Abstract
Inorganic polyphosphate (polyP) plays a significant role in increasing bacterial cell resistance to unfavorable environmental conditions and in regulating different biochemical processes. Using transmission electron microscopy of the polychlorinated biphenyl (PCB)-degrading bacterium Pseudomonas sp. strain B4 grown in defined medium with biphenyl as the sole carbon source, we observed large and abundant electron-dense granules at all stages of growth and following a shift from glucose to biphenyl or chlorobiphenyls. Using energy dispersive X-ray analysis and electron energy loss spectroscopy with an integrated energy-filtered transmission electron microscope, we demonstrated that these granules were mainly composed of phosphate. Using sensitive enzymatic methods to quantify cellular polyP, we confirmed that this polymer accumulates in PCB-degrading bacteria when they grow in the presence of biphenyl and chlorobiphenyls. Concomitant increases in the levels of the general stress protein GroEl and reactive oxygen species were also observed in chlorobiphenyl-grown cells, indicating that these bacteria adjust their physiology with a stress response when they are confronted with compounds that serve as carbon and energy sources and at the same time are chemical stressors.
Polyphosphate (polyP) is a ubiquitous linear polymer consisting of hundreds of orthophosphate residues (Pi) linked by high-energy phosphoanhydride bonds. The best-known enzymes involved in the metabolism of polyP in bacteria are the polyphosphate kinase (PPK) that catalyzes the reversible conversion of the terminal phosphate of ATP into polyP and the exopolyphosphatase that processively hydrolyzes the terminal residues of polyP to liberate Pi (15).
The involvement of polyP in the regulation of both enzyme activities and expression of large group of genes is the basis of survival for different bacteria, including pathogens, under stress conditions and of adaptation to the stationary growth phase (reviewed in reference 16). Mutant bacterial cells that lack polyP survive poorly during growth in the stationary phase and are less resistant to heat, oxidants, osmotic challenge, antibiotics, and UV radiation (6, 13, 24, 25, 35).
polyP accumulation in response to nutrient deprivation has also been reported in the genus Pseudomonas, and recent studies have demonstrated that PPK is essential in Pseudomonas aeruginosa not only for various forms of motility (26, 27) but also for biofilm development, quorum sensing, production of virulence factors, and virulence in the burned-mouse pathogenesis model (28).
Chlorinated biphenyls (CBs) and polychlorinated biphenyls (PCBs) belong to one of the most widely distributed classes of chlorinated chemicals in the environment (33, 34). The toxicities and carcinogenicities of some PCB congeners make them a serious environmental and health problem (14). For cleanup of large areas of PCB-contaminated soils and aquatic environments bioremediation seems to be a promising approach (22). Although many genetic, enzymological, and biochemical analyses of PCB-degradative pathways have provided the basis for the engineering of specific enzymes and genetically modified microorganisms in order to improve performance in bioremediation of PCBs, little is known about the physiological adjustments of PCB-degrading bacteria during growth with these kinds of organochlorine compounds.
Here we demonstrate that the PCB-degrading bacterium Pseudomonas sp. strain B4 accumulates much higher levels of polyP during exponential growth with biphenyl than when glucose is the sole carbon source. Following a shift from a defined medium with glucose to a medium with biphenyl or CBs as the single carbon source, numerous polyP granules accumulated in the cytoplasm. Additionally, induction of the general stress protein GroEl and oxygen reactive species (ROS) was observed, probably as a physiological adjustment to growth in the presence of these contaminating compounds, which appear to stress the cells.
MATERIALS AND METHODS
Chemicals.
Biphenyl was purchased from Merck (Hohenbrunn, Germany). 2-Chlorobiphenyl and 4-chlorobiphenyl were obtained from Accustandard, Inc. (New Haven, Conn.).
Bacterial strains and growth conditions.
The biphenyl-utilizing organisms Burkholderia fungorum strain LB400 and Pseudomonas sp. strain B4 were grown aerobically at 30°C on Luria-Bertani (LB) rich medium or M9 minimal salts medium (30) supplemented with 0.5% biphenyl, 1% glucose, 0.1% (wt/vol) 2-chlorobiphenyl, 0.1% (wt/vol) 4-chlorobiphenyl, or 0.05% (vol/vol) 3-chlorobiphenyl. For the shift experiments, Pseudomonas sp. strain B4 cells that were exponentially grown in M9 medium with 1% glucose as the sole carbon source were collected by centrifugation, washed twice with M9 medium, and finally resuspended in the same medium supplemented with biphenyl or another CB.
Electron microscopy.
Unstained cells from the different cultures were routinely examined for the presence of electron-dense bodies by transmission electron microscopy (9). Cells from the different cultures were mixed and dispersed in distilled water and then placed onto carbon-coated nickel grids. The drops containing the microorganisms were drained off with filter paper, and the preparations were air dried for 30 to 50 s. Electron microscopy was performed with a Philips Tecnai 12 electron microscope by using an accelerating voltage of 80 kV (Electron Microscopy Laboratory, Pontificia Universidad Católica de Chile).
EDAX analysis.
