Abstract
A random library of Escherichia coli MG1655 genomic fragments fused to a promoterless green fluorescent protein (GFP) gene was constructed and screened by differential fluorescence induction for promoters that are induced after exposure to a sublethal high hydrostatic pressure stress. This screening yielded three promoters of genes belonging to the heat shock regulon (dnaK, lon, clpPX), suggesting a role for heat shock proteins in protection against, and/or repair of, damage caused by high pressure. Several further observations provide additional support for this hypothesis: (i) the expression of rpoH, encoding the heat shock-specific sigma factor σ32, was also induced by high pressure; (ii) heat shock rendered E. coli significantly more resistant to subsequent high-pressure inactivation, and this heat shock-induced pressure resistance followed the same time course as the induction of heat shock genes; (iii) basal expression levels of GFP from heat shock promoters, and expression of several heat shock proteins as determined by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins extracted from pulse-labeled cells, was increased in three previously isolated pressure-resistant mutants of E. coli compared to wild-type levels.
One of the mechanisms that bacteria have developed to survive in unfavorable conditions is the ability to respond to stress situations. This stress response is mediated by a changed pattern of gene expression that typically results in an increased tolerance of the bacteria for the stress factor that triggered the response and usually also for a number of other stress factors (25, 26, 44). For the last 10 years stress responses to different types of stress have been intensively studied in various bacteria, and the genetic regulation and interaction between different stress regulons is well understood in model organisms such as Escherichia coli (2, 17, 51). On the other hand, the mechanisms of bacterial perception of most stresses remain ill defined.
In the field of food preservation, where stress is applied to inhibit or inactivate food-borne pathogens and spoilage organisms, the development of an adaptive stress tolerance response is highly relevant, particularly in so-called minimally processed and preserved foods, in which inhibition or inactivation relies on the synergistic combination of multiple stresses in low doses (13, 34, 45). The most commonly encountered stresses in foods are high or low temperature, high salt, and low pH, and the stress response to each of these factors has been characterized in some detail for important food-borne pathogens, such as Salmonella spp., E. coli O157:H7, and Listeria monocytogenes (6, 7). However, a number of new techniques for food preservation are emerging that make use of other types of stress. One of the most advanced of these is treatment by high hydrostatic pressure (HHP; 200 to 1,000 MPa) (14, 32), and research on the effects of pressure on the microbial stability and safety of foods has vastly expanded over the last few years (35, 48). At a more fundamental level, several studies have provided information on specific cellular targets of HHP in an attempt to identify the ultimate cause(s) of cellular inactivation. Many in vitro studies with purified proteins and with membrane vesicles have indicated that HHP can cause protein denaturation and affect membrane fluidity, effects which were predicted from thermodynamic principles (3, 18). Most in vivo observations on living cells exposed to HHP can be linked to these two effects: inhibition of key enzymes (43, 49); and inactivation of cellular structures and processes, including transcription (9), ribosomes (39), microtubules (33) and membrane proteins (46), and structural and functional disruption of the cell membrane (11, 42). Remarkable differences exist in the pressure sensitivity among bacterial species and even strains (1, 4), and several groups have reported the isolation of mutants with acquired HHP resistance (12, 16, 27). The bacterial response to high pressure has been studied mainly in deep-sea bacteria (20, 21, 29, 30, 59). However, because these works were conducted with piezotolerant and piezophilic bacteria under conditions that support growth (<100 MPa), it cannot be extrapolated to piezosensitive food-borne organisms exposed to pressure levels that cause inactivation (>100 MPa).
The first information on the cellular response of E. coli towards HHP was provided by Welch et al. (57), who demonstrated the induction of a specific set of proteins during growth of E. coli at pressures up to 100 MPa. Many of the induced proteins were identified as cold shock and heat shock proteins, including several chaperones, thus suggesting protein management as an important activity in cells growing at sublethal pressures. Expression of the lac promoter was also induced in E. coli at 30 MPa (31). Studies on the response of E. coli after brief exposure to an HHP shock at pressures that inhibit growth and protein synthesis have not been conducted to date but could provide insight into the nature of the cellular targets of high pressure and in high-pressure resistance mechanisms in E. coli. This information can be used to optimize HHP pasteurization processes of food. In this study we screened for promoters that are induced and proteins that are increasingly produced in E. coli subsequent to HHP shock and that provide several indications for a role of heat shock proteins in HHP resistance in E. coli.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli strain MG1655 (5, 15) and its pressure-resistant mutants LMM1010, LMM1020, and LMM1030 (16) were used in this study (Table 1). Overnight cultures were obtained by growth in Luria-Bertani (LB) broth (36) for 21 h at 37°C under well-aerated conditions. Ampicillin was added when necessary to a final concentration of 100 μg/ml (Ap100; Applichem, Darmstadt, Germany).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| E. coli | ||
| MG1655 | rph-1 F− lam− | 5, 15 |
| LMM1010 | HHP-resistant derivative of MG1655 | 16 |
| LMM1020 | HHP-resistant derivative of MG1655 | 16 |
| LMM1030 | HHP-resistant derivative of MG1655 | 16 |
| Plasmids | ||
| pFPV25 | Contains promoterless GFP | 53 |
| pAA210 | lon-gfp transcription fusion cloned in pFPV25 | This study |
| pAA211 | clpPX-gfp transcription fusion cloned in pFPV25 | This study |
| pAA212 | dnaK-gfp transcription fusion cloned in pFPV25 | This study |
| pAA213 | rpoH-gfp transcription fusion cloned in pFPV25 | This study |
Construction of a promoter trap library of MG1655.
