Abstract
The practical application of commercial malolactic starter cultures of Oenococcus oeni surviving direct inoculation in wine requires insight into mechanisms of ethanol toxicity and of acquired ethanol tolerance in this organism. Therefore, the site-specific location of proteins involved in ethanol adaptation, including cytoplasmic, membrane-associated, and integral membrane proteins, was investigated. Ethanol triggers alterations in protein patterns of O. oeni cells stressed with 12% ethanol for 1 h and those of cells grown in the presence of 8% ethanol. Levels of inosine-5′-monophosphate dehydrogenase and phosphogluconate dehydrogenase, which generate reduced nicotinamide nucleotides, were decreased during growth in the presence of ethanol, while glutathione reductase, which consumes NADPH, was induced, suggesting that maintenance of the redox balance plays an important role in ethanol adaptation. Phosphoenolpyruvate:mannose phosphotransferase system (PTS) components of mannose PTS, including the phosphocarrier protein HPr and EIIMan, were lacking in ethanol-adapted cells, providing strong evidence that mannose PTS is absent in ethanol-adapted cells, and this represents a metabolic advantage to O. oeni cells during malolactic fermentation. In cells grown in the presence of ethanol, a large increase in the number of membrane-associated proteins was observed. Interestingly, two of these proteins, dTDT-glucose-4,6-dehydratase and d-alanine:d-alanine ligase, are known to be involved in cell wall biosynthesis. Using a proteomic approach, we provide evidence for an active ethanol adaptation response of O. oeni at the cytoplasmic and membrane protein levels.
Malolactic bacteria are lactic acid bacteria that are able to carry out malolactic fermentation (MLF). The control of their activity is an important aspect of the technology of commercial wine production. MLF consists of the decarboxylation of l-malic acid to l-lactic acid, which decreases total acidity and improves the stability and quality of wine. Oenococcus oeni is recognized as the principal microorganism responsible for MLF under stress conditions, such as those prevailing in wine. However, inoculation of O. oeni starter cultures directly into wine leads to significant cell mortality and, consequently, failure of MLF. For an improved control of MLF in the wine industry, it is essential to understand the mechanisms involved in ethanol stress and tolerance (for a review of MLF, see reference 26).
We have previously examined the effects of ethanol in O. oeni cells and have shown that ethanol acts as a disordering agent of the O. oeni cytoplasmic membrane (7, 7a) and negatively affects metabolic activity (M. G. Da Silveira, F. M. Rombouts, and T. Abee, submitted for publication). The variety of inhibitory consequences of ethanol exposures makes assignment of primary targets problematic. In this paper, we examined not only the composition of cytoplasmic proteins of O. oeni cells but also that of membrane proteins in cells grown in the presence of ethanol. Membrane proteins may be associated with the membrane (extrinsic or peripheral membrane proteins) or be integrated in the membrane (intrinsic or integral membrane proteins). Integral membrane proteins contain one or more hydrophobic segments of the polypeptide chain (consisting predominantly of hydrophobic amino acids), which are able to span the membrane, sometimes repeatedly, and therefore are called membrane-anchoring domains (44). Membrane-associated proteins are generally bound to the membrane by protein-protein interaction (32) but may also include lipoproteins attached covalently to the membrane by acetylation.
One widely studied aspect of the ethanol-stress response is the modification of the cellular protein composition, including the so-called heat shock proteins (18, 28, 40). Recently Bourdineaud et al. (3) found that the O. oeni ftsH gene, encoding a protease belonging to the ATP binding cassette protein superfamily, was stress responsive, since its expression increased at high temperature and under osmotic shock. Expression of the O. oeni ftsH gene in Escherichia coli supplied resistance to wine toxicity, whereas no significant changes in ftsH gene expression were found when O. oeni was subjected to ethanol stress. In O. oeni the synthesis of Lo18, an 18-kDa protein, is markedly induced under a variety of stress conditions and during stationary growth phase (15), representing a general stress marker in this bacterium. Moreover, Lo18 was found to be peripherally associated with the cytoplasmic membrane, and it was suggested that it could be involved in the maintenance of membrane integrity (18). This prompted us to investigate also the ethanol-induced association of proteins with the cytoplasmic membrane in O. oeni cells.
It is now well established that the survival of microorganisms in a variety of potentially lethal conditions can be improved by preexposure to sublethal stress conditions of the same kind. O. oeni cells grown in the presence of 8% (vol/vol) ethanol were able to maintain the efficiency of the cytoplasmic membrane as a semipermeable barrier during ethanol challenge by adjusting their membrane fluidity at the lipid-water interface (Da Silveira, Golovina, Hoekstra, Rombouts, and Abee, submitted). This could be partially explained by the membrane composition shift observed during growth in the presence of ethanol, i.e., a decrease in the total amount of lipids. This result hints at an important role for the protein content of membranes during ethanol adaptation. However, the effect of growth in the presence of ethanol on the integral membrane proteins of O. oeni has never been established.
Proteomics is a very powerful tool for understanding how organisms respond to environment stresses, though the use of proteomic techniques with O. oeni is still limited. Radiolabeling with [35S]methionine has been used to study protein profiles in this bacterium under stress conditions (14, 15). Immunoblotting has been used to study the expression of specific proteins, e.g., malolactic enzyme (12, 23), [H+]ATPase (43), and Lo18 (16, 18, 43).
In this paper we provide evidence for an active ethanol-protective response in O. oeni. Subsets of cytoplasmic, membrane-associated, and integral membrane proteins were identified, and their role in adaptation to ethanol is discussed.
MATERIALS AND METHODS
Bacterial strain and growth conditions.
Oenococcus oeni GM (Microlife Technics, Sarasota, Fla.) was cultured at 30°C in FT80 medium (pH 4.5) (4) modified by the omission of Tween 80, containing 10 g of dl-malic acid per liter. Glucose and fructose were autoclaved separately and added to the medium just before inoculation, at a final concentration of 2 and 8 g per liter, respectively. Early-stationary-phase cultures were diluted 100-fold in fresh medium, incubated for 24 h, and then used to obtain 1% inoculated cultures. In the adaptation experiments, the culture medium was supplemented with 8% (vol/vol) ethanol.
Stress conditions.
