Abstract
Prior to registration of crop protection products (CPPs) their persistence in soil has to be determined under defined conditions. For this purpose, soils are collected in the field and stored for up to 3 months prior to the tests. During storage, stresses like drying may induce changes in microbiological soil characteristics (MSCs) and thus may influence CPP degradation rates. We investigated the influence of soil storage-related stress on the resistance and resilience of different MSCs by assessing the impact of a single severe drying-rewetting cycle and by monitoring recovery from this event for 34 days. The degradation and mineralization of the fungicide metalaxyl-M and the insecticide lufenuron were delayed by factors of 1.5 to 5.4 in the dried and rewetted soil compared to the degradation and mineralization in an undisturbed reference. The microbial biomass, as estimated by direct cell counting and from the soil DNA content, decreased on average by 51 and 24%, respectively. The bulk microbial activities, as determined by measuring substrate-induced respiration and fluorescein diacetate hydrolysis, increased after rewetting and recovered completely within 6 days after reequilibration. The effects on Bacteria, Archaea, and Pseudomonas were investigated by performing PCR amplification of 16S rRNA genes and reverse-transcribed 16S rRNA, followed by restriction fragment length polymorphism (RFLP) and terminal RFLP (T-RFLP) fingerprinting. Statistical analyses of RFLP and T-RFLP profiles indicated that specific groups in the microbial community were sensitive to the stress. In addition, evaluation of rRNA genes and rRNA as markers for monitoring the stress responses of microbial communities revealed overall similar sensitivities. We concluded that various structural and functional MSCs were not resistant to drying-rewetting stress and that resilience depended strongly on the parameter investigated.
Characterizing the stability and fate of crop protection products (CPPs) and their transformation intermediates in soil is an important component of the environmental safety assessment of these compounds (3, 23). These parameters are evaluated in small-scale laboratory studies under different controlled conditions by using defined natural soils (6, 36). For such CPP degradation studies, the use of fresh field soils is preferred over the use of stored soils (27). However, soil sampling in the field cannot be performed throughout the year due to seasonal or climatic conditions (e.g., there may be extensive drought periods in the summer or frozen or snow-covered soil during the winter). Therefore, soil storage under defined conditions is necessary, and data describing the impact of storage on microbial soil characteristics (MSCs) must be provided (39, 47). For maintenance of the biological CPP degradation potential of soils, indigenous microbial populations must be preserved. However, after removal of soils from the field, the microbial community structures and functions were shown to change when the soils were stored frozen (39; J. Schulze-Aurich and M. Lehmann, Int. Symp. Phys. Chem. Ecol. Seasonally Frozen Soils, p. 499-506, 1997) or were stored at the ambient temperature in a greenhouse (3). Soil moisture content is considered a critical factor for preserving MSCs. Drying-rewetting cycles may be a stress event that occurs in soils when they are stored in a greenhouse without regular watering and tight moisture control. Soil physical parameters, especially moisture content, also have a strong influence on CPP degradation and mineralization rates (11, 44) and have to be strictly controlled prior to CPP degradation studies (36).
The impact of drying and the impact of drying and then rewetting of soil on microbial communities have been investigated in numerous studies. In most of these investigations, bulk microbiological parameters were analyzed; these parameters included microbial cell number (5, 45), biomass carbon content (18, 35, 49), respiration (2, 34, 41, 45), enzyme activities (55), carbon and nitrogen mineralization (5, 18, 22), and substrate utilization (12, 18). Although useful for general microbiological characterization of soils, analyses at the whole-community level may be not sensitive enough to reveal changes in the structures or activities of small subgroups. Many CPPs are degraded by specialized microorganisms that have been shown to represent only small fractions of the entire microbial community (16).
There have been few studies in which the workers investigated the impact of soil drying and rewetting on CPP degradation in combination with MSCs (47; Schulze-Aurich and Lehmann, Int. Symp. Phys. Chem. Ecol. Seasonally Frozen Soils, 1997). In order to describe the impact of immediate stress and disturbance events on ecosystems, the concepts of resistance and resilience were developed (40). Resistance is defined as the magnitude of change after a system is disturbed (40), and resilience indicates the capacity of a system to recover after disturbance (24, 43). Recovery consists of two components, rate of recovery and degree of recovery. Use of these concepts to investigate the impact of stress on soil microbial communities may result in difficulties, since appropriate microbiological characteristics have to be identified. Different microbial parameters may also display various degrees of resistance and resilience (43), and magnitudes of resistance can only be determined on a relative scale (2).
Recently developed genetic fingerprinting techniques make it possible to investigate the impact of soil perturbations on microbial community structure (29, 48). Due to its high resolution power, terminal restriction fragment length polymorphism (T-RFLP) of 16S rRNA genes was successfully used to describe changes in microbial community structure in soil environments which had high levels of microbial species diversity (14, 31). In addition to analyses of rRNA genes, reverse transcription (RT) and amplification of rRNA may be used to investigate metabolically more active microbial groups (13, 17, 26). Genetic fingerprints derived from rRNA can be directly compared to fingerprints obtained from rRNA gene analyses, facilitating identification of active components of the microbial community.
In the present study we investigated the impact of a single severe drying-rewetting cycle on MSCs and the rates of degradation of two CPPs in soil. Our goals were to identify suitable microbiological markers for monitoring changes that may occur during soil storage and to describe the resistance and resilience of MSCs during a 34-day reequilibration period after the dried soil was rewetted. Bulk microbial biomass and activities were determined in order to analyze the impact of the stress event at the community level. To resolve the structure of microbial communities and specific groups, restriction fragment length polymorphism (RFLP) and T-RFLP fingerprinting analyses of 16S rRNA genes were performed. Genetic fingerprints of reverse-transcribed 16S rRNA were used to measure active components of bacterial communities and specific groups.
MATERIALS AND METHODS
Soil sampling and preequilibration.
The soil used for the dry stress experiment was Gartenacker, a silt loam grassland soil (39) that was harvested from a field site near Les Barges, Valais, Switzerland, in April 2001. This soil is routinely used for CPP degradation tests at Syngenta Crop Protection AG. Approximately 200 kg of soil (depth, 0 to 20 cm) was removed with a spade and put into open boxes for transport to the laboratory. The soil was passed through a 2-mm-pore-size sieve, and the moisture content was adjusted to 40% of the maximum water-holding capacity (WHC). The fresh soil was distributed into 300-ml Erlenmeyer flasks (for CPP degradation analyses) and into 1-liter bottles (for other analyses); the flasks and bottles contained 75 and 500 g (dry weight) of soil, respectively. The soil was preequilibrated in a climate room for 1 week under controlled conditions (darkness, 20°C). During the entire experiment a constant flow of water-saturated air was maintained in the bottles, and the soil moisture content was maintained at 40% of the WHC.
Application of the drying-rewetting stress.
After preequilibration in the climate chamber, a portion of the soil (test soil) was transferred from the bottles and Erlenmeyer flasks to a clean metal tray (200 by 100 cm), mixed, and spread in a 3- to 4-cm-thick layer. To induce drying stress, the soil was air dried for 14 days in the climate chamber in the dark at 20°C. Once per day the soil was carefully turned to ensure homogeneous drying to a minimum water content of 1% of the WHC. The remaining portion of the equilibrated soil was left undisturbed in the bottles and Erlenmeyer flasks and served as a reference during the experiment. After 14 days under drying stress conditions, the water content of the test soil was slowly readjusted to the original water content (40% of the WHC) by using sterile deionized water, which was dispersed with a garden water sprayer equipped with a uniform-distribution delivery head. The soil was carefully mixed and divided again among Erlenmeyer flasks and 1-liter bottles.
Sampling schedule.
