Abstract
Thlaspi goesingense is able to hyperaccumulate extremely high concentrations of Ni when grown in ultramafic soils. Recently it has been shown that rhizosphere bacteria may increase the heavy metal concentrations in hyperaccumulator plants significantly, whereas the role of endophytes has not been investigated yet. In this study the rhizosphere and shoot-associated (endophytic) bacteria colonizing T. goesingense were characterized in detail by using both cultivation and cultivation-independent techniques. Bacteria were identified by 16S rRNA sequence analysis, and isolates were further characterized regarding characteristics that may be relevant for a beneficial plant-microbe interaction—Ni tolerance, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase and siderophore production. In the rhizosphere a high percentage of bacteria belonging to the Holophaga/Acidobacterium division and α-Proteobacteria were found. In addition, high-G+C gram-positive bacteria, Verrucomicrobia, and microbes of the Cytophaga/Flexibacter/Bacteroides division colonized the rhizosphere. The community structure of shoot-associated bacteria was highly different. The majority of clones affiliated with the Proteobacteria, but also bacteria belonging to the Cytophaga/Flexibacter/Bacteroides division, the Holophaga/Acidobacterium division, and the low-G+C gram-positive bacteria, were frequently found. A high number of highly related Sphingomonas 16S rRNA gene sequences were detected, which were also obtained by the cultivation of endophytes. Rhizosphere isolates belonged mainly to the genera Methylobacterium, Rhodococcus, and Okibacterium, whereas the majority of endophytes showed high levels of similarity to Methylobacterium mesophilicum. Additionally, Sphingomonas spp. were abundant. Isolates were resistant to Ni concentrations between 5 and 12 mM; however, endophytes generally tolerated higher Ni levels than rhizosphere bacteria. Almost all bacteria were able to produce siderophores. Various strains, particularly endophytes, were able to grow on ACC as the sole nitrogen source.
Plants have acquired different mechanisms for growth in the presence of heavy metal concentrations usually considered phytotoxic. One strategy includes the accumulation of extremely large amounts of heavy metals. Hyperaccumulating plants are particularly interesting for phytoremediation technologies for the treatment of metal-polluted soils, sediments, and water resources (40). Several hundred plant species endemic to metalliferous soils have been identified as hyperaccumulators, of which 75% are able to hyperaccumulate Ni when growing in ultramafic soils (5). Thlaspi goesingense Hálácsy was first described as a hyperaccumulator by Reeves and Brooks (46), and plants grown in an ultramafic soil contained Ni concentrations as high as 12,400 μg g−1 of shoot dry biomass−1 (56).
Although a number of authors have addressed rhizosphere processes of hyperaccumulators, various questions still remain unanswered. Several studies indicated that rhizosphere acidification is not responsible for increased metal uptake (e.g., see references 36 and 39). So far, root architecture (e.g., see reference 58), effective root uptake systems (e.g., see reference 34), and partial depletion of labile metals in the rhizosphere (e.g., see reference 45) seem to be the most relevant rhizosphere processes involved in hyperaccumulation. The role of root exudates in metal mobilization is still unclear, and data reported so far are conflicting (45, 47, 57, 60). Recently it has been shown that rhizosphere microorganisms may play an important role, probably by increasing the availability of heavy metals for plant uptake (2, 59). Microbes are ubiquitous even in soils with high heavy metal concentrations. High numbers of Ni-resistant bacterial cells were found together with hyperaccumulator plants, whereas a decreased number of cells tolerating only lower Ni concentrations was associated with nonaccumulator plants grown in the same ultramafic soil or in nonrhizosphere soil (12, 49). Soil microorganisms may improve the metal solubility and availability by reducing the soil pH or by producing chelators and siderophores. Microbial siderophores prevent iron deficiency of the producing organism and of plants but may also be involved in the uptake of other metals (11, 17, 24). The production of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, an enzyme that has no function in bacteria but modulates ethylene levels in developing plants (21), may further contribute to the heavy metal tolerance of plants. It is well known that plants respond to environmental stresses by synthesizing “stress” ethylene (1). Several soil bacteria producing the enzyme ACC deaminase have been identified which reduce the amount of the precursor of ethylene, resulting in decreased ethylene biosynthesis in plants (6, 8, 20). In addition, plant-associated bacteria may produce phytohormones and provide nutrients to the plant. Besides rhizosphere microbes, endophytes, which colonize a niche similar to that of pathogens but do not cause damage to the plant, also have an intimate relationship with their host and have to tolerate high levels of heavy metal concentrations when colonizing hyperaccumulating plants. Furthermore, endophytic bacteria are known for their beneficial effects on plant growth and health (32, 53).
The aim of this study was to characterize rhizosphere and endophytic bacterial populations of T. goesingense growing in a serpentine soil in eastern Austria. Since only a minor percentage of naturally occurring microorganisms can be cultured, the plant-associated microflora was analyzed by using a cultivation-independent approach. Bacterial populations were characterized by terminal restriction fragment length polymorphism (T-RFLP), as well as by cloning and sequencing of 16S rRNA genes. Furthermore, Ni-resistant rhizosphere and endophytic bacteria were isolated, identified by partial 16S ribosomal DNA (rDNA) sequencing, and analyzed regarding their tolerance of Ni, as well as siderophore and ACC deaminase production.
MATERIALS AND METHODS
Field site description and sampling of plants and rhizosphere soil.
Plant and rhizosphere samples were obtained from a serpentine site in Redlschlag, Austria, previously described by Wenzel et al. (57). A total Ni concentration of 2,580 mg of Ni kg of soil−1 was reported, whereas the labile Ni concentrations in the bulk soil and in the rhizosphere of T. goesingense were 7.72 and 5.06 mg of Ni kg of soil−1, respectively (57). The sampling time was May 2002 at the flowering stage of the plants. Plant and soil material from the root zone was immediately transferred in polyethylene bags and transported to the laboratory in transportable coolers (4°C).
Isolation of rhizosphere and endophytic bacteria.
Soil tightly adhering to roots (2.5 g) was added to 25 ml of 1/10-strength tryptic soy broth and shaken for 2 h at 250 rpm at 28°C. Subsequently, the solution was left without shaking for 1 h in order to allow settling of soil particles. Various 10-fold dilutions were plated on 1/10-strength tryptic soy agar (TSA) (Merck) which was amended with cycloheximide (100 mg liter−1) in order to inhibit fungal growth and different concentrations of NiCl2 (5 or 10 mM). Plates were incubated for 7 days at 30°C. For the isolation of endophytes, plant material (2 g) was surface sterilized with 5% sodium hypochlorite for 5 min, rinsed four times with sterile, distilled water, dipped in 70% ethanol, and finally flamed. Plant surfaces were checked for their sterility by blotting them tightly on 1/10-strength TSA and incubating plates for 2 days at 30°C. No growth was observed. After flaming the plant material was macerated with 10 ml of 1/10-strength tryptic soy broth and subsequently plated on 1/10-strength TSA plates as described above. Viable cell counts were determined. Furthermore, from the rhizosphere and endosphere of five individual plants up to five Ni-resistant colonies of each morphology type were picked and further analyzed. In total, 50 rhizosphere isolates and 62 endophytic isolates were analyzed.
Isolation of rhizosphere and shoot-associated DNA.
Five plant samples were used for the isolation of rhizosphere and shoot-associated DNA. Rhizosphere DNA was isolated as previously described (50). Briefly, the bacteria of 0.5 g of soil were lysed with lysozyme and bead beating. Phenol-chloroform extraction was used to eliminate proteins, whereas humic acids were precipitated with potassium acetate. Nucleic acids were precipitated with ethanol, and the resulting DNA solution was further purified by passage through spin columns containing Sepharose.
For the isolation of bacterial DNA from shoots, plants (2 g) were surface sterilized as described above. Then, bacterial cells were dislodged from Thlaspi plant tissues prior to DNA extraction by using a slightly modified protocol published by Garbeva et al. (18). Disinfected sliced plant material was incubated in 50 ml of 0.9% sodium chloride with shaking for 1 h at room temperature. Cells were collected by centrifugation at 10,000 × g and 4°C and resuspended in 4 ml of TN150 buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl). Then, 0.3 g of 0.1-mm acid-washed glass beads (Sigma) were added to aliquots of 0.8 ml, and bead beating was performed twice for 1 min at full speed with an interval of 30 s (type MM2000, 220 V, 50 Hz; Retsch GmbH & Co. KG, Haam, Germany). After extraction with phenol and chloroform, nucleic acids were precipitated with a 0.1 volume of 3 M sodium acetate solution (pH 5.2) and a 0.7 volume of isopropanol for 20 min at −20°C. Nucleic acids were centrifuged for 10 min at 10,000 × g, washed, and dried. Finally, the DNA was resuspended in 60 μl of Tris-EDTA containing RNase (0.1 mg ml−1). It can be assumed that this DNA derives mainly from endophytes but may also contain nucleic acids from phylloplane bacteria.
