Abstract
In addition to the conventional pmoA gene (pmoA1) encoding the active site polypeptide of particulate methane monooxygenase, a novel pmoA gene copy (pmoA2) is widely distributed among type II methanotrophs (methane-oxidizing bacteria [MOB]) (M. Tchawa Yimga, P. F. Dunfield, P. Ricke, J. Heyer, and W. Liesack, Appl. Environ. Microbiol. 69:5593-5602, 2003). Here we report that the pmoA1 and pmoA2 gene copies in the type II MOB Methylocystis strain SC2 are each part of a complete pmoCAB gene cluster (pmoCAB1, pmoCAB2). A bacterial artificial chromosome (BAC) library of strain SC2 genomic DNA was constructed, and BAC clones carrying either pmoCAB1 or pmoCAB2 were identified. Comparative sequence analysis showed that these two gene clusters exhibit low levels of identity at both the DNA level (67.4 to 70.9%) and the derived protein level (59.3 to 65.6%). In contrast, the secondary structures predicted for PmoCAB1 and PmoCAB2, as well as the derived transmembrane-spanning regions, are nearly identical. This suggests that PmoCAB2 is, like PmoCAB1, a highly hydrophobic, membrane-associated protein. A total of 190 of the 203 amino acid residues representing a highly conserved consensus sequence of the currently known PmoCAB1 and AmoCAB sequence types could be identified in PmoCAB2. The amoCAB gene cluster encodes ammonia monooxygenase and is evolutionarily related to pmoCAB. Analysis of a set of amino acid residues that allowed differentiation between conventional PmoA and AmoA provided further support for the hypothesis that pmoCAB2 encodes a functional equivalent of PmoCAB1. In experiments in which we used 5′ rapid amplification of cDNA ends we identified transcriptional start sites 320 and 177 bp upstream of pmoC1 and pmoC2, respectively. Immediately upstream of the transcriptional start sites of both pmoCAB1 and pmoCAB2, sequence motifs similar to Escherichia coli σ70 promoters were identified.
Methane-oxidizing bacteria (MOB) (methanotrophs) are able to utilize methane (CH4) as a sole source of carbon and energy for growth (15). These bacteria play an important role in the global methane cycle by oxidizing CH4 released by methanogens in freshwater sediments and wetlands and thus mitigate the global warming effect of this greenhouse gas (6, 31). Phylogenies based on 16S rRNA genes show that MOB form distinct lineages in the gamma subclass of the class Proteobacteria (type I MOB) and the alpha subclass of the Proteobacteria (type II MOB) (3, 7, 8, 15, 18, 24). The two types of methanotrophs can be distinguished on the basis of biochemical and ultrastructural features (3, 15, 38).
The first step in CH4 oxidation, the conversion of methane to methanol, is carried out by a methane monooxygenase (MMO). This enzyme exists in two forms, a particulate, membrane-associated form (pMMO) and a soluble form (sMMO). The two forms of the enzyme differ in structure, in kinetic properties, and in the range of substrates which are utilized (23). Only a restricted number of MOB species possess sMMO, while almost all MOB possess pMMO. The only MOB lacking pMMO are Methylocella palustris (8) and Methylocella silvestris (10). In MOB that harbor both forms of MMO, sMMO is synthesized under copper-deficient conditions, while in the presence of even a minuscule amount of available Cu(II) (0.85 to 1.0 μmol/g [dry weight] of cells) only pMMO is synthesized (15, 27).
The pMMO gene cluster consists of three consecutive open reading frames (pmoC, pmoA, and pmoB) in both type I MOB (32, 35) and type II MOB (14). The pmo genes from Methylococcus capsulatus Bath are transcribed into a single 3.3-kb polycistronic mRNA (27). PmoA is presumed to contain the active site because it has been shown to be specifically labeled by [14C]acetylene, a suicide substrate for MMO (30, 40).
The type I MOB Methylococcus capsulatus Bath and Methylomicrobium album BG8 (32, 35), as well as the type II organisms Methylosinus trichosporium OB3b and Methylocystis sp. strain M (14), have been shown to contain duplicate copies of the pmo operon. The sequences of the duplicate pmoCAB gene clusters are nearly identical (e.g., there are 13 differences in 3,183 bp in M. capsulatus Bath).
However, the type II MOB Methylocystis strain SC2 has recently been shown to contain two very different pmoA-like genes (11). A 495-bp fragment of one gene (conventional pmoA, pmoA1) exhibited very high levels of sequence homology to pmoA genes of other type II MOB (encoding PmoA amino acid sequences identical to those of some other Methylocystis strains). The corresponding fragment of the second gene (novel pmoA, pmoA2) exhibited only 73% identity with pmoA1 at the nucleotide level and 68.5% identity (83% similarity) at the deduced amino acid level. Genes closely related to pmoA2 of strain SC2 are widely but not universally present in type II MOB (36). No pmoA2-like sequences were detected in five representative type I MOB tested. Comparative sequence analysis showed that all pmoA2-like sequences formed a coherent cluster that is clearly distinct from pmoA1 sequences of type I and type II MOB and from amoA sequences of the Nitrosomonas-Nitrosospira group. Reverse transcription-PCR provided evidence that pmoA2 was expressed in strain SC2 under standard laboratory growth conditions (36).
