Abstract
Investigation of the diversity of nirK and nirS in denitrifying bacteria revealed that salinity decreased the diversity in a nitrate-containing saline wastewater treatment system. The predominant nirS clone was related to nirS derived from marine bacteria, and the predominant nirK clone was related to nirK of the genus Alcaligenes.
Nitrogen removal from wastewater is accomplished by bacterial denitrification. Some types of industrial wastewater, such as metallurgic wastewater, contain large amounts of nitrate and saline (11). Although we have developed a nitrogen removal system for saline wastewater, nitrite often accumulates, particularly under relatively high-saline conditions (32). Therefore, it is important to understand the ecology of nitrite-reducing bacterial communities to determine stable operational conditions for denitrification processes.
A few researchers and we have studied microbial communities in anaerobic reactors for the denitrification of saline wastewater (13, 31, 32) and used an approach based on taxonomic analysis of the 16S rRNA gene to identify all of the bacterial species within a community. These studies suggested that members of the gamma subdivision of the class Proteobacteria are important in such wastewater treatment systems. However, 16S rRNA gene-based approaches are unable to detect denitrifying bacteria particularly in such systems because bacterial groups possessing denitrifying abilities are phylogenetically diverse (33). Furthermore, although aerobic halophilic species have been reported (7, 29), the microbial ecology of moderately halophilic denitrifying bacteria has hardly been reported.
Recently, primer sets specific for functional genes involved in denitrification, namely, nirK, nirS (3, 9), and nosZ (22), have been developed. Thus, PCR-based approaches revealed denitrifying microbial communities in activated sludge (9, 27), marine sediment (4, 5, 15), and soil (17, 19) on the basis of nirK and nirS and in marine sediment on the basis of nosZ (23, 24). These approaches have contributed to the improvement of the complete or partial sequences of nitrite reductase genes and nitrous oxide reductase genes in the database. We expect that the information on these genes detected from various environments will contribute to further studies on the ecophysiology of denitrifying communities.
In this study, we focused on the microbial ecology of nitrite-reducing bacteria in two series of metallurgic wastewater treatment systems (MWTSs) with different fluidity conditions; one of the MWTSs was composed of an anaerobic packed bed, and the other was composed of an anaerobic fluidized bed (31). The nirK and nirS heterogeneity in the anaerobic reactors of MWTSs was investigated by cloning, sequencing, and phylogenetic analysis to determine the actual denitrifying bacterial community. Furthermore, the anaerobic packed bed and the fluidized bed were compared for nirK and nirS diversity to investigate the influence of fluidity conditions on the denitrifying microbial community.
Sludge samples and isolates from MWTS.
Sludge samples were collected from two series of laboratory scale anaerobic-aerobic circulating bioreactors used as MWTSs, as described previously (31). These systems consisted of an anaerobic reactor (2 liters) and an aerobic fluidized reactor (1 liter). The anaerobic reactor of run 1 was packed with sponge cubic medium without mixing. The anaerobic reactor of run 2 was a completely mixed fluidized bed in which polyvinyl alcohol particles coated with activated carbon were used as a carrier. The metallurgic wastewater used in this study was from a factory that recovers precious metals from industrial waste. The wastewater was diluted with tap water, and acetic acid (approximately 3,000 mg liter−1) was added as a carbon source for denitrification (C/N ratio = 1.5) before being supplied to the anaerobic reactors. The composition of the inlet was NOx-N (nitrate and nitrite) (1,800 to 2,500 mg liter−1), NH4-N (320 to 1,690 mg liter−1), and saline (14,000 to 32,000 mg liter−1). For DNA extraction, sludge samples adhering and not adhering to bed materials were collected and suspended in TE buffer (1 mM EDTA, 10 mM Tris-HCl [pH 8.0]) and stored at −20°C. Some bacteria were isolated from runs 1 and 2 of MWTS by using Trypticase soy agar (Becton Dickinson, Mountain View, Calf.) medium under aerobic conditions, and their denitrification activity was tested. The isolates were identified by partial 16S rRNA gene sequence analysis (31).
DNA extraction, PCR amplification, and sequence analysis.