T-RFLP analysis.
Rhizosphere partial 16S rRNA gene sequences were amplified with a thermocycler (PTC-100; MJ Research, Inc.), using an initial denaturing step of 5 min at 95°C followed by 30 cycles of 30 s at 95°C, 1 min of annealing at 54°C, and a 2-min extension at 72°C. PCR mixtures (50 μl) contained 0.5 μl of extracted DNA, 1× reaction buffer (Gibco, BRL), 200 μM (each) dATP, dCTP, dGTP, and dTTP, 2 mM MgCl2, and 2.5 U of Taq DNA polymerase (Gibco BRL), and 0.2 μM concentrations of the primers 8f (5′-AGAGTTTGATCCTGGCTCAG-3′) (14), labeled with 6-carboxyfluorescein at the 5′ end, and 926r (31) (5′-CCGTCAATTCCTTT(AG)AGTTT-3′). For the analysis of endophytic bacteria, it was important to avoid the amplification of small-subunit rRNA genes derived from plant organelles. Therefore, the primer 799f [5′-AAC(AC)GGATTAGATACCC(GT)-3′] (10), which does not target the chloroplast 16S rRNA gene, in combination with primer pH (5′-AAGGAGGTGATCCAGCCGCA-3′) (14) was applied. Approximately 200 ng of fluorescently labeled PCR amplification products were digested with the restriction enzyme AluI (Gibco BRL). Aliquots of 0.75 μl were mixed with 1 μl of loading buffer (diluted five times in deionized formamide; Fluka) and 0.3 μl of the DNA fragment length standard (Rox 500; PE Applied Biosystems, Inc., Foster City, Calif.). Mixtures were denatured for 2 min at 92°C and immediately chilled on ice prior to electrophoretic separation on 5% polyacrylamide gels. Fluorescently labeled terminal restriction fragments were detected using an ABI 373A automated DNA sequencer (PE Applied Biosystems, Inc.) in the GeneScan mode. Lengths of labeled fragments were determined by comparison with the internal standard using the GeneScan 2.5 software package (PE Applied Biosystems Inc.).
Terminal restriction fragments (T-RFs) between 35 and 500 bp long with heights of ≥50 fluorescence units were included in the analysis. Profiles were normalized according to the method of Dunbar et al. (13).
Small-subunit rDNA clone libraries.
16S rRNA gene clone libraries were generated from total communities of DNAs isolated from the rhizosphere and endosphere (shoot-associated DNA). Rhizosphere 16S rRNA genes were amplified by PCR with the primers 8f and pH by using the conditions described above, whereas endophytic 16S rRNA genes were amplified by using the primer pair 799f and 1520r. Amplicons were ligated into the TpCR 4-TOPO vector (Invitrogen), and Escherichia coli DH5α-T1R (Invitrogen) was transformed with the ligation products according to the manufacturer's instructions. Sixty rhizosphere-associated colonies (obtained from the rhizosphere of plant I) and 80 shoot-associated colonies (obtained from shoots of plants III and V) were picked and resuspended directly in PCRs using the primers M13f and M13r and the conditions described above to amplify cloned inserts. PCR products were purified with the NucleoTraPCR kit (Macheroy-Nagel) according to the manufacturer's instructions and used as templates in sequencing reactions.
Theoretical terminal restriction fragment (T-RF) sizes of cloned sequences were determined after sequencing. However, since predicted and true T-RF sizes can differ up to several base pairs, clones were subjected to T-RFLP analysis in order to determine true T-RF lengths.
DNA isolation of bacterial isolates.
For the isolation of genomic DNA from isolates, bacteria were grown until late exponential phase in 5 ml of tryptic soy broth in a rotatory shaker at 28°C. Cells were harvested by centrifugation for 10 min at 10,000 × g at 4°C. After decanting the supernatant, 300-mg glass beads were added and DNA was isolated as described above.
PCR-RFLP analysis of the 16S rRNA gene and the 16S-23S rRNA IGS region.
Restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA gene was used to group isolates at the species level, whereas characterization of the 16S-23S rRNA intergenic spacer region (IGS) was applied to distinguish different strains of the same species. Small-subunit rRNA gene sequences were amplified as described above using the primer pair 8f and 1520r. The primers pHr (5′-TGCGGCTGGATCACCTCCTT-3′) and P23SR01 (5′-GGCTGCTTCTAAGCCAAC-3′) (38) were used for the amplification of the 16S-23S IGS. Aliquots of PCR product containing 200 ng of amplified DNA were digested with 5 units of the endonucleases and AluI (Gibco BRL) individually for 3 h at 37°C. The resulting DNA fragments were analyzed by gel electrophoresis in 2.5% agarose gels. A representative isolate of each IGS type was identified by partial 16S rRNA gene sequence analysis.
DNA sequencing.
For sequence analysis, 16S rRNA genes were PCR amplified under the conditions described above. PCR products were purified using the NucleoTraPCR kit (Macheroy-Nagel) according to the manufacturer's instructions and used as templates in sequencing reactions. Partial 16S rDNA sequencing was performed by applying the primer 518r (5′-ATTACCGCGGCTGCTGG-3′) (31) (rhizosphere bacteria and clones) and primer pH (endophytes and shoot-associated clones) by the dideoxy chain termination method (48) using an ABI 373A automated DNA sequencer and the ABI PRISM BigDye terminator cycle sequencing kit (PE Applied Biosystems, Inc.). Sequences were subjected to BLAST analysis (3) with the National Center for Biotechnology Information database. All sequences were submitted to the RDP-II Check Chimera program (37) in order to detect chimeric sequences.
Heavy metal tolerance and siderophore and ACC-deaminase production.
For determining the MIC of Ni, strains were cultivated on a phosphate-poor morpholinepropanesulfonic acid medium containing 0.1% glucose (43) or 0.5% glucose (strains iRIII19, iRIII6, iRIII7, and iRII17) instead of using the rather rich TSA medium in order to avoid complexation of Ni. MICs were determined by growing strains on morpholinepropanesulfonic acid medium with increasing concentrations (1, 3, 5, 10, 12, and 15 mM) of NiCl2. Siderophore production was measured by using a modified chrome azurol S agar plate assay according to the method of Milagres et al. (42). Strains that were able to grow on minimal BD medium (7) containing 0.7 g of ACC liter−1 as the sole N source were considered ACC-deaminase positive.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the National Center for Biotechnology Information (NCBI) database under accession no. AY357950 to AY358019, AY359822 to AY359851, AY364015 to AY364083, and AY369236 to AY369238.
RESULTS
T-RFLP analysis.
