5 | 738.3 | 84.9 | 1,060.7 | 213.9 | 190.7 | 554.4 | 4,499.1 | 524.1 | 0.9 | 0.7 | 0.6 |
| ASF492 | 69.9 | 966.4 | 89.9 | 424.0 | 10.3 | 42.5 | 104.8 | 113.1 | 75.5 | 0.1 | 0.0 | 0.3 |
| ASF500 | 30.1 | 154.2 | 7.4 | 81.0 | 6.1 | 8.7 | 16.7 | 11.8 | 8.9 | 0.3 | 0.1 | 0.2 |
| ASF502 | 675.2 | 3,632.5 | 130.5 | 2,388.8 | 209.1 | 289.1 | 1,022.8 | 787.6 | 298.2 | 7.7 | 1.2 | 5.7 |
| ASF519 | 82.1 | 324.2 | 24.6 | 3,597.2 | 1,357.4 | 296.2 | 4,027.7 | 2,847.9 | 826.6 | 11.4 | 0.8 | 2.1 |
ASF strain ratio was obtained by dividing the number of bacteria of each ASF strain by the number of ASF361 bacteria in the same section or sample. Sections L1 to L3 were proximal to distal sections of the colon, respectively, and F1 is a fecal sample. The ratios for the three different mice (mouse 1 [m1] to 3 [m3]) are shown.
Stability of ASF flora in RF mice.
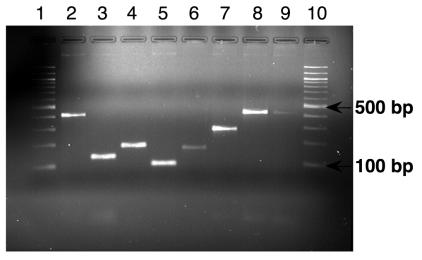
PCR of fecal pellet DNA from C57BL/6 RF mice with ASF primers yielded the correct-sized bands for all the ASF strains (Fig. 3). The PCR products were sequenced and found to be identical to the sequences of the ASF strains. This demonstrated the persistence of the ASF strains in the GI tracts of RF mice, which had been bred and maintained in an isolator with nonsterile food, water, and bedding for 3 years.
FIG. 3.
Specific PCR products obtained by amplification of fecal DNA extracted from C57BL/6 RF mice using the ASF-specific primers. These mice, which originally harbored the defined flora, were bred and maintained under nonsterile conditions for 3 years. Lanes 1 and 10, 100-bp ladder; lanes 2 to 9, RF fecal pellet DNA amplified with primers specific for ASF356 (417 bp), ASF360 (131 bp), ASF361 (182 bp), ASF457 (95 bp), ASF492 (167 bp), ASF500 (285 bp), ASF502 (427 bp), and ASF519 (429 bp) (expected sizes shown in parentheses), respectively.
DISCUSSION
Here, we present the defined-flora ASF mouse as a model system for the study of host-bacterium and interspecies interactions in the gut. Defined-flora models are invaluable in studying the effects of the microflora on the mammalian host in a manner not limited by the complexity and mouse-to-mouse variability that the full microbial complement entails (8). Recent studies have generally utilized the simplest defined-flora models, colonized with a single bacterial species, such as Bacteroides thetaiotaomicron (12) and E. coli (13), to assess the role of microflora in host development and colonization resistance to Salmonella infections, respectively. However, a more complex but easily quantifiable model system would be invaluable for many applications. The ASF model may be such a system for several reasons. First, mice colonized with the ASF strains have normal GI physiology, as shown by their resistance to opportunistic pathogens and the size of their ceca, which was similar to that of healthy mice. Second, the ASF flora is stable. Even when the defined-flora mice were maintained in clean conditions but with nonsterile food, water, and bedding over several years, allowing other bacteria to colonize the gut, we detected the ASF strains in the feces of the mice by PCR methods. Further, a recent study assessing the diversity of the mouse GI microbiota using clone libraries found several ASF strains in mice, indicating that they are stable members of the complex microbiota in the mouse GI tract (24). Third, ASF mice are widely available and with the increased routine use of QPCR, the model system is easily accessible. Since the constituents of the flora were chosen for both historical and functional reasons, they might not be ideal for all studies. In particular, several of the metabolic groups of intestinal bacteria are not represented in this model. However, additional bacterial strains can be added to this model to address various questions of interactions. Further, observations by studies using this model can serve as the basis for developing hypotheses to test in mice with a complex microflora. The choice of mouse strain in our study was influenced by its availability and prior use to test the pathogenesis of Helicobacter hepaticus in the development of IBD in a defined nonpathogenic microbial background (3). Since the mice lack both T and B cells, they afford some ability to separate the effects of bacterial interactions from specific immune responses of the host. Thus, they represent a powerful model for studying the role of the microecology of the gut in the pathogenesis of diseases, such as IBD.