The first samples for all analyses were obtained following the initial preequilibration phase before drying of the test soil, and this time was defined as day 0. Additional samples were obtained at the end of the drying phase (day 14) and 2, 6, and 34 days (days 16, 20, and 48) after rewetting of the test soil. The CPP metalaxyl-M was applied to reference and test soil samples on days 0, 20, and 48 and on days 20 and 48, respectively. The CPP lufenuron was applied on day 48 to reference and test soils. The experimental design allowed sampling of independent duplicate flasks (CPP degradation) and triplicate flasks (all other analyses).
CPP degradation.
The CPP degradation experiment was carried out in an open gas flow system according to Organisation for Economic Co-operation and Development guidelines (36). The system consisted of six interconnected flasks with a continuous flow of water-saturated air. At the head position, a gas-washing bottle saturated the incoming air with water. Then two 300-ml Erlenmeyer flasks (at the second and third positions) each contained 75 g (dry weight) of soil, and these flasks were followed by one empty flask (at the fourth position) and two flasks filled with 45 ml of 2 N NaOH (at fifth and sixth positions) to retain CO2 resulting from mineralization of the CPPs.
CPP application was performed by adding [phenyl-U-14C]metalaxyl-M (specific activity, 2.08 MBq/mg) or [dichlorophenyl-U-14C]lufenuron (specific activity, 1.90 MBq/mg) to the soil at a final concentration of 0.3 mg/kg (dry weight) of soil. This concentration corresponded to application of 0.2 kg ha−1.
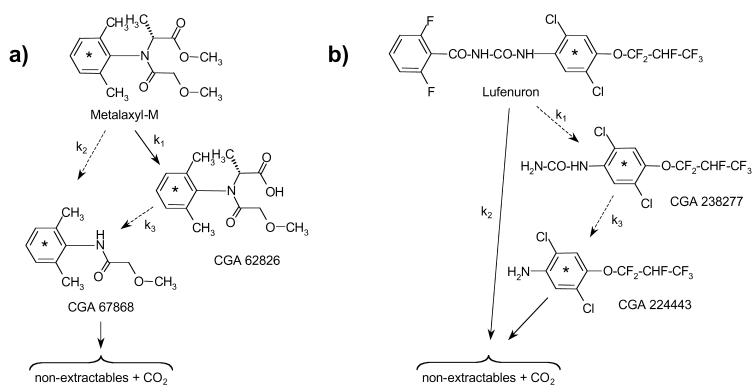
One-milliliter aliquots of the solution in each CO2 collection flask were transferred to scintillation vials, and 14CO2 was quantified by liquid scintillation counting (LSC) (Packard 2500 TR; Packard, Meriden, Conn.). To extract the parent compounds and metabolites, soils were transferred to 500-ml glass centrifuge tubes and extracted by horizontal shaking for 1.5 h at room temperature with 200 ml of acetonitrile-water (4/1, vol/vol). The suspensions were centrifuged at 2,000 × g for 20 min, and each supernatant was collected in a 500-ml flask. This extraction procedure was repeated three times, and the supernatants were pooled. One-milliliter aliquots of the pooled supernatants were used to quantify total extractable 14C by LSC. The pooled extracts were subsequently concentrated with a vacuum rotary evaporator at 30 to 40°C, and 100 μl was subjected to high-performance liquid chromatography analysis (Shimadzu LC-10 AD series instrument; Shimadzu Corporation, Kyoto, Japan) by using both a 14C detector (Berthold Radiomonitor beta detector; Berthold AG, Wildbad, Germany) and a UV detector (254 nm; Shimadzu SPD 6A). Two acetonitrile-water gradients, first a 15/85 (vol/vol) gradient and then a 50/50 (vol/vol) gradient, were used to separate the metabolites of metalaxyl-M and lufenuron, respectively. High-performance liquid chromatography was performed at a rate of 1.0 ml min−1 by using a C18-Nucleosil column (particle size, 5 μm; 250 by 4.6 mm; Macherey Nagel AG, Oensingen, Switzerland). For identification of compounds, the following four analytical reference standards were used: CGA 62826, CGA 67868, CGA 238277, and CGA 224443 (Fig. 1).
FIG. 1.
Chemical structures and initial degradation steps of the CPPs metalaxyl-M (a) and lufenuron (b). CGA 62826, CGA 67868, CGA 238277, and CGA 224443 are the major metabolites of metalaxyl-M and lufenuron monitored in this study. The degradation end products were nonextractables and CO2. The sixfold 14C-labeled aromatic rings (asterisks) and pseudo-first-order rate constants (k1 to k3) are indicated. Degradation steps observed exclusively in nonsterile soil are indicated by dashed arrows.
The extracted soil was air dried and ground to a fine powder with a planar mill (Fritsch GmbH, Idar-Oberstein, Germany). Bound 14C residues (nonextractables) were combusted in an oxidizing oven (Harvey OX500 biological oxidizer; R. J. Harvey Instrument, Hillsdale, N.J.). 14CO2 was collected in a 15-ml trap containing Scintillation Oxysolve C-400 absorption solution (Zinsser Analytik, Frankfurt, Germany) and was quantified by LSC.
Degradation of the CPPs metalaxyl-M and lufenuron has been investigated previously in detail by using different soils, and the degradation pathways shown in Fig. 1 were consistently observed (Syngenta Crop Protection AG, unpublished data). Metalaxyl-M is degraded in soil by cleavage of the methyl ester group, forming the metabolite CGA 62826 (Fig. 1a). A second metabolite, CGA 67868, is formed either directly from metalaxyl-M or from the metabolite CGA 62826 by N dealkylation. This reaction occurs only in nonsterile soil and is therefore assumed to be exclusively mediated by microorganisms. Significant amounts of other metabolites are not detectable. The end products of metalaxyl-M degradation are nonextractables and CO2. Degradation of lufenuron occurs by cleavage of the urea bond, resulting in metabolite CGA 238277 (Fig. 1b). A second metabolite, CGA 224443, is formed by release of the H2N-CO group. Degradation of lufenuron and formation of the two metabolites are detected only in nonsterile soil, indicating that microorganisms play a key role in the breakdown of this CPP. Significant accumulation of metabolites other than those described above has not been observed. The end products of the degradation processes are nonextractables and CO2.
Statistical analyses of CPP degradation data.
Degradation of parent compounds and metabolites was assumed to follow pseudo-first-order reaction kinetics (46). The degradation rates (k) and half-lives (ln2/k) of metalaxyl-M and lufenuron and the metabolites CGA 62826 and CGA 238277 were calculated with the computer program ModelMaker (Cherwell Scientific Publishing Ltd., Oxford, United Kingdom). Linear correlations between measured data and curves fitted according to the pseudo-first-order model were determined with ModelMaker. Mineralization of [phenyl-U-14C]metalaxyl-M and [dichlorophenyl-U-14C]lufenuron was assumed to follow zero-order reaction kinetics (46). Mineralization rates were calculated from linear regression of 14CO2 evolution curves by using Excel 98 (Microsoft Corporation, Redmond, Wash.).
Quantitative microbiological parameters.
For direct counting, cells were stained with the fluorescent dye 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma, Buchs, Switzerland) by using the protocol of Zarda et al. (54). Substrate-induced respiration (SIR) (4) was measured with the OxiTop OC 110 control system (Wissenschaftlich-Technische Werkstätten GmbH, Weilheim, Germany). The general microbial activity of soil microorganisms was estimated from hydrolysis of fluorescein diacetate (FDA) by using the procedure described by Schnürer and Rosswall (42). Dried soil (day 14) had to be rewetted before SIR and FDA hydrolysis were measured.
DNA and RNA extraction procedure.