Fragments of 1 to 3 kb of a partial Sau3AI digest of MG1655 chromosomal DNA were isolated from an agarose gel after electrophoresis with the High Pure PCR Product Purification kit (Roche Diagnostics Belgium, Vilvoorde, Belgium). This sized genomic DNA was ligated in the dephosphorylated BamHI site of pFPV25 (53), upstream of the promoterless gfp gene. After transformation of this ligation mixture to MG1655, approximately 15,000 independent clones were isolated on LB agar Ap100, purified, and stored at −80°C individually in microplate wells as well as in pools of 500 clones in cryo vials. Restriction analysis of ca. 500 random clones revealed less than 1% of clones with no insert.
Screening of the promoter trap library with DFI.
The differential fluorescence induction (DFI) technique has been described earlier by Valdivia and Falkow (53), but it was slightly customized for isolating HHP-induced promoters. Five microliters of a glycerol stock containing a pool of 500 random clones was inoculated in 4 ml of LB Ap100. Stationary-phase cultures of five such pools were combined 1:500 in 4 ml of fresh prewarmed LB Ap100, resulting in a pool of ca. 2,500 clones. This pool was grown further at 37°C until the (OD600) reached 0.6. Cells from 1 ml of this late-exponential culture were subsequently harvested by centrifugation (5 min at 6,000 × g) and resuspended in the same volume of fresh prewarmed LB. This culture was next sealed without air bubbles in a polyethylene bag and was pressurized to 150 MPa for 15 min in an 8-ml pressure vessel maintained at 20°C with an external cooling circuit (Resato, Roden, The Netherlands). After pressurization the culture was recovered and incubated at 37°C with aeration for an additional 4 h to allow green fluorescent protein (GFP) expression. After this expression period, cell sorting was performed by using a FACSCalibur apparatus (Becton Dickinson, Erembodegem, Belgium) fitted with an argon laser emitting at 488 nm. Bacteria were detected by forward scatter, side scatter, and fluorescence with logarithmic amplifiers. The flow rate for sorting bacteria was adjusted to 1,500 events/s. GFP-producing cells, representing ca. 17% of the total cell population, were divided into two fractions according to their fluorescence: the 80% least and the 20% most GFP-containing cells. Of each fraction, ca. 106 cells were sorted and grown overnight in LB Ap100 (positive selection). Subsequently these cultures were diluted 1:100 in fresh LB Ap100 and grown to late exponential phase as described above. The cells were harvested and resuspended in fresh LB as before, but this time the HHP treatment was omitted. After 4 h both cultures were sorted for the fractions of cells expressing less GFP than their induced counterparts (negative selection). The obtained fractions were grown overnight in LB Ap100 and went through positive and negative selection once again for enrichment of HHP-inducible clones. Afterwards cultures were plated out on LB agar Ap100, and 50 to 100 clones were individually checked for HHP induction as described below. In total, ca. 10,000 clones were screened in this fashion. Promoters inducible by HHP were sequenced.
Induction experiments.
Overnight cultures were diluted 1:100 in fresh prewarmed LB Ap100 and were further incubated until late exponential phase (OD600 = 0.6). Portions (4 ml) of this culture were then pelleted by centrifugation (5 min at 6,000 × g) and resuspended in the same volume of fresh prewarmed LB. For heat shock induction, 500 μl of this suspension was placed in a water bath at 50°C for 15 min. For HHP induction, 500 μl of suspension was sealed without air bubbles in a polyethylene bag and pressurized for 15 min in an 8-ml pressure vessel maintained at 20°C. Pressurization caused some adiabatic heating of the sample; however, this heating was less than 3°C at 150 MPa, which was the maximum inducing pressure used in this study. After induction, cultures were maintained at room temperature and used for the measurement of gfp induction or for determination of resistance to HHP inactivation.
GFP fluorescence measurements.
For induction experiments, 300-μl samples were transferred to microplate wells and placed in a Fluoroscan Ascent FL (Thermolabsystems, Brussels, Belgium). For measurement of basal (noninduced) expression, 300-μl cultures were grown overnight in a microplate at 37°C with orbital shaking (200 rpm). These cultures were subsequently diluted 1:100 in 300 μl of fresh prewarmed LB in a new microplate and were placed in the fluorescence reader. Fluorescence at 520 nm was then measured at 60-min intervals with intermittent shaking (every 5 min) at 37°C, using an excitation wavelength of 480 nm. Fluorescence data shown are representative results of at least four repetitions.
Determination of HHP resistance of heat-shocked cells.
