Abstract
We used genetic and biochemical methods to examine the genomic diversity of the enterobacterial plant pathogen Erwinia carotovora subsp. carotovora. The results obtained with each method showed that E. carotovora subsp. carotovora strains isolated from one ecological niche, potato plants, are surprisingly diverse compared to related pathogens. A comparison of 23 partial mdh sequences revealed a maximum pairwise difference of 10.49% and an average pairwise difference of 2.13%, values which are much greater than the maximum variation (1.81%) and average variation (0.75%) previously reported for Escherichia coli. Pulsed-field gel electrophoresis analysis of I-CeuI-digested genomic DNA revealed seven rrn operons in all E. carotovora subsp. carotovora strains examined except strain WPP17, which had only six copies. We identified 26 I-CeuI restriction fragment length polymorphism patterns and observed significant polymorphism in fragment sizes ranging from 100 to 450 kb for all strains. We detected large plasmids in two strains, including the model strain E. carotovora subsp. carotovora 71. The two least virulent strains had an unusual chromosomal structure, suggesting that a particular pulsotype is correlated with virulence. To compare chromosomal organization of multiple enterobacterial genomes, several genes were mapped onto I-CeuI fragments. We identified portions of the genome that appear to be conserved across enterobacteria and portions that have undergone genome rearrangements. We found that the least virulent strain, WPP17, failed to oxidize cellobiose and was missing several hrp and hrc genes. The unexpected variability among isolates obtained from clonal hosts in one region and in one season suggests that factors other than the host plant, potato, drive the evolution of this common environmental bacterium and key plant pathogen.
The plant-pathogenic species Erwinia carotovora is a complex taxon consisting of strains with a range of different phenotypic, biochemical, host range, and genetic characteristics. This species is well suited for studying the ecology, speciation, and pathogenicity of enterobacterial pathogens since it is widespread in the environment, it can infect numerous plant species (42), and many of its virulence genes have been identified, including genes encoding degradative enzymes, diverse regulatory systems, and the type III secretion system (TTSS) (42). E. carotovora is considered a broad-host-range soft rot pathogen because under laboratory conditions it can macerate plant tissues from numerous species. However, in nature, E. carotovora subspecies are associated with particular plant species, showing subspecies adaptation to specific hosts. None of the genetic differences responsible for host specialization in E. carotovora, which is one of the world's most economically important bacterial plant pathogens, have been identified yet.
The taxonomy of the soft rot Erwinia is in flux. Recently, Hauben et al. (16) proposed resurrecting the name Pectobacterium, but the majority of the Erwinia research community has not yet accepted this name because the data presented by Hauben et al. (16) are considered too weak to support transfer of the soft rot Erwinia to a new genus. Also, Gardan et al. (13) recently elevated three of the five E. carotovora subspecies to the species level and placed these new species in the genus Pectobacterium as well. The genome sequences of E. carotovora subsp. atroseptica (Pectobacterium atrosepticum) and Erwinia chrysanthemi (Pectobacterium chrysanthemi or Dickeya sp.) will be completed soon, and comparison of these sequences with available data should conclusively demonstrate into which genus the soft rot Erwinia should be placed. In this report, we use the original names.
Two of the original five subspecies, E. carotovora subsp. carotovora and E. carotovora subsp. atroseptica, are the major causal organisms of economically important potato diseases, including aerial stem rot, blackleg, and soft rot of potato tubers (28). In North America, E. carotovora subsp. atroseptica and blackleg are primarily found in the early spring, and overall, approximately 80% of the E. carotovora isolates from potato plants with aerial stem rot or tuber soft rot are E. carotovora subsp. carotovora (29, 45). E. carotovora subsp. carotovora has also been reported on numerous other hosts (19), including plants as diverse as celery (44) and sunflower (15).
Very little is known about the variability of E. carotovora in the field. Researchers used serogrouping and phage typing to examine the ecology and epidemiology of E. carotovora in the 1970s and 1980s with various degrees of success. The results were difficult to interpret since in some cases as many as 80% of the isolates obtained for study could not be typed with the available methods and were not classified further (26). However, the majority of the isolates from diseased plants were placed into 3 of the 40 identified serogroups (serogroups 3, 9, and 29) (9, 26). The serogrouping methods also showed that E. carotovora subsp. carotovora is more diverse than E. carotovora subsp. atroseptica, since E. carotovora subsp. atroseptica strains were members of only 4 of the 40 serogroups (serogroups 1, 18, 20, and 22), while E. carotovora subsp. carotovora isolates were members of 38 of the 40 serogroups (serogroups 2 to 21 and 23 to 40) (10). Recently, Avrova et al. (2) used amplified fragment length polymorphism fingerprinting to examine genetic diversity in E. carotovora strains and confirmed that E. carotovora subsp. carotovora is significantly more diverse than E. carotovora subsp. atroseptica.
The goals of this work were fourfold. Our first goal was to examine the intraspecies variation of recent E. carotovora subsp. carotovora isolates and of archived strains representing the different E. carotovora subsp. carotovora serogroups using sequence analysis of two housekeeping genes and biochemical assays. Our second goal was to quantify the relative virulence of the isolates in order to identify highly virulent and nonvirulent strains. Our third goal was to employ I-CeuI macrorestriction analysis of E. carotovora subsp. carotovora genomic DNAs by pulsed-field gel electrophoresis (PFGE) to determine if we could discriminate between virulent and nonvirulent strain types. Since there is a paucity of information on the organization and structure of the E. carotovora subsp. carotovora genome, our final goal was to construct a physical map of a representative virulent strain. To aid in data interpretation, the locations of virulence genes of E. carotovora subsp. carotovora and several housekeeping genes were mapped to serve as a guide for comparison to each other and to other enterobacterial pathogens, such as Escherichia, Salmonella, and Yersinia species.
MATERIALS AND METHODS
Strains, plasmids, and media.
The bacterial strains and plasmids used in this work are listed in Table 1. Strains representing 35 common E. carotovora subsp. carotovora serogroups were obtained from Arthur Kelman's culture collection (10). These strains were isolated from different geographical areas by various researchers and had been stored in nutrient broth containing dimethyl sulfoxide at −80°C since 1986. Most bacterial strains were routinely grown in Luria-Bertani medium (LB) at 37°C; the exceptions were E. carotovora subsp. atroseptica strains, which required a lower temperature (less than 33°C) for survival. When required, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 50 μg/ml; spectinomycin, 50 μg/ml; streptomycin, 50 μg/ml; and kanamycin, 50 μg/ml. Transformation, restriction endonuclease digestion, and other DNA techniques were basically performed as described by Sambrook and Russell (35). PCR-amplified fragments were cloned into the pGEMT-Easy vector by following the manufacturer's instructions (Promega, Madison, Wis.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, serogroups, or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (Δ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Clontech |
| E. carotovora | ||
| WPP1 to WPP27 | Wild-type strains isolated from diseased potato tubers and stems from fields in Wisconsin during the summer of 2001 | This study |
| Serogroups 2 to 17, 19, 21, 23, 24, 26 to 40 | Strains isolated in the United States, Canada, and the Netherlands representing 35 common serogroupsa | 10 |
| WPP49 | Cmr; ΔrrnA::cat derivative of WPP14 | This study |
| WPP51 | Cmr Spr/Smr; ΔrrnA::cat ΔrrnB::aadA derivative of WPP49 | This study |
| WPP53 | Cmr Spr/Smr; ΔrrnA::cat ΔrrnC::aadA derivative of WPP49 | This study |
| WPP54 | Cmr Spr/Smr; ΔrrnA::cat ΔrrnD::aadA derivative of WPP49 | This study |
| WPP55 | Cmr Spr/Smr; ΔrrnA::cat ΔrrnE::aadA derivative of WPP49 | This study |
| WPP57 | Cmr Kmr; ΔrrnA::cat ΔrrnF::kan derivative of WPP49 | This study |
| Plasmids | ||
| pGEMT-Easy | AmprlacZ′ cloning vector | Promega |
| pHP45ΩSp/Sm | Ampr Spr/Smr broad-host-range cloning vector | 12 |
| pKD3 | Kmr template plasmid carrying kan cassette | 8 |
| pKD4 | Cmr template plasmid carrying cat cassette | 8 |
| pTΔ23S | Ampr pGEMT-Easy carrying 2.1-kb I-CeuI-restricted 23S ribosomal DNA of WPP14 | This study |
| pTΔ23SKm | Ampr Kmr pTΔ23S derivative harboring 1.6-kb kan of pKD4 cloned into BamHI site | This study |
| pTΔ23SCm | Ampr Cmr pTΔ23S derivative harboring 1.1-kb cat gene of pKD3 cloned into BamHI site | This study |
| pTΔ23SΩSp | Ampr Spr/Smr pTΔ23S derivative harboring 2-kb ΩaadA gene of pHP45ΩSp cloned into BamHI site | This study |
For strain designations see reference 10.