DNA extraction was performed by using a bead beating procedure and the protocol of Bürgmann et al. (8). DNA pellets were resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8) at a concentration of 1 ml/g (dry weight) of extracted soil. The DNA concentration was determined by a fluorometric assay with PicoGreen (Molecular Probes, Eugene, Oreg.) as described by Bürgmann et al. (8). The DNA yield was expressed in micrograms of total DNA extracted per gram (dry weight) of soil. RNA extraction was carried out by using the procedure described by Bürgmann et al. (9). RNA pellets were resuspended in 30 μl (final volume) of RNase-free water and were stored at −80°C.
RT of 16S rRNA and PCR amplification.
RT of bacterial and archaeal 16S rRNA was performed by using the protocol of Bürgmann et al. (9) with avian myeloblastosis virus reverse transcriptase (Amersham Biosciences Europe GmbH, Dübendorf, Switzerland). Primer UNI-b-rev (Table 1) was used as a universal primer for RT. Controls without avian myeloblastosis virus reverse transcriptase were included to test for traces of DNA in the RNA extracts.
TABLE 1.
PCR primers and PCR conditions used
| Target group | PCR primer | Position in 16S rRNAa | Primer sequence (5′ → 3′)b | PCR conditions
|
Reference | |
|---|---|---|---|---|---|---|
| No. of cycles | Annealing temp (°C) | |||||
| Bacteria | 27F-(FAM) | 8-27 | AGAGTTTGATCMTGGCTCAG | 35 | 48 | 31 |
| 1378R | 1378-1401 | CGGTGTGTACAAGGCCCGGGAACG | ||||
| Archaea | Arch915for | 915-934 | AGGAATTGGCGGGGGAGCAC | 35 | 65 | 1 |
| UNI-b-rev | 1390-1407 | GACGGGCGGTGTGTRCAA | 7 | |||
| Pseudomonas | Ps-for | 292-311 | GGTCTGAGAGGATGATCAGT | 40 | 65 | 52 |
| Ps-rev | 1263-1280 | TTAGCTCCACCTCGCGGC | ||||
Escherichia coli numbering.
R = A or G; M = A or C.
Archaea- and Pseudomonas-specific PCRs with 16S rRNA gene fragments were performed in 100-μl mixtures as described by Pesaro and Widmer (38) and Widmer et al. (52), respectively (Table 1). PCR analyses of 16S rRNA genes of members of the bacterial domain were performed by using 50-μl mixtures and primers 27F (labeled with the fluorescent dye 6-carboxyfluorescein at the 5′ position) and 1378R (Microsynth, Balgach, Switzerland) (Table 1). The reaction mixtures contained 1.5 mM MgCl2, each primer at a concentration of 0.2 μM, each deoxynucleoside triphosphate (Qiagen, Basel, Switzerland) at a concentration of 0.2 mM, 0.6 mg of bovine serum albumin ml−1, 1 U of HotStar DNA polymerase (Qiagen), 1× PCR buffer (Qiagen), and 10 ng of soil DNA. The cycling conditions were as follows: one cycle of 15 min at 95°C to activate the HotStar DNA polymerase, followed by 35 cycles of denaturation at 94°C for 45 s, primer annealing at 48°C for 45 s, and DNA synthesis at 72°C for 2 min. The reaction was completed by a DNA synthesis step consisting of 5 min at 72°C.
RFLP analyses of Archaea and Pseudomonas.
Products obtained from Archaea- and Pseudomonas-specific PCR amplifications were subjected to RFLP analysis by using restriction endonuclease HaeIII as described by Widmer et al. (52). For restriction analysis of Archaea PCR products, 10 μl of a digestion product was separated by electrophoresis in 3% MetaPhor agarose (FMC BioProducts, Rockland, Maine) at 2.5 V cm−1 for 2.5 h and stained with ethidium bromide. Pseudomonas PCR products were resolved on 12% polyacrylamide gels that were electrophoresed for 4 h at 200 V and 35°C (DCode system; Bio-Rad Laboratories, Hercules, Calif.). To ensure high resolution of bands, the volumes that were loaded were less than 5 μl. Staining was performed with 50 ml of Sybr Green (1:5,000 in TAE buffer; Molecular Probes) by shaking the gels for 30 min in the dark.
T-RFLP analyses of Bacteria.
Bacteria-specific amplification products were purified as described by Widmer et al. (52) and were subsequently digested overnight at 37°C in 20-μl restriction digestion mixtures containing 3 U of MspI (Promega, Madison, Wis.) and 1× restriction enzyme buffer B (Promega). For T-RFLP analysis 12 μl of formamide and 0.2 μl of the internal size standard ROX500 (Applied Biosystems, Foster City, Calif.) were distributed in a 96-well plate, and 2 μl of restriction digest was added to each well. The DNA was denatured at 92°C for 2 min and chilled on ice for 5 min. Restriction fragments were separated with a genetic analyzer (ABI3100; Applied Biosystems) equipped with an array of 16 capillaries (length, 36 cm) filled with the POP-4 polymer (Applied Biosystems). The sizes of terminal restriction fragments (TRFs) were determined automatically by comparison to the internal standard by using GenScan 3.1 software (Applied Biosystems). Peak signals were converted into numeric data for fragment size and peak height by using the Genotyper 3.6 NT software (Applied Biosystems). If it was not possible to unambiguously determine the height of a specific peak (e.g., if there was a peak shoulder), the peak was omitted from the analysis for all samples. The peak heights were recorded and compiled in a data matrix for statistical analysis.
Statistical analyses of genetic fingerprinting data.
The intensities of all distinct bands in each RFLP pattern were quantified by using the lane tool and trace quantity option of the Quantity One software (Bio-Rad Laboratories). The band intensities were expressed as percentages of the sum of all of the intensities of the bands detected in a lane. This standardization procedure allowed us to objectively compare the relative band intensities in different lanes and gels. Two-sided t tests for pairwise comparisons of means of band intensities obtained from three independent replicate samples were perfromed by using Excel 98 (Microsoft Corporation). TRF heights were standardized by using the same procedure that was used for RFLP band intensities. In addition, for multivariate analyses, the heights of all TRFs belonging to the same length category were transformed to average 0 and standard deviation 1. This gave each TRF the same relative weight in statistical analyses. The data were then used for hierarchical cluster analysis (CA) based on pairwise Euclidean distances and single-linkage clustering and for principal-component analysis (PCA) with the Statistica v. 6 software package (StatSoft, Tulsa, Okla.). Analysis of variance (Statistica v. 6; StatSoft) was used to test treatment differences, time-treatment interactions, and temporal variations in TRF heights. The evenness of T-RFLP profiles was calculated by determining the Shannon evenness index of TRFs (33).
RESULTS
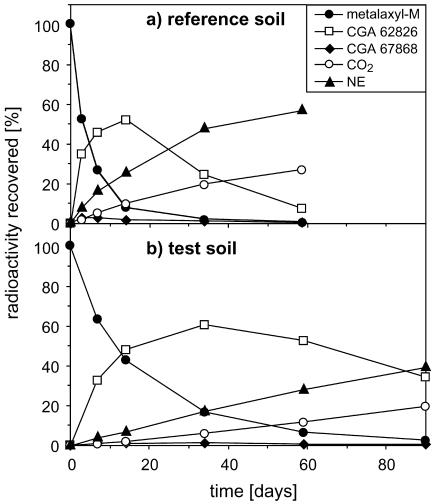
Degradation of metalaxyl-M and lufenuron. In the reference soil, the mean half-lives for metalaxyl-M were 3.2 days for the application at day 0, 3.4 days for the application at day 20, and 3.6 days for the application at day 48 (Table 2). In the test soil metalaxyl-M degradation was delayed (Fig. 2), and the half-lives were 16.6 days for the application at day 20 and 11.9 days for the application at day 48 (Table 2). In general, large amounts of the metalaxyl-M metabolite CGA 62826 were observed (Fig. 2). The mean half-lives of this metabolite were 11.0, 18.4, and 18.1 days for the three applications in the reference soil and 53.3 and 63.0 days for the two applications in the test soil (Table 2). The half-lives obtained in duplicate analyses differed by less than 3% of the larger value. The maximum quantities of the metabolite CGA 67868 detected were between 2.7 and 4.5% of the total radioactivity recovered for the reference soil and between 1.1 and 2.9% of the total radioactivity recovered for the test soil (Fig. 2). On application day 20 the mineralization rate for metalaxyl-M was 0.8% day−1 in the reference soil and 0.2% day−1 in the test soil. On application day 48 the mineralization rate was 0.5% day−1 in the reference soil and 0.2% day−1 in the test soil. Mass balance calculations revealed that between 92.8 and 105% of the total 14C applied (as [phenyl-U-14C]metalaxyl-M) was recovered as 14C-labled parent or 14C-labeled degradation products. The correlation coefficients (R2) of fitted curves for measured data for parent and metabolite degradation were more than 0.98 for all applications.