At different times after induction, heat-shocked and control cells, the latter obtained from the same culture but without pretreatment at 50°C, were sealed in polyethylene bags and pressurized together at 250 MPa as described above. Adiabatic heating of the samples at 250 MPa was less than 5°C. Serial dilutions were subsequently made from pressurized and nonpressurized samples and were plated on Tryptone Soy Agar (Oxoid, Basingstoke, United Kingdom) with a spiral plater (Spiral Systems Inc., Cincinnati, Ohio). Twenty-four hours later colonies on the plates were counted, and reduction factors (RF) were determined with the following equation: RF = (CFU per milliliter of nonpressurized sample)/(CFU per milliliter of pressurized sample).
Construction of plasmids.
Specific GFP transcriptional fusions were constructed in pFPV25 with the promoter regions of lon, clpPX, dnaK, and rpoH (σ32), which were obtained by PCR (Platinum Pfx DNA polymerase; Invitrogen, Merelbeke, Belgium) by using the following primers: 5′-CAGTGGATCCGTCGGTAATTGATGGTC-3′ and 5′-CAGTTCTAGATTATACGGGGATTTCAATGCG-3′ for lon; 5′-GACTGGATCCCTGTTGAAGCTGTACTGG-3′ and 5′-GACTTCTAGATTAGTTATCTCGTTCGCCGC-3′ for clpPX; 5′-TACGGGATCCCAATTTTACGTCTTGTCCTGC-3′ and 5′-TAGCTCTAGACTTAGCCCATCTAAACGTCTC-3′ for dnaK; and 5′-GATCTCTAGATTATGCCCGGATGTAGGA-3′ and 5′-GTAAGGATCCGCTGCGATTGTCATC-3′ for rpoH. All four PCR products and pFPV25 were cut with BamHI and XbaI to allow directional cloning of the promoters upstream of gfp, resulting in pAA210, pAA211, pAA212, and pAA213, respectively (Table 1). These constructs were subsequently confirmed by sequencing the promoter fragment and the gfp 5′ end and were transformed to MG1655, LMM1010, LMM1020, and LMM1030. All restriction enzymes were purchased from Roche Diagnostics Belgium.
Two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (2D-SDS-PAGE) analyses.
For analysis of protein synthesis, strains were grown aerobically at 37°C in M9 salts (36) supplemented with a solution of 0.04% glucose, 0.01 mM thiamine, 0.2 mM adenine, guanine, cytosine, and uracil bases and an amino acid mix as described by Wanner et al. (56). Overnight cultures in supplemented M9 medium were diluted 20-fold into 5 ml of the same medium, grown to an OD420 of 0.5, and diluted 10-fold into 10 ml of the same medium. These cultures were labeled for 5 min with 50 μCi of [35S]methionine (1,000 Ci/mmol) (Amersham, Uppsala, Sweden)/ml and chased for 3 min with 0.02 M unlabeled methionine at the onset of stationary phase.
Cell extracts were analyzed by two-dimensional polyacrylamide gels by the method of O'Farrell (41), with modifications (54). Protein spots were visualized by autoradiography and were identified by matching their x-y coordinates to the reference gel of the gene-protein database of E. coli (19, 50, 52, 55). Autoradiograms shown from two-dimensional analyses are representative results from two repetitions.
Sequencing.
Inserts in the pFPV25 plasmid were sequenced by MWG Biotech AG (Ebersberg, Germany), with 5′-GACAAGTGTTGGCCATGGAACAGGTAG-3′ in the 5′ region of gfp as the sequencing primer.
RESULTS
HHP induces heat shock genes.
DFI screening of ca. 10,000 clones of a gfp promoter probe library of MG1655, stressed by sublethal pressures, resulted in the isolation of a number of HHP-inducible clones. Sequencing revealed that three of these clones contained a heat shock-related promoter upstream of gfp from the genes encoding the Lon protease, the DnaK chaperone, and the ClpPX protease. The presence of functional heat shock promoters in these clones could be confirmed by heat shock induction of GFP expression.
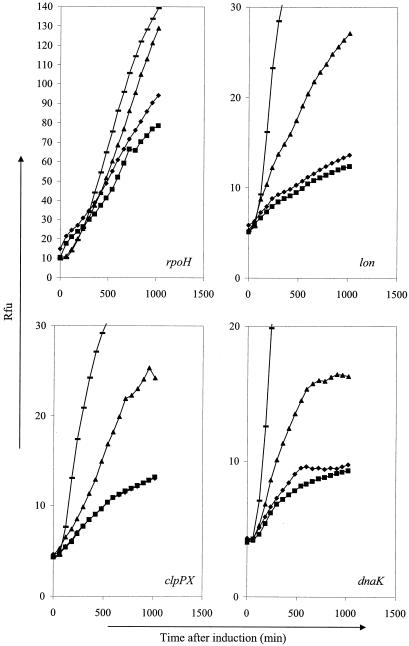
To confirm HHP shock induction of dnaK, lon, and clpPX, three new, precisely defined gfp transcription fusions were constructed with these genes. In addition, because the heat shock regulon is under the control of σ32, an additional gfp fusion was made with the rpoH gene encoding σ32. These constructs were confirmed by sequencing and were inducible by heat shock. Transcription of all four heat shock genes was also stimulated by pressure shock (Fig. 1). The induction of gene expression was dependent on the intensity of the pressure shock, with only a marginal increase when cells were treated at 75 MPa and a more substantial increase at 150 MPa. Pressures of 200 MPa and higher inactivated the majority of cells, thereby precluding measurement of gene induction under these conditions (data not shown).