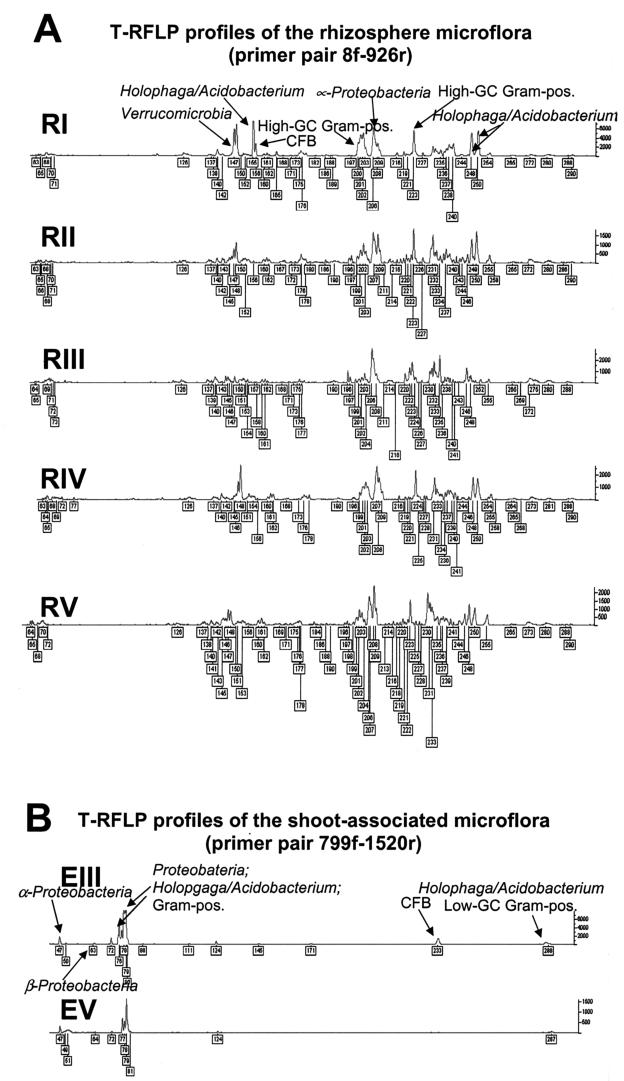
Community structures of plant-associated bacteria were determined by T-RFLP. Although plants were sampled several meters apart, similar T-RFLP patterns were obtained (Fig. 1). Chromatograms showed mostly T-RFs of the same size which occurred in several cases in different abundances. In normalized rhizosphere T-RFLP profiles, a total of 112 fragments with fluorescence intensities higher than 50 were detected, and 103 of them were found in at least 3 rhizosphere samples.
FIG. 1.
T-RFLP profiles produced by AluI digestion of 16S rRNA gene amplicons of rhizosphere bacteria (A) and of shoot-associated bacteria (B).
For the T-RFLP profiling of shoot-associated bacteria, only two samples were analyzed because insufficient amounts of bacterial DNA were obtained. However, community fingerprints were comparable (Fig. 1). Normalized sample profiles showed 15 T-RFs with more than 50 fluorescence units, and 10 of them were found in both samples.
Analysis of clone libraries.
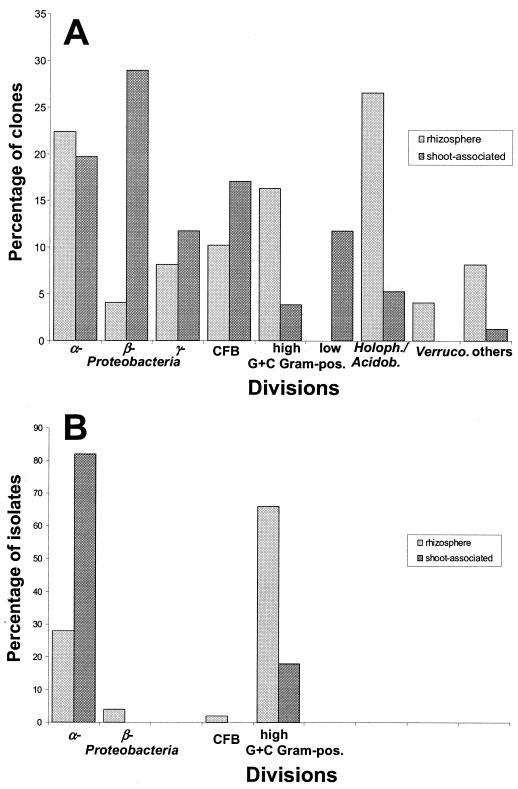
In order to identify the dominant bacteria associated with T. goesingense, 16S rRNA gene clone libraries were established and partial insert sequences were determined. From the rhizosphere, 47 nonchimeric 16S rRNA gene sequences were obtained, which were not evenly distributed among different bacterial divisions. Bacteria belonging to the Holophaga/Acidobacterium division accounted for 27% of the clones examined, whereas 22% could be classified as α-Proteobacteria. The remaining clones belonged to the high-G+C gram-positives (16%), the Cytophaga/Flexibacter/Bacteroides division (10%), or the γ- or β-Proteobacteria (8 and 4%) (Fig. 2). Three clones showed highest homology to sequences of the Gemmatimonadetes. Not a single sequence of low-G+C gram-positive bacteria was obtained. Most rhizosphere sequences showed at least 95% similarity to known sequences in the NCBI database, whereas three sequences were only moderately (90 to 94%) related to known 16S rRNA genes. From surface-sterilized shoots, 76 nonchimeric sequences were obtained. The majority of clones affiliated with the α-, β-, γ-, and δ-Proteobacteria (20, 29, 12, and 1%) and bacteria belonging to the Cytophaga/Flexibacter/Bacteroides division accounted for 17%. The remaining clones belonged to the low-G+C gram-positives (12%), the Holophaga/Acidobacterium division (5%), and the high-G+C gram-positive species (4%) (Fig. 2). Four shoot-associated 16S rRNA gene sequences showed only 90 to 93% homology to published sequences, whereas the remaining sequences were highly related (at least 95%) to NCBI entries.
FIG. 2.
Division-level phylogenetic diversity identified in 16S rDNA clone libraries (A) and the Ni-tolerant strain collection (B) of rhizosphere- and shoot-associated bacteria. CFB, Cytophaga/Flexibacter/Bacteroides division; Holoph./Acidobact., Holophaga/Acidobacterium division; Verruco., Verrucomicrobia.
Predicted and true T-RF sizes of clones generally differed by ±2 bp, and in rare cases differences of up to 5 bp were observed. True T-RF sizes were compared with T-RFLP community fingerprints in order to determine the identity of dominant peaks. In rhizosphere samples, for most peaks higher than 1,000 fluorescence units and several peaks higher than 500 fluorescence units, corresponding 16S rRNA gene sequences were found (Table 1). Results showed that the Holophaga/Acidobacterium division was highly abundant in the rhizosphere as well as Verrucomicrobia, α-Proteobacteria, high-G+C gram-positive species, and the Cytophaga/Flexibacter/Bacteroides division (Fig. 1). Due to the generally lower level of diversity of endophytes than of rhizosphere bacteria and the higher number of clones analyzed, all dominant T-RFLP peaks of the shoot-associated microflora could be identified by corresponding 16S rRNA gene sequences (Table 2). Comparison between clones and fingerprints indicated a dominance of mainly Proteobacteria; however, gram-positive species, the Cytophaga/Flexibacter/Bacteroides, and the Holophaga/Acidobacterium division were abundant as well (Fig. 1). A high number of 16S rRNA gene sequences with a T-RF size of 47 bp showed high homology to Sphingomonas sp. strain eh2, Sphingomonas sp. strain M3C203B-B, or clone Ape2_63 (again with high homology to Sphingomonas sp. strain eh2). However, all of these sequences were different but highly related to each other.
TABLE 1.