Cells of O. oeni cultured in the absence and in the presence of 8% (vol/vol) ethanol were harvested by centrifugation at exponential phase (20 and 40 h, respectively), suspended in the same medium containing 12% (vol/vol) ethanol, and incubated for 1 h at 30°C. Cells were recovered by centrifugation, washed twice with 50 mM potassium phosphate buffer (pH 7.0), and concentrated in the same buffer to an optical density at 600 nm of 5.
Extraction of total protein from O. oeni. Cells were disrupted by bead beating with an MSK cell homogenizer (B. Braun Biotech International, Melsungen, Germany) and zirconium beads (diameter, 0.1-mm; Biospec Products, Bartlesville, Okla.) six times for 1 min (with cooling on ice between treatments). Subsequently, proteins in the homogenate were analyzed by Western blotting and two-dimensional gel electrophoresis (2D-E). The protein concentration in cell extracts was determined by using the bicinchoninic acid assay (Sigma Chemical Co., St. Louis, Mo.).
Analysis of total protein by 2D-E. Total protein analysis was performed with a Multiphor 2D-E system (Pharmacia Biotech, Uppsala, Sweden) as described by Wouters et al. (47). Nonprotein impurities in the sample (e.g., lipids) interfered with separation and subsequent visualization of the 2D gels (data not shown), and therefore, protein samples were subjected to acetone precipitation prior to loading (13). Equal amounts of protein (120 μg) were separated on isoelectric-point gels at pI 4 to 7 and subsequently on homogeneous sodium dodecyl sulfate-12 to 14% polyacrylamide gels (Pharmacia Biotech). The gels were stained with Coomassie brilliant blue. The experiments were performed in triplicate, and representative gels are shown. The gels were analyzed by using PD-Quest software (Bio-Rad, Richmond, Calif.) and standardized by calculating the intensity of each spot as the percentage of the total intensity of the spots visualized on a gel, after which the induction or repression factors were calculated.
Determination of N-terminal amino acid sequences of total proteins. For determination of the N-terminal amino acid sequences of specific spots, protein samples (1.5 mg) were separated on the 2D-E gels under conditions identical to those used for running of the analytical gels. The proteins were blotted on a polyvinylidene difluoride membrane optimized for protein transfer (Amersham Life Science, Little Chalfont, Buckinghamshire, England) with a Trans-Blot unit in accordance with the instructions of the manufacturer (Bio-Rad), and stained with Coomassie brilliant blue. Protein spots were cut from the blot and subjected to consecutive Edman degradation and subsequent analysis with the model 476A protein sequencing system (Applied Biosystems, Foster City, Calif.) at the Sequence Center, University Utrecht (Utrecht, The Netherlands). By using BlastP and the O. oeni PSU-1 genome sequence (sequenced by Doe Joint Genome Institute, University of California; www.jgi.doe.gov) deposited in the ERGO database (http://ergo.integratedgenomics.com/ERGO), the derived N termini were analyzed for sequence similarities.
Membrane protein extraction from O. oeni. Cells (3 g [wet weight]) were resuspended in 12 ml of sample buffer (5 mM Mg acetate, 50 mM HEPES [pH 7.5], 100 mM K acetate [pH 7.5]), disrupted by a French pressure cell press (three times at 10,000 lb/in2), and centrifuged (10,000 × g, 10 min, 4°C) to eliminate cell debris. All steps were performed at 4°C, and one MiniComplete protease inhibitor tablet (Boehringer) was added to each of the solutions and buffers. The supernatant was recovered and centrifuged in a sucrose gradient in order to recover cytoplasmic membranes. Benzonase (125 U/liter) and sucrose (final concentration, 0.5 M) were added to the supernatant. Sucrose-gradient centrifugation was performed under conditions that stabilized ribosomes and consequently allowed ribosomal proteins to be removed easily: bottom layer, 3 ml of 2 M sucrose in sample buffer (5 mM Mg acetate, 50 mM HEPES [pH 7.5], 100 mM K acetate [pH 7.5], 8 mM β-mercaptoethanol); immediate up-layer, 3 ml of 1.5 M sucrose in sample buffer; middle layer, 8 ml of supernatant (0.5 M sucrose); and top layer, 2 ml of sample buffer without sucrose. After ultracentrifugation (99,700 × g, 45 min, 4°C), each membrane band was collected and diluted (1:1) in miliQ H2O and ultracentrifuged for 1 h at 99,700 × g. In order to remove remaining cytoplasmatic proteins, a series of washing steps were performed: membrane pellets were resuspended in 5 ml of 1 M Tris (pH 7.5), incubated for 30 min at 4°C, and ultracentrifuged (104,300 × g, 20 min, 4°C). The pellets were resuspended in 5 ml of 0.1 M sodium carbonate (pH 11), ultrasonicated for 5 min in an ice bath, incubated for 30 min on ice, and ultracentrifuged (104,300 × g, 20 min, 4°C). The last procedure was repeated, and the pellets were resuspended in 5 ml of miliQ H2O to remove the sodium carbonate. The protein suspension was ultracentrifuged (104,300 × g, 20 min, 4°C), and the pellet was resuspended in 1 ml of solubilization buffer (7 M urea, 2 M thiourea, 4% [wt/vol] 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 30 mM dithiothreitol [DTT], 0.5% [wt/vol] Pharmalyte [pH 4 to 7], 1 mM Pefabloc SC, 2 μM leupeptin). After ultracentrifugation at 104,300 × g for 20 min, the supernatant contained predominantly membrane-associated proteins, whereas the pellet contained hydrophobic proteins which were not soluble in the solubilization buffer.