QPCR has proven a robust and sensitive method for the quantification of bacterial species in complex environmental communities. QPCR assays developed for the quantification of the different ASF strains are specific for the strains, as shown both by a search against sequences in the GenBank database using BLAST and by amplifying the strain of interest in a complex background. The QPCR assays performed well in all regions of the GI tract, but the small intestine samples exhibited a high background. This was likely due to the higher number of mammalian cells, and hence mammalian DNA, which intercalated the fluorescent dye used to detect and quantify PCR amplification products. Dilution of these samples prior to quantification reduced the problem but led to a higher detection limit (∼104 to 105 copies versus 103 to 104 copies in other regions) and higher intra-assay variation (coefficient of variation, 0.08 to 1.67). Some of the cecal samples also had to be diluted. In contrast, the values obtained in the colon showed low intra-assay variation (coefficient of variation, 0.04 to 0.74).
Culture-based methods have previously been used to study the distribution of bacteria in the different parts of the GI tract. A comparison of total bacterial counts in feces with culture results indicated that 20 to 40% of fecal bacteria were culturable by the methods used (18). Thus, the culturability of intestinal bacteria is better than other environmental systems, where the value is typically around 0.01% (2, 14). Culture-based studies in mice (28) and humans (5) have indicated that there are about 107 bacteria/g in the esophagus, about 103 to 109 bacteria/g in the stomach, about 105 to 108 bacteria/g in the small intestine (predominantly lactobacilli), and about 1010 to 1012 bacteria/g in the large intestine (predominantly anaerobes). These numbers compare favorably with our results of a total of 106 to 107 bacteria/g in the esophagus and stomach, although we did not detect any bacteria in nonglandular stomach tissue, where the expected number and type of bacteria would be similar to those in the esophagus (28). Since nonglandular stomach tissue lacks a mucous layer and the bacteria are loosely attached, one of the possible factors leading to this result could be the loss of bacteria due to the wash that we subjected the stomach to in order to remove the ingesta. Given that the sensitivity of our assays was of the order of 10 bacteria/reaction, our lower limit of detection would be about 103 bacteria/g in nonglandular stomach tissue. In the small and large intestines, we observed about 106 to 108 and 108 to 1010 bacteria/g, respectively. As in the culture-based studies, we also observed a sharp thousandfold increase in bacterial numbers across the ileocecal valve. Thus, the QPCR-based data correspond well with the results of culture-based studies, and the ASF strains reach near natural levels in the defined-flora model.
The oxygen sensitivity of the individual ASF strains appears to be the predominant factor in their distribution along the GI tract and a good substitute for overall community patterns. The increasingly anaerobic nature of the GI tract from the stomach to the large intestine has recently been shown in live mice using noninvasive electron paramagnetic resonance methods (9). The esophagus and stomach are aerobic, with a decreasing gradient of oxygen along the small intestine, and the cecum and colon are anaerobic. Based on the results of previous culture-based studies (28), the aerotolerant Lactobacillus sp. strain ASF360 and ASF361 would be expected to be present at high numbers in the esophagus, stomach, and small intestine. Lactobacillus murinus ASF361 does follow this trend, with high numbers in these regions (106 to 108 cells/g [wet weight], 100% of esophageal bacteria, about 50% of small intestine bacteria). These high levels are maintained in the cecum, with a decrease over the length of the colon (104 to 107 cells/g [wet weight]). The anaerobic ASF strains would be expected to have high numbers in the cecum and colon (>109 cells/g) and low numbers (<105 cells/g) elsewhere, as seen by culture-based studies (6). Indeed, the distribution of Clostridium sp. strain ASF356, Flexistipes sp. strain ASF457, Eubacterium sp. strain ASF492, low-G+C-content strain ASF500, Clostridium sp. strain ASF502, and Bacteroides sp. strain ASF 519 follows this trend. There was a dramatic increase in the populations of the anaerobic ASF strains at the ileocecal valve, and the populations were maintained at high levels of 108 to 1010 cells/g (wet weight) in the cecum and colon. Thus, the patterns seen by QPCR correspond well with the results of culture-based studies but afford higher throughput and accuracy between samples. Furthermore, the oxygen sensitivity of the strains appears to explain the overall distribution of the ASF strains at the current spatial resolution of the assay.