DNA extraction from sludge samples was performed by the method of Smalla (26), with slight modifications. The isolates were added to TE buffer and heated at 95°C for 5 min to extract DNA. The oligonucleotide primer pairs nirK1F-nirK5R and nirS1F-nirS6R (3) were used in PCR amplifications performed with a model 9700 thermal cycler (Applied Biosystems, Foster City, Calif.). The presence of PCR products, whose sizes were approximately 514 and 890 bp for nirK and nirS, respectively, was confirmed by running 8 μl of the product on 2% agarose gels and then staining the agarose gels with ethidium bromide. The PCR products were purified by eluting the bands from the agarose gels with a Wizard SV gel and a PCR cleanup system (Promega Corp., Madison, Wis.). The eluted nirK or nirS PCR products were cloned with a ZERO Blunt TOPO PCR cloning kit (Invitrogen Corp., Carlsbad, Calif.) for nirK and a QIAGEN PCR cloning kit (QIAGEN, Hilden, Germany) for nirS in accordance with the manufacturer's instructions. The DNA insert was amplified and used as template DNA in a cycle sequencing reaction with a Big Dye Terminator cycle sequencing kit (Applied Biosystems) and a DYEnamic ET Terminator cycle sequencing kit (Amersham Biosciences, Freiburg, Germany) in accordance with the manufacturer's instructions. The nirK and nirS fragments were sequenced with an ABI PRISM 377 and ABI PRISM 3100-Avant DNA sequencing system (Applied Biosystems). Sequences with more than 97% similarity were considered to belong to the same operational taxonomic unit (OTU). The Shannon-Weaver index (H) (25) was calculated with the formula H = −Σ(pi)(log2pi), where pi is the proportion of each phylogenetic group to the total number of clones detected. Evenness (16) based on the Shannon-Weaver index was calculated with the formula E = H/log2S. A database search was conducted with BLAST (1) from the DDBJ (DNA Data Bank of Japan). The sequences determined in this study and those retrieved from the databases were aligned by using CLUSTAL W (28). Phylogenetic trees were constructed with CLUSTAL W and TreeView (18) by the neighbor-joining method (21).
Phylogenetic relationship of nirK and nirS.
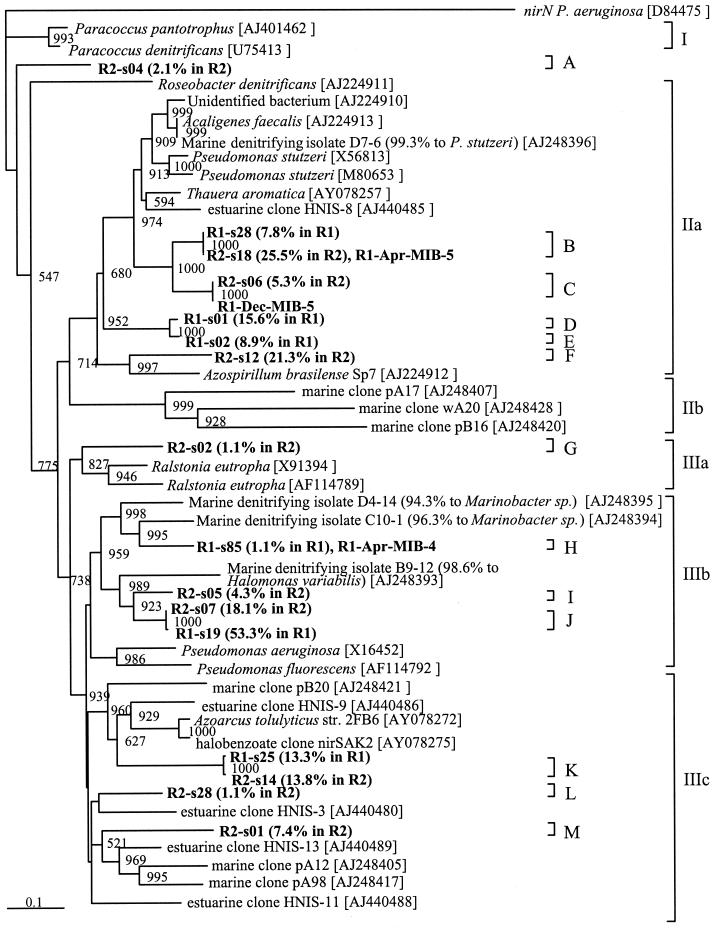
The phylogenetic tree based on the nirS sequence showed three major clusters (I to III) and 13 OTUs (A to M) (Fig. 1). The nucleotide sequence similarities between the nirS clones and those in the database ranged from 74.3 to 83.1%, and the deduced amino acid sequence similarities ranged from 73.3 to 88.9%. Most of the nirS clones obtained in this study exhibited less than 80% nucleotide sequence similarity to the nirS sequence in the database, suggesting that these nirS genes derived from novel denitrifying bacteria are indigenous to MWTSs.