Membrane protein analysis. Membrane-associated proteins of the supernatant were separated by 2D-GE essentially as described by O'Farrell (34). Isoelectric focusing (IEF) (first dimension) was performed using the IPGphor system and Immobiline DryStrip kit (Amersham Biosciences) with linear immobilized pH gradients (pH 4 to 7; 18 cm). The supernatants were diluted to a final volume of 360 μl with solubilization buffer and applied to the Immobiline gel strips by 12 h of in-gel rehydration. IEF was carried out sequentially at 100 V for 4 h, 300 V for 3 h, 3,000 V for 6 h, 5,500 V for 3 h, and 5,500 V for 20 h in a gradient mode at 20°C. For the second gel dimension, the gel strips were incubated for two intervals of 15 min each in equilibration solution (6 M urea, 30% glycerol, 2% [wt/vol] sodium dodecyl sulfate [SDS] in 0.05 M Tris-HCl buffer [pH 8.8]), with 1% (wt/vol) DTT added to reduce the proteins and then 4% (wt/vol) iodoacetamide added to carbamidomethylate them. The strips were transferred to 12 to 15% acrylamide gradient gels, and SDS-polyacrylamide gel electrophoresis (PAGE) was performed overnight at 125 V and 10°C using the ISO-DALT electrophoresis system (Amersham Biosciences). The 2D gels were stained with ruthenium II Tris according to the method of Rabilloud et al. (38). The separation of the hydrophobic proteins from the pellet was performed by 16-BAC-SDS-PAGE with a PROTEAN II system (Bio-Rad) as described by Hartinger et al. (17). The 16-BAC-SDS gels were stained with colloidal Coomassie brilliant blue G-250 according to the method of Neuhoff et al. (33).
Mass spectroscopy analysis of membrane proteins.
For mass spectrometry, proteins of the 2D gels were handled as described by Kruft et al. (22). Proteins of the 16-BAC-SDS gel were additionally carbamidomethylated before trypsin digestion. To reduce the cysteine residues, proteins were incubated in 50 μl of 20 mM DTT-25 mM NH4HCO3 for 30 min at 56°C. Excess liquid was removed, and the gel pieces were incubated in 70 μl of acetonitrile. When the gel pieces had shrunk, the acetonitrile was discarded and 50 μl of 55 mM iodacetamide-25 mM NH4HCO3 was added for 30 min in the dark. The gel pieces were dehydrated by acetonitrile and dried in a SpeedVak. Subsequently the gel pieces were incubated in 30 μl of digestion solution containing 2 ng of trypsin (Promega, Madison, Wis.)/μl. Mass measurements were performed by positive-ion matrix-assisted laser desorption ionization-time of flight mass spectrometry using a Bruker Reflex instrument equipped with delayed extraction and a nitrogen laser (337 nm). Zip-Tip elution was done with 1 μl of matrix solution (19 mg of α-cyano-4-hydroxycinnamic acid in 1 ml of 60% methanol-0.1% formic acid), which was placed directly on the matrix-assisted laser desorption ionization target. All spectra were recorded in the reflection mode with an acceleration voltage of 20 kV and a reflection voltage of 21.5 kV. Evaluation of the spectra was performed by using the BioTools package (Brüker Daltonik), and the identification was done by using the Mascot search engine (Matrix Science). Peptide sequencing was performed on a quadrupole time-of-flight mass spectrometry instrument (Q-tof II; Micromass) equipped with a nanospray ion source. Three microliters of Zip Tip-purified sample was filled into Au/PD-coated nanospray glass capillaries (Protana), which were placed orthogonally in front of the entrance hole of the Q-tof instrument. The generated sequences were compared to sequence entries in the O. oeni PSU-1 genome database (sequenced by Doe Joint Genome Institute, University of California; www.jgi.doe.gov). By using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/blast/blast.cgi), a search for short, nearly exact matches was performed.
Additional analytical methods.
Cellular dry weight was determined by filtering 80 ml of the cell suspensions through preweighed polyethylene filters, with a porosity of 0.22 μm, and dried at 100°C in an oven until a constant weight was reached. As a control the dry weight of the same volume of phosphate buffer was also determined. Protein was assayed by the method of Lowry et al. (27).
RESULTS
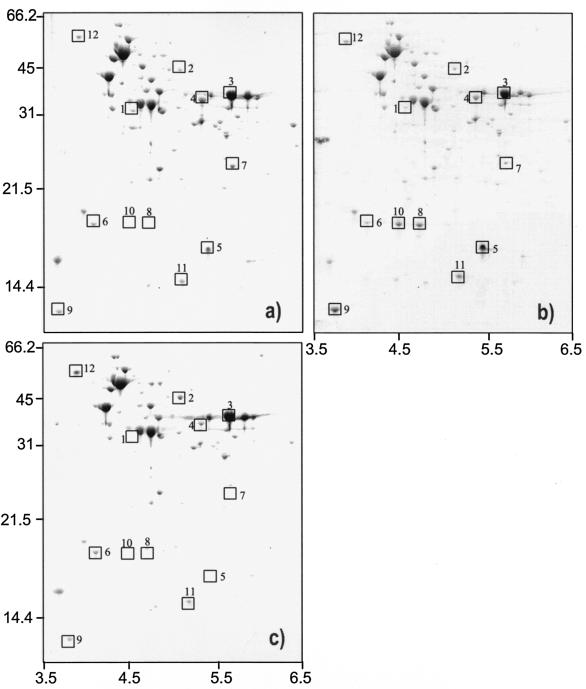
2D-E of total protein of O. oeni cells. Cell extracts from end-exponential cultures grown without or with 8% (vol/vol) ethanol and from cultures shocked with 12% ethanol for 1 h were separated using 2D-E. Analysis of the 2D-EF gels revealed a total of approximately 100 proteins, indicating low-efficiency staining. Combined with the protein identification protocol, which involves Coomassie staining after blotting, we decided to use Coomassie staining throughout the research. The analysis of the gels showed that both ethanol stress and adaptation changed the protein profiles of O. oeni cells (Fig. 1). The N-terminal sequences of a subset of proteins were determined, and the respective function was assigned from the annotation in the O. oeni genome sequence (Table 1). Stressing cells with 12% ethanol for 1 h promoted a decrease of several proteins, namely, spots 1, 3, and 4 (Fig. 1b; Table 2). These spots were identified as metal-dependent hydrolase, glyceraldehyde-3-phosphate dehydrogenase, and inosine-5′-monophosphate dehydrogenase (IMPDH), respectively (Table 2). The synthesis of some proteins within the region of 14 to 22 kDa and with a pI of 4.5 to 5.5 was also induced (Fig. 1b), including spot 5 (Table 2), identified as the phosphocarrier protein HPr. The proteins corresponding to spots 8 and 10 were present only when cells were exposed to 12% ethanol, suggesting that these proteins are typical ethanol stress-induced proteins. However, these two spots could not be identified, since the signals were too low to allow a proper assignment of the N-terminal amino acid sequence.