FIG. 1.
Induction of GFP expression by treatment with 75 MPa (♦), 150 MPa (▴), or 50°C (—) for 15 min. Noninduced control (▪) cells were kept at 20°C during pretreatment. Transcription was assayed from rpoH, lon, clpPX, and dnaK promoter fusions with gfp. Rfu, relative fluorescence units.
Heat shock-induced pressure resistance.
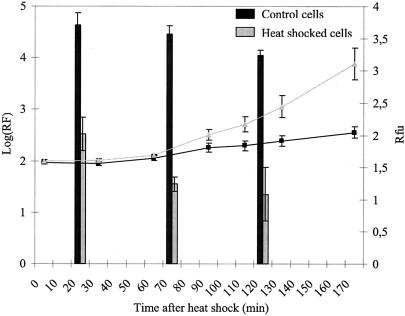
To investigate whether induction of heat shock genes results in increased pressure resistance, wild-type MG1655 cells were subjected to heat shock and subsequently were pressurized. Heat-shocked cells were strongly protected against HHP inactivation, showing hundreds-fold higher survival than non-heat-shocked cells upon pressurization at 250 MPa (Fig. 2). In addition, the increase in dnaK transcription after heat shock treatment paralleled the development of increased HHP resistance. (For Fig. 2 it should be noted that, due to intramolecular cyclization resulting in fluorochrome formation of the GFP protein, fluorescence considerably lags gfp expression. To estimate this lag time under our conditions, an arabinose-inducible promoter was cloned in pFPV25 upstream of gfp, and a 60-min lag time was observed between induction with arabinose and fluorescence detection [data not shown]). Much lower levels of dnaK induction and of HHP resistance occurred when the heat shock treatment was conducted in a buffer in the absence of nutrients (data not shown). Taken together, these results strongly suggest that the protective effect of heat shock on the HHP inactivation of E. coli is due to the induction of heat shock proteins. Conversely, we failed to increase heat or pressure resistance of E. coli by pressure shock (data not shown).
FIG. 2.
HHP reduction factors (250 MPa for 15 min) of heat-shocked and non-heat-shocked cells at different time intervals after pretreatment (bars). dnaK expression was assayed at the same time (line with black squares, control cells; line with gray triangles, heat-shocked cells) by pAA212. Rfu, relative fluorescence units.
HHP-resistant mutants of MG1655 display increased basal expression of heat shock proteins.
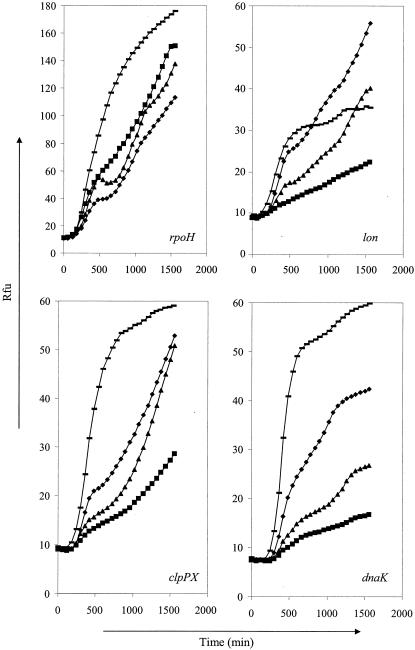
Basal expression levels of lon, clpPX, and dnaK during growth at 37°C in LB broth were strongly elevated in pressure-resistant mutants LMM1010 and LMM1030 and were slightly elevated in pressure-resistant mutant LMM1020 compared to the pressure resistance of parental strain MG1655 (Fig. 3). This difference in GFP expression reflected a true differential expression, because growth curves of the different strains were identical (except for LMM1030, which has a slightly lower cell count in stationary phase) and because it was confirmed by observation of individual cells by fluorescence microscopy (data not shown). Basal transcription of rpoH was higher only in LMM1030. However, because σ32 is subject to multiple posttranscriptional control mechanisms, the expression level of the rpoH gene is a poor indicator of σ32 activity. Nevertheless, the elevated transcription levels of lon, clpPX, and dnaK are suggestive of an increased σ32 activity in all the pressure-resistant strains.
FIG. 3.
Basal expression levels of rpoH, lon, clpPX, and dnaK promoter fusions with gfp in wild-type MG1655 (▪) and its pressure-resistant mutants LMM1010 (♦), LMM1020 (▴), and LMM1030 (—). Rfu, relative fluorescence units.