Phylogenetic assignment of rhizosphere clones corresponding to T-RFs bigger than 500 fluorescence unitsa
| Rhizosphere T-RF size (bp)b | Mean peak height (FUc) | Corresponding clone | Closest NCBI match/% homologyd | Phylogenetic group |
|---|---|---|---|---|
| 68 | 505 | cRI18d | Curtobacterium herbarum (AJ310413)/98 | High-GC gram-positives |
| cRI25d | Soil clone SC-I-19 (AJ252622)/96 | Holophaga/Acidobacterium | ||
| cRI43d | Clone LEE2 (AF392735)/96 | Holophaga/Acidobacterium | ||
| 70 | 615 | cRI42c | Pseudomonas sp. strain NZ011 (AY014803)/99 | Gammaproteobacteria |
| 137 | 520 | cRI40c | Uncult. Green Bay ferromang. bact. MNH4 (AF293002)/97 | Betaproteobacteria |
| 140 | 961 | cRI54d | Uncult. copper smelt. bact. D64 (AF337832)/97 | Alphaproteobacteria |
| cRI49d | Clone C17.43WL (AF431093)/100 | |||
| cRI47d | Soil clone DS-197 (AY289406)/99 | Alphaproteobacteria | ||
| 145 | 805 | cRI23c | Verrucomicrobiae clone pACH90 (AY297806)/97 | Verrucomicrobia |
| 147/148 | 2,444/3,416 | cRI35d | Uncult. Verrucomicrobia clone BAC-P37-1A (AY214910)/99 | Verrucomicrobia |
| 155 | 1,186 | cRI48d | Uncult. copper smeltery bact. D109 (AF337859)/96 | Holophaga/Acidobacterium |
| 156 | 558 | cRI5c | Uncult. PHOS-HE19 (AF314428)/90 | Cytophaga/Flexibacter/Bacteroides |
| 176 | 869 | cRI19d | Earthworm intestine clone 1w04 (AY154535)/98 | High-GC gram-positives |
| 199 | 1,049 | cRI34c | Uncult. clone NMS8.115WL (AF432672)/96 | Holophaga/Acidobacterium |
| cRI1a | Clone Riz1017 (AJ244321)/96 | Holophaga/Acidobacterium | ||
| 202 | 2,577 | cRI46d | Soil clone Tc135-228 (AY242765)/98 | High-GC gram-positives |
| 206 | 4,849 | cRI25c | Rhizobium leguminosarum bv. trifolii (AY196964)/99 | Alphaproteobacteria |
| cRI58c | Unident. eubact. (AF010068)/99 | Alphaproteobacteria | ||
| cRI28d | Eubact. dtb96 (AJ309970)/94 | Alphaproteobacteria | ||
| 208 | 3,432 | cRI38d | Rubrobacteridae clone glen99_5 (AY150874)/98 | High-GC gram-positives |
| cRI19c | Uncult. clone glen99_6 (AY150881)/96 | Alphaproteobacteria | ||
| cRI2a | Clone IAFR109 (AF270944)/99 | Alphaproteobacteria | ||
| 223 | 4,071 | cRI57d | Clone S54.26PG (AF432648)/99 | Gemmatimonedetes |
| 232 | 2,566 | cRI12c | Clone NMW.98 WL (AY043781)/98 | Betaproteobacteria |
| cRI21c | Rhizosphere clone RSC-II-36 (AJ252681)/98 | High-GC gram-positives | ||
| cRI6a | Clone C34.09SM (AF432645)/95 | High-GC gram-positives | ||
| cRI4d | Soil clone S055 (AY037588)/97 | Cytophaga/Flexibacter/Bacteroides | ||
| cRI30d | Pseudomonas fluorescens (AF375844)/99 | Gammaproteobacteria | ||
| 233 | 700 | cRI36d | Nocardioides OS4 (U61298)/98 | High-GC gram-positives |
| 238 | 1,146 | cRI37c | Uncult. copper smelt. bact. D92 (AF337839)/96 | Holophaga/Acidobacterium |
| 246 | 1,119 | cRI51d | Soil clone a13152 (AY102338)/95 | Holophaga/Acidobacterium |
| 248 | 2,752 | cRI8c, cRI27d | Soil clone S111 (AF013560)/95 | Holophaga/Acidobacterium |
| 250 | 2,705 | cRI45d | Clone 39p18 (AY281355)/99 | Holophaga/Acidobacterium |
Tentative phylogenetic placement and percent similarity values were determined by BLAST and are based on approximately 500 bp of the 16S rRNA gene sequence for each clone.
T-RFs bigger than 500 fluorescence units of the following sizes were not detected in the clone library: 146, 152, 160, 161, 162, 177, 178, 196, 197, 201, 203, 209, 220, 235, 236, 237, 240, 255, and 272 bp.
FU, fluorescence units.
Sequence accession numbers are given in parentheses. Uncult., uncultured; ferromang., ferromanganous; bact., bacteria; smelt., smelter.
TABLE 2.
Phylogenetic assignment of shoot-associated clones corresponding to T-RFsa
| T-RF size, group, or clone(s) | Closest NCBI match/% homologyb | T-RF size, group, or clone(s) | Closest NCBI match/% homologyb | |
|---|---|---|---|---|
| T-RF, 47 bp | ||||
| Alphaproteobacteria | ||||
| cEV17, cEV48, cEIII6, cEV10, cEV58 | Clone Ape2_63 (AB074640)/93-99 | |||
| cEIII37, cEIII40, cEIII48, cEIII58 | Sphingomonas sp. strain eh2 (AF548567)/98-99 | |||
| cEIII55 | Sphingomonas sp. strain M3C203B-B (AF395031)/98 | |||
| cEIII39 | Methylobact. sp. BF10 (Z23156)/99 | |||
| Holophaga Acidobacterium | ||||
| cEV16 | Uncult. Holophaga clone JG37-AG-144 (AJ519391)/95 | |||
| T-RF, 72 bp | ||||
| Betaproteobacteria | ||||
| cEV27 | Clone RB13C5 (AF407411)/98 | |||
| cEV41 | Uncult. sludge bact. (AF234753)/98 | |||
| T-RF, 77-80 bp | ||||
| Betaproteobacteria | ||||
| cEV13 | Clone FW66 (AF523953)/95 | |||
| cEV60 | Clone FW66 (AF23953)/96 | |||
| cEIII47 | Clone KD7-1 (AY218699)/97 | |||
| cEV57 | Blackwater bioreactor bact. BW32 (AF394175)/99 | |||
| cEV38 | Rape rhizosphere clone wr0070 (AJ295516)/92 | |||
| cEV32 | Clone UP9 (AY080915)/98 | |||
| cEV33 | Clone FW66 (AF523953)/96 | |||
| cEIII9 | Clone P30B-57 (AF414583)/96 | |||
| cEIII33 | Uncult. Green Bay ferromang. | |||
| Micronod. bact. (AF293006)/94 | ||||
| cEIII57 | Clone cvf62071 (AY100569)/94 | |||
| Gammaproteobacteria | ||||
| cEV46 | Uncult. γ-prot. MERTZ_0CM_353 (AF424152)/95 | |||
| cEV50 | Clone G-26 (AB094187)/99 | |||
| cEIII34 | Pseudomonas putida (AE016774)/99 | |||
| cEV18, cEV26 | Escherichia albertii LMG20976T (AJ508775)/99 | |||
| Low-GC gram-positives | ||||
| cEIII4 | Bacillus sp. strain LCSAOTU8 (AF506058)/99 | |||
| cEIII60 | Bacillus sp. strain 19494 (AJ315062)/99 | |||
| cEV34, cEIII35 | Bacillus sp. strain 433-D9 (AY266991)/99 | |||
| High-GC gram-positives | ||||
| cEV21 | Blastococcus sp. strain BC517 (AJ316572)/99 | |||
| cEV42 | Uncult. actinobact. clone Ape4_57 (AB074600)/95 | |||
| cEIII51 | Propionibacterium acnes (AB108482)/99 | |||
| Holophaga/Acidobacterium | ||||
| cEV11, cEV29 | Uncult. Holophaga clone JG37-AG-144 (AJ519391)/95 | |||
| T-RF, 233 bp | ||||
| Cytophaga/Flavobacterium/Bacteroides | ||||
| cEV20, cEV30, cEV53, cEIII8 | Clone PHOS-HE31 (AF314430)/96-97 | |||
| cEV25 | Clone esbio160215 (AY187354)/96 | |||
| cEIII42 | Bacterium CS57 (AY124340)/97 | |||
| cEV31 | Uncult. Flavobacterium sp. (AY1922048)/96 | |||
| cEV47 | Uncult. Flavobacterium OF70 (AY1777767)/96 | |||
| cEV39 | Soil clone S172 (AF507740)/99 | |||
| cEIII52 | Soil clone 760-2 (AF423292)/96 | |||
| T-RF, 287 bp | ||||
| Holophaga/Acidobacterium | ||||
| cEV45 | Soil clone C1192 (AF507640)/98 | |||
| Low-GC gram-positives | ||||
| cEV40 | Bacillus sp. strain 82352 (AF227852)/100 | |||
| cEV43 | Desulfitobacterium metallireductans (AF297871)/97 |
Tentative phylogenetic placement and percent similarity values were determined by BLAST and are based on approximately 500 bp of the 16S rRNA gene sequence for each clone.
Uncult., uncultured; bact., bacterium; ferromang., ferromanganous; micronod., micronodule; γ-prot., γ-proteobacterium; actinobact., actinobacterium.
Isolation and identification of plant-associated bacteria.