FIG. 1.
2D-E of total protein extracts of O. oeni cells grown in normal conditions (a), stressed with 12% ethanol for 1 h (b), and grown in the presence of 8% ethanol (c). Molecular masses (in kilodaltons) of marker bands (left side) and pI ranges (bottom) are indicated. Selected proteins are boxed and numbered (see also Tables 1 and 2).
TABLE 1.
List of cellular proteins of O. oenia
| Spot | N-terminal sequence | Mass (kDa) | Description |
|---|---|---|---|
| 1 | XELEFXXX | 36.2 | Metal-dependent hydrolase |
| MELEFLGT | |||
| 2 | XGPQQYDY | 48.6 | Glutathione reductase |
| MKNQQYDY | |||
| 3 | TVKIGINRFGRI | 37.1 | Glyceraldehyde 3-P dehydrogenase |
| TVKIGINGFGRI | |||
| 4 | TDFSNKYT | 40.1 | Inosine-5′-monophosphate dehydrogenase |
| TDFSNKYT | |||
| 5 | VSKEFTIT | 9 | Phosphocarrier protein HPr |
| VSKEFTIT | |||
| 6 | AKGLVAGI | 17.7 | General stress proteinb |
| AKGLVAGI | |||
| 7 | —d | ||
| 8 | XSELMXRXc | ||
| 9 | XLXXXKRVc | ||
| 10 | — | ||
| 11 | — | ||
| 12 | — |
For each spot (numbered according to Fig. 1), the N-terminal sequence, molecular mass, and possible function are indicated.
Closest hit, 37% similarity with L. monocytogenes.
No hits found.
—, signals were too low to give an assignment of a sequence.
TABLE 2.
Protein expression levels of O. oeni cellsa
| Spot | Protein | Ethanol stress | Ethanol adaptation |
|---|---|---|---|
| 1 | MDH | 0.6 | ND |
| 2 | GR | 1.1 | 3.6 |
| 3 | GPDH | 0.5 | 0.9 |
| 4 | IMPDH | 0.6 | 0.5 |
| 5 | HPr | 3 | ND |
Cells were stressed with 12% ethanol for 1 h, or adapted cells were grown in the presence of 8% ethanol (numbering is according to Fig. 1). Induction or repression factors were calculated by dividing the normalized values of the spots of stressed and adapted cells by the normalized value of the spots in the control. ND, not detected; MDH, metal-dependent hydrolase; GR, glutathione reductase; GPDH, glyceraldehyde-3-phosphate dehydrogenase; IMPDH, inosine-5′-monophosphate dehydrogenase; HPr, phosphocarrier protein.
Further analysis of 2D-EF gels revealed that a limited number of proteins were increased during growth in the presence of ethanol, mainly with those with high molecular weight (spots 2 and 12; Fig. 1). Spot 2 was identified as glutathione reductase (GR) and was threefold induced (Fig. 1; Table 2). Notably, some spots were not detected in these ethanol-adapted cells, including spots 1 and 5. Spot 5 corresponds to HPr, with a molecular mass, predicted from the HPr amino acid sequence, of 9.1 kDa; however, from the 2D-EF gel analysis we can infer a molecular mass of approximately 17 to 18 kDa. N-terminal sequence analysis revealed two sequences that differ only in the methionine of the first position (MVSKEFTI and XVSKEFTI).This suggests that HPr in O. oeni can be present in a dimeric form. Dimerization of HPr and the HPr-like protein, Crh, has been reported previously in gram-positive bacteria (11, 19).
Finally, the protein corresponding to spot 6 showed homology with a putative general stress protein in Listeria monocytogenes (Table 1); however, no induction was observed during ethanol shock or adaptation.
Integral membrane proteins of O. oeni cells.
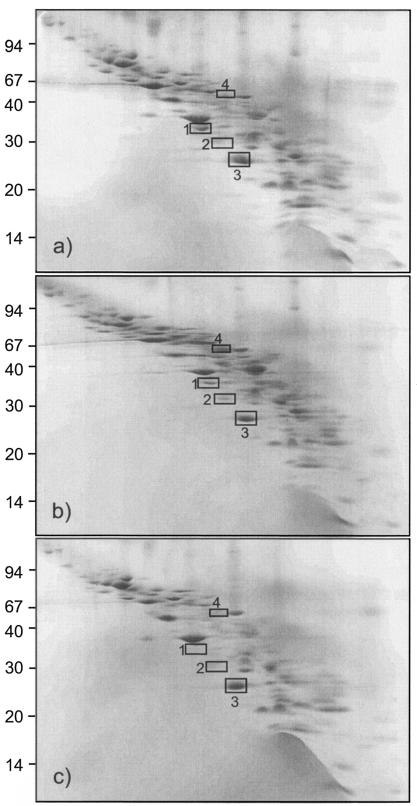
Integral membrane proteins contain one or more hydrophobic segments that are compatible with the hydrophobic interior of the lipid bilayer. The use of classical 2D-E for the analysis of these proteins is problematic since many of these proteins resolve poorly in the dimension of the pH gradient. This is partially due to the insolubility of membrane proteins in nonionic detergents, particularly at low ionic strength (despite the presence of a high concentration of urea). Moreover, because of their hydrophobicity, these proteins tend to precipitate before reaching their isoelectric points (17). 16-BAC-SDS-PAGE provides a good tool for the characterization of membrane proteins (17). The first dimension is electrophoresis towards the cathode at acidic pH in the presence of the cationic detergent 16-BAC. Analysis of the gels revealed that ethanol stress and adaptation resulted in minor changes in the membrane protein composition (Fig. 2). Q-tof tandem mass spectrometry analysis revealed that the mannose-specific IIAB component of the phosphoenolpyruvate:mannose phosphotransferase system (PTS) (EIIMan) corresponding to spot 1 (Fig. 2c; Table 3) was not present in ethanol-grown cells. Spot 2, which features high homology to a glutamine ABC transporter-ATP-binding protein, was decreased in ethanol-stressed cells. The protein corresponding to spot 3 was increased in ethanol-stressed cells. Q-tof analysis of this protein gave a good spectrum (EYVPALQEDLR), but no homologous sequences in the database of O. oeni were found. Even though several washing steps were done, we still found contamination with cytoplasmic proteins, since the analysis of spot 4 features high homology to PGDH. The level of this protein was induced in ethanol-stressed cells, whereas it was completely absent in ethanol-adapted cells.