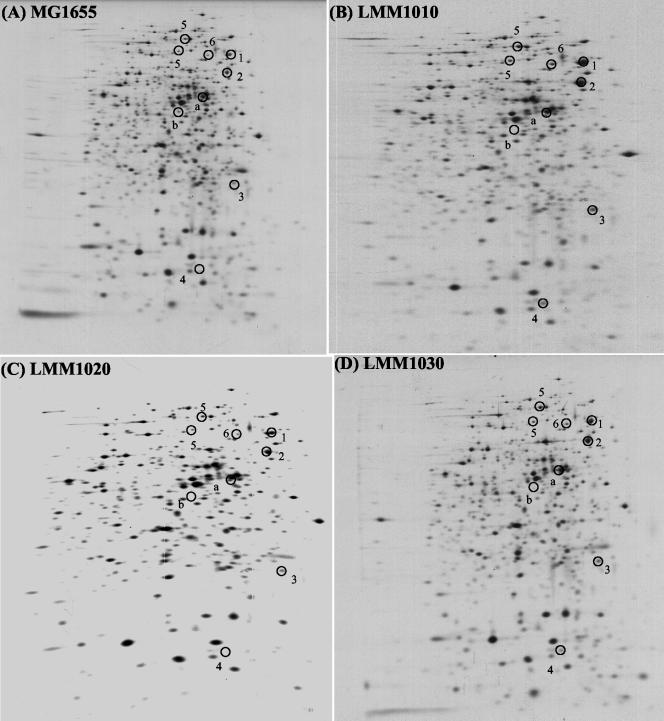
To complement these findings of increased heat shock gene expression with evidence at the protein level, protein synthesis was studied by 2D-SDS-PAGE analysis of pulse-labeled MG1655 and its HHP-resistant mutants (Fig. 4). Several heat shock proteins could be identified on the gels that were more strongly expressed in mutants LMM1010 and LMM1030 than in the parental strain: DnaK, GroEL, GroES, GrpE, ClpB, and HtpG. The levels of some, but not all, of these proteins also appeared to be elevated in mutant LMM1020, but less clearly so. These findings correspond well with results from the reporter fusion expression studies.
FIG. 4.
Two-dimensional autoradiograms of protein expression profiles in MG1655 (A), LMM1010 (B), LMM1020 (C), and LMM1030 (D) in late exponential phase at 37°C in supplemented M9 medium. Numbers mark heat shock proteins: 1, DnaK; 2, GroEL; 3, GrpE; 4, GroES; 5, ClpB; 6, HtpG. The letters a and b mark unidentified proteins with altered expression in the mutants. The presence of two ClpB spots is due to the presence of two translational starts on the clpB mRNA (50).
DISCUSSION
Because there are no reports, to our knowledge, on the genetics of bacterial HHP responses, we initiated a DFI (53) analysis of gene expression triggered by pressure treatment of E. coli MG1655. This analysis revealed that several heat shock genes were induced after exposure to sublethal pressures. These results resemble those of Welch et al. (57) who, by 2D-SDS-PAGE analyses, observed increased relative rates of synthesis of a set of heat shock proteins in anaerobic cultures of E. coli growing at 55 MPa compared to those of cultures grown at atmospheric pressure. However, the results in the latter study were obtained with cells growing at elevated pressure, whereas we studied gene expression in cells during recovery after exposure to a growth-inhibiting level of pressure.
Interestingly, HHP (150 MPa) induction of lon, clpPX, and dnaK transcription appears to be slower and weaker than heat shock (50°C) induction, although both treatments caused similar lag time before resumption of growth (data not shown). Therefore, the different patterns and levels of induction are likely to reflect subtle differences in cellular damage imposed by heat and HHP. In addition, the slow induction of heat shock proteins by HHP treatment may explain our failure to induce heat or pressure resistance by pressure shock (data not shown) in E. coli. A similar finding was reported by Fernandes et al. (10), who failed to induce pressure resistance in Saccharomyces cerevisiae by mild pressure pretreatment.
On the other hand, heat shock protein production upon temperature upshift occurred rapidly, and we could clearly demonstrate heat shock-induced pressure resistance in strain MG1655. This effect was reduced when induction was performed in the absence of nutrients (data not shown). Interestingly, for a specific E. coli isolate, Pagan and Mackey (42) observed pressure resistance in the absence of nutrients as a result of prolonged heat shock (45°C for 45 min) in phosphate buffer; however, this effect could be entirely attributed to plasma membrane stabilization. Taken together with the observation that pressure resistance observed in this study was correlated with the level of dnaK expression, our data strongly suggest that heat shock proteins prevent cellular damage and/or aid cell recovery. Heat shock-mediated HHP resistance has also been reported for S. cerevisiae, and further studies revealed the importance of heat shock proteins Hsc70 and Hsp104 in the pressure resistance of this yeast (22, 23, 24). In addition, the link between heat shock protein induction and HHP seems to extend to mammalian cells, because Elo et al. (8) observed induction of Hsp70 and Hsp90 in chondrocytic cells stressed by physiologic pressures of 30 MPa. More recently, cold shock also has been shown to affect HHP resistance in Staphylococcus aureus and L. monocytogenes (40, 58). To our knowledge, this study provides the first evidence of heat shock-induced HHP resistance in E. coli as well as a clue to the mechanism underlying this induced resistance.