Isolation of bacterial strains from potatoes.
E. carotovora subsp. carotovora strains WPP1 to WPP27 (Table 1) were isolated from potato samples essentially by the methods described by Schaad et al. (37). Cultures were incubated at room temperature throughout the isolation protocol to allow isolation of E. carotovora subsp. atroseptica. Potato stem and tuber samples were surface sanitized by soaking them in 10% bleach for approximately 3 min. A sterile scalpel was used to remove a section of a tuber or stem, and the sample was soaked in water for up to 5 min to allow the bacteria to stream from the plant tissue. One milliliter of this water was transferred to pectate enrichment medium (6). The pectate enrichment medium cultures were incubated without agitation at room temperature (20 to 25°C) for 3 days, and this was followed by streaking onto crystal violet pectate medium (CVP) (7). Individual colonies that formed pits in CVP were restreaked onto LB. If colonies with different morphologies were present on LB, bacteria from individual colonies were streaked to obtain pure cultures. One colony from LB was streaked onto CVP to determine if all of the resulting colonies were pectolytic. A single colony chosen from this CVP plate was streaked onto LB, and the resulting culture was then stored in 20% glycerol at −80°C.
Identification of E. carotovora subsp. carotovora strains.
All pectolytic bacterial strains isolated from the potato samples on CVP were presumed to be Erwinia strains. All WPP and Kelman strains were tested for growth at 37°C in LB containing 5% NaCl and for phosphatase activity (37), as well as for carbon source utilization as determined by the Microlog system (Biolog, Inc., Hayward, Calif.). Carbon source utilization studies were carried out by using GN2 plates as suggested by the manufacturer, except that the cultures were grown overnight at 30°C on LB agar prior to inoculation of GN2 plates and sodium thioglycolate was not used. Indole tests from two manufacturers (bioMériuex, Inc., Lyon, France, and Becton Dickinson Microbiology Systems, Sparks, Md.) were used.
All strains were also subjected to intergenic transcribed spacer (ITS)-PCR analysis as described by Toth et al. (41) to confirm the strain identification. To isolate DNA for ITS-PCR, bacterial cells were grown overnight on LB agar, scraped from the agar surface, and suspended in 500 μl of sterile water. The cells were boiled for 5 min, and cell debris was removed by centrifugation at 15,000 × g for 2 min. Two microliters of supernatant was used in each 20-μl ITS-PCR mixture containing 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 to 3.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 mM, 2.5 U of Taq polymerase (Promega), 100 pmol of primer G1, and 100 pmol of primer L2 (Table 2). E. carotovora subsp. carotovora was distinguished from other subspecies by PCR product sizes of 540, 575, and 740 bp (41).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Targeted region | Restriction enzyme site(s) |
|---|---|---|---|
| rrnSac-A | GAG CTC AGG GTG ACT GCG TAC C | rrn | SacI |
| rrnBSce-B | ATT ACC CTG TTA TCC CTA GGA TCC TGG AAG CGT AGC A | rrn | BamHI, I-SceI |
| rrnBSce-C | GGA TCC TAG GGA TAA CAG GGT AAT GAT GGC CAG GCT G | rrn | BamHI, I-SceI |
| rrnERI-D | GGT TAA GCC TCT CGG GGA ATT CGT AC | rrn | EcoRI |
| G1 | GAA GTC GTA ACA AGG | ITS region | |
| L2 | CAA GGC ATC CAC CGT | ITS region | |
| D34BP-F | CTG CAG GAT CCG TGT AGG CTG GAG CTG CTT C | cat or kan pKD3/4 | BamHI, PstI |
| D34BP-R | CTG CAG GAT CCC ATA TGA ATA TCC TCC TTA | cat or kan pKD3/4 | BamHI, PstI |
| acnA1 | GCC TCG CCG CCG CTG GTG GT | acnA | |
| acnA2 | CCG CGC ATC ATC ACT TCA TG | acnA | |
| hemGfor | GGT CAT ACG CAT AAT GAA CTG AAT C | hemG | |
| hemGrev | GCS AAT AMS CTS AAA GGR ACG | hemG | |
| hrpN2 | GTG TTG CCG GTA TCA CCC | hrp and hrc | |
| hrcC-bamF | ACA AGC GCA AAA GCT GGG G | hrp and hrc | |
| mdh2 | GCG CGT AAG CCG GGT ATG GA | mdh | |
| mdh4 | CGC GGC AGC CTG GCC CAT AG | mdh | |
| mobBforwd | TTA CTK AAG CAS STC ATC CC | mobB | |
| mobBrev | CCY TCA ACC AGM AYC ARA TC | mobB | |
| murIfor | AGG GCG AGA ATA CTA TCT CAC | murI | |
| murIrev | CMA CGA CRG GGA AAK TAA AAC | murI | |
| outH-Fwd | GCT GCT GGA AAT CAT GCT GG | outH | |
| outH-Rev | GTC TCC GGC GAC GTG TGC AC | outH | |
| pelB1 | CAG CAC AAA CAG CAC CAG CG | pelB | |
| pelB2 | GGG CCA CCG TTG TTG GTG CA | pelB | |
| purH-f1 | GAG TMM ACR CGG CTC ATY TG | purH | |
| purH-r1 | CCC RAC BAT GGT BCG CTC | purH | |
| rsmA-F1 | GGA TCC GGC AAG CAG GAT AG | rsmA | |
| rsmA-R1 | TGC GTC CCG CGA ACA CGA G | rsmA | |
| gmhB-f2 | GAT ATG GCB GCT TCT TAT ATG GTG | gmhB | |
| gmhB-r1 | CCA TAT AAG AAG CVG CCA TAT C | gmhB | |
| yieP-f1 | ACC TGG TGG ATG ACR GAA G | yieP | |
| yieP-r1 | ACC GAA TGA AAT AGC GWS GC | yieP |
DNA sequence analysis.
Primer sets were obtained from Integrated DNA Technologies, Inc., and were designed to anneal to conserved sequences in the malate dehydrogenase gene (mdh) and the aconitase gene (acnA) (Table 2). The primers were designed to amplify an approximately 500-bp sequence from each gene so that each strand of the DNA fragment could be sequenced with one reaction. The PCR-amplified fragments of mdh and acnA were purified with a PCR purification kit (Qiagen), and the sequencing reactions were performed with a Big-Dye Terminator kit (Perkin-Elmer). DNA sequences were analyzed and aligned by using Genetics Computer Group software (Genetics Computer Group, Madison, Wis.).
Virulence assays.
Virulence assays were performed with both potato plants and potato tubers. The relative virulence of E. carotovora subsp. carotovora strains for potato tubers was evaluated by measuring the amount of macerated tissue (24). For each experimental repetition, three to five surface-sanitized Yukon Gold potato tubers were inoculated with a solution containing 1 × 105 CFU of bacteria per ml and incubated at 100% relative humidity for 2 to 3 days at 28°C. After incubation, the tubers were cut open, and the macerated tissue was scooped from the tubers and weighed. Representative virulent and nonvirulent strains were also tested with 6-week-old Russet Norkotah potato plants grown from tissue culture plantlets by petiole inoculation by using an inoculum consisting of 5 × 104 CFU/ml per injection site. Virulence was expressed by the relative longitudinal lesion sizes after 2 days of incubation at room temperature.