TABLE 2.
Degradation of the CPPs metalaxyl-M and lufenuron in reference and test soils
| Application day | Compounda | Half-life (days)b
|
|||
|---|---|---|---|---|---|
| Metalaxyl-M
|
Lufenuron
|
||||
| Reference soil | Test soil | Reference soil | Test soil | ||
| 0 | Parent | 3.2 | NDc | ||
| CGA 62826 or CGA 238277 | 11.0 | ND | |||
| 20 | Parent | 3.4 | 16.6 | ND | ND |
| CGA 62826 or CGA 238277 | 18.4 | 53.3 | ND | ND | |
| 48 | Parent | 3.6 | 11.9 | 49.6 | 82.2 |
| CGA 62826 or CGA 238277 | 18.1 | 63 | 20.4 | 30.4 | |
CPP metabolites (CGA 62826 from metalaxyl-M degradation and CGA 238277 from lufenuron degradation) are shown in Fig. 1.
The degradation half-lives (DT50) were calculated by using the following equations: DT50 (parent) = In2/(k1 + k2) and DT50 (metabolite) = In2/k3, where k1, k2, and k3 are pseudo-first-order rate constants for parent compound and metabolite degradation as shown in Fig. 1.
ND, not determined.
FIG. 2.
Degradation of metalaxyl-M applied at day 48 in reference soil (a) and test soil (b). The data indicate the formation and degradation of the metabolites CGA 62826 and CGA 67868, as well as the formation of nonextractables (NE) and 14CO2. The data points are means of duplicate analyses.
The mean half-life of the lufenuron parent compound was 49.6 days for the application in reference soil on day 48, whereas in the test soil the half-life was 82.2 days (Table 2). The calculated mean half-lives of the lufenuron metabolite CGA 238277 were 20.4 days in the reference soil and 30.4 days in the test soil (Table 2). The differences between the half-lives for replicates were less than 10% of the larger value for both the parent and the metabolite. In general, only slow accumulation and decline of the metabolite CGA 224443 was observed. Forty-nine days after CPP application, 16.2 and 15.3% of the recovered radioactivity had accumulated as the metabolite CGA 224443 in the reference and test soils, respectively. After 120 days, this metabolite accounted for 4 and 10.5% of the radioactivity recovered for the reference and test soils, respectively. Mineralization of lufenuron occurred at a rate of 0.09% day−1 in the reference soil, and the rate was threefold lower in the test soil. Mass balance calculations revealed that between 96.4 and 104.7% of the total 14C applied (as [dichlorophenyl-U-14C]lufenuron) was recovered as 14C-labeled parent or 14C-labeled degradation products. The correlation coefficients (R2) of fitted curves for measured data for parent and metabolite degradation were more than 0.98.
Bulk microbiological parameters.
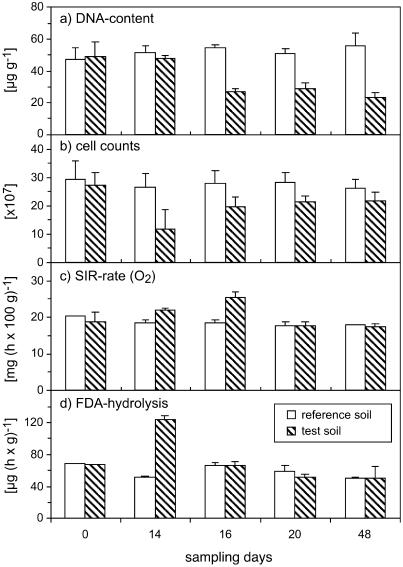
The DNA yields from the test soil were approximately the same at the beginning (day 0) and at the end (day 14) of the drying phase (Fig. 3). After the soil was rewetted on day 14, the DNA content decreased by 43% on day 16 (P < 0.001). Between day 16 and day 48, the DNA persisted at this reduced level and did not recover. The DNA yields for the reference soil, which had remained in the climate room under controlled conditions, showed only small, insignificant fluctuations (P = 0.05). The DNA contents of the reference soil between day 16 and day 48 were significantly higher than those of the corresponding test soil samples (P < 0.001 at day 16; P < 0.01 at day 20; P < 0.05 at day 48).
FIG. 3.
Effects of drying-rewetting stress and reequilibration on soil DNA content (a), direct cell counts (b), SIR rates (c), and FDA hydrolysis (d). The data for reference soil samples and test soil samples are means for three independent replications; the error bars indicate standard deviations.
The direct cell count as determined by DAPI staining was 29.5 × 107 cells/g (dry weight) of soil on day 0 for the reference soil, and the level remained constant throughout the experiment (Fig. 3). On day 14 (the end of the drying phase), the cell count was 56% lower for the dried soil than for the corresponding reference soil. On day 16, the difference between the reference and test soils was 29%, and the differences were 25 and 17% on days 20 and 48, respectively (significant time-treatment interaction; P < 0.05). All differences between reference soil and test soils were highly significant (P < 0.001).
The SIR rates at the end of the drying period (day 14) were 20% greater than the rates for the corresponding reference soil (P < 0.05) (Fig. 3). After rewetting, there was a 38% increase on day 16 (P < 0.001), and this was followed by a decrease to reference soil levels on day 20. The respiration rates of the reference soil decreased gradually by 12% from day 0 until day 48 (P < 0.001) (Fig. 3), but no significant differences between the reference and test soils were detected except for the differences described above.
When hydrolysis of FDA was examined, there were some fluctuations in the reference soil, but no clear trend was identified over the course of the experiment (Fig. 3). The maximum value in the reference soil (68.7 μg/h/g [dry weight] of soil) was observed on day 0, and the minimum value, observed on day 48, was 27% lower. In the test soil, a strong (80%) increase in the FDA hydrolysis rates occurred on day 14 (P < 0.001). After rewetting, the rates decreased again, and no significant differences were detected between the reference and test soils from day 16 until the end of the experiment at day 48.
RFLP analyses of Archaea.
A comparison of the 16S rRNA gene RFLP patterns for the domain Archaea revealed distinct differences between the reference soil and the test soil. These differences were associated with two bands migrating at 360 and 220 bp in the gel (Fig. 4a). The intensity of the band at 360 bp for the dried soil on day 14 was 60% of the intensity on day 0. After rewetting, a further decrease to 41% of the day 0 value was detected on day 16. Until day 48, the intensity of this band remained constant and was significantly lower than that observed for the corresponding reference soil (P < 0.01). The faint band migrating at 220 bp was visible only for the reference soil and for the day 0 test soil samples. The Archaea RFLP pattern obtained after RT-PCR amplification of 16S rRNA revealed changes similar to the changes in the pattern based on a 16S rRNA gene analysis. In the dried soil (day 14), the intensity of the band at 360 bp declined to 54% of the initial value (Fig. 4b). Two days after rewetting (day 16), the intensity markedly decreased, to 23% of the day 0 value. The band intensity then remained constant until day 48 and was significantly lower than the intensities detected in reference samples (P < 0.01). In contrast to the RFLP pattern based on rRNA genes, the band at 220 bp was very faint in RFLP patterns obtained from rRNA analyses and could not be unambiguously quantified.