Factors other than oxygen sensitivity may also play a role in regulation of population abundance. Mice are coprophagic, and ingestion of fecal matter may account for the transient populations detected, especially in the upper GI tract. This, for example, may explain the low levels of anaerobic populations (105 to 107 bacteria/g) in the stomachs and small intestines of all mice. However, the existence of anaerobic microniches cannot be discounted at this point and may be a more likely explanation for the presence of ASF500 in the esophagus. This strain is EOS and was the only anaerobe to be present in the esophagus in all three mice. Competition due to high niche overlap may explain why one of the lactobacilli was present only at trace amounts in the guts of all three mice. Lactobacillus sp. strain ASF360 was detected only in very low numbers (<103 cells/g [wet weight]) in a few regions of the GI tract. However, we were able to detect ASF360 by PCR in fecal pellet samples from C57BL/6 RF mice (Fig. 3), indicating that it was a stable colonizer of formerly gnotobiotic strains of mice. To determine the reason for this discrepancy and to further differentiate the distribution of the ASF strains, more detailed studies with finer resolution will need to be performed.
Intermouse variation of the total bacterial numbers per gram was relatively moderate. Higher levels of bacteria were seen in the stomach and small intestine of mouse 3, while the stomach of mouse 1 had fewer anaerobic ASF strains than the other two mice. Fecal matter consumed prior to the dissection is a possible cause for this increase in mouse 3, since the levels of bacteria were also high in the stomach of mouse 3. Individual strains showed low intermouse variability over the length of the GI tract, with the exception of Clostridium sp. strain ASF502 and Bacteroides sp. strain ASF519. ASF502 exhibited high intermouse variation in the small intestine, with a higher number in the small intestine of mouse 3 than in mouse 1 and mouse 2. This mirrored the total bacterial distribution discussed above, and coprophagy is also a likely cause for this difference. ASF519 showed high intermouse variation in the cecum. The numbers of ASF519 in mouse 2 were 2 to 3 orders of magnitude higher than those in mouse 1 and mouse 3. This was confirmed by repeating the quantification. However, the numbers of ASF519 in the colon were comparable for all three mice. Overall, the intermouse variation is within an order of magnitude, and the few differences that were observed between the three mice could be due to differences in feeding or coprophagy. Moreover, the colon, which is often the region of interest in studying the microecology of the gut, showed little intermouse variation for the different ASF strains.
The lack of accessibility of the gut for easy sampling has resulted in the frequent use of fecal matter as a substitute for the colonic flora. However, the utility of this approximation may not be suitable for all purposes, since the qualitative and quantitative correspondence of the fecal bacterial distribution to the colonic biota has not been studied in detail (23). Zoetendal et al. (35) provided evidence that the human colonic mucosal flora and fecal flora are significantly different on the basis of molecular community fingerprints obtained by denaturant gradient gel electrophoresis. To the best of our knowledge, our study is the first to use quantitative molecular methods to compare the quantitative distribution of bacteria in fecal matter and the colon. Our results show that even with the limited diversity of the ASF model, the microflora of the colon is poorly reflected in fecal matter. The distribution of ASF strains in feces was similar for all three mice, and all the ASF strains (except ASF360) were present in feces varying within an order of magnitude. The colonic distribution, however, varied over 3 orders of magnitude, with anaerobes exceeding the aerotolerant ASF361 by a thousandfold. In contrast, the levels of ASF361 in feces were comparable to the levels of the anaerobes. Thus, our results, in conjunction with the study of Zoetendal et al. (35), suggest that the fecal microflora differs quantitatively from the colonic microflora and that the fecal flora may be an inappropriate substitute for some questions of GI ecology.