Here we show that both pmoA1 and pmoA2 are part of complete pmoCAB gene clusters in Methylocystis strain SC2. Although the deduced amino acid sequences of PmoCAB2 are very different from those of PmoCAB1, the putative secondary structure and regions of transmembrane-spanning helices seem to be highly conserved in the two PmoCAB variants. The biochemical equivalent of PmoCAB1 is the particulate methane hydroxylase (pMH), which is the main component of functionally active pMMO. The pMH complex consists of the following three subunits: α (45 kDa, PmoB1), β (27 kDa, PmoA1), and γ (23 kDa, PmoC1) (26, 40). The three polypeptides associate by noncovalent bonds and form a single complex with a stoichiometry of 1:1:1 (αβγ) (23). Since our data were derived from an analysis of pmo genes, here we mainly refer to PmoCAB1 and PmoCAB2 rather than to pMH.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The conditions used for growth of Methylocystis strain SC2 were adapted from the conditions described by Heyer et al. (18, 19). For extraction of high-molecular-weight (HMW) DNA, cells were grown in liquid cultures in medium 10 containing NaNO3 instead of NH4Cl as the nitrogen source. The cultures were grown for 3 to 5 days at 30°C under a headspace containing 20% (vol/vol) CH4, 5% (vol/vol) CO2, and 75% (vol/vol) air. Cells were pelleted by centrifugation at 6,000 × g for 30 min at 4°C and washed once with washing buffer (WB) (20 mM Tris-HCl, 50 mM EDTA; pH 8.0). The cell biomass was immediately used for extraction of DNA. Cells of Escherichia coli strain DH10B were grown overnight in Luria-Bertani medium in a liquid culture at 37°C.
Preparation of HMW DNA.
For extraction and further treatment of HMW DNA from strain SC2, whole cells were embedded in agarose at a final density of 3 × 109 cells ml−1 by using protocols described previously (12, 34). The embedded cells were treated with lysis buffer (100 mM Tris-HCl, 100 mM EDTA, 1.5 M NaCl, 1% [wt/vol] hexadecyltrimethylammonium bromide, 2% [wt/vol] sodium dodecyl sulfate; pH 8.0) (9) for 20 h at 37°C, rinsed with 25 ml of WB, and incubated in 5 ml of proteinase K reaction buffer (100 mM EDTA, 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg of proteinase K per ml; pH 8.0) at 50°C for 14 h. To remove residual proteins, agar plugs were washed four times for 1 h in WB. The DNA-containing agarose plugs were stored in 0.5 M EDTA (pH 8.0) at 4°C until they were used.
Construction of BAC library.
Cloning of HMW DNA into a bacterial artificial chromosome (BAC) vector (pIndigoBAC-5 Cloning-Ready; Epicentre, Madison, Wis.) required partial digestion with restriction endonuclease HindIII. To obtain the maximum percentage of genomic DNA fragments in the desired size range, a time series of partial restriction digestions (29) was performed for each extract of HMW DNA prior to final mass digestion. To enable subsequent agarase digestion, the center part of a 1% pulse-field agarose gel (Bio-Rad, Madison, Wis.) was replaced by 1% low-melting-point agarose (Bio-Rad). DNA size fractionation was performed in relation to a low-range PFG marker (New England Biolabs, Beverly, Mass.) with a contour-clamped homogeneous electric field apparatus (CHEF-DR III; Bio-Rad) by using three distinct separation steps, as described by Osoegawa et al. (28).
Ethidium bromide-stained gel slices containing both electrophoresed HMW DNA and the PFG marker allowed us to cut slices from the unstained agarose gel containing genomic DNA that was approximately 50 to 150 kb long. HMW DNA was extracted immediately by agarase digestion with β-agarase I as described by the manufacturer (New England Biolabs). To prevent shearing of HMW DNA, subsequent steps were carried out without removing the digested agarose.
Eluted HMW DNA was ligated with 100 ng of pIndigoBAC-5. The molar ratio of insert to vector DNA was approximately 1:10. Ligation was performed in a 100-μl (total volume) mixture with 800 U of T4 ligase (New England Biolabs) for 24 h. The reaction was stopped by heat inactivation at 70°C for 30 min. The ligation mixture was spotted onto the middle of a microdialysis filter (pore size, 0.025 μm; Millipore, Bedford, Mass.) and was desalted by floating the filter on sterile 20% (vol/vol) polyethylene glycol for 3 h. Subsequently, E. coli cells were transformed with desalted DNA (ElectroMax DH10B competent cells; Invitrogen Life Technologies; Carlsbad, Calif.) at 2.5 kV, 25 μF, and 100 Ω in a precooled 1-mm electroporation cuvette. A total of 960 single clones were picked and transferred into 96-well microtiter plates.
PCR-based screening for BAC clones carrying pmo genes.
For identification of clones carrying either pmoA1 or pmoA2, 10-μl aliquots of BAC clone-positive E. coli cells, which were grown in 12 wells (one row) of a 96-well microtiter plate, were combined. An aliquot (20 μl) of each row pool was combined to generate plate pools. Pooled cells were lysed by boiling for 15 min, and the cell debris was subsequently pelleted by centrifugation with a microcentrifuge at 8,000 × g for 5 min. Microtiter plates that contained at least one clone carrying target DNA were identified by PCR-based amplification of pmoA genes by using 1 μl of the supernatant of each plate pool as the template. Row pools of test-positive plates were screened in the same way. Finally, clones of single rows that tested positive were analyzed separately.