Energy-dispersive spectroscopy of chemical elements in bacteria was performed with an EDAX-PV 9800 energy-dispersive microanalyzer at an accelerating voltage of 120 kV (8). The electron beam was focused on the location at which the elemental composition was to be determined. Due to the interaction between the primary electron and the sample, X-ray signals were collected with the energy-dispersive X-ray (EDAX) analysis spectrometer, which was connected to the electron microscope.
EELS and element mapping.
Electron energy loss spectroscopy (EELS) analysis (17) was performed with a Zeiss CEM 902 integrated energy-filtered transmission electron microscope. The microscope was operated in the electron spectroscopic imaging (ESI) mode for element mapping, and parallel EELS was performed for spectrum registration with the aid of ESI-Vision software (Soft Imaging Systems, Münster, Germany). Aperture settings described by Lünsdorf et al. (17) were used. Commercial hydroxyapatite was used as the internal phosphate standard.
Purification and analysis of proteins.
Purified recombinant His6-PPK was prepared by using Escherichia coli strain NR 100 as described previously (1), and this preparation was used in the polyP assay described below. The protein concentration was determined by the method of Bradford (Coomassie Plus protein assay reagent; Pierce, Rockford, Ill.).
Western immunoblotting.
The total protein fractions corresponding to the different cells in the shift experiments were separated by sodium dodecyl sulfate- polyacrylamide gel electrophoresis and electrotransferred to a polyvinylidene difluoride membrane as described previously (23). For the antigen-antibody reaction, the membrane containing the transferred proteins was treated with an anti-Acidithiobacillus ferroxidans GroEL polyclonal antibody (1:1,000 dilution) as the primary antibody and with monoclonal anti-rabbit antibodies conjugated with peroxidase (Amersham, Little Chalfont, United Kingdom) as the secondary antibodies (1:5,000 dilution). A colorimetric method was used to develop Western blots as recommended by Promega (Madison, Wis.).
polyP quantification.
polyP was quantified by using a two-step conversion of polyP into ATP by PPK and quantification of ATP by luciferase to generate light (2). First, polyP was extracted from cell extracts by using Glassmilk, and then it was assayed by using the reverse reaction of E. coli PPK with excess ADP. Finally, the ATP content was determined by using the luciferase (Boehringer, Mannheim, Germany) reaction, and the luminescence was measured with a luminometer (BioScan Lumi/96). The concentration of polyP was expressed in terms of Pi residues.
Analysis of polyP by gel electrophoresis.
polyP samples extracted with Glassmilk were prepared for gel electrophoresis by the method of Robinson et al. (29). Polyacrylamide gel electrophoresis was performed as previously described (18) with Tris-borate-EDTA buffer (pH 8.3), using 18% urea gels and a Protean IIxi cell system (Bio-Rad) for 90 min at 400 V. The gels were stained with toluidine blue (0.05%) in 25% methanol. The estimated size range of the polyP was determined by comparison with polyP standards having chain lengths of 45 and 75 P residues (Sigma) and PolyP750 (chain length, ∼750 P residues), synthesized in vitro as described previously (2, 4).
In vivo detection of ROS.
Overproduction of ROS in cells exponentially grown under different conditions was detected by using the oxidative stress-sensitive probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) (7). The acetyl groups in this compound are removed by membrane esterases to form 2′,7′-dichlorodihydrofluorescein (DCFH) when the probe is taken up by living cells. DCFH is not fluorescent but is highly sensitive to ROS; it is oxidized by these active species to the highly fluorescent compound 2′,7′-dichlorofluorescein (12). DCFH can be oxidized by several reactive species, including RO2·, RO·, OH·, HOCl·, and ONOO−, but only longer-lived radicals contribute to the increase in fluorescence (10). For our experiments, DCFH-DA was added at a final concentration of 5 μM from a 2 mM stock solution in ethanol to cells exponentially grown with glucose, biphenyl, 2-chlorobiphenyl, or 4-chlorobiphenyl as the sole carbon source. The cells were incubated at 30°C for 1 h in the dark. The samples were handled to avoid light, and fluorescence was measured with a spectrofluorometer (Fluoromax-2; Instruments S.A, Inc.).
RESULTS
Formation of electron-dense granules by growth of cells with biphenyl and CBs.