The role of the microbiota in the host remains difficult to assess largely due to the complexity of the gut ecosystem. Thus, the characterization of defined-flora mice colonized with the ASF flora, along with the highly specific and sensitive assays developed for accurate quantification of the ASF, should provide a useful tool for the study of a wide range of questions in GI microecology. Examples of questions that may be addressed with this model system include the population dynamics that drive colonization resistance to pathogens and understanding the role of the normal microbiota in the prognosis of diseases such as IBD and the spread of antibiotic resistance.
Acknowledgments
This work was supported in part by NIH grant AI-50952 (to J.G.F., D.B.S., and M.F.P.).
We thank Jeff Bajko for help with the dissections, Yan Feng for help with cloning, and Nancy Taylor and Roger Orcutt for their expertise and helpful suggestions for strain culture.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahill, R. J., C. J. Foltz, J. G. Fox, C. A. Dangler, F. Powrie, and D. B. Schauer. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 65:3126-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dewhirst, F. E., C. C. Chien, B. J. Paster, R. L. Ericson, R. P. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaldson, R. M., and P. P. Toskes. 1989. The relation of enteric bacterial populations to gastrointestinal function and disease, p. 107-114. In J. S. Sleisenger (ed.), Gastrointestinal disease: pathophysiology, diagnosis, management. Saunders, Philadelphia, Pa.
- 6.Dubos, R. J., D. C. Savage, and R. W. Schaedler. 1967. The indigenous flora of the gastrointestinal tract. Dis. Colon Rectum 10:23-34. [DOI] [PubMed] [Google Scholar]
- 7.Falk, P. G., L. V. Hooper, T. Midtvedt, and J. I. Gordon. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, G., R. A. Shankar, M. Chzhan, A. Samouilov, P. Kuppusamy, and J. L. Zweier. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. USA 96:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentges, D. J., A. J. Stein, S. W. Casey, and J. U. Que. 1985. Protective role of intestinal flora against infection with Pseudomonas aeruginosa in mice: influence of antibiotics on colonization resistance. Infect. Immun. 47:118-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 12.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 13.Hudault, S., J. Guignot, and A. L. Servin. 2001. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut 49:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy, M. J., and P. A. Volz. 1985. Ecology of Candida albicans gut colonization: inhibition of Candida adhesion, colonization, and dissemination from the gastrointestinal tract by bacterial antagonism. Infect. Immun. 49:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. S. M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 18.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orcutt, R. P., F. J. Gianni, and R. J. Judge. 1987. Development of an “Altered Schaedler Flora” for NCI gnotobiotic rodents. Microecol. Ther. 17:59. [Google Scholar]
- 22.Orcutt, R. P. 2002. Culturing members of the Altered Schaedler Flora from stock vials. Taconic Technical Library. [Online.] http://www.taconic.com/library/Culturing_ASF.htm.
- 23.Rumney, C. J., and I. R. Rowland. 1992. In vivo and in vitro models of the human colonic flora. Crit. Rev. Food Sci. Nutr. 31:299-331. [DOI] [PubMed] [Google Scholar]
- 24.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 25.Sartor, R. B. 1997. The influence of normal microbial flora on the development of chronic mucosal inflammation. Res. Immunol. 148:567-576. [DOI] [PubMed] [Google Scholar]
- 26.Savage, D. C. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107-133. [DOI] [PubMed] [Google Scholar]
- 27.Savage, D. C. 1986. Gastrointestinal microflora in mammalian nutrition. Annu. Rev. Nutr. 6:155-178. [DOI] [PubMed] [Google Scholar]
- 28.Savage, D. C., R. Dubos, and R. W. Schaedler. 1968. The gastrointestinal epithelium and its autochthonous bacterial flora. J. Exp. Med. 127:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaedler, R. W., R. Dubos, and R. Costello. 1965. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122:77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer, S. M., and T. E. Nash. 2000. The role of normal flora in Giardia lamblia infections in mice. J. Infect. Dis. 181:1510-1512. [DOI] [PubMed] [Google Scholar]
- 31.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vollaard, E. J., and H. A. Clasener. 1994. Colonization resistance. Antimicrob. Agents Chemother. 38:409-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoetendal, E. G., K. Ben-Amor, A. D. Akkermans, T. Abee, and W. M. de Vos. 2001. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst. Appl. Microbiol. 24:405-410. [DOI] [PubMed] [Google Scholar]
- 35.Zoetendal, E. G., A. von Wright, T. Vilpponen-Salmela, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]