Primers A189f and 682b and an annealing temperature of 50°C were used to specifically detect pmoA1, while primers PmoA206f and PmoA703b and an annealing temperature of 66°C were used to screen for pmoA2 (Table 1). Each reaction mixture contained 1 μl of template, 10 μl of 10× reaction buffer, 1.5 mM Mg2+, each deoxynucleoside triphosphate at a concentration of 200 μM, 2.5 U of Taq polymerase (Promega, Madison, Wis.), and each primer at a concentration of 0.25 μM. Amplifications were performed in 100-μl (total volume) mixtures in 0.2-ml reaction tubes by using a DNA thermal cycler (model 2400; PE Applied Biosystems, Weiterstadt, Germany). The thermal profile was as follows: initial denaturation for 3 min at 94°C, followed by 30 cycles consisting of denaturation at 94°C for 40 s, primer annealing for 45 s at the temperatures indicated above, and elongation at 72°C for 75 s. The final elongation step was extended to 7 min. Aliquots of the amplicons were checked by electrophoresis on 1% agarose gels. Amplicons of the expected size were purified and sequenced to verify identification of test-positive clones. To ensure that complete pmo operons were located on the selected clones, the 5′ and 3′ termini of the (cloned) inserts were sequenced by using vector-specific primers. The data obtained were compared with sequences of known pmoB and pmoC genes.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Position | Sequence (5′→3′)a | Direction |
|---|---|---|---|
| Primers for screening | |||
| A189fb | 1486-1504c | GGN GAC TGG GAC TTC TGG | Forward |
| PmoA206fd | 1486-1521e | GGNGACTGGGACTTCTGGATCGACTTCAAGGATCG | Forward |
| 682bd | 2026-2009c | GAA SGC NGA GAA GAA SGC | Reverse |
| PmoA703bb | 2026-1992e | GAASGCNGAGAAGAASGCGGCGACCGGAACGACGT | Reverse |
| RACE primers | |||
| NovC40r | 236-216e | GAT CGT ATC GGT CTC GCC GGC | Reverse |
| NovC100r | 293-273e | GCC GAC GTA GAA GAC CAG CAT | Reverse |
| NovC313r | 511-492e | CAG CCG ATG AGC GTC AGG AT | Reverse |
| ConvC-60r | 269-289c | TGC GCG TGA CCG GTA TCG CTC | Reverse |
| ConvC40r | 372-352c | CAG CTT CCG TGC CAG CAG CAG | Reverse |
| ConvC100r | 445-425c | TCG TAA ATG CGA ACG ACC AGA | Reverse |
| RACE kit primersf | |||
| Oligod(T)Anchored | −38-−1c,e | GAC CAC GCG TAT CGA TGT CGA C(T)16V | Forward |
| Anchor primer | −38-−17c,e | GAC CAC GCG TAT CGA TGT CGA C | Forward |
N = A, C, G, or T; S = C or G; V = A, C, or G.
Data from reference 20.
Primer position in relation to the transcriptional start site of pmoCAB1.
Data from reference 36.
Primer position in relation to the transcriptional start site of pmoCAB2.
Obtained from Roche Diagnostics (Mannheim, Germany).
Two clones that carried either pmoA1 (clone SC2-VII-C1) or pmoA2 (clone SC2-IIX-C7) were selected for further analysis.
DNA sequencing of BAC clones.
The primary structures of the pmoCAB1 (clone SC2-VII-C1) and pmoCAB2 (clone SC2-IIX-C7) gene clusters and their flanking regions were determined by direct sequence analysis of BAC DNA by using a primer walking approach and oligonucleotide primers designed for accurate sequencing. The known partial sequences of pmoA1 and pmoA2 (11) were used as the starting points to formulate novel oligonucleotide primers. BAC DNA was extracted with a QIAGEN Plasmid MAXI kit (Qiagen, Hilden, Germany) used according to the manufacturer's instructions. Each sequencing reaction mixture contained 13 μl (approximately 200 to 800 ng) of BAC DNA, 30 pmol of primer, and 6 μl of BigDye terminator mixture (PE Applied Biosystems). After initial denaturation at 94°C for 3 min, the following profile was repeated 99 times: rapid thermal ramp to 94°C and denaturation for 30 s, primer annealing at 50°C for 20 s, and elongation at 60°C for 4 min. After completion of the reaction, cycle sequencing products were purified with AutoSeq G-50 columns (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). Sequences determined by primer walking were edited manually and assembled by using EditSeq and Seqman II (DNAstar Inc., Madison, Wis.). In addition, the pmoA2-carrying clone SC2-IIX-C7 was fully sequenced by a shotgun cloning approach with a minimal sequence quality of phred 30.
Isolation of total RNA.
Cells of strain SC2 were grown in batch cultures to an optical density at 600 nm of 0.5 to 0.7 (mid-exponential growth phase). Expression of pmo genes was promoted by ensuring that the gas phase contained 10% CH4 approximately 20 h before RNA extraction. Total RNA was stabilized before cell lysis by application of the RNAprotect bacterial reagent (Qiagen). Aliquots containing 1× 109 cells were lysed by mechanical disruption (bead beating at 2,500 rpm for 90 s), and RNA was extracted with an RNEasy mini kit (Qiagen). The manufacturer's protocol was modified slightly. Instead of the recommended on-column DNase digestion, which failed to remove DNA completely, the RNA preparations were treated with RNase-free DNase (RQ1; Promega) for 40 min at 37°C. The concentration of nucleic acids was determined by photometric measurement at 260 nm.
Identification of transcriptional start sites of pmoCAB gene clusters.
cDNA was produced by using the ThermoScript reverse transcription-PCR system (Invitrogen) and a primer (Table 1) with target specificity for either pmoC1 (ConvC100r) or pmoC2 (NovC313r). ConvC100r and NovC313r were selected for analysis from a set of newly designed primers because they exhibited the best performance in cDNA synthesis. Aliquots (0.5 to 1 μg) of DNA-free RNA were used as templates and were transcribed at 56°C for 1 h. The cDNA was purified with a Qiaquick PCR purification kit (Qiagen).
The transcriptional start sites were determined by rapid amplification of cDNA ends (RACE) by using a 5′/3′RACE kit (Roche, Mannheim, Germany) according to the manufacturer's protocol. The oligonucleotide primers used are shown in Table 1. Briefly, a poly(A) tail was synthesized on the 3′ end of the first-strand cDNA by using terminal transferase. The use of an anchored oligo(dT) primer and a primer targeting a region of pmoC (for pmoC1, primer ConvC40r; for pmoC2, primer NovC100r) allowed us to amplify tailed cDNA.
A second PCR was performed with a nested pmoC-targeting primer (for pmoC1, primer ConvC-60r; for pmoC2, primer NovC40r) and an anchored forward primer; a 1:50 dilution of the first PCR product was used as the template. First- and second-round PCR were carried out as described above with an annealing temperature of 61°C. PCR products were purified with a QIAquick PCR purification kit and were sequenced by using the BigDye terminator mixture (PE Applied Biosystems) as described by the manufacturer. The starting point of the poly(A) sequence indicated the 3′ end of the cDNA, which corresponded to the putative transcriptional start site.