The average numbers of CFU g of rhizosphere soil−1 formed by rhizosphere bacteria on 1/10-strength TSA amended with 5 and 10 mM NiCl2 were 2.9 × 105 and 2.8 × 104. Endophytes formed 2.2 × 103 and 5.6 × 102 CFU g of shoot weight−1 on Ni-amended plates. Seven morphology types were found among rhizosphere isolates, whereas endophytes showed three types. Several colonies of each morphology type were picked, resulting in 50 Ni-resistant colonies formed by rhizosphere bacteria and 62 endophytes which were further analyzed. Rhizosphere bacteria could be grouped in 17 16S rRNA types and 25 IGS PCR-RFLP types (Table 3), whereas endophytes consisted of 15 16S rRNA types and 22 IGS PCR-RFLP types (Table 4). Isolates were further characterized by partial sequencing of the 16S rRNA gene. The majority of rhizosphere strains belonged to the high-G+C gram-positives and the α-Proteobacteria. One isolate fell into the Cytophaga/Flexibacter/Bacteroides division. Three genera were highly abundant among rhizosphere isolates—Methylobacterium (26%), Rhodococcus (30%), and Okibacterium (26%) (Table 3). Endophytes were mainly the α-Proteobacteria and high-G+C gram-positive bacteria. The majority of isolates (42%) showed high level of similarity to Methylobacterium mesophilicum or clone D9, which is highly related to that species. In addition, Sphingomonas sp. strains were highly abundant (37%). Remaining endophytes showed high homology to the genera Rhodococcus, Curtobacterium, and Plantibacter (Table 4). All isolates except isolate iRIII15 showed at least 97% homology to known 16S rRNA genes.
TABLE 3.
Sequence analysis of partial 16S rDNA (approximately 500 bp) of isolates obtained from the rhizosphere of T. goesingense and analysis of phenotypic properties
| Representative isolate (IGS typea) | 16S rDNA type | Closest relativeb | % Homology | MIC of Ni (mM) | ACC deaminase | Siderophore productiond |
|---|---|---|---|---|---|---|
| Proteobacteria | ||||||
| iRII1 (1) | RII1 | Methylobacterium mesophilicum (AJ400919) | 99 | 10 | + | ++ |
| iRIV2 (1) | RIV2 | Methylobacterium mesophilicum (AJ400919) | 99 | 10 | − | ++ |
| iRV3 (3) | RV3 | Methylobacterium mesophilicum (AJ400919) | 98 | 12 | − | ++ |
| iRIV1 (1) | RIV1 | Methylobacterium extorquens (AF531770) | 100 | 10 | − | + |
| iRIII1 (3) | RIII1 | Methylobacterium extorquens (AF531770) | 100 | 5 | − | ++ |
| iRII2 (2) | RII2 | Methylobacterium extorquens (L20847) | 97 | 10 | − | + |
| iRIV3 (1) | RIV2 | Methylobacterium sp. strain V3 (AF324201) | 99 | 5 | − | ++ |
| iRV5 (1) | RV3 | Clone D9 (AY268275) | 97 | 5 | − | + |
| iRV1 (1) | RV1 | Uncult. α-proteobact. (AJ413602) | 97 | 5 | − | + |
| iRIII19 (2) | RIII19 | Burkholderia terricola (AY040362) | 98 | 1 | − | + |
| Gram-positives | ||||||
| iRIII6 (2) | RIII6 | Uncult. bact. WR169 (AJ233561) | 97 | 1 | − | ++ |
| iRIII7 (2) | RIII6 | Uncult. bact. WR169 (AJ233561) | 97 | 3 | − | ++ |
| iRIII11 (7) | RIII11 | Okibacterium fritillariae (AB042097) | 99 | 5*c | − | + |
| iRII14 (2) | RII14 | Okibacterium fritillariae (AB042097) | 99 | 10 | − | + |
| iRII17 (1) | RII14 | Okibacterium fritillariae (AB042097) | 99 | 1 | − | +++ |
| iRV16 (2) | RV16 | Okibacterium fritillariae (AB042097) | 98 | 5 | + | ++ |
| iRIV11 (1) | RIV11 | Okibacterium fritillariae (AB042097) | 99 | 5 | + | − |
| iRIV4 (2) | RIV4 | Rhodococcus fascians (AJ011329) | 99 | 12 | + | + |
| iRIV6 (4) | RIV4 | Rhodococcus fascians (AJ011329) | 99 | 5 | − | ++ |
| iRIV10 (1) | RIV4 | Rhodococcus fascians (AJ011329) | 99 | 10 | − | ++ |
| iRV10 (4) | RV8 | Rhodococcus fascians (AJ011329) | 99 | 5 | − | ++ |
| iRV8 (3) | RV8 | Rhodococcus spP-wp0233 (AY188941) | 99 | 12 | + | ++ |
| iRV15 (1) | RV8 | Rhodococcus spP-wp0233 (AY188941) | 99 | 5 | − | ++ |
| iRIV12 (1) | RIV12 | Microbacterium sp. strain VA8728_00 (AF306834) | 99 | 5* | − | − |
| Cytophaga/Flexibacter/ Bacteroides | ||||||
| iRIII15 (1) | RIII15 | Potato root bacterium RC-III-57 (AJ252723) | 94 | 5 | − | + |
Abundance of an IGS type among all isolates analyzed.
Uncult., uncultured; bact., bacterium; α-proteobact., α-proteobacterium. Sequence accession numbers are given in parentheses.
*, MIC test was performed on 1/10-strength TSA because strain did not grow in minimal medium. Real MIC may be lower as part of the heavy metal may have been complexed in this rich medium.
Siderophore production: +, moderate, ++, high; +++, very high.
TABLE 4.
Sequence analysis of partial 16S rDNA (approximately 500 bp) of endophytic isolates obtained from T. goesingense and analysis of phenotypic properties
| Representative isolate (IGS typea) | 16S rDNA type | Closest relativeb | % Homology | MIC of Ni (mM) | ACC deaminasec | Siderophore prodn.d |
|---|---|---|---|---|---|---|
| Proteobacteria | ||||||
| iEI1 (1) | EI1 | M. mesophilicum (AJ400919) | 98 | 12 | − | + |
| iEI6 (1) | EI1 | M. mesophilicum (AJ400919) | 100 | 12 | − | ++ |
| iEIII3 (5) | EIII3 | M. mesophilicum (AJ400919) | 100 | 12 | − | ++ |
| iEIII5 (2) | EIII3 | M. mesophilicum (AJ400919) | 100 | 10 | − | ++ |
| iEIV1 (3) | EIV1 | M. mesophilicum (AJ400919) | 100 | 12 | − | ++ |
| iEV8 (1) | EV1 | M. mesophilicum (AJ400919) | 99 | 5 | − | + |
| iEIV8 (1) | EIV8 | M. extorquens (AF531770) | 100 | 10 | − | + |
| iEII3 (1) | EII1 | Methylobacterium sp. strain V3 (AF324201) | 98 | 10 | − | ++ |
| iEIII4 (1) | EIII4 | Clone D9 (AY268275) | 98 | 12 | + | ++ |
| iEIV4 (1) | EIV1 | Clone D9 (AY268275) | 98 | 10 | − | ++ |
| iEV1 (8) | EV1 | Clone D9 (AY268275) | 99 | 12 | + | ++ |
| iEII1 (2) | EII1 | Clone D9 (AY268275) | 97 | 10 | + | ++ |
| iEV10 (1) | EV10 | Clone D9 (AY268275) | 98 | 5 | + | ++ |
| iEII15 (8) | EII15 | Sphingomonas sp. strain pfB27 (AY336556) | 98 | 5 | ++ | + |
| iEII17 (2) | EII15 | Sphingomonas sp. strain M3C203B-B (AF395031) | 98 | 10 | − | + |
| iEIV10 (6) | EIV10 | Sphingomonas sp. strain M3C203B-B (AF395031) | 100 | 5 | − | + |
| iEIII15 (7) | EIII15 | Sphingomonas sp. strain ch2 (AF548567) | 99 | 10 | − | + |
| Gram positives | ||||||
| iEI7 (3) | EI7 | Curtobacterium sp. strain VKM Ac-2062 (AB042097) | 98 | 5 | ++ | ++ |
| iEII7 (2) | EII7 | Curtobacterium sp. strain VKM Ac-2062 (AB042097) | 97 | 10 | − | ++ |
| iEII8 (3) | EII7 | Plantibacter flavus (AY275509) | 99 | 5 | − | ++ |
| iEI10 (2) | EI10 | Rhodococcus sp. strain 5/1 (AF181689) | 100 | 5 | + | ++ |
| iEIII14 (1) | EIII14 | Rhodococcus sp. strain 5/1 (AF181689) | 99 | 5 | + | + |
Abundance of an IGS type among all isolates analyzed.