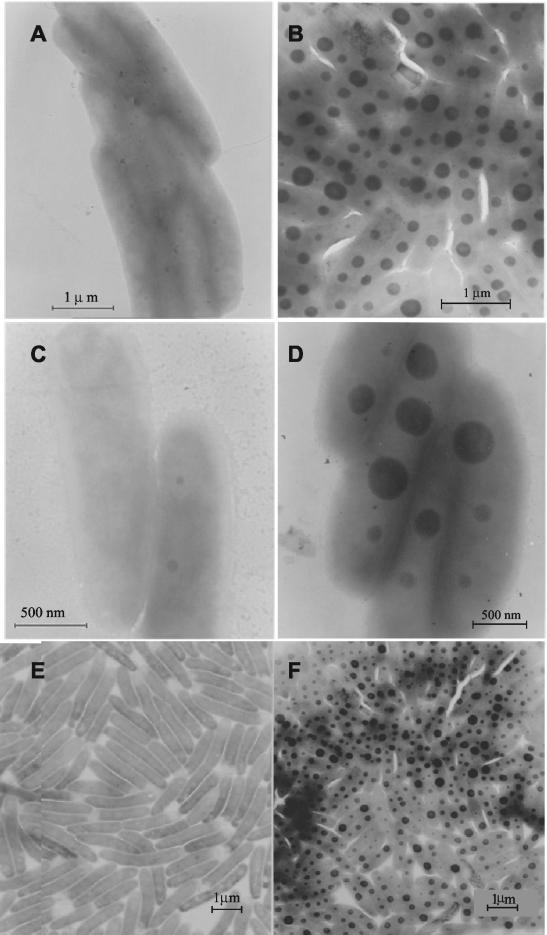
polyP have been detected as electron-dense bodies in several microorganisms (9). Both Pseudomonas sp. strain B4 and the PCB-degrading reference strain B. fungorum LB400 were found to contain electron-dense bodies in the cytoplasm, and the number and size of these bodies depended on the growth medium and the phase of growth. In all of the experiments performed, the results for the two microorganisms were the same. Figure 1 shows electron micrographs of Pseudomonas sp. strain B4 exponentially grown on M9 medium with either glucose (Fig. 1A) or biphenyl (Fig. 1B) as the sole carbon source. With unstained microorganisms like those used in this analysis, it is not possible to discern a well-defined outline of a bacterial cell. However, this method clearly showed that 80 to 90% of the cells collected in the exponential phase contained numerous large electron-dense granules when they were grown with biphenyl, in contrast to the less than 10% of the cells that contained very few much smaller granules when the organisms were grown with glucose. However, more than 90% of the cells collected in the stationary phase contained visible electron-dense granules when they were grown with either of the two carbon sources (data not shown). The granules were abundant and in many cells occupied 20 to 30% of the cell contour area (Fig. 1B). The largest granules had diameters of ∼200 to 300 nm, and generally all cells contained three or more granules (Fig. 1B). The same phenomenon was observed with B. fungorum LB400 cells (Fig. 1C and D). Thus, the PCB-degrading bacteria Pseudomonas sp. strain B4 and B. fungorum LB400 accumulated large amounts of large electron-dense granules when they were grown with biphenyl at all stages of growth; when they were grown with glucose, they accumulated granules only when the cells entered the stationary phase.
FIG. 1.
Electron micrographs of exponentially grown cells of Pseudomonas sp. strain B4 (A, B, E, and F) and B. fungorum LB400 (C and D). Grids containing the unstained cells were prepared as described in Materials and Methods. Cells were grown to the exponential phase with glucose (A and C) or biphenyl (B and D) as the sole carbon source. A sample of exponentially growing cells of Pseudomonas sp. strain B4 in a defined medium supplemented with glucose (E) was shifted to a medium containing 4-chlorobiphenyl as the sole carbon source (F).
When exponentially grown cells of Pseudomonas sp. strain B4 were shifted from a medium with glucose to the same medium containing CBs, such as 2-, 3-, and 4-chlorobiphenyls, as the sole carbon sources, after 4 h we observed massive accumulation of granules in all cases (as shown for cells grown with 4-chlorobiphenyl in Fig. 1E and F). After cells were shifted from glucose to biphenyl, a dramatic accumulation of electron-dense granules also occurred 4 h after the shift (data not shown). Neither Pseudomonas sp. strain B4 nor B. fungorum LB400 contained electron-dense granules in any of the growth phases when they were grown in LB medium (data not shown).
Elemental analysis of electron-dense granules by EDAX and EELS.
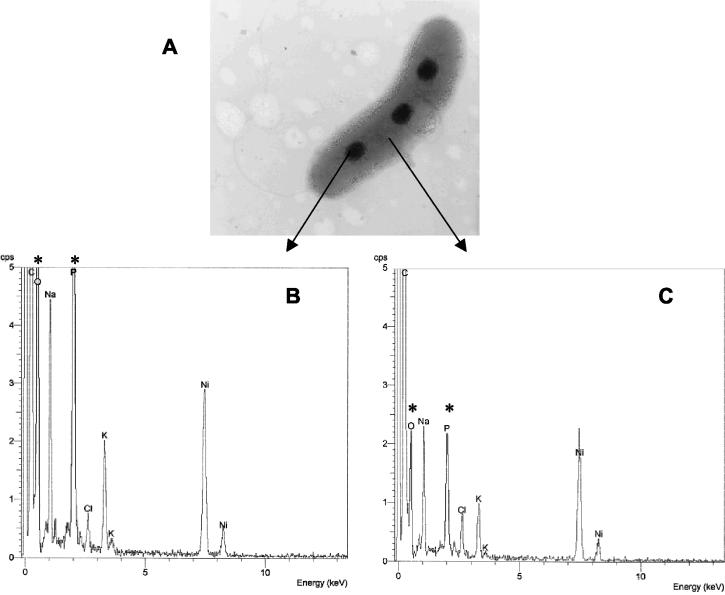
EDAX analysis revealed that the electron-dense granules contained large amounts of phosphorus and oxygen (Fig. 2A and B) compared with the amounts in other cytoplasmic regions of the cells (Fig. 2A and C). The EDAX data on the chemical composition of the electron-dense granules from Pseudomonas sp. strain B4 were similar to those for intracellular polyP granules from Acinetobacter strain 210A (3) and Desulfovibrio gigas (11).