FIG. 2.
2D separation of hydrophobic membrane proteins by 16-BAC-SDS-PAGE from O. oeni cells grown under normal conditions (a), stressed with 12% ethanol for 1 h (b), and grown in the presence of 8% ethanol (c). Molecular masses (in kilodaltons) of marker bands (left side) are indicated. Analyzed proteins are boxed and numbered (see also Table 3).
TABLE 3.
Putative membrane proteins of O. oenia
| Spot | Sequence of peptide analyzed | Mass (kDa) | Description |
|---|---|---|---|
| 1 | ALVLFENPEDALK | 35.5 | PTS system, mannose-specific IIAB (EIIman) |
| 2 | AQQFVDQVQNH | 27.2 | Glutamine ABC transporter, ATP-binding protein |
| 3 | EYVPALQEDLRc | ||
| 4 | LIPSFTIEDFVK | 53 | Phosphogluconateb dehydrogenase (PGDH) |
For each spot (numbered according to Fig. 2), sequences of short single peptides, the molecular mass, and the possible function are indicated.
Cytoplasmic protein.
No hits found.
Membrane-associated proteins of O. oeni cells.
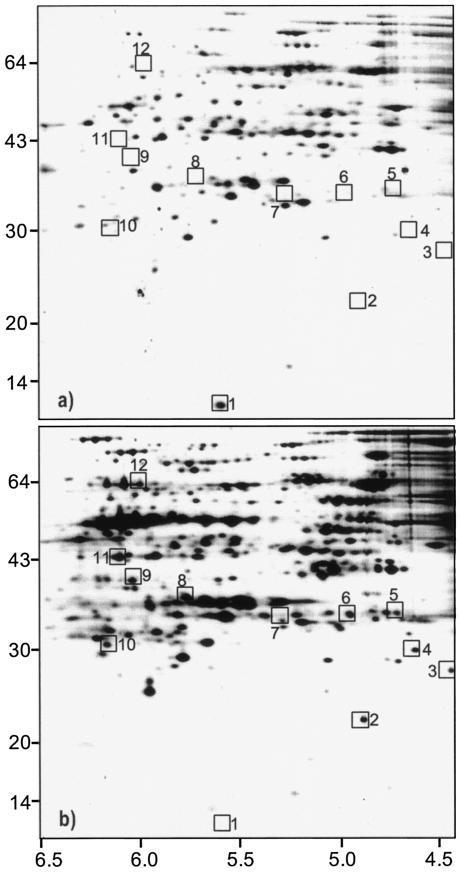
Fractions with membrane-associated proteins were isolated after sodium carbonate extraction. According to the prior treatment of the membranes, these fractions contain proteins which were attached to the membrane by protein-protein interaction, including the so-called lipoproteins, which are attached covalently to the membrane by acetylation. O. oeni cells grown in the presence of 8% (vol/vol) ethanol showed additional protein spots of putative membrane-associated proteins compared to control cells (Fig. 3). From the 2D gels, 12 protein spots were selected for analysis by mass spectroscopy and shown to include putative dTDT-glucose-4,6-dehydratase, d-alanine:d-alanine ligase, phosphomethylpyrimidine kinase, CMP kinase, and a protein belonging to the decarboxylase family (Table 4). All selected spots were present only in ethanol-grown cells (Fig. 3), except spot 1, which was absent in these cells.
FIG. 3.
2D-E of membrane-associated proteins of O. oeni cells grown in the absence (a) and in the presence (b) of 8% ethanol. Proteins were extracted from the same amount of cells. The same samples were used to extract integral membrane proteins (see above and Materials and Methods). Molecular masses (in kilodaltons) of marker bands (left side) and pI ranges (bottom) are indicated. Selected proteins are boxed and numbered (see also Table 4).
TABLE 4.
List of membrane-associated proteins of O. oenia
| Spot | Mass (kDa) | Description | Closest hit (% similarity) |
|---|---|---|---|
| 3 | 22.1 | Decarboxylase family protein | E. faecalis (62)b |
| 7 | 29.2 | Phosphomethylpirimidine kinase | E. faecalis (69) |
| 9 | 37.5 | dTDP-glucose 4,6-dehydratase | L. monocytogenes (82) |
| 10 | 25.0 | Cytidine monophosphate kinase | L. monocytogenes (74) |
| 11 | 42.1 | d-alanine-d-alanine ligase | L. mesenteroides (70)c |
For each spot (numbering according to Fig. 3), the molecular mass, the possible function, and the closest hits are indicated.
Enterococcus faecalis.
Leuconostoc mesenteroides.
DISCUSSION
Performance of microorganisms under ethanol stress conditions, such as those prevailing in wine, requires specific cellular features, including modification of the protein composition to allow survival under such adverse conditions. We examined the proteome of O. oeni, a lactic acid bacterium of enological interest, the genome of which has been sequenced recently by Miles (Doe Joint Genome Institute, University of California; see above URL). Our main focus was the characterization of proteins involved in ethanol adaptation. To understand the physiological relevance of the site-specific location of these proteins, cytoplasmic, membrane-associated, and integral membrane proteins were investigated. Ethanol triggered alterations in protein patterns of O. oeni cells. From the 28 proteins analyzed, 14 were identified as proteins with assigned function involved in a variety of cellular processes. The possible role of these proteins in ethanol adaptation is discussed.
GR catalyzes the regeneration of glutathione from glutathione disulfide at the expense of NADPH and was more than threefold induced during ethanol adaptation. The first report of GR in lactic acid bacteria appeared in 1995, by Pebay et al. (35), providing new data concerning lactic acid bacterium responses to oxidative stress. Since the principal thiol-disulfide redox buffer in the cytoplasm of bacteria is thought to be glutathione (46), GR is probably important for maintaining the redox potential at a low level for protection of thiol-containing proteins against oxidative stress. One of the known toxic effects of acetaldehyde accumulation as a consequence of ethanol oxidation comes from the reaction of the aldehyde group with thiol compounds, such as cysteine and glutathione (Rawat, 1975, and Lieberthal, 1979; reviewed in reference 20). Moreover, it has been shown that ethanol increases dramatically the production of reactive oxygen species in yeast, which cause severe oxidative damage to proteins, lipid, and DNA (31). Thus, the increase of GR observed in O. oeni ethanol-adapted cells suggests that the cells have to cope with oxidative damage during ethanol adaptation.