Computational analysis.
Homology searches were carried out with standard software tools and the BLAST server of the National Center for Biotechnology Information by using the BLAST algorithm (1) with a nonredundant DNA database, as well as with a nonredundant protein database compiled for this study from SWISSPROT, TREMBL, and PIR.
Putative factor-independent terminators were calculated by using the program TERMINATOR of the Genetics Computer Group software package (4). Similarity and identity values for deduced amino acid sequences were calculated by using the stretcher program (http://bioweb.pasteur.fr/seqanal/interfaces/stretcher.html) based on global pairwise alignments (25) computed with a BLOSSUM65 (17) substitution matrix. Alternatively, similarity and identity values were calculated with the ARB program package (developed by O. Strunk and W. Ludwig of the Technical University of Munich [http://www.arb-home.de]), based on manually edited multiple alignments.
Hydrophobicity plots of PmoCAB1 and PmoCAB2 were calculated by using the comparison tool of the Protein Hydrophilicity/Hydrophobicity Search and Comparison Server (http://bioinformatics.weizmann.ac.il/hydroph/) of the Weizmann Institute of Science, Rehovot, Israel. The locations of putative transmembrane-spanning regions were computed by using the toppred transmembrane topology prediction program (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html).
Nucleotide sequence accession numbers.
The sequences of the pmoCAB gene clusters plus flanking regions from Methylocystis strain SC2 have been deposited in the EMBL, DDBJ, and GenBank databases under accession numbers AJ584611 and BX649604 for pmoCAB1 and pmoCAB2, respectively. The information on the flanking regions of pmoCAB2 includes the annotation results for the fully sequenced 18.3-kb insert of clone SC2-IIX-C7.
RESULTS AND DISCUSSION
Methylocystis strain SC2 BAC clones carrying pmo genes.
Gilbert et al. (14) reported that in accordance with previous studies (32), cloning of pmo operons in multicopy vectors, such as pUC plasmids, is possible only in overlapping fragments because parts of the genes might be toxic to E. coli. Analysis of a pmo operon sequence by cloning of overlapping fragments involves the risk that sequence data obtained originate from different, multiple pmo operons. This problem is negligible if the sequences of multiple pmo operons are virtually identical, as is the case for the duplicate pmo operons of M. trichosporium OB3b (14). However, reconstruction of the primary operon structure from a genome containing multiple pmo operons with divergent sequences could be much more problematic. Thus, in order to avoid cloning of overlapping pmo fragments and to obtain full-length pmo operons on the cloned fragments, we used a single-copy BAC vector to construct a genomic library of strain SC2.
Using a coordinated PCR-based screening approach, we identified six and seven clones carrying pmoA1 and pmoA2, respectively, in a set containing 960 BAC clones. To ensure that complete pmo operons were located on the BAC clones, the sequences of the terminal regions of the inserts were compared to known pmoC and pmoB sequences. One clone that carried pmoCAB1 (clone SC2-VII-C1) and another clone that carried pmoCAB2 (clone SC2-IIX-C7) were selected for further analysis. The complete sequences of the two pmo gene clusters plus their flanking regions were determined by a primer walking strategy. In addition, the complete sequence of the pmoCAB2 cluster was confirmed by applying shotgun sequencing to clone SC2-IIX-C7. Comparison of the sequences determined for the pmoCAB2 operon and flanking regions either by primer walking or by shotgun cloning did not reveal any ambiguous nucleotide positions.
Structural organization of pmoCAB1 and pmoCAB2.
Comparative sequence analysis revealed that pmoA1 was present in a defined pmoCAB gene cluster. This gene arrangement agrees well with the structural organization reported previously for pmo operons of type I MOB (32, 35) and type II MOB (14). In autotrophic nitrifiers, the amo genes are also arranged in the order amoCAB. The amoCAB cluster encodes ammonia monooxygenase (AMO) and is believed to be an evolutionary homolog of pmoCAB (20, 21, 23). pmoA2 also corresponds to a defined pmoCAB gene cluster, but the primary structure of this cluster is clearly distinct from that of pmoCAB1 (see below).
pmoCAB1.
The pmoCAB1 genes have putative lengths of 771, 759, and 1,263 bp, respectively. The intergenic sequence regions cover 276 bp (pmoC1-pmoA1) and 161 bp (pmoA1-pmoB1), respectively. Putative Shine-Dalgarno sequence motifs very similar to the E. coli Shine-Dalgarno consensus sequence (5′-TAAGGAG-3′) (33) could be identified in the upstream region of each of the three pmo1 genes.
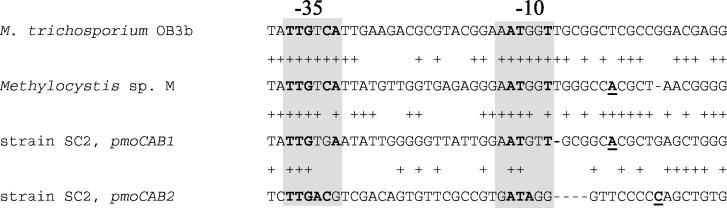
By using a 5′ RACE approach, the putative transcriptional start site was determined to be located 320 bp upstream of pmoC1 (designated position A1). Upstream of position A1, putative σ70 promoter-like sequence motifs were identified. PCR amplification of cDNA by using primer ConvC100r and a primer targeting a region directly upstream or downstream of the predicted start site confirmed the data obtained by 5′ RACE. Only primers targeting the cDNA downstream of position A1 resulted in PCR products (data not shown). The putative −35 and −10 hexamer promoter motifs agreed with the corresponding E. coli consensus sequences at four and three nucleotide positions, respectively (16). The putative pmo1 promoter in strain SC2 also exhibited high levels of similarity in both the −35 and −10 regions to the promoter sequences predicted for the pmo operons of M. trichosporium OB3b and Methylocystis sp. strain M (14) (five identical nucleotide sequence positions for both the −35 and −10 regions) (Fig. 1). Detection of putative σ70 pmo promoters in the latter two organisms led to the conclusion that pmo operons are recognized by σ70 and are negatively regulated under low-copper conditions (14).