FIG. 2.
EDAX spectra of Pseudomonas sp. strain B4 grown in a defined medium supplemented with biphenyl as the sole carbon source. (A) Electron micrograph of a single unstained cell used to analyze the chemical composition of different areas (indicated by arrows). (B) Spectrum obtained from an electron-dense body. (C) Spectrum of a cytoplasmic area. The asterisks indicate Ka peak intensities of phosphorus and oxygen.
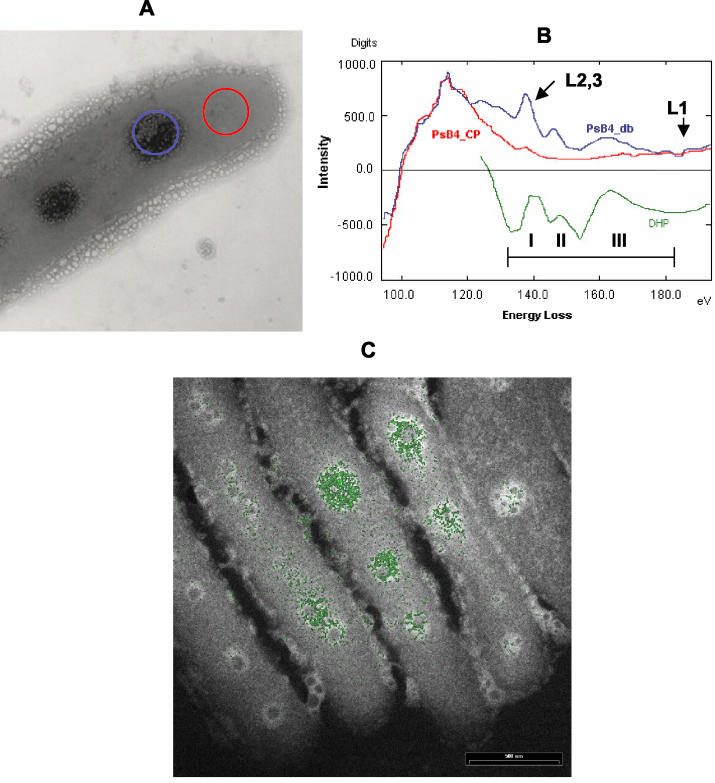
To obtain more detailed information about the polyP-like inclusions, the ultrastructure was examined by scanning electron microscopy coupled to EELS analysis with an integrated energy-filtered transmission electron microscope. Figure 3A shows the electron energy loss spectrum for a cell area containing an electron-dense granule and a cytoplasmic area without such granules. As shown in Fig. 3B, the electron energy loss spectrum of the cell area containing the granules was similar to that of hydroxyapatite, which was used as a phosphate reference after background subtraction and spectrum filtering. The ionization energy onset of the PL2,3 edge is at 135 eV for all three spectra and is followed by characteristic energy loss near edge structure features in the 50-eV-higher energy loss region (Fig. 3B). Although all three main maxima (peaks I, II, and III) are present, the maximum of peak III is lower for the polyP granules than for the hydroxyapatite reference. This indicates that there are differences in the electronic vicinity of the phosphorus atoms in the two compounds (i.e., the phosphorus acid anhydride character of polyP and the nonanhydride character of the calcium hydroxyapatite). By operating the microscope in the ESI mode it was possible to map the phosphorus distribution in the cell area. As shown in Fig. 3C, the distribution of phosphorus, obtained by ESI analysis, exactly matched the electron-dense polyP areas. Thus, by using electron microscopic microanalyses we demonstrated that the electron-dense granules that accumulated in PCB-degrading bacteria during growth with biphenyl or CBs as the sole carbon sources were mainly composed of phosphate and most likely were polyP granules.
FIG. 3.
EELS spectrum of electron-dense bodies present in unstained Pseudomonas sp. strain B4. (A) Electron micrograph of an unstained cell grown in a defined medium supplemented with biphenyl as the sole carbon source. The electron-dense granule analyzed is indicated by a blue circle, and the cytoplasmic reference area analyzed is indicated by a red circle. (B) EELS spectra (after background subtraction) of the electron-dense body (PsB4_db) (blue line) and the cytoplasmic area (PsB4_CP) (red line). The spectrum of hydroxyapatite was used as the phosphate reference standard (DHP) (green line). (C) Phosphate distribution image (green), obtained by ESI superimposed with the electron micrograph negative image of Pseudomonas sp. strain B4 cells.
Enzymatic determination of polyP in cells grown under different conditions.
To determine whether the amazingly large increase in electron-dense granules composed of phosphate during growth with different CBs was indeed due to greater accumulation of polyP, we measured the polyP content under all the conditions analyzed during the electron microscopy observations. By using cell extracts, polyP was enzymatically quantified as described in Materials and Methods.