FIG. 1.
Phylogenetic tree constructed by the neighbor-joining method on the basis of partial nirS sequences cloned from run 1 (R1), run 2 (R2), or MWTS isolates (MIB). The nirN sequence of Pseudomonas aeruginosa was used as an outgroup to root the tree. A single sequence from each system was chosen as a representative of a cluster with more than 97% similarity. Bootstrap numbers indicate the value of 1,000 replicate trees supporting order (8), and values of less than 500 were omitted. Scale bar = 10% nucleotide substitution. The values in parentheses are the percent distributions of the clones within the nirS population.
Cluster II included five OTUs of nirS clones from MWTSs, and these nirS clones were associated with the nirS gene of Pseudomonas stutzeri. The fractions of the nirS clone affiliated with cluster II relative to all of the nirS clones detected were 32.3 and 52.1% in runs 1 and 2, respectively. OTUs B to E were most similar to the nirS gene of marine denitrifying isolate D7-6 (accession no. AJ248396) described by Braker et al. (5). The nirS genes of isolates R1-Apr-MIB-5 and R1-Dec-MIB-5, which were affiliated with the genus Pseudomonas (31), were included in OTUs B and C, respectively. OTUs D and E were unique to run 1, and their fractions were 15.6 and 8.9%, respectively. All of the nirS clones of MWTSs did not belong to cluster IIb, including the nirS clones found in a marine environment by Braker et al. (5). For cluster IIIb, the most abundant nirS clones from run 1 were affiliated with OTU J and accounted for 53.3 and 18.1% of all of the nirS clones detected in runs 1 and 2, respectively. OTU J was the most similar to the nirS gene of marine denitrifying isolate B9-12 (accession no. AJ248393) (98.6% similarity to Halomonas variabilis) found by Braker et al. (5). The nirS clones similar to the nirS gene of Marinobacter sp. were found, although their fraction was small (OTU H). The sequences of these clones were almost the same as the nirS sequence of isolate R1-Apr-MIB-4. Cluster IIIc consisted mainly of nirS clones of unknown denitrifying bacteria. OTU K was similar to the nirS gene of Azoarcus sp. at similarities of 13.3 and 13.8% to all of the nirS clones detected in runs 1 and 2, respectively. Clusters IIa and IIIb, whose fractions were large relative to all of the detected nirS clones, were related to the marine bacteria isolated by Braker et al. (5). The NaCl concentration of the influent appropriately diluted for MWTSs was 1.5 to 3.5%, which is similar to that of seawater. Many nirS clones similar to the nirS gene of marine bacteria were detected from MWTSs regardless of the differences in composition except that of saline, suggesting that a microbial community possessing nirS is affected by salinity. Although the physiology of moderately halophilic bacteria under anaerobic conditions has hardly been reported, Halomonas elongata (30), Halomonas desiderata (2), and Bacillus halodenitrificans (6) are well-known denitrifying bacteria. It was reported that H. elongata can reduce both nitrate and nitrite and that B. halodenitrificans is so unusually tolerant to nitrite as to grow in 0.58 M NaNO2.