HR assays.
Hypersensitive response (HR) assays (3) were performed with tobacco leaves (Nicotiana tabacum L. cv. Xanthi NN). Mid-log-phase cultures of E. carotovora subsp. carotovora strains grown at 28°C were washed twice with sterile deionized water, and the optical density at 600 nm was adjusted to 1.4. Fully expanded tobacco leaves were infiltrated with a suspension containing 2× 108 CFU of bacteria per ml and kept at room temperature. The plants were examined for the HR 24 h after inoculation.
PFGE analysis.
The conventional PFGE protocol for enteric bacteria has been described elsewhere (33, 40). Briefly, cells were grown overnight on LB agar, washed once in sterile water, and suspended in 1 ml of T100E100 (100 mM Tris, 100 mM EDTA; pH 8.0) at an optical density at 600 nm of 1.4. One hundred microliters of the cell suspension was mixed with 0.2 mg of proteinase K (Sigma Chemical Co., St. Louis, Mo.) and added to 100 μl of molten 1.5% SeaKem Gold agarose (FMC BioProducts, Rockland, Maine) in plug buffer (T100E100, 1% sodium dodecyl sulfate). The resultant mixture was dispensed into 75-μl plug molds and allowed to solidify. The plugs were then removed from the molds and incubated in 1 ml of T100E100 supplemented with 1% sodium dodecyl sulfate and 0.1 mg of proteinase K at 55°C for 4 to 16 h. The plugs were washed six times with TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0) and stored at 4°C in TE buffer until they were used. The bacterial DNA in the agarose plugs was digested with 2 U of I-CeuI (New England Biolabs, Inc.) in a 50-μl reaction solution containing the appropriate buffer for 2 h at 37°C. Electrophoresis was performed with a Pulsaphor Plus system with a hexagonal electrode array (Pharmacia, Uppsala, Sweden) by following the manufacturer's instructions. The digested DNA was separated by electrophoresis on 1% agarose-0.5× TBE (Tris-borate-EDTA)-10 mM thiourea (Acros Organics, Inc.) gels at 12°C for 20 h at 5 V/cm with the switch time ramping from 25 to 45 s. ProMega lambda ladder (Promega) was used as the size marker, and the DNA was stained with ethidium bromide and visualized with a UV light transilluminator.
DNA hybridization.
Southern blot analysis was performed essentially by the recommended procedure (Millipore Co.) and the procedure described by Sambrook and Russell (35). I-CeuI-digested chromosomal DNAs were transferred to nylon membranes (Millipore Co.) and hybridized with 32P-labeled probes prepared by using a random priming kit (Amersham Pharmacia Biotech). Twelve gene probes were PCR amplified by using primers listed in Table 2.
Construction of I-CeuI-restricted Δrrn mutants of E. carotovora subsp. carotovora WPP14.
To construct a physical map of the E. carotovora subsp. carotovora genome, several rrn operons were deleted from strain WPP14. To accomplish this, an I-CeuI deletion mutant allele was PCR amplified by using a crossover PCR strategy described by Link et al. (21) and Yang et al. (46). Oligonucleotide primers rrnSac-A, rrnBSce-B, rrnBSce-C, and rrnERI-D (Table 2) were used to precisely delete the 26-bp recognition site of I-CeuI and to introduce a BamHI site and 24-bp barcode into the E. carotovora subsp. carotovora 23S rRNA gene. The final 2.1-kb PCR product was cloned into pGEMT-Easy (Promega) to generate plasmid pTΔ23S, and then the antibiotic resistance gene cassettes cat, kan, and aadA (Table 2) were cloned into the BamHI site of pTΔ23S, producing plasmids pTΔ23SCm, pTΔ23SKm, and pTΔ23SΩSp, respectively (Table 1). These plasmids were subsequently electrotransformed, and the mutant alleles were marker exchanged into wild-type E. carotovora subsp. carotovora WPP14 or other WPP14 mutant derivatives by using the methods described by Ried and Collmer (34). All deletion mutants were further confirmed by I-CeuI-PFGE analysis.
Nucleotide sequence accession numbers.
The E. carotovora subsp. carotovora mdh and acnA partial gene sequences reported here have been deposited in the GenBank sequence database under accession numbers AY428968 to AY429013.
RESULTS AND DISCUSSION
Identity and diversity of E. carotovora subsp. carotovora potato isolates.
Strains isolated in 2001 from diseased potatoes with aerial stem rot and tuber soft rot were all E. carotovora based on the ITS-PCR assay developed by Toth et al. (41). We did not serotype the strains isolated in 2001 since the reagents are not widely available. Rather, we decided to use DNA-based methods to characterize the archived serotyped strains.
All strains formed pits on CVP, were able to grow at 37°C, and were salt tolerant and phosphatase negative, all of which are typical biochemical and physiological characteristics of E. carotovora subsp. carotovora. All of the Kelman isolates were indole negative, as expected (Table 3). However, one-third of the E. carotovora subsp. carotovora WPP isolates were indole positive. This was surprising since the indole assay is recommended for differentiation of E. carotovora from Erwinia chrysanthemi, species which are described as indole negative and indole positive, respectively (37) (Table 3). The indole assay results were confirmed for a subset of strains by using a kit obtained from a second manufacturer.
TABLE 3.
Representative biochemical test results for 62 E. carotovora subsp. carotovora strains
| Test | % of strains positive
|
|
|---|---|---|
| WPP strains (n = 27) | Kelman strains (n = 35) | |
| Acetic acid | 0 | 94 |
| Cellobiosea | 96 | 100 |
| Citric acid | 96 | 91 |
| d-Galacturonic acid | 67 | 85 |
| d-Galacturonic acid lactone | 44 | 74 |
| d-Gluconic acid | 30 | 77 |
| d-Glucuronic acid | 0 | 6 |
| d-Melibiose | 100 | 91 |
| dl-Lactic acid | 0 | 20 |
| d-Psicose | 4 | 97 |
| d-Raffinose | 100 | 94 |
| Glucuronamide | 0 | 11 |
| Glucose-1-phosphate | 26 | 69 |
| Formic acid | 33 | 91 |
| Inosine | 0 | 51 |
| l-Glutamic acid | 0 | 20 |
| l-Serine | 89 | 100 |
| Lactulose | 0 | 14 |
| Monomethyl succinate | 0 | 74 |
| Succinamic acid | 0 | 42 |
| Succinic acid | 89 | 94 |
| Thymidine | 0 | 97 |
| Tween 80 | 0 | 71 |
| Uridine | 0 | 82 |
| Indole production | 39 | 0 |
WPP17 was the only strain that did not oxidize cellobiose.
Although the atypical indole-positive E. carotovora subsp. carotovora WPP strains were unexpected, Seo et al. (38) also reported that approximately one-third of E. carotovora subsp. carotovora isolates from Asia were indole positive. None of the isolates in the Kelman collection were indole positive, and we were unable to find other reports of indole-positive E. carotovora strains. It is possible that indole-positive E. carotovora-like strains either were characterized as atypical by Kelman and other colleagues or were not present in regions where these workers collected strains. Because of this unexpected result, numerous methods were used to characterize the WPP strains, including biochemical assays, ITS-PCR (41), and construction of phylogenetic trees by using gene sequences (the phylogenetic data will be reported elsewhere). In all cases, all of the WPP strains were clearly E. carotovora strains.
Sixty-two E. carotovora subsp. carotovora strains, including members of 35 serogroups obtained from A. Kelman's collection, were also analyzed by independently inoculating three Biolog plates to determine if the strains oxidized the individually available carbon sources. Of the 95 carbon sources, 12 and 22 were differentially oxidized by the WPP and Kelman strains, respectively (Table 3). Notably, WPP17 was the only strain that did not oxidize cellobiose. The Kelman strains oxidized a wider variety of carbon sources than the WPP strains oxidized, probably because they were collected from a wider geographical region over several years and are a more diverse group of strains.