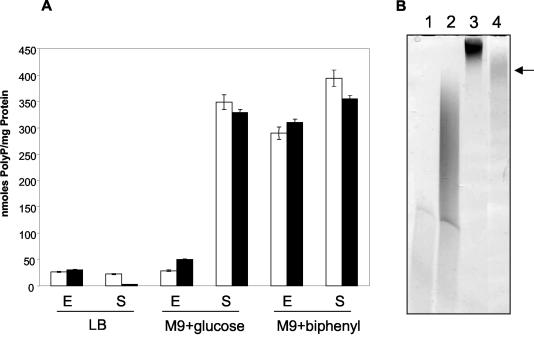
As shown in Fig. 4A, the polyP contents of Pseudomonas sp. strain B4 during growth under different conditions exactly matched the pattern observed for the appearance of electron-dense granules in the cells. Greater accumulations of polyP were observed in cells grown in the presence of biphenyl at all stages of growth and in glucose-grown cells only in the stationary phase. The values were more than 10-fold higher than those seen during exponential growth of cells with glucose as the sole carbon source or in LB medium at all stages of growth. Identical results were obtained when B. fungorum LB400 cells were used (Fig. 4A). These results indicated that the accumulation of electron-dense bodies during growth in the presence of biphenyl was due to greater accumulation of polyP. The same phenomenon was observed when CBs were tested (data not shown). The polyP synthesized under these conditions had an average chain length of more than 75 residues and less than 750 residues as determined by polyacrylamide gel electrophoresis (Fig. 4B).
FIG. 4.
Influence of the growth phase and different carbon sources on polyP accumulation by cells of Pseudomonas sp. strain B4 and B. fungorum LB400. (A) Cells of Pseudomonas sp. strain B4 (open bars) or B. fungorum LB400 (solid bars) were grown in LB medium or in M9 medium supplemented with glucose or biphenyl as the sole carbon source. Aliquots of the cells were analyzed to determine the polyP contents during the exponential (E) or stationary (S) phase of growth. The error bars indicate the standard deviations based on three different experimental values. (B) Polyacrylamide gel electrophoresis of polyP purified from Pseudomonas sp. strain B4 cells grown with biphenyl (lane 4, arrow). Lanes 1 to 3 contained polyP standards (lane 1, 45 residues; lane 2, 75 residues; lane 3, 750 residues).
polyP accumulation and stress.
As polyP plays a significant role in increasing the resistance of bacterial cells to unfavorable and stressful environmental conditions, it was of interest to find out if these microorganisms were stressed while they metabolized CBs. Organochlorine compounds, including PCBs, are known to increase oxidative stress in several biological systems (31). In living microorganisms that have been subjected to environmental stresses, oxidative stress is caused by both overproduction of ROS and depletion of antioxidants. Therefore, we measured the possible oxidative stress in cells exponentially grown under different conditions by using the oxidative stress-sensitive probe DCFH-DA (7). Table 1 shows that exponentially growing cells of Pseudomonas sp. strain B4 grown with glucose had a minor background level of fluorescence generated by ROS. A 10-fold increase in fluorescence was seen in biphenyl-grown cells, whereas 40- and 20-fold increases were seen in cells grown in 2-chlorobiphenyl and 4-chlorobiphenyl, respectively. These results provided clear evidence that an oxidative stress was generated when the microorganism was grown with these compounds as the sole carbon sources.
TABLE 1.
Growth with biphenyl and CBs greatly enhances generation of ROSa
| Carbon source | Growth (FU/mg)c |
|---|---|
| Glucose (1%) | 16,837 |
| Biphenyl (2 mM) | 174,075 |
| 4-Chlorobiphenyl (2 mM) | 606,597 |
| 2-Chlorobiphenyl (2 mM) | 319,027 |
| Glucose (1%) + H2O2 (50 mM)b | 1,384,377 |
DCFH-DA (5 μM) was added to Pseudomonas sp. strain B4 cells growing exponentially with different carbon sources, and the preparations were incubated for 1 h in the dark.
As a positive control, cells grown with glucose were incubated for the same time in the presence of 50 mM hydrogen peroxide.
Fluorescence was measured as described in Materials and Methods and was expressed in fluorescence units (FU).
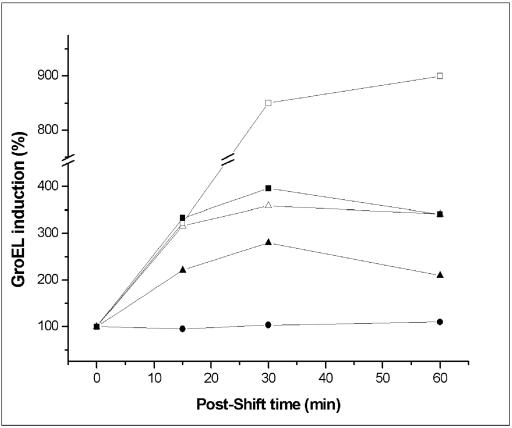
It was of interest to determine if a more general stress response could take place. To do this, we determined the levels of the protein GroEL during shift experiments in the presence of chloroaromatic compounds. As shown in Fig. 5, the GroEL levels increased in PCB-degrading bacterial cells when they were shifted from growth with glucose to growth with different CBs. Although maximum induction of GroEL (approximately eightfold) occurred when control cells were heat shocked at 42°C, during the shift to 4-chlorobiphenyl the stress protein was induced fourfold compared to control cells maintained with glucose as the sole carbon source and showed a transient response, which is typical of heat shock responses. The induction of GroEL was greater in all the cells exposed to the CBs tested than in the cells exposed to biphenyl. Thus, CBs not only induced the synthesis of polyP and ROS but most likely generated a heat shock-like response in PCB-degrading bacteria.