FIG. 1.
Sequence alignment of experimentally determined (Methylocystis sp. strain M, pmoCAB1 and pmoCAB2 of Methylocystis strain SC2) and computationally predicted (M. trichosporium OB3b) promoter regions of pmo operons. The alignment was computed by using a ClustalW algorithm. The shading indicates the −35 and −10 sequence motifs. Nucleotides identical to the corresponding nucleotides of the E. coli σ70 promoter consensus sequences (−35 sequence, TTGACA; −10 sequence, TATAAT) are indicated by boldface type, and the putative transcriptional start sites (positions A1 and C1) are underlined.
A factor-independent terminator was identified 121 bp downstream of pmoB1. In addition, a factor-independent terminator was predicted to occur 172 bp upstream of position A1, probably terminating an operon that is localized upstream of pmoCAB1 (the sequence of an acid phosphatase from Mesorhizobium loti exhibited the best E value in a tBLASTx analysis [4e-34]).
pmoCAB2.
The pmoCAB2 genes have putative lengths of 777, 771, and 1,290 bp, respectively. They are separated by two intergenic sequence regions that are 229 bp (pmoC2-pmoA2) and 95 bp (pmoA2-pmoB2) long. As in the pmo1 operon, putative Shine-Dalgarno sequences were identified upstream of the start codons of all three pmo2 genes.
The putative transcriptional start site of pmoCAB2 was determined by 5′ RACE to be located 177 bp (at a position designated C1) upstream of pmoC2. As in the pmo1 operon, PCR amplification of pmo2 cDNA with a primer set consisting of NovC100r and a primer targeting the promoter region directly upstream or downstream of position C1 confirmed that C1 was the putative start site (data not shown). However, it should be mentioned that when cDNA synthesis was carried out at 37°C instead of 56°C, partial polyadenylation occurred 40 bp downstream of position C1. This may indicate that there is an interruption of cDNA synthesis due to mRNA secondary structure elements.
The promoter sequences predicted for pmoCAB2 matched the −35 and −10 hexamer sequence motifs of the E. coli σ70 consensus sequence at five and three nucleotide positions, respectively. The comparison of the sequence motifs predicted for pmoCAB1 and pmoCAB2 revealed a lower degree of identity (Fig. 1).
A factor-independent terminator was identified 75 bp downstream of pmoB2. In addition, a putative factor-independent terminator was identified 368 bp upstream of position C1. This indicated that the 3′ end of an unidentified operon (characterized by ORPHEUS [13] as a hypothetical protein) is localized upstream of the pmo2 operon.
Comparative molecular analysis of pmoCAB1 and pmoCAB2.
(i) Identity and similarity values. In silico translation of pmoCAB1 resulted in derived amino acid sequences with 256 residues (PmoC1), 252 residues (PmoA1), and 420 residues (PmoB1). The corresponding data for pmoCAB2 were 258 residues (PmoC2), 256 residues (PmoA2), and 429 residues (PmoB2). Despite the fact that N-terminal amino acids were predicted to constitute leader sequences (26), all derived amino acids were included in a further comparative analysis. The pmo operons determined for M. trichosporium OB3b and Methylocystis sp. strain M (14), as well as one of the two nearly identical pmo operons of the type I MOB M. capsulatus Bath (32, 35), were used for comparison (Table 2). Comparative analysis of the amino acid sequences deduced from pmoC and pmoB revealed that PmoC1 and PmoC2, as well as PmoB1 and PmoB2, have identity and similarity values which are in the same range as the values for partial PmoA1 and PmoA2 sequences (11). The levels of identity between polypeptides predicted for PmoCAB2 of strain SC2 and polypeptides predicted for the conventional PmoCAB proteins of type II MOB ranged from 59.3 to 65.6%, while the levels of similarity were in the range from 74.7 to 80.1%. Comparison of PmoCAB2 with PmoCAB of the type I MOB M. capsulatus Bath resulted in moderately lower levels of identity and similarity for PmoC and PmoA and clearly lower values for PmoB.
TABLE 2.
Comparison of pmo genes at the DNA and amino acids levels
| Organisms |
pmoC
|
pmoA
|
pmoB
|
|||
|---|---|---|---|---|---|---|
| % DNA identity | % Amino acid similarity (identity) | % DNA identity | % Amino acid similarity (identity) | % DNA identity | % Amino acid similarity (identity) | |
| Strain SC2 pmo2-strain SC2 pmo1 | 69.0 | 74.8 (59.3) | 70.9 | 78.9 (65.6) | 67.4 | 75.4 (63.5) |
| Strain SC2 pmo2-Methylocystis sp. strain M | 71.7 | 77.1 (64.0) | 71.1 | 79.7 (65.2) | 67.0 | 74.7 (61.4) |
| Strain SC2 pmo2-M. trichosporium OB3b | 71.6 | 77.1 (63.6) | 70.7 | 80.1 (64.5) | 69.2 | 75.8 (64.1) |
| Strain SC2 pmo2-M. capsulatus Bath | 67.5 | 71.6 (57.6) | 65.5 | 74.6 (57.0) | 60.5 | 67.1 (46.9) |
| Strain SC2 pmo1-Methylocystis sp. strain M | 83.9 | 91.8 (85.9) | 91.7 | 97.6 (95.2) | 86.1 | 94.5 (89.5) |
| Strain SC2 pmo1-M. trichosporium OB3b | 82.8 | 91.0 (80.5) | 85.2 | 94.0 (87.3) | 78.8 | 88.9 (81.2) |
| Strain SC2 pmo1-M. capsulatus Bath | 68.8 | 72.9 (59.2) | 66.6 | 76.2 (58.3) | 59.2 | 61.2 (45.6) |
| Methylocystis sp. strain M-M. trichosporium OB3b | 85.5 | 93.0 (85.9) | 84.2 | 94.4 (86.5) | 80.5 | 86.8 (78.7) |
| Methylocystis sp. strain M-M. capsulatus Bath | 78.7 | 74.4 (60.3) | 66.4 | 75.4 (57.5) | 61.2 | 60.9 (45.2) |
| M. trichosporium OB3b-M. capsulatus Bath | 69.2 | 73.3 (69.2) | 67.4 | 76.2 (57.9) | 60.6 | 61.8 (44.0) |
(ii) Putative secondary structures.