Because induction of heat shock proteins apparently protects wild-type E. coli strain MG1655, we further investigated whether they also play a role in the pressure resistance of three independently isolated pressure-resistant mutants of MG1655 (16). As shown in Fig. 3, all three mutants displayed increased expression of lon, clpPX, and dnaK without any induction by heat or pressure. Overall, mutant LMM1030 had the highest expression levels, followed by mutant LMM1010 and finally mutant LMM1020. In addition, it appeared from the subsequent two-dimensional analysis of protein synthesis that mutants LMM1010 and LMM1030 had higher constitutive levels of several heat shock proteins, including DnaK, GroEL, GroES, GrpE, ClpB, and HtpG (Fig. 4). For mutant LMM1020 this was not the case, and this could be a reflection of the lower transcription rate of the heat shock genes in this strain. Previous observations showed that these mutants were also more resistant (although not always statistically significant) to heat inactivation at 58 to 60°C (16), and this agrees well with the increased heat shock protein expression observed in this study. Comparison of protein expression patterns also revealed differences in proteins other than these heat shock proteins, but we were unable to identify the majority of these protein spots because they were not assigned in the E. coli gene-protein database (19, 50, 52, 55).
Interestingly, some studies observed a correlation between heat and pressure resistance among natural isolates of E. coli (1, 4). Other studies, in which HHP-resistant mutants of E. coli (16) and of L. monocytogenes (27) were isolated after selection, also found that HHP resistance coincided with increased heat resistance. In E. coli O157:H7, the natural variation in pressure resistance was suggested to be correlated to the activity of the stationary-phase sigma factor (47). In L. monocytogenes, pressure resistance was caused by a mutation in CtsR, a negative regulator of class III heat shock genes, and was accompanied by enhanced expression of these genes and by enhanced heat resistance (28). Judging from the data shown in Fig. 3, one might consider overexpression of σ32 to be at the basis of high pressure resistance in the MG1655 mutants, particularly in LMM1030. It should be taken into account, however, that σ32 mRNA is not readily accessible for the translational apparatus due to secondary structures that can be resolved by heat (37, 38). Consequently, because these mutants are pressure resistant without heat shock induction, an additional mechanism must be active. Moreover, episomal expression of σ32 mRNA from a tac promoter had no effect on pressure resistance in MG1655 (data not shown). Attempts are presently being undertaken to isolate the determinant responsible for this altered expression of heat shock genes.
Our findings suggest a causative relationship between the expression of heat shock proteins and pressure resistance in E. coli, emphasizing proteome alteration as the primary target of inactivation by pressure. More specifically, heat shock protein-based pressure resistance can occur in two different forms: transient pressure resistance, induced by heat shock, and acquired pressure resistance, achieved by selection after repeated exposures to high pressure. These results may have important consequences for the use of high-pressure treatment, particularly in combination with mild heat treatment, for food preservation. It should be noted, however, that our screening of the promoter trap library with the DFI technique was not exhaustive, and HHP induction of other genes or pathways might prove relevant in the future.
Acknowledgments
This work was supported by a fellowship from the Flemish Institute for the Promotion of Scientific Technical Research (IWT) to A.A. and by research grants from the K. U. Leuven Research Fund (OT/01/35) and from the Fund for Scientific Research Flanders (G.0195.02).
We thank J. Vanderleyden (Centre for Microbial and Plant Genetics, K. U. Leuven) for providing access to his FACS facilities.