The sequences of the mdh and acnA housekeeping genes were determined to estimate the genetic depth of E. carotovora subsp. carotovora. The mdh gene has been used for analysis of the relationships among other enteric species (4, 5, 30, 32). A comparison of 23 WPP mdh sequences revealed a maximum pairwise difference of 10.49% and an average pairwise difference of 2.13%, values which are much greater than the maximum variation (1.81%) and average variation (0.75%) previously reported for E. coli (4, 30). In addition, the levels of identity for the acnA sequences from the WPP strains ranged from 80 to 100% (Table 4), and the average difference was 8.97%.
TABLE 4.
Intraspecies comparison of DNA sequence identity as determined by the BLAST algorithm for diverse E. coli, Salmonella, and E. carotovora subsp. carotovora strains for the malate dehydrogenase gene (mdh) and the aconitase gene (acnA)a
| Taxon | % Identity (no. of strains) for:
|
|
|---|---|---|
| mdh | acnA | |
| E. coli | 99-100 (50) | 95-100 (30) |
| Salmonella | 95-100 (30) | 90-100 (4) |
| E. carotovora subsp. carotovora WPP strains | 90-100 (23) | 80-100 (23) |
See reference 1.
This finding suggests that there is considerable genetic diversity in E. carotovora subsp. carotovora strains isolated from the same region in the same season from the same host species and that the genomes of two different E. carotovora subsp. carotovora potato isolates may be more different than the genomes of E. coli K-12 and E. coli O157:H7. Because of this surprising level of diversity, we decided to use methods that allowed us to examine the entire genome to determine if the variation seen in the two individual genes was reflected throughout the bacterial chromosome.
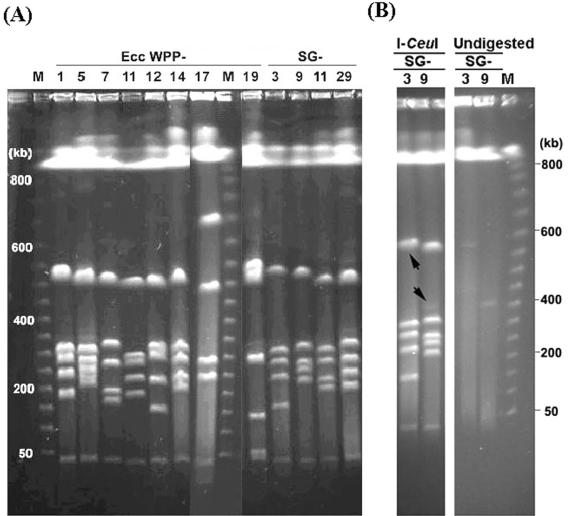
E. carotovora subsp. carotovora strains had diverse I-CeuI PFGE restriction fragment length polymorphism (RFLP) pulsotypes.
To examine gross genome-level variation between E. carotovora subsp. carotovora strains, we used I-CeuI digestion followed by PFGE. I-CeuI is encoded by a class I mobile intron and specifically targets a 26-bp sequence in the 23S rRNA gene of the rrn operon in all enteric bacteria examined so far (22, 27). Thus, the I-CeuI fragments represent the number and genomic distribution of rrn operons. Most of the Enterobacteriaceae chromosomes reported to date contain seven rRNA operons, and some of the genes flanking the rRNA operons are highly conserved across genera (23, 39). With the exception of WPP17, which had six I-CeuI fragments and thus is likely to have only six rRNA operons, all of the E. carotovora subsp. carotovora WPP strains and strains belonging to 16 representative serogroups isolated from potato contained, as expected, seven I-CeuI fragments.
The gene sequence divergence of the E. carotovora subsp. carotovora strains was paralleled by the heterogeneous large-scale organizational patterns of the genomes. There were at least 16 and 10 I-CeuI RFLP patterns (or pulsotypes) among the 27 WPP strains and 16 Kelman strains (each belonging to a different serogroup), respectively. In comparison, in Salmonella species, a high degree of structural conservation is observed, and the I-CeuI pulsotypes of 32 Salmonella strains representing eight subgenera are virtually indistinguishable (23). In each E. carotovora subsp. carotovora genome, there were six fragments smaller than 700 kb whose total size was 1.8 to 2.2 Mb, and a 40-kb fragment was present in all strains (Fig. 1A). If the size of the E. carotovora subsp. carotovora genome were similar to the size of the 5-Mb E. carotovora subsp. atroseptica SCRI1043 genome (http://www.sanger.ac.uk/Projects/E_carotovora/), as estimated by current genome sequencing data, these six fragments would represent approximately 40% of the total genome. All strains except WPP17 and WPP19 had four I-CeuI fragments that were between 100 and 450 kb long, and there was extensive polymorphism in the sizes of these fragments (Fig. 1A). In contrast, the size of the smallest fragment was conserved across all strains, including WPP17 and WPP19.
FIG. 1.
Genomic fingerprints of E. carotovora subsp. carotovora as determined by PFGE. (A) I-CeuI genomic cleavage patterns of representative E. carotovora subsp. carotovora strains. Strains WPP17 and WPP19 had unique chromosomal structures compared to the chromosomal structures of other E. carotovora subsp. carotovora strains. (B) Detection of large plasmids in E. carotovora subsp. carotovora 71 (serogroup 3) and E. carotovora subsp. carotovora 63 (serogroup 9). Undigested genomic DNA was compared to I-CeuI-digested DNA of the same strains. In the left panel, the arrows indicate the positions of the putative plasmids. Electrophoresis was performed by using pulse times of 25 to 45 s at 200 V for 20 h at 12°C. Lanes M contained markers. Ecc, E. carotovora subsp. carotovora; SG, serogroup.
Two previously identified serogroups contained an extrachromosomal element.
In addition to the seven I-CeuI-digested fragments, a faint band at 400 kb was consistently detected in E. carotovora subsp. carotovora 63 (serogroup 9), even after prolonged incubation to eliminate the possibility of incomplete digestion. When undigested genomic DNAs of all strains were examined, bands were present at 550 and 400 kb for the model strain E. carotovora subsp. carotovora 71 (serogroup 3) and for E. carotovora subsp. carotovora 63 (serogroup 9), respectively (Fig. 1B), indicating that large plasmids were present. An additional 32 strains were examined, but none were found to contain large plasmids. Because plasmids appear to be rare in E. carotovora subsp. carotovora, it is likely that biological differences among most E. carotovora subsp. carotovora strains and serogroups are due to insertions and deletions (indels) or mutations rather than to genes carried on plasmids. Because the plasmids are not common in E. carotovora subsp. carotovora, we did not characterize them further here.
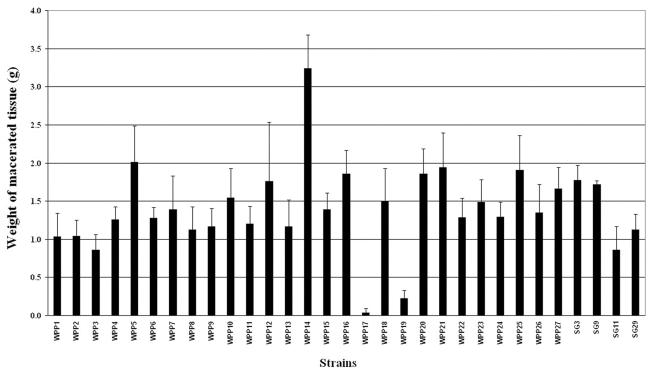
Two strains with unusual PFGE pulsotypes were the least virulent strains for potato tubers and petioles.