FIG. 5.
GroEL is induced in PCB-degrading bacteria during growth with different CBs. Pseudomonas sp. strain B4 cells were shifted from a defined medium supplemented with glucose to a medium supplemented with biphenyl (▴), 2-chlorobiphenyl (▵), or 4-chlorobiphenyl (▪) as the sole carbon source. Control cells were maintained in the presence of glucose (•) or were subjected to heat shock at 42°C (□) for different times. The growth of the shifted cells was determined by measuring the optical density at 600 nm during the experiment. The only cells that grew during the experiment were the control cells grown with glucose. Cell extracts from the cells shifted to the different conditions were used to determine the levels of GroEL by Western blotting after separation of the total proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Quantitation of the GroEL bands after Western blotting was done by using an image analysis program (Scion Image).
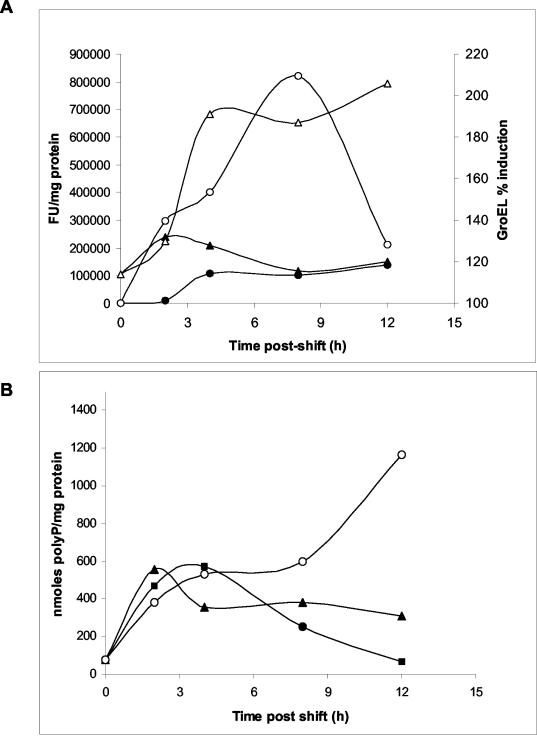
To monitor these three responses simultaneously, we determined the time course changes in polyP, GroEL, and ROS in the same cells when they were shifted to different substrates (Fig. 6). Shortly after the shift (after 2 h), there was no great difference in ROS between the cells shifted to glucose and the cells shifted to 2-chlorobiphenyl, but the GroEL level was much higher in the cells shifted to 2-chlorobiphenyl (Fig. 5 and 6A). This great difference in induction of GroEL levels in cells shifted to 2-chlorobiphenyl remained or even increased during the remaining time (up to 12 h). This prolonged synthesis of GroEL suggests that there is slow and continuous accumulation of abnormal proteins which most likely trigger a heat shock-like response. In the present case, the abnormal proteins were generated by production of ROS as a consequence of metabolism of the 2-chlorobiphenyl substrate. Figure 6A shows a similar situation with the ROS levels, since these levels were between six- and sevenfold higher in the cells shifted to 2-chlorobiphenyl.
FIG. 6.
Time course of production of ROS, GroEL, and polyP in Pseudomonas sp. strain B4. (A) Exponentially grown cells were shifted from a defined medium supplemented with glucose to a medium supplemented with 2-chlorobiphenyl (▵ and ○) as the sole carbon source. Control cells were maintained in a medium containing glucose (▴ and •). GroEL levels (• and ○) and ROS production (▵ and ▴) were measured as described in the text. (B) polyP content of cells shifted from a defined medium supplemented with glucose to a medium supplemented with 2-chlorobiphenyl (○), glycerol (▴), or glucose (▪) as the sole carbon source. FU, fluorescence units.
Since simply changing substrates in the cell could also induce a stress response, as a control we shifted the cells from glucose to another carbon source, such as glycerol. Shortly after the shift (up to 4 h after the shift) there were increases in the polyP levels with all the substrates employed (Fig. 6B), probably reflecting initial adaptation of the cells to the medium change. However, at later times after the shift (4 to 12 h) there was a marked increase in the level of polyP when the cells were shifted to 2-chlorobiphenyl compared with the level in cells shifted to glucose or glycerol, in which the level of polyP slowly decreased to a level that was 6- to 10-fold lower than that in cells grown with 2-chlorobiphenyl. This clearly indicates that the effect seen with 2-chlorobiphenyl was not simply due to a change in the substrate.
DISCUSSION
Bacterial survival and stress resistance studies are important for many aspects of bioremediation and biocontrol. For adaptation to stress, cells must coordinate major changes in the rates of transcription, translation, and replication, as well as changes in the genes expressed. polyP could provide activated phosphates or could coordinate an adaptive response by binding metals and/or specific proteins. Here, we describe the massive accumulations of polyP that occur in PCB-degrading bacteria during the exponential phase of growth in the presence of biphenyl and some CBs. The accumulations of polyP under these conditions are remarkable because they are so large and are sustained compared to those in E. coli, which does not form visible polyP granules under common growth conditions (15). The levels that were more than 400 nmol/mg of protein in the exponential phase of cells growing in a medium containing biphenyl and in the stationary phase of glucose-grown cells were roughly 100-fold greater than those seen in E. coli. The amounts of polyP accumulated by Pseudomonas sp. strain B4 were even larger than the amount reported previously (150 nmol/mg) for another microorganism accumulating polyP granules, Vibrio cholerae, during the exponential phase of growth in a rich medium (20). It is likely that the polyP accumulations observed in Pseudomonas sp. strain B4 and B. fungorum LB400 contribute not only to the energy supply but also to enhanced survival and stress resistance in the presence of CBs.