IMPDH catalyzes the conversion of IMP to XMP with the concomitant reduction of NAD+ to NADH. This reaction is the rate-limiting step in guanine nucleotide biosynthesis (6). We observed that the synthesis of IMPDH was inhibited in ethanol-adapted cells. Conceivably this inhibition will result in depletion of guanine nucleotides in these cells. The physiological effect remains to be elucidated.
In lactic acid bacteria the transport and phosphorylation of glucose is carried out by the PTS (5). The phosphocarrier protein HPr, a general non-sugar-specific component of energy-coupling proteins, and the sugar-specific EII complex (EIIMan) of the PTS (36) were not present in ethanol-adapted cells, suggesting that the mannose PTS is absent in these O. oeni cells. It is now well established that in Bacillus subtilis and other low-GC-content gram-positive bacteria, the dominant catabolic repression (CR) pathway involves HPr and a transcription regulator, CcpA (for a review, see reference 41). HPr plays a role in CR as a corepressor of CcpA when phosphorylated at Ser-46 (9). It has also been shown that mutations rendering the EIIMan complex inactive have a pleiotropic effect on an inducible fructose PTS activity (2), resulting in an increased rate of phosphoenolpyruvate-dependent phosphorylation of fructose (5). Thus, our results strongly suggest that adaptation to ethanol is associated with a relief of CR exerted by glucose over other carbon sources in O. oeni cells. This represents a metabolic advantage to O. oeni during MLF, since glucose is known to induce inhibition of malolactic enzyme activity via NADH accumulation during glucose metabolism (30). Moreover, phosphomethylpyrimidine (HMP-P) kinase, which catalyzes the stepwise phosphorylation of HMP-P into HMP-PP (21), a heterocyclic intermediate in the de novo synthesis of thiamine diphosphate (ThDP), was increased in ethanol-adapted cells. The only ThDP-dependent enzyme known in nonaerobic bacteria is xylulose 5-phosphate (X5P) phosphoketolase, a central enzyme of the pentose phosphate pathway (PKP) of heterofermentative lactic acid bacteria (39, 45). Since the X5P concentration is expected to be low in ethanol-adapted cells (PGDH is almost absent), a possible explanation for the putative increased activity of X5P phosphoketolase in these cells is that pentoses, e.g., xylose, are taken up, converted into X5P, and then fueled into the PKP pathway. However, this hypothesis presupposes that mechanisms involved in CR are offset (25) in ethanol-adapted O. oeni cells, as discussed above. These findings are in line with our observation that ATP synthesis and malic acid degradation are much more efficient in ethanol-adapted cells (Da Silveira, Rombouts, and Abee, submitted).
The presence of 8% ethanol in the growth medium promoted a shutdown in the synthesis of several integral membrane proteins. In contrast, additional protein spots which may be associated with the membrane were found in ethanol-grown cells compared to results for control cells, suggesting that cytoplasmic proteins can assume the function of stabilizing the O. oeni cytoplasmic membrane in response to the ethanol-induced membrane disordering (7a). It was previously described that the membrane association of Lo18, a general stress marker for O. oeni that is peripherally associated with the cytoplasmic membrane, was increased significantly upon temperature upshift (8). Torok et al. (42) presented evidence that the soluble chaperonin GroEL from E. coli can associate with model lipid membranes and that binding was apparently governed by the composition of the host lipid bilayer, suggesting lipid-protein interactions. It is conceivable that ethanol-induced hydrophobicity of cytoplasmic proteins (37) may increase the affinity of these proteins for membranes, which may offer an explanation for the high level of membrane-associated proteins displayed in ethanol-adapted cells. Interestingly, two of these associated proteins were assigned with functions that can be associated with the biosynthesis of the cell wall: dTDT-glucose-4,6-dehydratase (4,6-dehydratase), which catalyzes the irreversible conversion of dDTD-glucose to dTDT-4-keto-6-deoxyglucose (29), and d-alanine:d-alanine ligase, which is an ATP-dependent enzyme that promotes dipeptide formation of d-Ala-d-Ala (1). The 6-deoxysugars are principle components of bacterial lipopolysaccharides (24), and in many prokaryotes d-Ala-d-Ala is incorporated into UDP-muramyl pentapeptide, which ultimately is used to produce peptidoglycan polymers (10).
Formerly, we reported that the protective effect of growth in the presence of ethanol is, to a large extent, based on modification of the physicochemical state of the membrane, i.e., O. oeni cells adjust their membrane permeability by decreasing fluidity at the lipid-water interface. Moreover, ethanol-adapted cells displayed increased metabolic capacity, i.e., they were revealed to be much more efficient in degrading malic acid and generating ATP than control cells (Da Silveira, Rombouts, and Abee, submitted). In this study, using a proteomic approach, we provide evidence for an active ethanol adaptation response of O. oeni, including the modification of cytoplasmic and membrane protein profiles, reflecting the diversity of physiological responses. Three possible sites for cellular adaptation to ethanol are proposed. (i) Cell wall: dTDT-glucose-4,6-dehydratase and d-alanine:d-alanine ligase, which were increased in ethanol-adapted cells, are known to be involved in lipopolysaccharide and peptidoglycan biosynthesis, respectively, suggesting that the cell wall is modulated during ethanol adaptation. (ii) Cytoplasmic membrane: the observed association between proteins and membranes during growth in the presence of ethanol may constitute a general mechanism that preserves membrane integrity during ethanol stress. (iii) Metabolic pathways: amounts of IMPDH and PGDH, which generate reduced nicotinamide nucleotides, are reduced during growth in the presence of ethanol, while GR, which consumes NADPH, is increased, suggesting that maintaining the redox balance plays an important role in ethanol adaptation. These results are in agreement with previous observations that ethanol-adapted cells contain very low levels of NAD(P)H (Da Silveira, Rombouts, and Abee, submitted). Furthermore, two components of mannose PTS, i.e., HPr and EIIMan, were lacking in ethanol-adapted cells, providing strong evidence that mannose PTS is absent in ethanol-adapted cells, which represents a metabolic advantage to O. oeni cells during MLF. Finally, the increased level of HMP-P kinase involved in the de novo synthesis of ThDP, which is a coenzyme of X5P phosphoketolase, a central enzyme of the PKP pathway, provides further evidence that ethanol-adapted cells are metabolically more active than control cells. Besides the above-mentioned proteins, which are involved in a variety of cellular processes, a putative general stress response protein and a protein involved in oxidative stress (GR) were also identified.