FIG. 4.
RFLP patterns obtained from 16S rRNA gene and 16S rRNA analyses of the domain Archaea (a and b) and the genus Pseudomonas (c and d) for the reference and test soils. Bands described in the text are indicated by arrowheads. The bands which indicate that there are significant differences between reference and test soils are indicated by solid arrowheads. For clear identification, the fragment sizes (in base pairs) of bands are indicated. The numbers at the bottom indicate sampling days ([d]). The data for only one of three replicates are presented. A 1-kb marker (lane M) was used as a DNA size standard.
RFLP analyses of Pseudomonas.
The RFLP patterns based on 16S rRNA genes of the genus Pseudomonas revealed no statistically significant changes for the replicates throughout the experiment (Fig. 4c). The band patterns derived from 16S rRNA, however, revealed several clear changes in response to drying and rewetting (Fig. 4d). On day 16, the intensity of the band located at 550 bp was 66% of the value at day 0. On day 20 the signal intensity declined further, to 51% of the value at day 0, and remained constant until day 48. In the reference soil, the intensity of this band remained constant throughout the experiment. Differences in the intensities of the band at 550 bp between the reference and test soils were statistically significant (P < 0.05). Additional differences between the reference and test soils were associated with three bands located at 320, 225, and 125 bp. These bands were very faint and not consistently detected in the reference soil, but they became dominant components of the patterns obtained from rewetted soil.
T-RFLP analyses of the domain Bacteria.
Fifty-seven different TRFs were identified in electropherograms obtained from all rRNA gene and rRNA analyses. On average, 43 and 44 TRFs were detected in rRNA gene fingerprints for the reference and rewetted test soils, respectively (Table 3). In the rRNA fingerprints, on average, 36 and 38 TRFs were identified. The numbers of TRFs obtained for reference and test soil samples in the rRNA gene and rRNA analyses were not significantly different.
TABLE 3.
TRF richness and evenness for analysis of the domain Bacteria in reference and test soils
| Day | TRF richness (mean ± SD; n = 3)
|
TRF evenness (mean ± SD; n = 3)
|
||||||
|---|---|---|---|---|---|---|---|---|
| rRNA genes
|
rRNA
|
rRNA genes
|
rRNA
|
|||||
| Reference soil | Test soil | Reference soil | Test soil | Reference soil | Test soil | Reference soil | Test soil | |
| 0 | 43.7 ± 0.6 | 41.3 ± 0.6 | 36.3 ± 0.6 | 36.3 ± 0.6 | 0.922 ± 0.001 | 0.923 ± 0.002 | 0.901 ± 0.001 | 0.901 ± 0.006 |
| 14 | 44.0 ± 0.0 | 43.7 ± 1.5 | 34.7 ± 0.6 | 29.7 ± 0.6 | 0.919 ± 0.000 | 0.918 ± 0.009 | 0.887 ± 0.010 | 0.896 ± 0.001 |
| 16 | 42.0 ± 0.0 | 45.0 ± 0.0 | 36.3 ± 1.2 | 35.0 ± 1.0 | 0.917 ± 0.001 | 0.889 ± 0.004 | 0.883 ± 0.007 | 0.868 ± 0.010 |
| 20 | 44.0 ± 0.0 | 42.7 ± 0.6 | 37.0 ± 0.0 | 38.3 ± 1.2 | 0.918 ± 0.000 | 0.900 ± 0.002 | 0.894 ± 0.007 | 0.866 ± 0.005 |
| 48 | 41.0 ± 1.0 | 43.3 ± 0.6 | 34.3 ± 0.6 | 39.0 ± 1.7 | 0.916 ± 0.005 | 0.906 ± 0.005 | 0.887 ± 0.002 | 0.875 ± 0.012 |
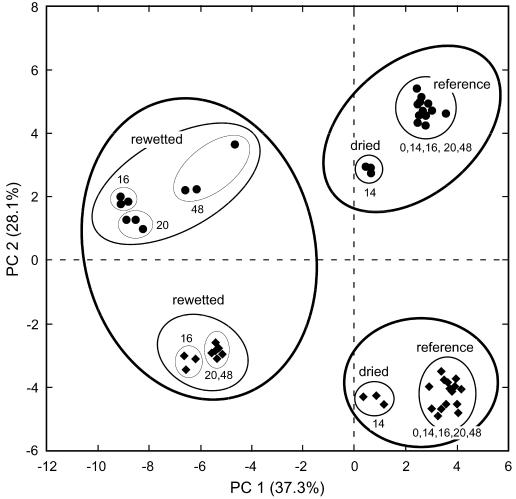
CA (data not shown) revealed that there was distinct clustering of soil treatments (reference, dried, and rewetted) and nucleic acid type (rRNA genes and rRNA) (Fig. 5). The PCA was entirely consistent with the CA and revealed that there were clear separations of reference soil from test soil and the rRNA gene-based analyses from rRNA-based analyses (Fig. 5). The first two principal components (PC 1 and PC 2) accounted for 65.4% of the variance. Soil treatments (reference, dried, and rewetted) were mainly separated on PC 1, while nucleic acid types (rRNA genes and rRNA) were mainly separated on PC 2. The data for reference soil samples from the rRNA gene- and rRNA-based analyses formed very tight clusters, and each cluster contained the data points for the five sampling days. Samples from test soils were clearly separated from reference soil samples. For the rRNA gene analyses of rewetted soil, three distinct subclusters containing the triplicate samples from days 16, 20, and 48 could be distinguished. For the rRNA analyses of rewetted soil, two closely related subclusters resulting from day 16 data and from day 20 and 48 data were detected. The triplicate samples from rRNA gene and rRNA analyses of the dried soil were located between the corresponding reference and rewetted soil samples; however, they were closer to the reference data points. The two reference soil clusters from the rRNA gene and rRNA analyses were clearly farther from each other (mean relative distance on PC 2, 8.95) than the corresponding clusters with the samples from rewetted soil (mean relative distance on PC 2, 4.88) (P < 0.001).
FIG. 5.
PCA of T-RFLP data obtained from 16S rRNA gene (•) and rRNA (♦) analyses of the domain Bacteria for the reference and test soils. Each data point represents a single analysis of an independent sample. The lines around data points indicate the groups that formed clusters in the hierarchical CA. The different types of lines indicate the different hierarchic clustering levels identified, as follows: thick lines, three main clusters at the highest branching level; medium lines, subclusters at the second branching level; thin lines, subclusters at the third branching level. The numbers next to the subclusters indicate the sampling days included. The values in parentheses indicate the percentages of variance accounted for by the first two principal components, PC 1 and PC 2.
Nine TRFs (21%) that were found in all reference soil rRNA gene analyses were not found in all rRNA gene analyses of rewetted soil. On the other hand, 11 TRFs (25%) that were not detected in the reference soil rRNA gene analyses appeared in the rRNA gene analyses of rewetted soil. Twenty-nine TRFs were found in the T-RFLP profiles for both reference and rewetted soils. In the rRNA analyses of rewetted test soil, six TRFs (17%) that were present in the reference soil were not detected, whereas 10 TRFs (27%) that were not present in the reference soil appeared in rRNA analyses of rewetted soil. Twenty-seven TRFs were detected in both reference and rewetted soil samples. The TRF evenness for fingerprints of rRNA gene analyses was on average 0.919 for reference soil and 0.898 for rewetted soil (Table 3) (P < 0.001). The evenness for rRNA-derived fingerprints was 0.892 for reference soil samples and 0.870 for the samples from rewetted soil (P < 0.001).