Regarding the physiological function of polyP as a regulatory factor for gene expression in E. coli, it has been shown that mutant E. coli cells that are unable to accumulate polyP have reduced resistance to heat shock and to oxidative stress caused by H2O2 (32). On the other hand, organochlorine compounds, including PCBs, are known to increase oxidative stress in several biological systems (31). We found clear evidence of generation of oxidative stress in Pseudomonas sp. strain B4 cells grown in the presence of biphenyl and CBs. At present, we do not know how this oxidative stress correlates with the concomitant massive increase in polyP levels under these conditions. In E. coli it has been found that polyP controls rpoS and recA expression at the transcriptional level, thereby affecting the expression of many stress-inducible and stationary-phase-inducible genes, including some genes related to oxidative stress and thermotolerance (32).
Our finding that there are increased levels of polyP and GroEL during growth in the presence of biphenyl and CBs not only indicates that the pollutant compounds used as carbon sources induce a chemical stress but is consistent with the behavior of mutant E. coli cells that are unable to accumulate polyP, which are less thermotolerant (15). In this regard, several stress shock proteins, including DnaK and GroEL, have been reported to be newly synthesized in Pseudomonas sp. strain DJ-12 when the organism is subjected to lethal stress conditions, such as the presence of 4-chlorobiphenyl and biphenyl and heat shock (21). It has been found that in Pseudomonas putida the pollutant toluene not only serves as a carbon and energy source but also is a chemical inducer of stress (36). Also, the stress shock proteins, which contribute to the resistance to the cytotoxic effects of the phenoxy herbicide 2,4-dichlorophenoxyacetic acid, were induced at different 2,4-dichlorophenoxyacetic acid concentrations in exponentially growing cultures of Burkholderia sp. strain YK-2 (5). Based on the similarities among polyP, ROS, and heat shock protein accumulations during several kinds of stress conditions, it might be assumed that CBs generate a stress response in bacteria when the organisms use these compounds as sole sources of carbon. Additionally, increased polyP synthesis requires enhanced phosphate uptake, a fact that should be taken into account when bioremediation of PCBs is undertaken.
Recently, it was shown that Burkholderia cepacia AM19 grown under low-pH conditions exhibited enhanced polyP accumulation, possibly representing a widespread microbial response to stressful low external pH values (19). polyP may therefore play an important role in the physiological adaptation of microorganisms during growth and development and in their responses to starvation and environmental stresses.
Acknowledgments
This research was supported by projects ICM P-99-031-F and ICGEB, CRP/CHI00-04, contract 01/001. F.P.C. was the recipient of a DAAD Ph.D. scholarship.
We are very grateful to Arthur Kornberg for kindly providing E. coli strain NR 100, to Kenneth Timmis and Bernd Hofer for their kind gift of Pseudomonas sp. strain B4 and B. fungorum LB400, and to Ricardo B. Maccioni for providing the reagent DCFH-DA. We also thank Alejandro Munizaga for helpful collaboration during the electron microscopy sessions.
REFERENCES
- 1.Ahn, K., and A. Kornberg. 1990. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265:11734-11739. [PubMed] [Google Scholar]
- 2.Ault-Riché, D., C. D. Fraley, C.-M. Tzeng, and A. Kornberg. 1998. A novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonting, C. F. C., M. Rigoulet, B. Guerin, and P. Canioni. 1993. The elemental composition dynamics of large polyphosphate granules in Acetobacter strain 210A. Arch. Microbiol. 159:428-434. [Google Scholar]
- 4.Cardona, S. T., F. P. Chávez, and C. A. Jerez. 2002. The exopolyphosphatase gene from Sulfolobus sofataricus: characterization of the first gene found to be involved in polyphosphate metabolism in Archaea. Appl. Environ. Microbiol. 68:4812-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, Y. S., S. H. Park, C. K. Kim, and K. H. Oh. 2000. Induction of stress shock proteins DnaK and GroEL by phenoxyherbicide 2,4-D in Burkholderia sp. YK-2 isolated from rice field. Curr. Microbiol. 41:33-38. [DOI] [PubMed] [Google Scholar]
- 6.Crooke, E., M. Akiyama, N. N. Rao, and A. Kornberg. 1994. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 269:6290-6295. [PubMed] [Google Scholar]
- 7.Crow, J. P. 1997. Dichlorodihydrofluorescein and dihydrorhodamine 123 are sensitive indicators of peroxynitrite in vitro: implications for intracellular measurement of reactive nitrogen and oxygen species. Nitric Oxide Biol. Chem. 1:145-157. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg, J., H. Gonzalez, T. E. Jensen, and W. A. Corpe. 2001. Quantitative analysis of the elemental composition and the mass of bacterial polyphosphate bodies using STEM EDX. Microbios 106:177-188. [PubMed] [Google Scholar]