The relative virulence of each of the E. carotovora subsp. carotovora WPP strains was quantified by a potato tuber assay. In the related species E. chrysanthemi, multiple virulence genes, including genes encoding pectic enzymes, the TTSS, and an antimicrobial peptide degradation system, are required for full virulence on potato tubers (25). The TTSS and antimicrobial peptide degradation probably contribute to virulence by inhibiting plant defenses. Thus, this assay is likely to measure the contributions of multiple types of virulence genes. In this assay, strain WPP17 macerated the least tuber tissue, and the amount of maceration observed with WPP19 was significantly less than the amounts of maceration observed with the other strains (Fig. 2). On the other hand, WPP14 was consistently the most virulent strain when several independent replicates were examined (Fig. 2). These data were supported by the results of a potato leaf petiole assay, in which the relative lesion lengths for WPP1, WPP14, WPP17, and WPP19 after inoculation were concordant with the degrees of tuber maceration (data not shown). Since WPP17 and WPP19 also had unusual I-CeuI pulsotypes, these data suggest that rearrangements leading to these pulsotypes may also be responsible for the reduced virulence of these strains. Extensive mutagenesis of E. carotovora genes encoding plant cell wall-degrading enzymes has not been completed. However, in the related species E. chrysanthemi, deletion of individual pectic enzymes had little effect either on pitting on CVP or in plant maceration assays, and several genes with overlapping functions had to be deleted before significant reductions were seen (34). Thus, the reduced virulence of WPP17 and WPP19 is likely to be due to the absence of either numerous pectic enzymes or other unrelated virulence proteins.
FIG. 2.
Relative virulence of E. carotovora subsp. carotovora strains on potato tubers. Tubers were inoculated separately with 105 CFU of different strains per ml. Decayed tissue was weighed after 48 h of incubation at 28°C with high humidity. The errors bars indicate standard deviations of three independent replicates. SG, serogroup.
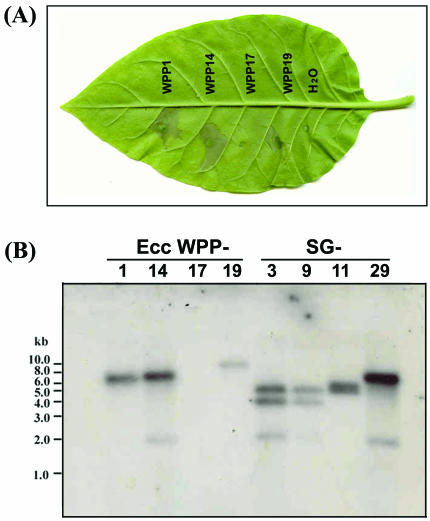
WPP17 was HR negative and was missing at least eight hrp and hrc genes.
All E. carotovora subsp. carotovora strains were tested on tobacco leaves for HR elicitation. Of the 62 E. carotovora subsp. carotovora strains examined, only WPP17 failed to elicit an HR, indicating an absence of a functional TTSS (Fig. 3A). A Southern blot probed with a DNA fragment comprised of a 6-kb region containing hrpN, hrpV, hrpT, hrcC, hrpG, hrpF, hrpE, and hrpD (31) confirmed that these genes, which are required for a functional TTSS, were not present in WPP17. As shown in Fig. 3B, the probe did not hybridize to WPP17 chromosomal DNA under low-stringency conditions, which would allow a detectable signal with 60% DNA sequence similarity. This DNA hybridization experiment and the PFGE data also revealed that strain WPP14 and E. carotovora subsp. carotovora 380 (serogroup 29) have identical I-CeuI pulsotypes and hrp gene RFLPs, suggesting that these strains are closely related even though they were isolated in Wisconsin in 2001 and in Oregon prior to 1986, respectively. This suggests that although there are multiple E. carotovora subsp. carotovora pulsotypes, strains with identical pulsotypes may persist and may be widespread in nature.
FIG. 3.
Phenotypic and genotypic analysis of a naturally occurring E. carotovora subsp. carotovora strain deficient in TTSS. (A) HR assay with tobacco (N. tabacum cv. Xanthi). The bacteria were infiltrated into tobacco leaves at a level of 2× 108 CFU/ml. Deionized sterile water was used as a negative control. (B) Southern analysis of genomic DNA isolated from representative E. carotovora subsp. carotovora strains and digested with EcoRI. The probe used for the blot was PCR amplified by using primers hrpN2 and hrc-bamF, which generated a 6.0-kb product spanning eight hrp and hrc genes. Ecc, E. carotovora subsp. carotovora; SG, serogroup.
Currently, we do not know how common hrp-deficient strains, like WPP17, are in nature or if deletion of the hrp and hrc genes occurred prior to culturing of this strain. However, if an hrp and hrc deletion happened during isolation of WPP17, such a deletion could also happen in nature and it may not be a rare event, considering the small size of the strain collection. In fact, natural deletion of a type III secretion gene cluster in Salmonella spp. (14) and a natural HR-deficient Erwinia pyrifoliae strain have been reported (20). However, the inability of the E. pyrifoliae strain to cause HR was not due to the absence of the hrp genes. Rather, a single-base mutation of the regulatory gene, hrpL, was responsible for the absence of HR. In E. pyrifoliae, no difference in growth rate was observed in the HR-negative strain when it was coinoculated with an HR-positive strain at different ratios, suggesting that the HR-positive strain aids the growth of the HR-negative variant in host plant tissue (20). WPP17 was isolated from the same tuber as WPP16 and WPP18, two virulent strains able to elicit the HR, suggesting that growth of WPP17 could have been aided by these more virulent strains.
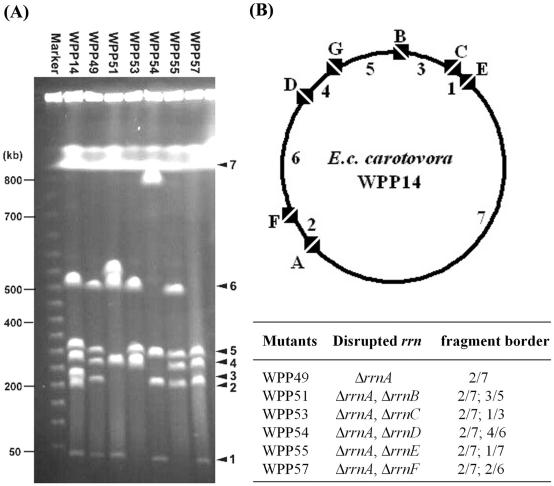
Construction of I-CeuI physical map of E. carotovora subsp. carotovora WPP14.
In general, in enterobacterial species, large genomic inversions due to recombination at rrn operons are rare. Thus, it is possible to construct physical maps of the rrn operons with partial I-CeuI digests (for example, see references 22 and 27). We chose to make a physical map of WPP14, which is a highly virulent strain that lacks plasmids and is very similar to a representative strain, E. carotovora subsp. carotovora 380 belonging to serogroup 29, which is commonly found on diseased potatoes. We were not successful in deducing the E. carotovora subsp. carotovora genome map with either partial digestions or multiple enzyme digestions due to the lack of suitable restriction enzymes and ambiguous results from poorly resolved or overlapping fragments. Therefore, the order and organization of each I-CeuI fragment from WPP14 were determined by allelic exchange to delete the I-CeuI site in the 23S ribosomal DNA genes. Successful deletion of an I-CeuI site was visualized simply on a PFGE gel, as indicated by a larger DNA fragment resulting from fusion of two adjacent I-CeuI fragments and the disappearance of the two original DNA fragments (Fig. 4A). By using this approach, six 23S ribosomal DNA genes were inactivated by constructing both single and double mutants, and the physical map was constructed as shown in Fig. 4B. Since the results for multiple mutations were consistent, it appears that at least in E. carotovora subsp. carotovora WPP14, homologous recombination between rrn operons is not common in cultured E. carotovora subsp. carotovora cells.
FIG. 4.
Construction of an I-CeuI physical map. (A) I-CeuI cleavage patterns of genomic DNA of WPP14 and I-CeuI-restricted derivatives. (B) Circular genome map based on restriction data in panel A. The mapped I-CeuI sites localized seven rrn operons, which were designated rrnA through rrnG, and the flanking junction I-CeuI fragments were numbered 1 through 7 in ascending order (the smallest fragment was fragment 1).