Analysis of variance revealed that 91% of all TRFs obtained from rRNA gene analyses had significantly different peak heights when the data for the reference and rewetted soil samples were compared (P < 0.05). Analogous calculations for rRNA-based analyses resulted in significantly different heights for 83% of the TRFs (P < 0.05). In addition, time-treatment interactions were examined to assess the temporal changes that PCA revealed after rewetting of the test soil. When sampling days 16, 20, and 48 were considered, the data for 59% of the TRFs revealed significant time-treatment interactions for rRNA gene-based analyses (P < 0.05); the level of significance for 38% of these TRFs was <0.001. For rRNA-based analyses, 40% of the TRFs exhibited significant time-treatment interactions (P < 0.05), and 9% had a significance level of <0.001.
DISCUSSION
Degradation of metalaxyl-M and lufenuron.
Degradation and mineralization of the widely used CPPs metalaxyl-M and lufenuron were used as parameters to evaluate the practical importance of drying-rewetting stress in soil. In addition, these compounds were used as sensors to test specific catabolic processes in soil. The drying and rewetting procedure used resulted in a significant decrease in the CPP degradation potential of the indigenous microbial soil communities (Fig. 2 and Table 2). In agreement with these results, drying and rewetting of an agricultural soil caused a significant reduction in the degradation and mineralization of the insecticide Carbofuran (44). Decomposition of plant material was found to be retarded in soil that had been dried for 7 days and subsequently rewetted (32). In contrast, no effects of soil drying and rewetting on mineralization of atrazine and trichloroacetic acid were observed (D. Rugbjerg and A. Helveg, 6th Danish Plant Prot. Conf. Pestic. Environ., p. 23-31, 1989). These studies may not be directly comparable, since the compounds used for testing microbial degradation activity have different physical and chemical properties and may also be degraded by different microbial groups.
During our experiment, metalaxyl-M was applied to the reference soil on days 0, 20, and 48 and to the test soil on days 20 and 48. The observed half-lives in the reference soil suggested that there was a tendency for decreasing CPP degradation potential over time. In a comparison of the degradation of a thiocarbamate herbicide in a freshly collected soil to the degradation of a thiocarbamate herbicide stored for 90 days at 20°C and 40% of the WHC, Anderson (3) observed a 2.3-fold increase in the half-life of the parent compound in the stored soil. The slowly decreasing CPP degradation potential in the stored soil was attributed to nutrient depletion in the absence of a plant cover and subsequent dormancy of the soil microorganisms (3). Apparently, these effects were less pronounced in the soil used in the present study. The reduced metalaxyl-M half-lives in the test soil on days 20 and 48 indicated that recovery of the CPP degradation potential to reference soil levels did not occur until day 48.
The metalaxyl-M degradation and mineralization rates were three- to fourfold lower in the test soil. For lufenuron the decreases were between 1.5-fold (metabolite degradation) and 3-fold (mineralization). In comparison, the DNA content and the microscopic cell counts decreased only 1.3- and 2-fold, respectively, suggesting that the delay in CPP degradation was not entirely explained by the decrease in microbial biomass. Due to cell death during drying and rewetting, specialized catabolic functions may have been lost, which were not replaced or were only partially replaced by the surviving microorganisms. Alternatively, soil conditions (e.g., nutrient availability) were altered by the stress, and this may have resulted in decreased activities of CPP-degrading organisms.
Bulk microbiological biomass.
The results obtained for DNA content and direct cell counts indicated that there were pronounced average decreases in microbial biomass (51 and 24%, respectively) after soil drying and rewetting (Fig. 3). A similar trend for these two parameters was observed for the same soil (Gartenacker) when it was subjected to freezing and thawing (39). However, the magnitudes of the effects after drying and rewetting exceeded those after freezing and thawing by factors of 2.1 for DNA content and 1.1 for direct cell counts. This indicated that drying and rewetting represented a considerably harsher stress than freezing and thawing. In the soil subjected to drying-rewetting stress, the DNA content remained constant during drying and was 50% of the reference value after rewetting. This change appears to indicate that the DNA of microorganisms that had been lysed during the drying phase was not degraded until the soil was rewetted. This moisture effect is in agreement with the finding that degradation of purified plasmid DNA was strongly delayed in a soil that was at 10% of the WHC compared to the degradation in a soil that was at 40% of the WHC (53). The lowest cell counts were obtained in the dry soil just before rewetting (day 14). This observation may be explained by cell lysis during the drying phase from day 0 to day 14 and possibly also by increased proportions of spores that may have formed as an adaptation to the decreasing water content. Due to decreased permeability for DAPI, spores are less detectable by the cell fixation and staining procedure used (20). Fungal mycelial length and bacterial numbers, volumes, and surface areas were shown to decrease when moist soils were air dried (50), demonstrating that microbial adaptation and damage were induced by a decreasing soil moisture content.
Bulk microbiological activities.
In contrast to CPP degradation activity, the SIR and FDA hydrolysis values were not significantly different from the reference values after 6 and 34 days of reequilibration (Fig. 3). It has been shown that the number of active bacteria can increase after soil drying and rewetting (10). Mamilov and Dilly (34) found that the ratio of SIR to microbial C (determined by the chloroform fumigation-extraction method) increased by a factor of 1.3 after repeated soil drying and rewetting. In our experiment, the activity per cell, as calculated from the ratio of SIR to the number of cells, increased by a factor of 1.2 after 6 and 34 days of reequilibration. In agreement with Mamilov and Dilly (32), we concluded that the microbial fraction that responded to the addition of substrate increased after rewetting. Our data further indicate that changes in specific activities (e.g., CPP degradation) are not necessarily reflected by bulk microbial activities.
Microbiological community structure and activity.
T-RFLP analyses of rRNA genes from the domain Bacteria revealed a clear stepwise difference in the community structure between the reference soil and the dried soil and between the dried soil and the rewetted soil (Fig. 5). The data agree with shifts in community composition that were observed after several drying and rewetting cycles of an oak forest soil (19). When the same procedure was applied to a grassland soil comparable to the soil used in the present study, no changes in bacterial community composition were induced, as determined by T-RFLP analyses (19). It has been hypothesized that the moisture regimen to which a soil is naturally exposed may be responsible for the resistance of microbial communities to drying and rewetting stress (55). The mean annual precipitation in Les Barges (the origin of Gartenacker soil) is approximately 1,000 mm. The microbial communities at this location may be not exposed to severe drying and rewetting events as frequently as the grassland soil used in the study of Fierer et al. is (19), which originated from a region with annual rainfall of only 500 mm. The distinct groups formed by rRNA samples in PCA and CA analyses and the separation from the corresponding rRNA gene samples indicated that there was a significant difference between the pool of rRNA genes and the pool of rRNA present in the soil (Fig. 5). This may reflect not only differences in the extraction and detection methods used but also the presence both inactive and highly active bacterial groups at the same time in the soil (13, 17, 26). The distance between the PCA clusters of rRNA gene and rRNA samples decreased in the following order: reference soil-dried soil-rewetted soil. This may be an indication that bacteria that survived the stress treatment became more uniformly active.
The use of rRNA analyses to characterize active microbial communities is based on the assumption that active or growing cells have increased levels of intracellular rRNA, whereas inactive or dormant cells contain less rRNA. Kerkhof and Kemp (28) found that there were clear, although nonlinear, correlations between 16S rRNA contents and growth rates and also that there were rapid decreases in intracellular rRNA levels after cessation of in vitro growth for various pure cultures of marine proteobacteria. They concluded that under steady-state conditions, species-specific growth rates may be determined by measuring cellular rRNA content. On the other hand, in pure-culture experiments with a marine Vibrio strain, the ribosome contents decreased with a half-life of 79 h after carbon starvation (21). These findings from RNA analyses of pure in vitro cultures indicate that active bacteria contain increased levels of rRNA. However, different rRNA stabilities were observed after cessation of growth. In rhizosphere microbial communities a total RNA turnover rate of 20% day−1 was calculated by using an isotopic tracer approach (37). These results indicate that the total RNA turnover in an undisturbed rhizosphere system may be relatively slow. Therefore, it cannot be assumed that only the active microbial community components are detected when rRNA is analyzed.