Sequence accession numbers are given in parentheses.
Growth on ACC: −, none; +, good; ++, very good.
Siderophore production. +, high; ++, very high.
Nickel resistance and siderophore and ACC deaminase production.
Representative isolates of each IGS type were analyzed regarding characteristics that may be relevant for the survival and interaction of T. goesingense and the associated microflora. Sixteen strains out of twenty-five tested tolerated a maximum of 5 mM Ni, whereas the remaining isolates tolerated 10 to 12 mM Ni. Five rhizosphere strains belonging to different phylogenetic groups showed ACC deaminase activity. Except isolates iRIV11 and iRV12, all rhizosphere bacteria analyzed were able to produce siderophores (Table 3). For 8 endophytes out of 22 strains, the MIC was 5 mM; the remaining isolates were able to tolerate 10 to 12 mM Ni. Eight isolates—again belonging to different genera—showed growth on ACC as sole N source. All endophytes were able to produce siderophores (Table 4).
DISCUSSION
Cultivation-independent analysis.
Hyperaccumulating plants accumulate extremely large amounts of heavy metals in their shoots and therefore provide a specific environment for bacterial endophytes. Furthermore, the rhizosphere of hyperaccumulators typically contains large concentrations of easily bioaccessible heavy metal concentrations (39), providing a niche for heavy metal-resistant bacteria. A cultivation-independent approach was chosen in order to identify the dominant bacterial species colonizing the rhizosphere and shoots of the hyperaccumulator T. goesingense when grown in an ultramafic soil containing high Ni concentrations. Emphasis was put on the analysis of bacterial endophytes, since they have intimate contact with the plant, must be able to tolerate high Ni concentrations, and have also been reported to confer plant beneficial effects. Yet endophytes rarely have been approached by cultivation-independent methods. Flowering plants were investigated because this stage of plant development is considered to be characterized by high levels of physiological activity and hyperaccumulation of Ni; however, it may be that the plant-associated bacterial populations fluctuate with plant growth stage and season.
The rhizosphere of T. goesingense hosted a broad diversity of bacteria that were able to live in this extreme environment. High abundance was encountered for α-Proteobacteria, high-G+C gram-positives, and bacteria showing closest similarity to the Holophaga/Acidobacterium division. This last phylum contains very few known cultivated species but has been found to be highly abundant in many soils worldwide (28, 35, 50). The ecological function of Acidobacterium bacteria in soils and rhizospheres is largely unknown; however, this study suggests that several strains belonging to this division are able to tolerate high Ni concentrations. A comparison between T-RFs of clones and T-RFLP profiles confirmed the dominance of α-Proteobacteria, high-GC gram-positives, and the Holophaga/Acidobacterium division in the rhizosphere of T. goesingense but also suggested a high abundance of Verrucomicrobia. Community fingerprinting by T-RFLP analysis indicated a lower level of diversity of shoot endophytes compared to rhizosphere bacteria; however, a different primer pair (799f and 1520r for endophytes, 8f and 926r for rhizosphere bacteria) was used for the amplification of bacterial 16S rRNA genes in order to avoid the amplification of plant-derived small-subunit rRNA genes. The 799f primer was particularly designed for the analysis of endophytes and was found to be highly specific for bacterial sequences except those of Verrucomicrobia, cyanobacteria, spirochetes, and all Obsidian Pool candidate divisions originally detected in a Yellowstone Hot Spring (10). T-RFLP analysis of rhizosphere samples using the 799f and 1520r primer pair resulted in a smaller number of T-RFs than when the 8f and 926r primer pair was applied (results not shown), indicating that the primer pair applied may have underestimated the diversity of bacteria associated with shoots. Cloning and sequencing of endophytic 16S rRNA genes showed that a high number of different divisions, genera, and species are able to colonize T. goesingense shoots despite extremely high Ni concentrations. Six divisions were represented in the clone library, with the majority (67%) of clones clustering with the α-,β-, and γ-Proteobacteria. This is in agreement with previous studies showing that the interior of plants is preferentially colonized by Proteobacteria (10, 18, 51). Striking was the high abundance of sequences showing closest similarity to the genus Sphingomonas (Sphingomonas sp. strain eh2, Sphingomonas sp. strain M3C203B-B, and clone Ape2_63) (Table 3). Related or identical endophytic Ni-resistant Sphingomonas strains were also isolated from T. goesingense shoots. Similarly, heavy metal-resistant Sphingomonas spp. had been isolated from roots and shoots of the Zn hyperaccumulator Thlaspi caerulescens (33). Furthermore, Abou-Shanab et al. (2) reported that a Ni-resistant Sphingomonas macrogoltabidus strain, which was obtained from the rhizosphere of the Ni hyperaccumulator Alyssum murale, increased the uptake of the heavy metal by the plant when bacteria were inoculated onto surface-sterilized seeds before growth in nonsterile soil. However, the mechanism of this effect is still unclear.
Although it has been reported that endophytes represent a subset of rhizosphere bacteria (19, 30, 41), this study clearly indicates that the plant apoplast (including eventually the phylloplane) and the rhizosphere are colonized by highly different bacterial populations, which can be explained by the fact that both habitats provide distinct growth conditions for microorganisms. Low-G+C gram-positives belonging to the genus Bacillus were found to be associated with T. goesingense shoots; however, no low-G+C gram-positives were found in the rhizosphere 16S rRNA gene clone libraries. Similarly, the gram-positive bacteria associated with the surface and interior of maize roots were affiliated closely with Bacillus and Paenibacillus (10). High-G+C gram-positive bacteria were frequently detected in the rhizosphere, but they represented only a minority among endophytes. For the first time, endophytic bacteria belonging to the Holophaga/Acidobacterium division were found, although they were more abundant in the rhizosphere than in association with shoots.
Cultivation-independent analysis indicated the presence of several novel species and genera in association with T. goesingense, since their 16S rRNA gene sequences showed low homology to known genes. In general, T-RFLP community profiles and clone libraries matched well, and clones corresponding to the dominant T-RFLP peaks were found. T-RFs that were not found in the clone library may represent pseudo-T-RFs, additional secondary T-RFs that derive from partially single-stranded amplicons (15). Alternatively, the analysis of more clones may be required to correlate all peaks with specific 16S rRNA gene sequences. From the cultivation-independent analysis it is not clear whether all detected bacteria are actually resistant to Ni or whether the heavy metal can be located in different microhabitats than the bacteria. In soil the heavy metal may be bound to particular particles or concentrated in specific pores, whereas some bacteria may be present at Ni-free sites. In T. goesingense about 70% of total leaf Ni was found to be localized in the apoplast or bound to cell wall material (27), suggesting that bacterial endophytes are confronted with high Ni concentrations.
Characterization of cultivated plant-associated bacteria.
Cultivation-independent analysis generally does not give any information on the function of individual community members. Therefore, Ni-resistant bacteria associated with T. goesingense were isolated, identified by partial 16S rDNA sequencing, and characterized regarding traits that may support the hyperaccumulation or stress tolerance of the associated plant. The rhizosphere as well as the interior of shoots hosted a large number of different culturable bacterial strains that showed high Ni resistance. In general, endophytes were able to tolerate larger concentrations of Ni than rhizosphere bacteria, indicating an adaptation of the endophytic microflora to the large heavy metal concentrations present in T. goesingense shoots. Almost all strains obtained were able to produce siderophores in the absence of iron. Siderophores may be important for the mobilization of the heavy metal in the rhizosphere. They show high affinity for ferric iron but also form complexes with bivalent heavy metal ions (16) that can be assimilated by the plant. Furthermore, heavy metals have been shown to stimulate the production of bacterial siderophores (54). In addition, bacterial siderophores can alleviate heavy metal toxicity by increasing the supply of iron to the plant (8, 9). Among endophytes, siderophore production may be a common phenotype, since almost all strains isolated from surface sterilized and aseptically peeled potato shoots were able to produce siderophores (52). Endophytes have to compete with plant cells for Fe supply, and therefore, siderophore production may be highly important for endophytic growth. However, the formation of siderophore-Ni complexes in the apoplast may also lead to decreased Ni toxicity. Similar effects were previously observed for some plant species, where root exudates contributed to reduced metal toxicity; e.g., of Al in maize plants (26) and of Ni in Thlaspi arvense (45, 47).