E. carotovora subsp. carotovora has undergone significant genomic rearrangements compared with related enterobacteria.
Based on the extraordinary similarity in genome structure among diverse E. coli and Salmonella strains and even between E. coli and Salmonella (22, 36), a comparable level of similarity was expected for E. carotovora subsp. carotovora strains. However, the I-CeuI RFLPs differed greatly between some strains, particularly for fragments between 100 and 450 kb long (Fig. 1). We wanted to determine if fragments in the same relative location (e.g., the third largest fragment in each strain) encoded homologous genes. To do this, we mapped several conserved genes to specific E. carotovora subsp. carotovora I-CeuI fragments. Most of the E. coli, Salmonella, and Yersinia rrn operons are flanked by homologous genes (17), and the housekeeping genes on each I-CeuI fragment between the rrn flanking genes tend to be colinear within enterobacterial species. Thus, it is likely that most E. carotovora subsp. carotovora I-CeuI fragments with homologous flanking genes also carry other colinear homologous genes. Six of these genes, murH, hemG, murI, mobB, yieP, and gmhB, were mapped in several E. carotovora subsp. carotovora strains by DNA hybridization to Southern blots of the I-CeuI-digested fragments.
Six additional genes or loci, including rsmA, acnA, mdh, outH, and a fragment that contained eight contiguous hrp and hrc genes, were also mapped to the I-CeuI fragments. The acnA, mdh, and rsmA genes were chosen as additional representatives of genes conserved among enterobacteria. We chose the hrp and hrc, outH, and pelB genes as representative virulence genes that might be present on different chromosome fragments in different E. carotovora subsp. carotovora strains. The results for E. carotovora subsp. carotovora strains from both the WPP and Kelman strain collections representing the four E. carotovora subsp. carotovora map types observed among the multiple strains examined are compared to each other and to the results for other enterobacteria in Table 5 and Fig. 5.
TABLE 5.
Summary of the genes mapped on I-CeuI genomic fingerprints of E. carotovora subsp. carotovora strains and comparison to genes in other closely related human pathogensa
| Fragment | Genes
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
E. carotovora subsp. carotovora
|
E. coli
|
Salmonella
|
Yersinia
|
|||||||
| WPP1b | WPP14b | WPP17c | WPP19 | K-12 | EDL933 | LT2 | CT18 | CO92c | KIM | |
| 1d | purH | purH | purH | purH | purH | purH | purH | purH | purH | purH |
| 2e | rsmA | rsmA | rsmA | hemG | hemG | hemG | hemG | hemG | hemG | hemG |
| 3e | hemG, mobB | hemG, mobB | hemG, murI | rsmA | murI, mobB | murI, mobB | murI, mobB | murI, mobB | murI, yieP, oriCg | murI |
| 4f | murI | ? | mobB | murI | yieP, oriC | yieP, oriC, TTSS | yieP, oriC | yieP, oriC | yieP, gmhB, oriCg | |
| 5h | ? | murI | ? | gmhB | gmhB | gmhB | rsmA, mdh, TTSS | rsmA, mdh | rsmA, mdh, TTSS | |
| 6 | gmhB | gmhB | gmhB | gmhB, mobB | rsmA, mdh | rsmA, mdh, TTSS | rsmA, mdh | gmhB | gmhB, mobB, TTSS | mobB |
| 7i | acnA, yieP, mdh, outH, pelB, TTSS | acnA, yieP, mdh, outH, pelB, TTSS | acnA, yieP, mdh, outH, pelB, [TTSS] | acnA, yieP, mdh, outH, pelB, TTSS | acnA | acnA | acnA, TTSS | acnA, TTSS | acnA | acnA |
Seven I-CeuI fragments were designated fragments 1 through 7 in accordance to the diagram shown in Fig. 5; the smallest fragment was fragment 1. Gene locations for E. coli, Salmonella, and Yersinia were determined from genome sequence data. Among the gene probes used, the purH, hemG, mobB, murI, yieP, and gmhB genes are housekeeping genes flanking rrn operons in most enterobacteria examined to date; mdh and acnA are the two housekeeping genes subjected to sequence analysis; and outH, rsmA, pelB, and TTSS are important virulence factors in E. carotovora subsp. carotovora. Brackets indicate the absence of the genes. A question mark indicates that no gene has been assigned to the fragment.
The DNA hybridization results for E. carotovora subsp. carotovora 193 (serogroup 11) and E. carotovora subsp. carotovora 63 (serogroup 9) were similar to those for WPP1, and the DNA hybridization results for E. carotovora subsp. carotovora 71 (serogroup 3) and E. carotovora subsp. carotovora 380 (serogroup 29) were similar to those for strain WPP14.
E. carotovora subsp. carotovora WPP17 and Yersinia pestis CO92 contain six rRNA operons instead of the seven copies commonly found in enterobacteria.
purH is on fragment 1 (40 to 50 kb) in all enteric species examined.
The gene location of hemG and murI is conserved in E. coli, Salmonella, and Yersinia but not in E. carotovora subsp. carotovora. The rsmA gene is on the same fragment as mdh in E. coli, Salmonella, and Yersinia but not in E. carotovora subsp. carotovora.
The location of yieP is conserved on fragment 4 in multiple species, including E. coli, Salmonella sp., and Y. pestis KIM. This gene is not located on fragment 4 in Y. pestis CO92, which has only six rRNA operons, or in any of the E. carotovora subsp. carotovora strains.
The Y. pestis oriC is based on DNA sequence characteristics, not experimental data.
For a better comparison of the genes mapped on similar-size fragments, fragment 5 of WPP17 and fragment 4 of Y. pestis CO92 were displaced by one row.
acnA was mapped on the largest fragment in all enteric species examined. The locations of three housekeeping genes and three virulence genes are highly conserved in E. carotovora subsp. carotovora. Note that WPP17 lacked several TTSS genes.
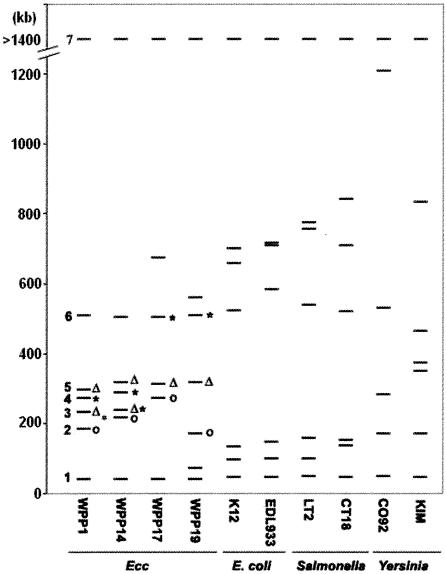
FIG. 5.
Diagrammatic representation of I-CeuI fingerprints of E. carotovora subsp. carotovora and related enteric bacteria. The fragment sizes for E. coli, Salmonella, and Yersinia were calculated from genome sequence data. Fragments were numbered 1 through 7, and the smallest fragment was fragment 1. Fragments 3, 4, and 5 of WPP19 (indicated by an open circle, an open triangle, and an asterisk, respectively) were used as probes to determine the homologous regions in other E. carotovora subsp. carotovora strains. Hybridizing fragments from the other WPP strains are indicated by the same symbols used to indicate the WPP19 probe fragments. Ecc, E. carotovora subsp. carotovora.
All of the genes mapped on I-CeuI fragments of four archived Kelman strains, E. carotovora subsp. carotovora 71 (serogroup 3), E. carotovora subsp. carotovora 63 (serogroup 9), E. carotovora subsp. carotovora 193 (serogroup 11), and E. carotovora subsp. carotovora 380 (serogroup 29), were located on similar-size fragments in either WPP1 or WPP14 (Table 5). Based on the results of comparative genomics of other enterobacteria, it is likely that other genes on the similar-size fragments in these strains are colinear and that the size differences between homologous fragments from different species are due to indels. The size differences among strains for homologous fragments 2, 3, 4, and 5 are up to 50 kb for each set of homologous fragments, suggesting that numerous gene islands are present in the E. carotovora subsp. carotovora population. So far, none of these gene islands have been identified, and their functions remain unknown.
As expected, the E. carotovora subsp. carotovora purH gene was on the smallest I-CeuI fragment, as it is in related members of the Enterobacteriaceae. In contrast to the fragments that were between 100 and 450 kb long, the size of the smallest fragment varied little among E. carotovora subsp. carotovora strains and among other enterobacteria. The genes on this fragment are well conserved among diverse enterobacterial species; thus, in addition to purH, we would expect to find several other conserved genes and we would not expect to find large indels on this fragment in E. carotovora subsp. carotovora.