Large portions of the TRF fragments detected completely disappeared from the reference soil samples, whereas many TRFs were found exclusively in samples from rewetted soil. This finding may reflect the fact that some bacterial groups were strongly affected by the stress event, while others profited from the changed conditions after drying and rewetting and became active or started to grow.
It may be surprising that the average TRF richness did not change for either the rRNA gene or rRNA analyses after the test soil was rewetted (Table 3). Apparently, the decrease in biomass, as indicated by the DNA contents and the cell counts, was not accompanied by a decrease in the number of bacterial subgroups detected. TRF evenness, however, was reduced after stress for rRNA gene-based analyses, as well as for rRNA-based analyses, suggesting that there was a change in the dominance of certain TRFs (Table 3). However, it has been shown that due to detection biases genetic fingerprints do not accurately represent the entire microbial community richness (15, 30). Therefore, it is important to note that microbial community indices calculated from genetic fingerprints should be interpreted with caution. These indices should be used only for relative comparisons of different samples and not as absolute characteristics of the microbial communities investigated.
Certain groups belonging to the domain Archaea appeared to be particularly sensitive to drying and rewetting, as revealed by the decreasing intensities of specific bands in the rRNA gene and rRNA analyses (Fig. 4). Similar changes in Archaea RFLP fingerprints were observed when soil was frozen and thawed, and these changes were confirmed by gene cloning and independent PCR detection with new specific primers (39). These consistent findings indicated that the affected archaeal groups may represent microorganisms that are not well adapted to rapid environmental changes. This characteristic may make these organisms suitable indicators for monitoring microbiological soil characteristics in stored soils, where changes in MSCs are not desirable. Changes due to drying and rewetting for the genus Pseudomonas were more clearly detected by rRNA analyses. The intensities of three bands were increased in the rewetted soil, suggesting that certain Pseudomonas spp. were better adapted to the conditions after rewetting. The intensity of one band decreased, indicating that organisms represented by this band may have been less successful in coping with the altered conditions in the soil during and after the stress.
Resistance and resilience of microbiological parameters.
All microbiological parameters determined indicated that MSCs of Gartenacker soil were not resistant to drying-rewetting stress. SIR and FDA hydrolysis suggested that there was complete resilience of MSCs within less than 1 week after rewetting. This finding is in agreement with previous observations that bulk microbial activities are able to completely recover after transient stress. Based on measurements of the soil respiratory quotient, Anan'eva et al. (2) observed complete resilience of dried and rewetted soil within 7 days. After heat stress, short-term decomposition of grass shoot residues revealed resilience within 13 days (25). In the present study, direct cell counts recovered partially, while CPP degradation, DNA content, and community structure and activity data indicated that the MSCs were severely altered by the stress and no substantial recovery occurred within 34 days of reequilibration. Westergaard et al. (51) investigated the effects on microbial communities after a single application of the antibiotic tylosin to soil using plating experiments, substrate utilization profiles, and denaturing gradient gel electrophoresis analyses. Similar to our results, molecular biological tools were the most sensitive tools and detected permanent changes in the microbial community structure even after disappearance of the antibiotic. T-RFLP analysis revealed significant time-treatment interactions due to temporal changes in TRF heights in the rewetted soil. This analysis indicated that microbial structures and activities were still changing during the reequilibration phase, and thus a stable state was not reached after 34 days of reequilibration.
Conclusions.
Our results suggested that different MSCs are suitable for monitoring the impact of drying-rewetting stress on soil. The DNA content, in particular, may be used as a reliable and simple parameter to monitor bulk microbial changes. Bacteria, Pseudomonas, and Archaea were comparably sensitive to drying-rewetting stress. T-RFLP analyses of Bacteria suggested that rRNA genes and rRNA reacted equally sensitively to drying-rewetting stress. rRNA analyses revealed greater changes than rRNA gene analyses only for Pseudomonas. We therefore suggest that there should be case-by-case selection of different microbial groups and marker molecules when the effects of different stresses on MSCs are monitored.
Acknowledgments
We thank Sophie Ferbach for excellent technical assistance with the CPP degradation experiments. William V. Sigler is acknowledged for constructive discussions and helpful comments on the manuscript. We are grateful to Doris Hermann for assistance with T-RFLP analyses and to Helmut Bürgmann for stimulating discussions during this work.
Manuel Pesaro was supported by funds from Syngenta Crop Protection AG, Basel, Switzerland.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anan'eva, N. D., E. V. Blagodatskaya, and T. S. Demkina. 2002. Estimating the resistance of soil microbial complexes to natural and anthropogenic impacts. Eurasian Soil Sci. 35:514-521. [Google Scholar]
- 3.Anderson, J. P. E. 1987. Handling and storage of soils for pesticide experiments, p. 45-60. In L. Sommerville and M. P. Greaves (ed.), Pesticide effects on soil microflora. Taylor and Francis, London.
- 4.Anderson, J. P. E., and K. H. Domsch. 1978. A physiological method for the quantitative measurement of microbial biomass in soil. Soil Biol. Biochem. 10:215-221. [Google Scholar]
- 5.Bloem, J., P. C. Deruiter, G. J. Koopman, G. Lebbink, and L. Brussaard. 1992. Microbial numbers and activity in dried and rewetted arable soil under integrated and conventional management. Soil Biol. Biochem. 24:655-665. [Google Scholar]
- 6.Blumhorst, M. R. 1996. Experimental parameters used to study pesticide degradation in soil. Weed Technol. 10:169-173. [Google Scholar]
- 7.Bundt, M., F. Widmer, M. Pesaro, J. Zeyer, and P. Blaser. 2001. Preferential flow paths: biological ′hot spots' in soils. Soil Biol. Biochem. 6:729-738. [Google Scholar]
- 8.Bürgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 9.Bürgmann, H., F. Widmer, W. V. Sigler, and J. Zeyer. 2003. mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil. Appl. Environ. Microbiol. 69:1928-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen, H., M. Hansen, and J. Sorensen. 1999. Counting and size classification of active soil bacteria by fluorescence in situ hybridization with an rRNA oligonucleotide probe. Appl. Environ. Microbiol. 65:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupples, A. M., G. K. Sims, R. P. Hultgren, and S. E. Hart. 2000. Effect of soil conditions on the degradation of cloransulam-methyl. J. Environ. Qual. 29:786-794. [Google Scholar]
- 12.Degens, B. P. 1998. Decreases in microbial functional diversity do not result in corresponding changes in decomposition under different moisture conditions. Soil Biol. Biochem. 30:1989-2000. [Google Scholar]
- 13.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Fantroussi, S. 2000. Enrichment and molecular characterization of a bacterial culture that degrades methoxy-methyl urea herbicides and their aniline derivates. Appl. Environ. Microbiol. 66:5110-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felske, A., A. Wolterink, R. Van Lis, W. M. De Vos, and A. D. L. Akkermans. 2000. Response of a soil bacterial community to grassland succession as monitored by 16S rRNA levels of the predominant ribotypes. Appl. Environ. Microbiol. 66:3998-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fierer, N., and J. P. Schimel. 2002. Effects of drying-rewetting frequency on soil carbon and nitrogen transformations. Soil Biol. Biochem. 34:777-787. [Google Scholar]
- 19.Fierer, N., J. P. Schimel, and P. A. Holden. 2003. Influence of drying-rewetting frequency on soil bacterial community structure. Microb. Ecol. 45:63-71. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, K., D. Hahn, W. Hönerlage, F. Schönholzer, and J. Zeyer. 1995. In situ detection of spores and vegetative cells of Bacillus megaterium in soil by whole cell hybridization. Syst. Appl. Microbiol. 18:265-273. [Google Scholar]
- 21.Flärdh, K., P. S. Cohen, and S. Kjelleberg. 1992. Ribosomes exist in large excess over the apparent demand for protein synthesis during carbon starvation in marine Vibrio sp. strain CCUG 15956. J. Bacteriol. 174:6780-6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franzluebbers, A. J. 1999. Potential C and N mineralization and microbial biomass from intact and increasingly disturbed soils of varying texture. Soil Biol. Biochem. 31:1083-1090. [Google Scholar]
- 23.Gerber, H. R., J. P. E. Anderson, B. Bügel-Mogensen, D. Castle, K. H. Domsch, H.-P. Malkomes, L. Sommerville, D. J. Arnold, H. Van De Werf, R. Verbeken, and J. W. Vonk. 1989. 1989 Revision of recommended laboratory tests for assessing side-effects of pesticides on the soil microflora. Toxicol. Environ. Chem. 30:249-261. [Google Scholar]
- 24.Greenland, D. J., and I. Szabolcs. 1994. Soil resilience and sustainable land use. CAB International, Wallingford, United Kingdom.