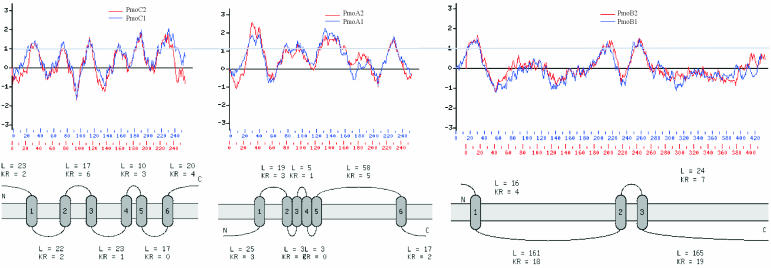
The polypeptides predicted for both PmoCAB1 and PmoCAB2 are highly hydrophobic and contain several transmembrane-spanning regions (Fig. 2 and 3). All N-terminal sequences were predicted to be located in the cytosol. Comparison with putative transmembrane helices of PmoCAB subunits from M. trichosporium OB3b, Methylocystis sp. strain M, and M. capsulatus Bath revealed very high levels of structural similarity. The high degree of structural conservation of PmoCAB2 suggests that it clearly resembles PmoCAB1 in terms of its physicochemical properties. This allowed us to conclude that, like pmoCAB1, pmoCAB2 encodes a membrane-associated protein (Fig. 3). As described by Gilbert and coworkers (14), the first PmoB transmembrane-spanning region (PmoB [helix 1]) (Fig. 3) has to be regarded hypothetically only, because the N-terminal region of PmoB is considered to constitute a leader sequence (26, 37, 40).
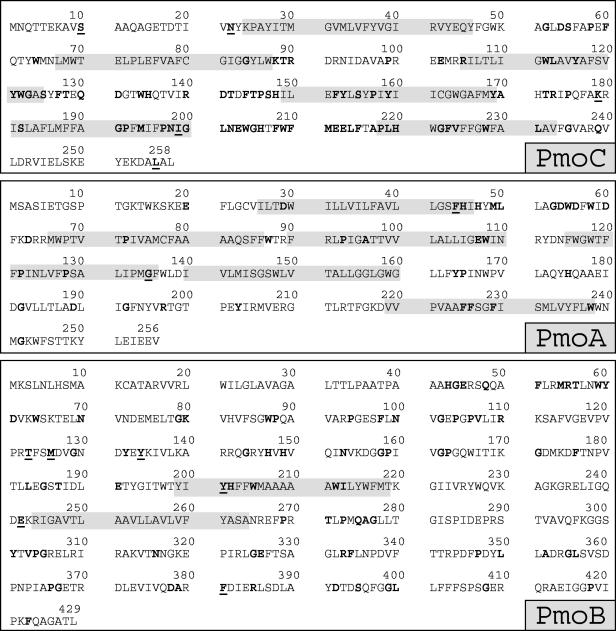
FIG. 2.
Primary structures of derived PmoCAB2 expressed in one-letter code. Predicted transmembrane helices are indicated by shading. Residues that are highly conserved in PmoCAB1 and AmoCAB (consensus sequence) are indicated by boldface type (37). Amino acids that are located at conserved positions but differ from the amino acids in the consensus sequence determined by Tukhavatullin et al. (37) are underlined. The N-terminal helix of PmoB, as predicted by toppred and shown in Fig. 3, is not indicated because the residues are thought to constitute a leader sequence (26, 37).
FIG. 3.
Predicted topologies of derived PmoCAB1 and PmoCAB2 from Methylocystis strain SC2. (Top) Hydrophobicity plot of PmoCAB1 versus PmoCAB2. The y axis indicates the relative hydrophobicity value at a given position in PmoCAB. (Bottom) Schematic presentation of predicted transmembrane helices of PmoCAB2. N-terminal sequences are predicted to be located in the cytosol. Exact locations of transmembrane-spanning regions are shown in Fig. 2.
(iii) Highly conserved amino acid residues.
Recently, Tukhvatullin et al. (37) described a structural and functional model for pMH from M. capsulatus Bath. This model was predicted based on amino acid residues found to be almost completely conserved in a data set consisting of 3 sequence types for PmoB1 and 12 sequence types for AmoB, 112 sequence types for PmoA1 and 349 sequence types for AmoA, and 5 sequence types for PmoC1 and 5 sequence types for AmoC. Because of the high degree of conservation, these amino acid positions are believed to include various functionally important residues. Of the 85 residues determined by Tukhvatullin et al. (37) to be highly conserved in PmoB1 and AmoB (α-peptide), 79 were also identified in PmoB2. Likewise, PmoA2 (β-peptide) and PmoC2 (γ-peptide) possessed 34 of 36 and 77 of 82 highly conserved residues, respectively.
In total, the PmoCAB2 sequence had 13 deviations from the consensus sequence. PmoCAB1 of strain SC2 differed at only two positions (P86→ A97 and I249→ L254 of PmoC1; the numbering of the consensus sequence is that of Tukhvatullin et al. [37], who did not consider putative leader sequences). However, it should be noted that of the 13 residues at which PmoCAB2 differed from the PmoCAB1-AmoCAB consensus sequence, only a single residue is thought to have a different chemical property than the corresponding residue of the consensus sequence (R180 of PmoC2, compared with the consensus residue G167 in PmoC1 of M. capsulatus Bath). Each of the other 12 residues belonged to the same amino acid similarity group (as defined empirically by the BLOSUM65 matrix [17]) as its counterpart in the consensus sequence. Thus, the similarity value for the consensus sequence of PmoCAB1 and AmoCAB and the corresponding positions in PmoCAB2 (>99%) provides support for the assumption that pmoCAB2 encodes a pMMO-like (or AMO-like) enzyme.