Heavy metals can induce ethylene production in plants (55), and an excess of stress ethylene can inhibit plant growth (1). It has been reported that ACC-utilizing bacteria promote plant growth in heavy metal-contaminated soils (6, 8, 9). Consequently, bacterial ACC deaminase production may also support growth of T. goesingense in this extreme environment. Furthermore, the observed stimulated root elongation by ACC deaminase-producing bacteria (6, 20) may result in enhanced Ni uptake in T. goesingense. Our results showed that the capacity to utilize ACC is not restricted to specific phylogenetic groups, which is in agreement with the findings of Belimov et al. (6). It is evident from this study that the ability to produce ACC deaminase is strain dependent. Interestingly, a higher percentage of endophytes (36%) than of rhizosphere bacteria (20%) showed ACC deaminase activity. This may be explained by the more intimate relationship of endophytes with the plant and the better availability of ACC that may favor the growth of utilizing bacteria. However, these percentages are based on a rather small number of strains, and further testing would be required in order to verify this assumption.
Similar to the results obtained by cultivation-independent analysis, analysis of isolates showed that the rhizosphere and shoots of T. goesingense hosted different microbial populations, although the genera Methylobacterium and Rhodococcus were isolated from both niches. Sphingomonas spp. were exclusively found in association with the interior of shoots. The rhizosphere hosted a higher percentage of high-G+C gram-positive bacteria and a smaller amount of α-Proteobacteria than the endosphere of T. goesingense, which is in agreement with the cultivation-independent analysis. Bacillus spp. were not isolated, although they were abundant in 16S rRNA gene clone libraries of shoot-associated bacteria and are generally considered culturable. These strains may be sensitive to Ni, may not be able to grow under the conditions used, or may eventually have entered a viable-but-nonculturable state. The high abundance of highly Ni-resistant pink-pigmented Methylobacterium spp. in the rhizosphere and particularly in the interior of T. goesingense shoots was striking. Most endophytic methylobacteria were affiliated with Methylobacterium mesophilicum and showed very high homology to a Zn-resistant strain isolated from a Japanese soil (see accession no. AJ400919), whereas Methylobacterium extorquens was more abundant in the rhizosphere. Similarly, Lodewyckx et al. (33) isolated a large number of heavy-metal-tolerating methylobacteria from shoots of the Zn hyperaccumulator T. caerulescens, but they were not found in association with roots. Methylobacterium spp. have been reported to interact with plants (4, 22) and even have been detected within cells of the Scoth pine (44). There is evidence that several pink-pigmented Methylobacterium strains produce plant hormones, such as cytokinins (22, 23) and indole acetic acid (25). Genome sequencing of an M. extorquens strain revealed a number of open reading frames with significant homology to Rhizobium and Agrobacterium genes involved in the interaction with plants (29), suggesting an intimate relationship with plants. Since some methylobacteria associated with T. goesingense showed ACC deaminase activity and produced siderophores, they may have exhibited a beneficial effect on plant growth.
Conclusions.
Our results demonstrate that highly diverse microbial communities live in association with the Ni hyperaccumulator T. goesingense and can withstand the high heavy metal concentrations. Only a minor proportion of the bacteria identified by cultivation-independent analysis could be isolated on TSA medium amended with Ni, but nevertheless, the uncultured microflora may exhibit beneficial traits. Functional analysis of isolates revealed several characteristics that potentially support the uptake of the heavy metal by the plant and the reduction of stress symptoms. Whiting et al. (59) showed that soil bacteria increased bioavailable Zn significantly, leading to a fourfold-enhanced accumulation of the heavy metal by T. caerulescens. Similarly, Abou-Shanab et al. (2) reported the importance of bacteria for Ni uptake in the hyperaccumulator A. murale. Nevertheless, the mechanisms by which plant-associated bacteria, in particular endophytes, can support the hyperaccumulation process are not fully understood. Modification of metal speciation by endophytic siderophores and of the bioavailability of such metal species should be considered in experiments establishing metal uptake kinetics of hyperaccumulator plants. Such experiments are typically conducted in hydroponic cultures without consideration of rhizosphere and endophytic microbes. Future work will address the effect of selected bacteria on plant growth and the uptake of heavy metals by the plant as well as the mechanisms involved.
Acknowledgments
This project was financed by the Austrian Science Foundation (Fonds zur Förderung der wissenschaftlichen Forschung). R. Trifonova received a fellowship funded by the Austrian Exchange Service (ÖAD).
We thank Alexandra Weilharter for her excellent assistance with sequence analysis.
REFERENCES
- 1.Abeles, F. B., P. W. Morgan, and M. E. Saltveit, Jr. 1992. Regulation of ethylene production by internal, environmental and stress factors, p. 56-119. In Ethylene in plant biology, 2nd ed. Academic Press, Inc., San Diego, Calif.
- 2.Abou-Shanab, R. A., J. S. Angle, T. A. Delorme, R. L. Chaney, P. van Berkum, H. Moawad, K. Ghanem, and H. A. Ghozlan. 2003. Rhizobacterial effects on nickel extraction from soil and uptake by Alyssum murale. New Phytol. 158:219-224. [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäfer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-PLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araújo, W. L., J. Marcon, W. Maccheroni, Jr., J. D. van Elsas, J. W. L. van Vuurde, and J. L. Azevedo. 2002. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl. Environ. Microbiol. 68:4906-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, A. J. M., S. P. McGrath, R. D. Reeves, and J. A. C. Smith. 2000. Metal hyperaccumulator plants: a review of the ecology and physiology of a biological resource for phytoremediation of metal-polluted soils, p. 85-107. In N. Terry and E. Banuelos (ed.), Phytoremediation of contaminated soil and water. Lewis Publishers, London, United Kingdom.
- 6.Belimov, A. A., V. I. Safronova, T. A. Sergeyeva, T. N. Egorova, V. A. Matveyeva, V. E. Tsyganov, A. Y. Borisov, I. A. Tikhonovich, C. Kluge, A. Preisfeld, K. J. Dietz, and V. V. Stepanok. 2001. Characterization of plant growth-promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 47:642-652. [DOI] [PubMed] [Google Scholar]
- 7.Brown, C. M., and M. J. Dilworth. 1975. Ammonia assimilation by Rhizobium cultures and bacteroids. J. Gen. Microbiol. 122:61-67. [DOI] [PubMed] [Google Scholar]
- 8.Burd, G. I., D. G. Dixon, and B. R. Glick. 1998. A plant growth-promoting bacterium that decreases nickel toxicity in seedlings. Appl. Environ. Microbiol. 64:3663-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd, G. I., D. G. Dixon, and B. R. Glick. 2000. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can. J. Microbiol. 46:237-245. [DOI] [PubMed] [Google Scholar]
- 10.Chelius, M. K., and E. W. Triplett. 2001. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 41:252-263. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Y., E. Jurkevitch, E. Bar-Ness, and Y. Hadar. 1994. Stability constants and pseudobactin complexes with transition metals. Soil Sci. Soc. Am. J. 58:390-396. [Google Scholar]
- 12.Delorme, T. A., J. V. Gagliardi, J. S. Angle, and R. L. Chaney. 2001. Influence of the zinc hyperaccumulator Thlaspi caerulescens J. & C. Presl. and the nonmetal accumulator Trifolium pratense L. on soil microbial populations. Can. J. Microbiol. 47:773-776. [DOI] [PubMed] [Google Scholar]
- 13.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes: characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evers, A., R. D. Hancock, A. E. Martell, and R. J. Motekaitis. 1989. Metal ion recognition in ligands with negatively charged oxygen donor groups. Complexation of Fe(III), Ga(III), In(III), Al(III) and other highly charged metal ions. Inorg. Chem. 28:2189-2195. [Google Scholar]