- 25.Griffiths, B. S., M. Bonkowski, J. Roy, and K. Ritz. 2001. Functional stability, substrate utilisation and biological indicators of soils following environmental impacts. Appl. Soil Ecol. 16:49-61. [Google Scholar]
- 26.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International Organization for Standardization. 1993. Soil quality—sampling. Part 6: guidance on the collection, handling and storage of soil for the assessment of aerobic microbial processes in the laboratory 10381-6. International Organization for Standardization, Geneva, Switzerland.
- 28.Kerkhof, L., and P. Kemp. 1999. Small ribosomal RNA content in marine proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 29.Kozdrój, J., and J. D. van Elsas. 2001. Structural diversity of microorganisms in chemically perturbed soil assessed by molecular and cytochemical approaches. J. Microbiol. Methods 43:197-212. [DOI] [PubMed] [Google Scholar]
- 30.Lueders, T., and M. W. Friedrich. 2003. Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl. Environ. Microbiol. 69:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukow, T., P. F. Dunfield, and W. Liesack. 2000. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 32:241-247. [DOI] [PubMed] [Google Scholar]
- 32.Magid, J., C. Kjaergaard, A. Gorissen, and P. J. Kuikman. 1999. Drying and rewetting of a loamy sand soil did not increase the turnover of native organic matter, but retarded the decomposition of added 14C-labelled plant material. Soil Biol. Biochem. 31:595-602. [Google Scholar]
- 33.Magurran, A. E. 1988. Ecological diversity and its measurements. Croom Helm Limited, London, United Kingdom.
- 34.Mamilov, A. S., and O. A. Dilly. 2002. Soil microbial eco-physiology as affected by short-term variations in environmental conditions. Soil Biol. Biochem. 34:1283-1290. [Google Scholar]
- 35.Min, S. S., P. C. Brookes, and D. S. Jenkinson. 1987. Soil respiration and the measurement of microbial biomass-C by the fumigation technique in fresh and in air-dried soil. Soil Biol. Biochem. 19:153-158. [Google Scholar]
- 36.Organisation for Economic Co-operation and Development. 2002. OECD guideline for the testing of chemicals: aerobic and anaerobic transformation in soil 307. Organisation for Economic Co-operation and Development, Paris, France.
- 37.Ostle, N., A. S. Whiteley, M. J. Bailey, D. Sleep, P. Ineson, and M. Manefield. 2003. Active microbial RNA turnover in a grassland soil estimated using a 13CO2 spike. Soil Biol. Biochem. 35:877-885. [Google Scholar]
- 38.Pesaro, M., and F. Widmer. 2002. Identification of novel Crenarchaeota and Euryarchaeota clusters associated with different depth layers of a forest soil. FEMS Microbiol. Ecol. 42:89-98. [DOI] [PubMed] [Google Scholar]
- 39.Pesaro, M., F. Widmer, G. Nicollier, and J. Zeyer. 2003. Effects of freeze-thaw stress during soil storage on microbial communities and methidathion degradation. Soil Biol. Biochem. 35:1049-1061. [Google Scholar]
- 40.Pimm, S. L. 1984. The complexity and stability of ecosystems. Nature 307:321-326. [Google Scholar]
- 41.Powlson, D. S., and D. S. Jenkinson. 1976. The effects of biocidal treatments on metabolism in soil. II. Gamma irradiation, autoclaving, air-drying and fumigation. Soil Biol. Biochem. 8:179-188. [Google Scholar]
- 42.Schnürer, J., and T. Rosswall. 1982. Fluorescein diacetate hydrolysis as a measure of total microbial activity in soil and litter. Appl. Environ. Microbiol. 43:1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seybold, C. A., J. E. Herrick, and J. J. Brejda. 1999. Soil resilience: a fundamental component of soil quality. Soil Sci. 164:224-234. [Google Scholar]
- 44.Shelton, D. R., and T. B. Parkin. 1991. Effect of moisture on sorption and biodegradation of carbofuran in soil. J. Agric. Food Chem. 39:2063-2068. [Google Scholar]
- 45.Soulides, D. A., and F. E. Allison. 1961. Effects of drying and freezing soils on carbon dioxide production, available mineral nutrients, aggregation and bacterial population. Soil Sci. 91:291-298. [Google Scholar]
- 46.Spanggord, R. J., G. R. Gordon, R. I. Starr, and D. J. Elias. 1996. Aerobic biodegradation of [14C]3-chloro-P-toluidine hydrochloride in a loam soil. Environ. Toxicol. Chem. 15:1664-1670. [Google Scholar]
- 47.Stenberg, B., M. Johansson, M. Pell, K. Sjöhdahl-Svensson, J. Stenström, and L. Torstensson. 1998. Microbial biomass and activities in soil as affected by frozen and cold storage. Soil Biol. Biochem. 30:393-402. [Google Scholar]
- 48.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Ovreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 49.Van Gestel, M., R. Merckx, and K. Vlassak. 1993. Microbial biomass responses to soil drying and rewetting—the fate of fast-growing and slow-growing microorganisms in soils from different climates. Soil Biol. Biochem. 25:109-123. [Google Scholar]
- 50.West, A. W., G. P. Sparling, and W. D. Grant. 1987. Relationships between mycelial and bacterial populations in stored, air-dried and glucose-amended arable and grassland soils. Soil Biol. Biochem. 19:599-605. [Google Scholar]
- 51.Westergaard, K., A. K. Muller, S. Christensen, J. Bloem, and S. J. Sorensen. 2001. Effects of tylosin as a disturbance on the soil microbial community. Soil Biol. Biochem. 33:2061-2071. [Google Scholar]
- 52.Widmer, F., R. J. Seidler, P. M. Gillevet, L. S. Watrud, and G. D. Di Giovanni. 1998. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl. Environ. Microbiol. 64:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Widmer, F., R. J. Seidler, and L. S. Watrud. 1996. Sensitive detection of transgenic plant marker gene persistence in soil microcosms. Mol. Ecol. 5:603-613. [Google Scholar]
- 54.Zarda, B., D. Hahn, A. Chatzinotas, W. Schönhuber, A. Neef, R. I. Amann, and J. Zeyer. 1997. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch. Microbiol. 168:185-192. [Google Scholar]
- 55.Zelles, L., P. Adrian, Q. Y. Bai, K. Stepper, M. V. Adrian, K. Fischer, A. Maier, and A. Ziegler. 1991. Microbial activity measured in soils stored under different temperature and humidity conditions. Soil Biol. Biochem. 23:955-962. [Google Scholar]