While the structure of pMMO and its content of metal centers is still under discussion (2, 5, 22), it is widely accepted that pMMO requires copper as a cofactor for both functional activity and structural integrity (2, 5). Tukhvatullin et al. (37) analyzed the set of highly conserved amino acid residues described above to examine their possible roles in the formation of complexes with metal ions. This analysis took into account the physicochemical characteristics of particular amino acid residues, as well as experimental data for pMMO and pMH obtained by electron spin resonance, electron nuclear double resonance, Mössbauer, and atomic absorption spectroscopy. The analysis aimed to elucidate elements of pMMO-AMO structure and to reveal amino acid residues which are involved in the formation of the active sites. Thirty-nine residues which have the potential to act as ligands were identified. Thirty-eight of these residues could also be identified in PmoCAB2. The only exception, Y26 (PmoA1 and AmoA), is replaced in PmoA2 by F (PmoA2 in Fig. 2 at position F44). Y26 is thought to be involved in formation of the catalytic center only in a subset of the structural-functional models predicted. However, in the majority of suggested models, Y26 is replaced as a ligand by other residues. Thus, the replacement of tyrosine by phenylalanine does not necessarily have an impact on the functional properties of PmoA2.
Among the PmoCAB1 and AmoCAB sequences, the largest data set is available for PmoA1 and AmoA, which include the active sites of the enzymes. To define the putative cellular function of PmoCAB2 more precisely, we identified in 315 PmoA1 and 919 AmoA sequences a set of 18 amino acid positions that can be used to differentiate between conventional pMMO and AMO (Table 3). Analysis of these signature positions in 40 PmoA2 sequences clearly showed that PmoA2 agrees with PmoA1 at all but one position.
TABLE 3.
Amino acid residues enabling differentiation of AmoA, PmoA1, and PmoA2
| Positiona | Residue in:
|
||
|---|---|---|---|
| AmoAb | PmoA1 | PmoA2 | |
| 142 | Mc | 1 | L |
| 145 | T | s | S |
| 147 | n (d)d | S (A) | S (A) |
| 157e | (a) | (i) | (I) |
| 171 | F (F) | I (I) | l (i) |
| 172 | G | a | A |
| 179 | V (I) | e (d) | E (D) |
| 185 | L | m | l |
| 190 | (f) | (i) | (I) |
| 204 | v | i | I |
| 206 | l | m | M |
| 207 | I | v | v (I) |
| 209 | (d) | (h) | (H) |
| 211 | S | t | T |
| 217 | G | k | K |
| 218 | H (H) | d (d) | D (D) |
| 219 | T (A) | v (i) | V (I) |
| 221 | V (I) | p (a) | P (A) |
The positions are the positions in PmoA2, as shown in Fig. 2.
Identification of signature residues indicative of either AmoA or PmoA was based on 919 AmoA sequences, 315 PmoA1 sequences, and 40 PmoA2 sequences.
Amino acid residues are indicated by the one-letter codes. Uppercase letters indicate residues which are conserved in at least 95% of the reference data set, while lowercase letters indicate residues which are conserved in at least 80% of the sequences compared.
The letters in parentheses represent amino acid similarity groups, as follows: A, PAGST; D, QNEDBZ; H, HKR; I, LIVM; and F, FYW.
A highly conserved amino acid residue could not be identified for positions 157, 190, and 209. However, the majority of residues identified for each individual data set at these positions belonged to the same amino acid similarity group.
Taken together, the analysis of a set of amino acid residues that are highly conserved among all pMMO and AMO and of a set that can be used to differentiate between the two enzymes clearly suggests that pmoCAB2 encodes a functional equivalent to conventional pMMO. Calculation of the ratio of nonsynonymous nucleotide substitutions to synonymous nucleotide substitutions in a phylogeny (36, 39) suggested that in recent evolutionary history pmoA2 was subject to strong purifying selection and therefore has an important cellular function. However, the analyses also suggested that pmoA2 may have been subject to diversifying selection forces acting at particular times or on particular codons. The elevated number of deviations in the PmoCAB2 sequence from the consensus sequence of PmoCAB1 and AmoCAB supports the hypothesis that there have been diversifying selection forces and may suggest that the novel pMMO-like enzyme has a different functional role in strain SC2 than the conventional pMMO has. To obtain further evidence for this hypothesis, we are currently assessing in cultivation studies the response of the level of pmoCAB2 expression to various environmental conditions (e.g., different concentrations of methane and competitive substrates) and trying to generate mutants defective in either pmoCAB1 or pmoCAB2. The pmoCAB2 operon is expressed under standard laboratory growth conditions (36; this study), but its level of expression is clearly lower than that of pmoCAB1 (36). This observation may be a further indication that the novel enzyme has a different functional role than the conventional pMMO has.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB 395 to W. Liesack) and by the Competence Network Göttingen project Genome Research on Bacteria (GenoMik) financed by the German Federal Ministry of Education and Research (BMBF).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Basu, P., B. Katterle, K. K. Andersson, and H. Dalton. 2003. The membrane-associated form of methane mono-oxygenase from Methylococcus capsulatus (Bath) is a copper/iron protein. Biochem. J. 369:417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman, J. P., L. Jimenez, I. Rosario, T. C. Hazen, and G. S. Sayler. 1993. Characterization of the methanotrophic bacterial community present in a trichloroethylene-contaminated subsurface groundwater site. Appl. Environ. Microbiol. 59:2380-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brendel, V., and E. N. Trifonov. 1984. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 12:4411-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, D. W., R. C. Kunz, E. S. Boyd, J. D. Semrau, W. E. Antholine, J. I. Han, J. A. Zahn, J. M. Boyd, A. M. de la Mora, and A. A. DiSpirito. 2003. The membrane-associated methane monooxygenase (pMMO) and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol. 185:5755-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedysh, S. N., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, W. Liesack, and J. M. Tiedje. 2002. Methylocapsa acidiphila gen. nov., sp. nov., a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bogs. Int. J. Syst. Evol. Microbiol. 52:251-261. [DOI] [PubMed] [Google Scholar]