REFERENCES
- 1.Alpas, H., N. Kalchayanand, F. Bozoglu, A. Sikes, C. P. Dunne, and B. Ray. 1999. Variation in resistance to hydrostatic pressure among strains of food-borne pathogens. Appl. Environ. Microbiol. 65:4248-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 3.Balny, C., P. Masson, and K. Heremans. 2002. High pressure effects on biological macromolecules: from structural changes to alteration of cellular processes. Biochim. Biophys. Acta 1595:3-10. [DOI] [PubMed] [Google Scholar]
- 4.Benito, A., G. Ventoura, M. Casadei, T. Robinson, and B. Mackey. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 65:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duche, O., F. Tremoulet, P. Glaser, and J. Labadie. 2002. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elo, M. A., R. K. Sironen, K. Kaarniranta, S. Auriola, H. J. Helminen, and M. J. Lammi. 2000. Differential regulation of stress proteins by high hydrostatic pressure, heat shock, and unbalanced calcium homeostasis in chondrocytic cells. J. Cell Biochem. 79:610-619. [DOI] [PubMed] [Google Scholar]
- 9.Erijman, L., and R. M. Clegg. 1998. Reversible stalling of transcription elongation complexes by high pressure. Biophys. J. 75:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes, P. M., A. D. Panek, and E. Kurtenbach. 1997. Effect of hydrostatic pressure of a mutant of Saccharomyces cerevisiae deleted in the trehalose-6-phosphate synthase gene. FEMS Microbiol. Lett. 152:17-21. [DOI] [PubMed] [Google Scholar]
- 11.Ganzle, M. G., and R. F. Vogel. 2001. On-line fluorescence determination of pressure mediated outer membrane damage in Escherichia coli. Syst. Appl. Microbiol. 24:477-485. [DOI] [PubMed] [Google Scholar]
- 12.Gao, X., J. Li, and K. C. Ruan. 2001. Barotolerant Escherichia coli induced by high hydrostatic pressure. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao 33:77-81. [PubMed] [Google Scholar]
- 13.Gould, G. W. 1996. Methods for preservation and extension of shelf life. Int. J. Food Microbiol. 33:51-64. [DOI] [PubMed] [Google Scholar]
- 14.Gould, G. W. 2000. Preservation: past, present and future. Br. Med. Bull. 56:84-96. [DOI] [PubMed] [Google Scholar]
- 15.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in Escherichia coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 16.Hauben, K. J., D. H. Bartlett, C. C. Soontjens, K. Cornelis, E. Y. Wuytack, and C. W. Michiels. 1997. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 63:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter σS-selective? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 18.Heremans, K., and L. Smeller. 1998. Protein structure and dynamics at high pressure. Biochim. Biophys. Acta 1386:353-370. [DOI] [PubMed] [Google Scholar]
- 19.Hoogland, C., J.-C. Sanchez, L. Tonella, P. A. Binz, A. Bairoch, D. F. Hochstrasser, and R. D. Appel. 2000. The 1999 SWISS-2DPAGE database update. Nucleic Acids Res. 28:286-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikegami, A., K. Nakasone, C. Kato, Y. Nakamura, I. Yoshikawa, R. Usami, and K. Horikoshi. 2000. Glutamine synthetase gene expression at elevated hydrostatic pressure in a deep-sea piezophilic Shewanella violacea. FEMS Microbiol. Lett. 192:91-95. [DOI] [PubMed] [Google Scholar]
- 21.Iwahashi, H., H. Shimizu, M. Odani, and Y. Komatsu. 2003. Piezophysiology of genome wide gene expression levels in the yeast Saccharomyces cerevisiae. Extremophiles 7:291-298. [DOI] [PubMed] [Google Scholar]
- 22.Iwahashi, H., K. Obuchi, S. Fujii, and Y. Komatsu. 1997. Effect of temperature on the role of Hsp104 and trehalose in barotolerance of Saccharomyces cerevisiae. FEBS Lett. 416:1-5. [DOI] [PubMed] [Google Scholar]
- 23.Iwahashi, H., S. Nwaka, and K. Obuchi. 2001. Contribution of Hsc70 to barotolerance in the yeast Saccharomyces cerevisiae. Extremophiles 5:417-421. [DOI] [PubMed] [Google Scholar]
- 24.Iwahashi, H., S. C. Kaul, K. Obuchi, and Y. Komatsu. 1991. Induction of barotolerance by heat shock treatment in yeast. FEMS Microbiol. Lett. 64:325-328. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins, D. E., J. E. Schultz, and A. Matin. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170:3910-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins, D. E., S. A. Chaisson, and A. Matin. 1990. Starvation-induced cross protection against osmotic challenge in Escherichia coli. J. Bacteriol. 172:2779-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karatzas, K. A., and M. H. Bennik. 2002. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl. Environ. Microbiol. 68:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karatzas, K. A., J. A. Wouters, C. G. Gahan, C. Hill, T. Abee, and M. H. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 29.Kato, C., and M. H. Qureshi. 1999. Pressure response in deep-sea piezophilic bacteria. J. Mol. Microbiol. Biotechnol. 1:87-92. [PubMed] [Google Scholar]
- 30.Kato, C., M. Smorawinska, L. Li, and K. Horikoshi. 1997. Comparison of the gene expression of aspartate beta-D-semialdehyde dehydrogenase at elevated hydrostatic pressure in deep-sea bacteria. J. Biochem. 121:717-723. [DOI] [PubMed] [Google Scholar]
- 31.Kato, C., T. Sato, M. Smorawinska, and K. Horikoshi. 1994. High pressure conditions stimulate expression of chloramphenicol acetyltransferase regulated by the lac promoter in Escherichia coli. FEMS Microbiol. Lett. 122:91-96. [DOI] [PubMed] [Google Scholar]