In this paper we present evidence that the physiological significance of ethanol adaptation in O. oenis cells is reflected in its proteome. In addition to the introductory global view of the O. oeni proteome when cells are grown in different environmental conditions, the various protocols described here provide new tools to achieve deeper insight into the protein composition and functionality of the O. oeni membrane.
Acknowledgments
We thank J. Wehland (GBF, Braunschweig, Germany) for making it possible to perform membrane protein analysis in his laboratory. We thank Jeroen Wouters (NIZO, Ede, Holland) and the proteomics group (Henrike Wemekamp-Kamphuis, Jasper Kieboom, and Willem van Schaik) for discussion and support throughout this work. We also thank Airidas Dapkavicius for the layout of the images.
This work was supported by the Portuguese Foundation for Science and Technology, under the program PRAXIS XXI.
REFERENCES
- 1.Arthur, M., P. E. Reynolds, F. Depardieu, S. Evers, S. Dutka-Malen, R. Quintiliani, and P. Courvalin. 1996. Mechanisms of glycopeptide resistance in enterococci. J. Infect. 32:10-16. [DOI] [PubMed] [Google Scholar]
- 2.Bourassa, S., and C. Vadeboncoeur. 1992. Expression of an inducible enzyme II fructose and activation of a cryptic enzyme II glucose in glucose-grown cells of spontaneous mutants of Streptococcus salivarius lacking the low-molecular-mass of IIIman, a component of the phosphoenolpyruvate:mannose phosphotransferase system. J. Gen. Microbiol. 138:769-777. [DOI] [PubMed] [Google Scholar]
- 3.Bourdineaud, J.-P., B. Nehmé, S. Tesse, and A. Lonvaud-Funel. 2003. The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl. Environ. Microbiol. 69:2511-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavin, J. F., P. Schmitt, A. Arias, J. Lin, and C. Divies. 1988. Plasmid profiles in Leuconostoc species. Microbiol. Aliment. Nutr. 6:55-62. [Google Scholar]
- 5.Chaillou, S., W. P. Postma, and P. H. Pouwels. 2001. Contribution of the phosphoenolpyruvate:mannose phosphotransferase system to carbon catabolite repression in Lactobacillus pentosus. Microbiology 127:671-679. [DOI] [PubMed] [Google Scholar]
- 6.Collart, F. R., and E. Huberman. 1988. Cloning and sequence analysis of the human and Chinese hamster inosine-5′-monophosphate dehydrogenase cDNAs. J. Biol. Chem. 263:15769-15772. [PubMed] [Google Scholar]
- 7.Da Silveira, M. G., M. V. San Romão, M. C. Loureiro-Dias, F. M. Rombouts, and T. Abee. 2002. Flow cytometric assessment of membrane integrity of ethanol-stressed Oenococcus oeni cells. Appl. Environ. Microbiol. 68:6087-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Da Silveira, M. G., E. A. Golovina, F. A. Hoekstra, F. M. Rombouts, and T. Abee. 2003. Membrane fluidity adjustments in ethanol-stressed Oenococcus oeni cells. Appl. Environ. Microbiol. 69:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delmas, F., F. Pierre, F. Coucheney, C. Divies, and J. Guzzo. 2001. Biochemical and physiological studies of the small heat shock protein Lo18 from the lactic acid bacterium Oenococcus oeni. J. Mol. Microbiol. Biotechnol. 3:601-610. [PubMed] [Google Scholar]
- 9.Deutscher, J., E. Kuster, U. Bergstedt, Charrier, V., and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolitic activity to carbon catabolite repression in Gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 10.Evers, S., B. Casadewall, M. Charles, S. Dutka-Malen, M. Galimand, and P. Courvalin. 1996. Evolution of structure and substrate specificity in D-alanine:D-alanine ligases and related enzymes. J. Mol. E vol. 42:706-711. [DOI] [PubMed] [Google Scholar]
- 11.Favier, A., B. Brutscher, M. Blackledge, A. Galinier, J. Deutscher, F. Penin, and D. Marion. 2002. Solution structure and dynamics of Crh, the Bacillus subtilis catabolite repression HPr. J. Mol. Biol. 317:131-144. [DOI] [PubMed] [Google Scholar]
- 12.Galland, D., R. Tourdot-Marechal, M. Abraham, K. S. Chu, and J. Guzzo. 2003. Absence of malolactic activity is a characteristic of H+-ATPase-deficient mutants of the lactic acid bacterium Oenococcus oeni. Appl. Environ. Microbiol. 69:1973-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guy, G. R., R. Philip, and Y. H. Tan. 1994. Analysis of cellular phosphoproteins by two-dimensional gel electrophoresis: applications for cell signaling in normal and cancer cells. Electrophoresis 15:417-440. [DOI] [PubMed] [Google Scholar]
- 14.Guzzo, J., J. F. Cavin, and C. Diviès. 1994. Induction of stress proteins in Leuconostoc oenos to perform direct inoculation of wine. Biotechnol. Lett. 16:1089-1094. [Google Scholar]
- 15.Guzzo, J., J. F. Delmas, F. Pierre, M.-P. Jobin, B. Samyn, J. Van Beeumen, J.-F. Cavin, and C. Diviès. 1997. A small heat shock protein from Leuconostoc oenos induced by multiple stresses and during stationary growth phase. Lett. Appl. Microbiol. 24:393-396. [DOI] [PubMed] [Google Scholar]
- 16.Guzzo, J., M.-P. Jobin, and C. Diviès. 1998. Increase of sulfite tolerance in Oenococcus oeni by means of acidic adaptation. FEMS Microbiol. Lett. 160:43-47. [Google Scholar]
- 17.Hartinger, J., K. Stenius, D. Högemann, and J. Reinhard. 1996. 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal. Biochem. 240:116-133. [DOI] [PubMed] [Google Scholar]
- 18.Jobin, M.-P., F. Delmas, D. Garmyn, C. Divies, and J. Guzzo. 1997. Molecular characterization of the gene encoding an 18-kilodalton small heat shock protein associated with the membrane of Leuconostoc oenos. Appl. Environ. Microbiol. 63:609-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, B. E., V. Dossonnet, E. Kuster, W. Hillen, J. Deutscher, and R. E. Klevit. 1997. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 272:26530-26535. [DOI] [PubMed] [Google Scholar]