- 17.Fekete, F. A., and L. L. Barton. 1992. Effects of iron(III) analogs on growth and pseudobactin synthesis of a chromium-tolerant Pseudomonas isolate. Biol. Metals 4:211-216. [DOI] [PubMed] [Google Scholar]
- 18.Garbeva, P., L. S. van Overbeek, J. W. L. van Vuurde, and J. D. van Elsas. 2001. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb. Ecol. 41:369-383. [DOI] [PubMed] [Google Scholar]
- 19.Germida, J. J., S. D. Siciliano, J. R. de Freitas, and A. M. Seib. 1998. Diversity of root-associated bacteria associated with field-grown canola (Brassica napus L.) and wheat (Triticum aestivum L.). FEMS Microbiol. Ecol. 26:43-50. [Google Scholar]
- 20.Glick, B. R., C. B. Jacobson, M. M. K. Schwarze, and J. J. Pasternak. 1994. 1-Aminocyclopropane-1-carboxylic acid deaminase mutants of the plant growth promoting rhizobacterium Pseudomonas putida GR12-2 do not stimulate canola root elongation. Can. J. Microbiol. 40:911-915. [Google Scholar]
- 21.Glick, B. R., D. M. Penrose, and J. Li. 1998. A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol. 190:63-68. [DOI] [PubMed] [Google Scholar]
- 22.Holland, M. A., and J. C. Polacco. 1994. PPFMs and other covert contaminants: is there more to plant physiology than just plant? Annu. Rev. Plant Physiol. Plant Mol. Biol. 45:197-209. [Google Scholar]
- 23.Holland, M. A. 1997. Occam's razor applied to hormonology. Are cytokinins produced by plants? Plant Physiol. 115:865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, X., and G. L. Boyer. 1996. Siderophore-mediated aluminum uptake by Bacillus megaterium ATCC 19213. Appl. Environ. Microbiol. 62:4044-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanova, E. G., N. V. Doronina, A. O. Shepeliakovskaia, A. G. Laman, F. A. Brovko, and Y. A. Trotsenko. 2001. Aerobic methylobacteria are capable of synthesizing auxins. Microbiology 70:392-397. [PubMed] [Google Scholar]
- 26.Kochian, L. V. 1995. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46:237-260. [Google Scholar]
- 27.Krämer, U., I. J. Pickering, R. C. Prince, I. Raskin, and D. E. Salt. 2000. Subcellular localization and speciation of nickel in hyperaccumulator and non-accumulator Thlaspi species. Plant Physiol. 122:1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S. M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lidstrom, M. E., and L. Chistoserdova. 2002. Plants in the pink: cytokinin production by Methylobacterium. J. Bacteriol. 184:1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilley, A. K., J. C. Fry, M. J. Bailey, and M. J. Day. 1996. Comparison of aerobic heterotrophic taxa isolated from four root domains of mature sugar beet (Beta vulgaris). FEMS Microbiol. Ecol. 21:231-242. [Google Scholar]
- 31.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodewyckx, C., J. Vangronsfeld, F. Porteous, E. R. B. Moore, S. Taghavi, M. Mergeay, and D. van der Lelie. 2002. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 21:563-606. [Google Scholar]
- 33.Lodewyckx, C., M. Mergeay, J. Vangronsfeld, H. Clijsters, and D. van der Lelie. 2002b. Isolation, characterization, and identification of bacteria associated with the zinc hyperaccumulator Thlaspi caerulescens subsp. calaminaria. Int. J. Phytorem. 4:101-105. [DOI] [PubMed] [Google Scholar]
- 34.Lombi, E., F. J. Zhao, S. P. McGrath, S. D. Young, and S. A. Sacchi. 2001. Physiological evidence for a high-affinity cadmium transporter highly expressed in a Thlaspi caerulescens ecotype. New Phytol. 149:53-60. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 36.Luo, Y. M., P. Christie, and A. J. M. Baker. 2000. Soil solution Zn and pH dynamics in non-rhizosphere soil and in the rhizosphere soil of Thlaspi caerulescens grown in a Zn/Cd-contaminated soil. Chemosphere 41:161-164. [DOI] [PubMed] [Google Scholar]
- 37.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massol-Deya, A. A., D. A. Odelson, R. F. Hickey, and J. M. Tiedje. 1995. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA). In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, chapter 3.3.2, p. 1-8. Kluwer Academic Publishers., Dortrecht, The Netherlands.
- 39.McGrath, S. P., Z. G. Shen, and F. J. Zhao. 1997. Heavy metal uptake and chemical changes in the rhizosphere of Thlaspi caerulescens and Thlaspi ochroluceum grown in contaminated soils. Plant Soil 232:207-214. [Google Scholar]
- 40.McGrath, S. P., and F. J. Zhao. 2003. Phytoextraction of metals and metalloids from soils. Curr. Opin. Biotechnol. 14:1-6. [DOI] [PubMed] [Google Scholar]
- 41.McInroy, J. A., and J. W. Kloepper. 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 173:337-342. [Google Scholar]
- 42.Milagres, A. M. F., A. Machuca, and D. Napoleão. 1999. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 37:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirttila, A. M., H. Laukkanen, H. Pospiech, R. Myllylä, and A. Hohtola. 2000. Detection of intracellular bacteria in the buds of Scotch pine (Pinus sylvestris L.) by in situ hybridization. Appl. Environ. Microbiol. 66:3073-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puschenreiter, M., S. Wieczorek, O. Horak, and W. W. Wenzel. 2003. Chemical changes in the rhizosphere of hyperaccumulator and metal-excluder Thlaspi species. J. Plant Nutr. Soil Sci., 579-584. 166:
- 46.Reeves, R. D., and R. R. Brooks. 1983. European species of Thlaspi L. (Cruciferae) as indicators of nickel and zinc. J. Geochem. Explor. 18:275-283. [Google Scholar]
- 47.Salt, D. E., N. Kato, U. Krämer, R. D. Smith, and I. Raskin. 2000. The role of root exudates in nickel hyperaccumulation and tolerance in accumulator and nonaccumulator species of Thlaspi, p. 189-200. In N. Terry and G. Banuelos (ed.), Phytoremediation of contaminated soil and water. CRC Press LLC, Boca Raton, Fla.
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with the chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlegel, H. G., J. P. Cosson, and A. J. M. Baker. 1991. Nickel-hyperaccumulating plants provide a niche for nickel-resistant bacteria. Bot. Acta 104:18-25. [Google Scholar]
- 50.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirchmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2002. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39:23-32. [DOI] [PubMed] [Google Scholar]
- 52.Sessitsch, A., B. Reiter, and G. Berg. Endophytic bacterial communities of field-grown potato plants and their plant growth-promoting and antagonistic abilities. Can. J. Microbiol., in press. [DOI] [PubMed]
- 53.Sturz, A. V., B. R. Christie, and J. Nowak. 2000. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 19:1-30. [Google Scholar]
- 54.van der Lelie, D., P. Corbisier, L. Diels, A. Gilis, C. Lodewyckx, M. Mergeay, S. Taghavi, N. Spelmans, and J. Vangronsveld. 1999. The role of bacteria in the phytoremediation of heavy metals. p. 265-281. In N. Terry and G. Banuelos (ed.), Phytoremediation of contaminated soil and water. Lewis Publishers, London, United Kingdom.
- 55.Weckx, J., J. Vangronsveld, and H. Clijster. 1993. Heavy metal induction of ethylene production and stress enzymes. I. Kinetics of response, p. 238-239. In J. C. Pech, A. Latche, and C. Balaque (ed.), Cellular and molecular aspects of the plant hormone ethylene. Kluwer Academic Publishers, Dortrecht, The Netherlands.
- 56.Wenzel, W. W., and F. Jockwer. 1999. Accumulation of heavy metals in plants grown on mineralized soils of the Austrian Alps. Environ. Pollut. 104:145-155. [Google Scholar]
- 57.Wenzel, W. W., M. Bunkowski, M. Puschenreiter, and O. Horak. 2003. Rhizosphere characteristics of indigenously growing nickel hyperaccumulator and excludor plants on serpentine soil. Environ. Pollut. 123:131-138. [DOI] [PubMed] [Google Scholar]
- 58.Whiting, S. N., J. R. Leake, S. P. McGrath, and A. J. M. Baker. 2000. Positive responses to Zn and Cd by roots of the Zn and Cd hyperaccumulator Thlaspi caerulescens. N. Phytol. 145:199-210. [Google Scholar]
- 59.Whiting, S. N., M. P. de Souza, and N. Terry. 2001. Rhizosphere bacteria mobilize Zn for hyperaccumulation by Thlaspi caerulescens. Environ. Sci. Technol. 35:3144-3150. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, F. J., R. E. Hamon, and M. J. McLaughlin. 2001. Root exudates of the hyperaccumulator Thlaspi caerulescens do not enhance metal mobilization. New Phytol. 151:613-620. [DOI] [PubMed] [Google Scholar]