- 9.Gonzalez, H., and T. E. Jensen. 1998. Nickel sequestering by polyphosphate bodies in Staphylococcus aureus. Microbios 93:179-185. [PubMed] [Google Scholar]
- 10.He, Y. Y., M. Klisch, and D.-P. Häder. 2002. Adaptation of cyanobacteria to UV-B stress correlated with oxidative stress and oxidative damage. Photochem. Photobiol. 76:188-196. [DOI] [PubMed] [Google Scholar]
- 11.Hensgens, C. M. H., H. Santos, C. Zhang, W. H. Kruizinga, and T. A. Hansen. 1996. Electron-dense granules in Desulfovibrio gigas do not consist of inorganic triphosphate but of a glucose pentakis(diphosphate). Eur. J. Biochem. 242:327-331. [DOI] [PubMed] [Google Scholar]
- 12.Ischiropoulos, H., A. Gow, S. R. Thom, N. W. Kooy, J. A. Royall, and J. P. Crow. 1999. Detection of reactive nitrogen species using 2,7-dichlorodihydrofluorescein and dihydrorhodamine 123. Methods Enzymol. 301:367-373. [DOI] [PubMed] [Google Scholar]
- 13.Kim, K. S., N. N. Rao, C. D. Fraley, and A. Kornberg. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. 99:7675-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimbrough, R. D. 1995. Polychlorinated biphenyls (PCBs) and human health: an update. Crit. Rev. Toxicol. 25:133-163. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 16.Kulaev, I., and T. Kulakovskaya. 2000. Polyphosphate and phosphate pump. Annu. Rev. Microbiol. 54:709-734. [DOI] [PubMed] [Google Scholar]
- 17.Lünsdorf, H., C. Strömpl, A. M. Osborn, A. Bennasar, E. R. B. Moore, W.-R. Abraham, and N. T. Kenneth. 2000. Approach to analyze interactions of microorganisms, hydrophobic substrates, biofilms and to study initial events in microbiogeological processes. Methods Enzymol. 336:317-331. [DOI] [PubMed] [Google Scholar]
- 18.McGrath, J. W., and J. P. Quinn. 2000. Intracellular accumulation of polyphosphate by the yeast Candida humicola G-1 in response to acid pH. Appl. Environ. Microbiol. 66:4068-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullan, A., J. P. Quinn, and J. W. McGrath. 2002. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low pH conditions. Microb. Ecol. 44:69-77. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa, N., C. M. Tzeng, C. D. Fraley, and A. Kornberg. 2000. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182:6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, S. H., K. H. Oh, and C. K. Kim. 2001. Adaptive and cross-protective responses of Pseudomonas sp. DJ-12 to several aromatics and other stress shocks. Curr. Microbiol. 43:176-181. [DOI] [PubMed] [Google Scholar]
- 22.Pieper, D. H., and W. Reineke. 2000. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 11:262-270. [DOI] [PubMed] [Google Scholar]
- 23.Ramírez, P., H. Toledo, N. Guiliani, and C. A. Jerez. 2002. An exported rhodanese-like protein is induced during growth of Acidithiobacillus ferrooxidans in metal sulfides and different sulfur compounds. Appl. Environ. Microbiol. 68:1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, N. N., Liu, S., and A. Kornberg. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid, M. H., N. N. Rao, and A. Kornberg. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson, N. A., J. E. Clark, and H. G. Wood. 1987. Polyphosphate kinase from Propionibacterium shermanii—demonstration that polyphosphates are primers and determination of the size of the synthesized polyphosphate. J. Biol. Chem. 262:5216-5222. [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Schilderman, P. A., L. M. Maas, D. M. Pachen, T. M. de Kok, J. C. Kleinjans, and F. J. van Schooten. 2000. Induction of DNA adducts by several polychlorinated biphenyls. Environ. Mol. Mutagen. 36:79-86. [DOI] [PubMed] [Google Scholar]
- 32.Shiba, T., K. Tsutsumi, K. Ishige, and T. Noguchi. 2000. Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry (Moscow) 65:375-384. [PubMed] [Google Scholar]
- 33.Tiedje, J. M., J. F. Quensen, J. Chee-Sanford, J. P. Schimel, and S. A. Boyd. 1993. Microbial reductive dechlorination of PCBs. Biodegradation 4:231-240. [DOI] [PubMed] [Google Scholar]
- 34.Timmis, K. N., R. J. Steffan, and R. Unterman. 1994. Designing microorganisms for the treatment of toxic wastes. Annu. Rev. Microbiol. 48:525-557. [DOI] [PubMed] [Google Scholar]
- 35.Tsutsumi, K., M. Munekata, and T. Shiba. 2000. Involvement of inorganic polyphosphate in expression of SOS genes. Biochim. Biophys. Acta 493:73-81. [DOI] [PubMed] [Google Scholar]
- 36.Varcellone-Smith, P., and D. S. Herson. 1997. Toluene elicits a carbon starvation response in Pseudomonas putida mt-2 containing the TOL plasmid pWW0. Appl. Environ. Microbiol. 63:1925-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]