The outH, pelB, and hrp and hrc genes were on the same I-CeuI fragment in all strains examined, while the location of the rsmA gene was different in different strains. The mobB and murI genes and the rsmA and mdh genes were not on the same fragment in any E. carotovora subsp. carotovora strain, unlike the situation in related members of the Enterobacteriaceae, suggesting that at least one chromosome rearrangement occurred after divergence of the last common ancestor of E. carotovora subsp. carotovora and other Enterobacteriaceae (Table 5).
This analysis also revealed additional genomic rearrangements among E. carotovora subsp. carotovora strains. For example, hemG and mobB are not on the same I-CeuI fragment in WPP17 and WPP19, unlike in all other E. carotovora subsp. carotovora strains examined. Also, the murI gene is on the fourth largest I-CeuI fragment of WPP1 and on the fifth largest fragment of WPP14, which could be due to either a rearrangement or indels.
Because the I-CeuI fragment sizes and DNA hybridization results for WPP19 were different from the fragment sizes and hybridization results for other strains, several I-CeuI fragments of WPP19 were end labeled and used as probes to determine which fragments in the other E. carotovora subsp. carotovora strains were homologous. As shown in Fig. 5, fragment 3 from WPP19, which contained rsmA, was similar to fragment 2 of other E. carotovora subsp. carotovora strains, which also contained rsmA. Fragment 4 from WPP19, which contained murI, hybridized to fragment 3 in WPP14, which contained hemG, as well as to fragment 5 in WPP14, which contained murI. WPP19 fragment 5 hybridized to both fragment 3 and fragment 4 of WPP1 and WPP14. Together, these results suggest that multiple rearrangements have occurred in WPP19 compared with the other E. carotovora subsp. carotovora strains. Once an E. carotovora subsp. carotovora genome sequence becomes available and it is possible to design gene microarrays, similar I-CeuI fragment probes could be used to map numerous genes in diverse strains and better pinpoint where chromosomal rearrangements have occurred and which genes may have been lost during these rearrangements in strains with low virulence, such as WPP19 and WPP17.
The significant changes in the lengths of the I-CeuI fragments and the different locations of housekeeping genes mapped to WPP19 and WPP17 I-CeuI fragments are likely to be due to chromosomal recombination (Table 5) and are similar to the insertion sequence element-associated rearrangements which Wei et al. (43) observed in their comparison of the E. coli and Shigella flexneri genomes. To our knowledge, intraspecies variation in the number of rrn operons in Enterobacteriaceae has only been reported for Yersinia and was demonstrated to be a result of recombination between large tandem repeats rather than recombination between rRNA operons (11). Other events, including inversions between rrn operons, are also possible in E. carotovora subsp. carotovora but would not be detected by our approach, unless the orientation of each rrn operon and the location of oriC and the termination of replication (TER) in multiple strains were obtained.
Curiously, although fragments carrying homologous genes varied in length, the total sizes of fragments 1 to 6 were nearly identical in all E. carotovora subsp. carotovora strains examined, including WPP17 and WPP19, suggesting that there is a limit on genome size and that when one gene island is inserted into one of these six fragments, another gene island must be deleted. If both the E. carotovora subsp. carotovora origin (oriC) and the TER, which are generally directly opposite each other in enterobacterial genomes, are on fragment 7, this could explain the apparent total-size constraint on fragments 1 to 6. The yieP gene, a conserved gene that borders an rrn operon, and oriC are on the fourth largest I-CeuI fragment in E. coli and Salmonella enterica. In contrast, the yieP gene is on the largest fragment in all of the E. carotovora subsp. carotovora strains. Our data suggest that single chromosomal rearrangement may have placed both yieP and oriC on the largest I-CeuI fragment in E. carotovora subsp. carotovora, and thus, both oriC and TER may be on the same I-CeuI fragment.
Synthesis of genetic and phenotypic data.
This study was the first attempt to examine genomic diversity in E. carotovora. Several of our strains were isolated from the same region in the same season and from the same host species, yet we found considerable genomic diversity. Our results suggest that the genomes of two different E. carotovora subsp. carotovora potato isolates from the same field may differ more than the genomes of E. coli K-12 and E. coli O157:H7 differ. We originally hypothesized that we would find little genomic variation since the host plants from which we isolated these strains, potatoes, are clonally propagated and widely planted in 2-year rotations in this region. Thus, compared to many other host-pathogen interactions, there is little pressure for strain variation from the host, and one might expect to find a clonal pathogen population well adapted to this host species. Since we obtained strains with significant genomic differences from single fields and even from single infected plants, it is likely that the pressure for E. carotovora subsp. carotovora genome diversification is from other parts of its life cycle. If this is true, then the gene islands present in the different E. carotovora subsp. carotovora strains may play a larger role in other parts of the E. carotovora subsp. carotovora life cycle than they play in virulence in potato, and to understand their functions, we must learn more about the effect of E. carotovora subsp. carotovora genes on how and where E. carotovora subsp. carotovora survives in the environment.
The virulence of the two strains with unusual genomes, WPP17 and WPP19, for potato was significantly less than the virulence of the other strains, but we do not know if these two strains are less fit in other environmental niches. One of these two strains, WPP17, appeared to be missing important factors for association with host plants. This strain does not appear to have genes required for a functional TTSS, which contributes to bacterial growth in host plants for many gram-negative bacterial plant pathogens (3). WPP17 is also unable to oxidize cellobiose in Microlog assays. Cellobiose is part of the cellulose degradation pathway (18); thus, WPP17 should not be able to access as much carbon released from plant cell wall degradation during infection as other E. carotovora subsp. carotovora strains access. The presumed loss of the TTSS and cellobiose degradation may partially account for the relatively low virulence of this strain. However, a previously described E. carotovora subsp. carotovora TTSS mutant showed only a slight reduction in virulence (31), and a large reduction in virulence would not be expected for a cellobiose-deficient mutant, suggesting that other virulence genes are also missing or not expressed in WPP17.
The absence of hrp and hrc genes and cellobiose degradation in WPP17 is probably due to gene loss during intrachromosomal recombination since preliminary phylogenetic trees constructed with E. carotovora subsp. carotovora housekeeping genes showed that WPP17 is in the same clade as several virulent E. carotovora subsp. carotovora strains (data not shown). It seems likely that multiple recombination events and deletions occurred in WPP17 since the region containing mobB, which is on fragment 3 in the majority of the strains examined, is on WPP17 fragment 5 and several hrp and hrc (TTSS) genes have been deleted from fragment 7. In contrast, the missing virulence functions in WPP19, which is pectolytic, able to elicit the HR, and able to oxidize cellobiose, are unknown.
The results presented here provide a basis for comparing the rrn genomic skeletons of multiple closely related genomes and enhance information about the correlations of chromosomal structures with E. carotovora subsp. carotovora diversity and pathogenicity. Elucidation of the physical map and gene locations for multiple E. carotovora subsp. carotovora strains is also a step forward in terms of comparative genome analysis among the Enterobacteriaceae and should aid in E. carotovora subsp. carotovora genome sequencing projects. Our observations should serve as a guide to determine which E. carotovora subsp. carotovora strains are most representative of E. carotovora subsp. carotovora in general by providing a baseline with which to compare the genome structure of E. carotovora subsp. carotovora with the genome structures of other E. carotovora subspecies, by showing which strains have large plasmids, and by providing information on how many rrn loci to expect.
Acknowledgments
This work was supported by Hatch grant 4605, by the U.S. Department of Agriculture (Agricultural Research Service CRIS project 5325-42000-040-00D), and by the Department of Plant Pathology and the University of Wisconsin—Madison College of Agricultural and Life Sciences.