- 8.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 9.Dedysh, S. N., N. S. Panikov, W. Liesack, R. Großkopf, J. Zhou, and J. M. Tiedje. 1998. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science 282:281-284. [DOI] [PubMed] [Google Scholar]
- 10.Dunfield, P. F., V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, and S. N. Dedysh. 2003. Methylocella silvestris sp. nov., a novel methanotrophic bacterium isolated from an acidic forest Cambisol. Int. J. Syst. Evol. Microbiol. 53:1231-1239. [DOI] [PubMed] [Google Scholar]
- 11.Dunfield, P. F., M. Tchawa Yimga, S. N. Dedysh, U. Berger, W. Liesack, and J. Heyer. 2002. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol. Ecol. 41:17-26. [DOI] [PubMed] [Google Scholar]
- 12.Flanagan, J. L., L. Ventra, and A. S. Weiss. 1989. Rapid method for preparation and cleavage of bacterial DNA for pulsed-field gel electrophoresis. Nucleic Acids Res. 17:814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frishman, D., A. Mironov, H. W. Mewes, and M. Gelfand. 1998. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 26:2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert, B., I. R. McDonald, R. Finch, G. P. Stafford, A. K. Nielsen, and J. C. Murrell. 2000. Molecular analysis of the pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl. Environ. Microbiol. 66:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyer, J., V. F. Galchenko, and P. F. Dunfield. 2002. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology 148:2831-2846. [DOI] [PubMed] [Google Scholar]
- 19.Heyer, J., Y. R. Malashenko, U. Berger, and E. N. Budkova. 1984. Verbreitung methanotropher Bakterien. Z. Allg. Mikrobiol. 24:725-744. [Google Scholar]
- 20.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 21.Klotz, M. G., J. Alzerreca, and J. M. Norton. 1997. A gene encoding a membrane protein exists upstream of the amoA/amoB genes in ammonia oxidizing bacteria: a third member of the amo operon? FEMS Microbiol. Lett. 150:65-73. [DOI] [PubMed] [Google Scholar]
- 22.Miyaji, A., T. Kamachi, and I. Okura. 2002. Improvement of the purification method for retaining the activity of the particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biotechnol. Lett. 24:1883-1887. [Google Scholar]
- 23.Murrell, J. C., B. Gilbert, and I. R. McDonald. 2000. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 173:325-332. [DOI] [PubMed] [Google Scholar]
- 24.Murrell, J. C., and S. Radajewski. 2000. Cultivation-independent techniques for studying methanotroph ecology. Res. Microbiol. 151:807-814. [DOI] [PubMed] [Google Scholar]
- 25.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear space. Comput. Applic. Biosci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, H. H., S. J. Elliott, J. H. Yip, and S. I. Chan. 1998. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. Isolation and characterization. J. Biol. Chem. 273:7957-7966. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen, A. K., K. Gerdes, and J. C. Murrell. 1997. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol. 25:399-409. [DOI] [PubMed] [Google Scholar]
- 28.Osoegawa, K., P. Y. Woon, B. Zhao, E. Frengen, M. Tateno, J. J. Catanese, and P. J. de Jong. 1998. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Peterson, D. G., J. P. Tomkins, D. A. Frisch, R. A. Wing, and A. H. Paterson. 2000. Journal of Agricultural Genomics, vol. 5. Construction of plant bacterial artificial chromosome (BAC) libraries: an illustrated guide. CABI Publishing, Cambridge, Mass.
- 30.Prior, S. D., and H. Dalton. 1985. The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath). J. Gen. Microbiol. 131:155-163. [Google Scholar]
- 31.Reeburgh, W. S., S. C. Whalen, and M. J. Alperin. 1993. The role of methylotrophy in the global methane budget, p. 1-14. In J. C. Murrell and D. P. Kelly (ed.), Microbial growth on C1 compounds. Intercept Ltd., Andover, United Kingdom.
- 32.Semrau, J. D., A. Chistoserdov, J. Lebron, A. Costello, J. Davagnino, E. Kenna, A. J. Holmes, R. Finch, J. C. Murrell, and M. E. Lidstrom. 1995. Particulate methane monooxygenase genes in methanotrophs. J. Bacteriol. 177:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to non-sense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobral, B. W., and A. G. Atherly. 1989. A rapid and cost-effective method for preparing genomic DNA from gram-negative bacteria in agarose plugs for pulsed-field gel electrophoresis. BioTechniques 7:938. [PubMed] [Google Scholar]
- 35.Stolyar, S., A. M. Costello, T. L. Peeples, and M. E. Lidstrom. 1999. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology 145:1235-1244. [DOI] [PubMed] [Google Scholar]
- 36.Tchawa Yimga, M., P. F. Dunfield, P. Ricke, J. Heyer, and W. Liesack. 2003. Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl. Environ. Microbiol. 69:5593-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tukhvatullin, I., A., R. I. Gvozdev, and K. K. Anderson. 2001. Structural and functional model of methane hydroxylase of membrane-bound methane monooxygenase from Methylococcus capsulatus (Bath). Russ. Chem. Bull. 50:1867-1876. [Google Scholar]
- 38.Whittenbury, R., K. C. Phillips, and J. F. Wilkinson. 1970. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 61:205-218. [DOI] [PubMed] [Google Scholar]
- 39.Yang, Z., and J. P. Bielawski. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahn, J. A., and A. A. DiSpirito. 1996. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol. 178:1018-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]