- 20.Jones, R. P. 1989. Biological principles of the effects of ethanol: a review. Enzyme Microb. Technol. 10:130-152. [Google Scholar]
- 21.Kawasaki, T., T. Nakata, and Y. Nose. 1968. Genetic mapping with a thiamine-requiring auxotroph of Escherichia coli K-11 defective in thiamine phosphate pyrophosphorylase. J. Bacteriol. 95:1233-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruft, V., H. Eubel, L. Jansch, W. Werhahn, H. P. Braun. 2001. Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 117:1694-1710. [PMC free article] [PubMed] [Google Scholar]
- 23.Labarre, C., J. F. Cavin, C. Divies, and J. Guzzo. 1998. Using specific polyclonal antibodies to study the malolactic enzyme from Leuconostoc oenos and other lactic acid bacteria. Lett. Appl. Microbiol. 26:293-296. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H. W., and J. S. Thorson. 1994. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol. 48:223-256. [DOI] [PubMed] [Google Scholar]
- 25.Lokman, B. C., M. Heerikhuisen, R. J. Leer, A. van den Broek, Y. Borsboom, S. Chaillou, P. W. Postma, and P. H. Pouwels. 1997. Regulation of expression of the Lactobacillus pentosus xylAB operon. J. Bacteriol. 179:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonvaud-Funel, A. 1999. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Leewenhoek 76:317-331. [PubMed] [Google Scholar]
- 27.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 28.Mantis, N. J., and S. C. Winans. 1992. Characterization of the Agrobacterium tumefaciens heat shock response: evidence for a σ32-like sigma factor. J. Bacteriol. 174:991-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marolda, C. L., and M. A. Valvano. 1995. Genetic analysis of dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rft cluster and correct location of the rftE gene. J. Bacteriol. 177:5539-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda, M., A. Ramos, M. Veiga-da-Cunha, M. C. Loureiro-Dias, and H. Santos. 1997. Biochemical basis for glucose-induced inhibition of malolactic fermentation in Leuconostoc oenos. J. Bacteriol. 179:5347-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moradas-Ferreira, P., V. Costa, P. Piper, and W. Mager. 1996. The molecular defences against reactive oxygen species in yeast. Mol. Microbiol. 19:651-658. [DOI] [PubMed] [Google Scholar]
- 32.Mouritsen, O. G., and M. Bloom. 1984. Mattress model of lipid-protein interaction in membranes. Biophys. J. 46:121-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 34.O'Farrel, P. H. 1975. High-resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 35.Pebay, M., A. C. Holl, J. M. Simonet, and B. Decaris. 1995. Characterization of the gor gene of the lactic acid bacterium Streptococcus thermophilus CNRZ368. Res. Microbiol. 126:371-383. [DOI] [PubMed] [Google Scholar]
- 36.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Privalov, P. L., and S. L. Gill. 1988. Stability of protein structure and hydrophobic interactions. Adv. Protein Chem. 39:191-217. [DOI] [PubMed] [Google Scholar]
- 38.Rabilloud, T., J. M. Strub, S. Luche, A. van Dorsselaer, and J. Lunardi. 2001. A comparison between Sypro Ruby and ruthenium II Tris(bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics 1:699-704. [DOI] [PubMed] [Google Scholar]
- 39.Schorken, U., and G. A. Sprenger. 1998. Thiamin-dependent enzymes as catalysts in chemoenzymatic syntheses. Biochim. Biophys. Acta 1385:229-243. [DOI] [PubMed] [Google Scholar]
- 40.Soto, T., J. Fernandez, J. Vicente-Soler, J. Cansado, and M. Gacto. 1999. Accumulation of trehalose by overexpression of tps1, coding for trehalose-6-phosphate synthase, causes increased resistance to multiple stresses in the fission yeast Schizosaccharomyces pombe. Appl. Environ. Microbiol. 65:2020-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Titgemeywer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Leewenhoek 82:52-71. [PubMed] [Google Scholar]
- 42.Torok, Z., I. Horvath, P. Goloubinoff, E. Kovacs, A. Glatz, G. Balogh, and L. Vigh. 1997. Evidence for a lipochaperonin: association of active protein-folding GroESL oligomers with lipids can stabilize membranes under heat shock conditions. Proc. Natl. Acad. Sci. USA. 94:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tourdot-Marechal, R., L. C. Fortier, J. Guzzo, B. Lee, and C. Divies. 1999. Acid sensitivity of neomycin-resistant mutants of Oenococcus oeni: a relationship between reduction of ATPase activity and lack of malolactic activity. FEMS Microbiol Lett. 178:319-326. [DOI] [PubMed] [Google Scholar]
- 44.Van Renswoude, J., and C. Kempf. 1984. Purification of integral membrane proteins. Methods Enzymol. 104:329-339. [DOI] [PubMed] [Google Scholar]
- 45.Veiga-da-Cunha, M., H. Santos, and E. Van Schaftingen. 1993. Pathway and regulation of erythritol formation in Leuconostoc oenos. J. Bacteriol. 175:3941-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.William, A. P., èslund, F. Holmgren, A., and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in Escherichia coli cytoplasm. J. Chem. 272:15661-15667. [DOI] [PubMed] [Google Scholar]
- 47.Wouters, J. A., B. Jeynov, F. M. Rombouts, W. M. de Vos, O. P. Kuipers, and T. Abee. 1999. Analysis of the role of 7 kDa cold-shock proteins of Lactococcus lactis MG1363 in cryoprotection. Microbiology 125:3185-3194. [DOI] [PubMed] [Google Scholar]