- 32.Knorr, D. 1999. Novel approaches in food-processing technology: new technologies for preserving foods and modifying function. Curr. Opin. Biotechnol. 10:485-491. [DOI] [PubMed] [Google Scholar]
- 33.Kobori, H., M. Sato, A. Tameike, K. Hamada, S. Shimada, and M. Osumi. 1995. Ultrastructural effects of pressure stress to the nucleus in Saccharomyces cerevisiae: a study by immunoelectron microscopy using frozen thin sections. FEMS Microbiol. Lett. 132:253-258. [DOI] [PubMed] [Google Scholar]
- 34.Leistner, L. 2000. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 55:181-186. [DOI] [PubMed] [Google Scholar]
- 35.Linton, M., and M. F. Patterson. 2000. High pressure processing of foods for microbiological safety and quality (a short review). Acta Microbiol. Immunol. Hung. 47:175-182. [DOI] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Morita, M., M. Kanemori, H. Yanagi, and T. Yura. 1999. Heat-induced synthesis of σ32 in Escherichia coli: structural and functional dissection of rpoH mRNA secondary structure. J. Bacteriol. 181:401-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morita, M. T., Y. Tanaka, T. S. Kodama, Y. Kyogoku, H. Yanagi, and T. Yura. 1999. Translational induction of heat shock transcription factor σ32: evidence for a built-in RNA thermosensor. Genes Dev. 13:655-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niven, G. W., C. A. Miles, and B. M. Mackey. 1999. The effects of hydrostatic pressure on ribosome conformation in Escherichia coli: an in vivo study using differential scanning calorimetry. Microbiology 145:419-425. [DOI] [PubMed] [Google Scholar]
- 40.Noma, S., and I. Hayakawa. 2003. Barotolerance of Staphylococcus aureus is increased by incubation at below 0°C prior to hydrostatic pressure treatment. Int. J. Food Microbiol. 80:261-264. [DOI] [PubMed] [Google Scholar]
- 41.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 42.Pagan, R., and B. Mackey. 2000. Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: differences between exponential- and stationary-phase cells and variation among strains. Appl. Environ. Microbiol. 66:2829-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pagan, R., S. Jordan, A. Benito, and B. Mackey. 2001. Enhanced acid sensitivity of pressure-damaged Escherichia coli O157 cells. Appl. Environ. Microbiol. 67:1983-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pichereau, V., A. Hartke, and Y. Auffray. 2000. Starvation and osmotic stress induced multiresistances. Influence of extracellular compounds. Int. J. Food Microbiol. 55:19-25. [DOI] [PubMed] [Google Scholar]
- 45.Raso, J., and G. V. Barbosa-Canovas. 2003. Nonthermal preservation of foods using combined processing techniques. Crit. Rev. Food Sci. Nutr. 43:265-285. [DOI] [PubMed] [Google Scholar]
- 46.Ritz, M., M. Freulet, N. Orange, and M. Federighi. 2000. Effects of high hydrostatic pressure on membrane proteins of Salmonella typhimurium. Int. J. Food Microbiol. 55:115-119. [DOI] [PubMed] [Google Scholar]
- 47.Robey, M., A. Benito, R. H. Hutson, C. Pascual, S. F. Park, and B. M. Mackey. 2001. Variation in resistance to high hydrostatic pressure and rpoS heterogeneity in natural isolates of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:4901-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.San Martin, M. F., G. V. Barbosa-Canovas, and B. G. Swanson. 2002. Food processing by high hydrostatic pressure. Crit. Rev. Food Sci. Nutr. 42:627-645. [DOI] [PubMed] [Google Scholar]
- 49.Simpson, R. K., and A. Gilmour. 1997. The effect of high hydrostatic pressure on the activity of intracellular enzymes of Listeria monocytogenes. Lett. Appl. Microbiol. 25:48-53. [DOI] [PubMed] [Google Scholar]
- 50.Squires, C. L., S. Pedersen, B. M. Ross, and C. Squires. 1991. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 173:4254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storz, G., and R. Hengge-Aronis. 2000. Bacterial stress responses. ASM Press, Washington, D.C.
- 52.Tonella, L., B. J. Walsh, J.-C. Sanchez, K. Ou, M. R. Wilkins, M. Tyler, S. Frutiger, A. A. Gooley, I. Pescaru, R. D. Appel, J. X. Yan, A. Bairoch, C. Hoogland, F. S. Morch, G. J. Hughes, K. L. Williams, and D. F. Hochstrasser. 1998. ′98 Escherichia coli SWISS-2DPAGE database update. Electrophoresis 19:1960-1971. [DOI] [PubMed] [Google Scholar]
- 53.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 54.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanBogelen, R. A., E. R. Olson, B. L. Wanner, and F. C. Neidhardt. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 178:4344-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wanner, B. L., R. Kodaira, and F. C. Neidhardt. 1977. Physiological regulation of a decontrolled lac operon. J. Bacteriol. 130:212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welch, T. J., A. Farewell, F. C. Neidhardt, and D. H. Bartlett. 1993. Stress response of Escherichia coli to elevated hydrostatic pressure. J. Bacteriol. 175:7170-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wemekamp-Kamphuis, H. H., A. K. Karatzas, J. A. Wouters, and T. Abee. 2002. Enhanced levels of cold shock proteins in Listeria monocytogenes LO28 upon exposure to low temperature and high hydrostatic pressure. Appl. Environ. Microbiol. 68:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yayanos, A. A. 1995. Microbiology to 10,500 meters in the deep sea. Annu. Rev. Microbiol. 49:777-805. [DOI] [PubMed] [Google Scholar]