We thank Robert Goodman and Jo Handelsman for kindly allowing us to use their equipment and facilities, Nicole Perna for help with interpretation of data, and Caitilyn Allen for reviewing the manuscript. We are grateful to Andy Witherell for assistance with laboratory techniques and to Tzu-Pi Huang, Paul Rabadeaux, and Michael Hibbing for performing some of the DNA sequencing and strain characterization.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Avrova, A. O., L. J. Hyman, R. L. Toth, and I. K. Toth. 2002. Application of amplified fragment length polymorphism fingerprinting for taxonomy and identification of the soft rot bacteria Erwinia carotovora and Erwinia chrysanthemi. Appl. Environ. Microbiol. 68:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, D. W., A. J. Bogdanove, S. V. Beer, and A. Collmer. 1994. Erwinia chrysanthemi hrp genes and their involvement in soft rot pathogenesis and elicitation of the hypersensitive response. Mol. Plant-Microbe Interact. 7:573-581. [DOI] [PubMed] [Google Scholar]
- 4.Boyd, E. F., K. Nelson, F. S. Wang, T. S. Whittam, and R. K. Selander. 1994. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. 91:1280-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collmer, A., C. Schoedel, D. L. Roeder, J. L. Ried, and J. F. Rissler. 1985. Molecular cloning in Escherichia coli of Erwinia chrysanthemi genes encoding multiple forms of pectate lyase. J. Bacteriol. 161:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cother, E. J., A. B. Blakeney, and S. J. Lamb. 1980. Laboratory-scale preparation of sodium polypectate for use in selective media for pectolytic Erwinia spp. Plant Dis. 64:1086-1087. [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Boer, S. H., R. J. Copeman, and H. Vruggink. 1979. Serogroups of Erwinia carotovora potato strains determined with diffusible somatic antigens. Phytopathology 69:316-319. [Google Scholar]
- 10.De Boer, S. H., and M. Sasser. 1986. Differentiation of Erwinia carotovora ssp. carotovora and E. carotovora ssp. atroseptica on the basis of cellular fatty acid composition. Can. J. Microbiol. 32:796-800. [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett, 3rd, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 13.Gardan, L., C. Gouy, R. Christen, and R. Samson. 2003. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int. J. Syst. Evol. Microbiol. 53:381-391. [DOI] [PubMed] [Google Scholar]
- 14.Ginocchio, C. C., K. Rahn, R. C. Clarke, and J. E. Galan. 1997. Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect. Immun. 65:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gudmestad, N. C., G. A. Secor, P. Nolte, and M. L. Straley. 1984. Erwinia carotovora as a stalk rot pathogen of sunflower in North Dakota [Helianthus annuus]. Plant Dis. 68:189-192. [Google Scholar]
- 16.Hauben, L., E. R. B. Moore, L. Vauterin, M. Steenackers, J. Mergaert, L. Verdonck, and J. Swings. 1998. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 21:384-397. [DOI] [PubMed] [Google Scholar]
- 17.Helm, R. A., and S. Maloy. 2001. Rapid approach to determine rrn arrangement in Salmonella serovars. Appl. Environ. Microbiol. 67:3295-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram, L. O., and J. B. Doran. 1995. Conversion of cellulosic materials to ethanol. FEMS Microbiol. Rev. 16:235-241. [Google Scholar]
- 19.Jabuonski, R. E., A. Takatsu, and F. J. B. Reifschneider. 1986. Survey and identification of Erwinia spp. of different host plants and regions of Brazil. Fitopatol. Bras. 11:185-195. [Google Scholar]
- 20.Jock, S., W. S. Kim, M. A. Barny, and K. Geider. 2003. Molecular characterization of natural Erwinia pyrifoliae strains deficient in hypersensitive response. Appl. Environ. Microbiol. 69:679-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S. L., and K. E. Sanderson. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J. Bacteriol. 177:3355-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, S. L., A. B. Schryvers, K. E. Sanderson, and R. N. Johnston. 1999. Bacterial phylogenetic clusters revealed by genome structure. J. Bacteriol. 181:6747-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lojkowska, E., and A. Kelman. 1994. Comparison of the effectiveness of different methods of screening for bacterial soft rot resistance of potato tubers. Am. Potato J. 71:99-113. [Google Scholar]
- 25.Lopez-Solanilla, E., A. Llama-Palacios, A. Collmer, F. Garcia-Olmedo, and P. Rodriguez-Palenzuela. 2001. Relative effects on virulence of mutations in the sap, pel, and hrp loci of Erwinia chrysanthemi. Mol. Plant-Microbe Interact. 14:386-393. [DOI] [PubMed] [Google Scholar]
- 26.Maher, E. A., S. H. De Boer, and A. Kelman. 1986. Serogroups of Erwinia carotovora involved in systemic infection of potato plants and infestation of progeny tubers. Am. Potato J. 63:1-11. [Google Scholar]
- 27.Marshall, P., and C. Lemieux. 1992. The I-CeuI endonuclease recognizes a sequence of 19 base pairs and preferentially cleaves the coding strand of the Chlamydomonas moewusii chloroplast large subunit rRNA gene. Nucleic Acids Res. 20:6401-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perombelon, M. C. M. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 51:1-12. [Google Scholar]
- 29.Powelson, M. L. 1980. Seasonal incidence and cause of black leg and a stem soft rot of potatoes in Oregon. Am. Potato J. 57:301-306. [Google Scholar]
- 30.Pupo, G. M., D. K. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rantakari, A., O. Virtaharju, S. Vahamiko, S. Taira, E. T. Palva, H. T. Saarilahti, and M. Romantschuk. 2001. Type III secretion contributes to the pathogenesis of the soft-rot pathogen Erwinia carotovora: partial characterization of the hrp gene cluster. Mol. Plant-Microbe Interact. 14:962-968. [DOI] [PubMed] [Google Scholar]
- 32.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 33.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ried, J. L., and A. Collmer. 1987. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Sanderson, K. E., and S. L. Liu. 1998. Chromosomal rearrangements in enteric bacteria. Electrophoresis 19:569-572. [DOI] [PubMed] [Google Scholar]
- 37.Schaad, N. W., J. B. Jones, and W. Chun. 2001. Laboratory guide for identification of plant pathogenic bacteria, 3rd ed. APS Press, The American Phytopathological Society, St. Paul, Minn.
- 38.Seo, S. T., N. Furuya, C. K. Lim, Y. Takanami, and K. Tsuchiya. 2002. Phenotypic and genetic diversity of Erwinia carotovora ssp. carotovora strains from Asia. Phytopathol. Z. 150:120-127. [Google Scholar]
- 39.Shu, S., E. Setianingrum, L. Zhao, Z. Li, H. Xu, Y. Kawamura, and T. Ezaki. 2000. I-CeuI fragment analysis of the Shigella species: evidence for large-scale chromosome rearrangement in S. dysenteriae and S. flexneri. FEMS Microbiol. Lett. 182:93-98. [DOI] [PubMed] [Google Scholar]
- 40.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toth, I. K., A. O. Avrova, and L. J. Hyman. 2001. Rapid identification and differentiation of the soft rot erwinias by 16S-23S intergenic transcribed spacer-PCR and restriction fragment length polymorphism analyses. Appl. Environ. Microbiol. 67:4070-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toth, I. K., K. S. Bell, M. C. Holeva, and P. R. J. Birch. 2003. Soft rot erwiniae: from genes to genomes. Mol. Plant Pathol. 4:17-30. [DOI] [PubMed] [Google Scholar]
- 43.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett, 3rd, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janecky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wimalajeewa, D. L. S. 1976. Studies on bacterial soft rot [Erwinia carotovora carotovora] of celery in Victoria. Aust. J. Exp. Agric. Anim. Husb. 16:915-920. [Google Scholar]
- 45.Xu, G. W., and D. C. Gross. 1986. Field evaluations of the interactions among fluorescent pseudomonads, Erwinia carotovora, and potato yields. Phytopathology 76:423-430. [Google Scholar]
- 46.Yang, C. H., M. Gavilanes-Ruiz, Y. Okinaka, R. Vedel, I. Berthuy, M. Boccara, J. W. Chen, N. T. Perna, and N. T. Keen. 2002. hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant-Microbe Interact. 15:472-480. [DOI] [PubMed] [Google Scholar]