It is said that nirS phylogenetic trees generally show the same clustering as 16S rRNA gene phylogenetic trees (5). The nirS genes of the bacteria isolated from MWTSs, which are affiliated with Pseudomonas sp. (R1-MIB-Apr-5, Dec-5) and Marinobacter sp. (R1-MIB-Apr-4), branched with the same clusters including the same species isolated from a marine environment. However, no nitrite reductase genes have been detected from the isolated bacterium identified as Halomonas sp. by partial 16S rRNA gene sequence analysis (31), although nirS clone analysis detected clones similar to the nirS gene of Halomonas sp. (OTUs I and J). One possible explanation for these results is that the nitrite reductase genes of isolated Halomonas sp. might have some mismatch to the primers used in this study whereas uncultured bacteria had no mismatch to the primers used. Our previous study revealed that the group of bacteria belonging to the gamma subdivision of the class Proteobacteria including Halomonas sp. is dominant in MWTSs (31). Simultaneous detection of the 16S rRNA and the functional gene will make it possible to combine the phylogenetic information of each gene.
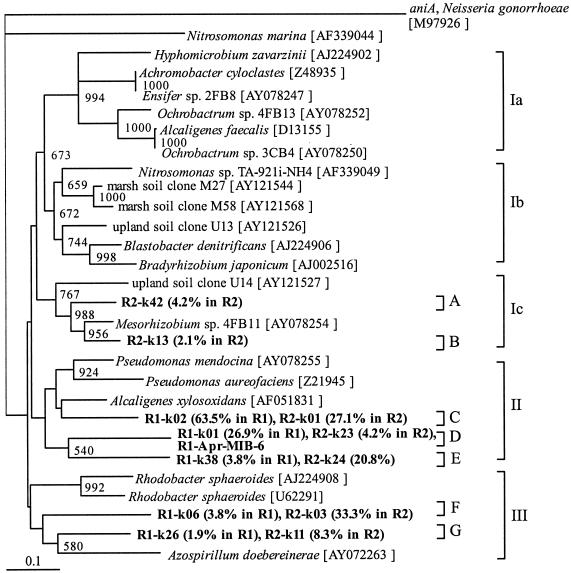
For nirK, the number of clusters detected was lower than that for nirS; there were three major clusters and seven OTUs (Fig. 2). According to the BLAST search, the nucleotide sequence similarities between the nirK clones and those in the database ranged from 74.6 to 91.1% and the deduced amino acid sequence similarities ranged from 70.8 to 97.5%. In contrast to nirS clones, all of the nirK clones did not cluster with any marine clones. The fractions of clones including OTUs D, E, and F, which were not similar to any nirK sequence in the database, were high relative to all of the nirK genes detected, suggesting that those clones are inherent in MWTSs. For both reactors, most of the nirK clones were affiliated with cluster II, including OTUs C, D, and E, similar to the nirK gene of Alcaligenes xylosoxidans (accession no. AF051831). In particular, the percentages of nirK clones affiliated with OTU C relative to all of the clones were 63.5 and 27.1% in runs 1 and 2, respectively. The nirK gene of isolate R1-Apr-MIB-6, which clustered in the genus Alcaligenes in the phylogenetic tree on the basis of the partial 16S rRNA gene sequence, was included in OTU D. The fraction of cluster III, which consisted of the nirK of Rhodobacter sp. and Azospirillum sp., included the most abundant OTU in run 2 (F), and the fractions of cluster III were 5.7 and 41.6%, respectively. The nirK clones affiliated with cluster Ic, which were similar to the nirK gene of Mesorhizobium sp., were derived from run 2 only.
FIG. 2.
Phylogenetic tree constructed by the neighbor-joining method on the basis of partial nirK sequences cloned from run 1 (R1) or run 2 (R2) of MWTS. The aniA sequence of Neisseria gonorrhoeae was used as an outgroup to root the tree. A single sequence from each system was chosen as a representative of a cluster with more than 97% similarity. Bootstrap numbers indicate the value of 1,000 replicate trees supporting order (8), and values of less than 500 were omitted. Scale bar = 10% nucleotide substitution. The values in parentheses are the percent distributions of the clones within the nirK population.
It was mentioned that great care should be taken to detect nirK fragments from environments with high salinity because the nirK primer set used in this study shows no sequence homology to the nirK gene of an archaean denitrifier, which has been recently sequenced (12). PCR amplification was carried out with primers specific for the archaean 16S rRNA gene to examine whether archaea exist in MWTSs, resulting in the lack of PCR products from these systems (data not shown). Therefore, there is no possibility of underestimating nirK diversity in MWTSs, which is consistent with the fact that halophilic archaea generally inhabit a hypersaline environment with more than 2.5 M NaCl. Nevertheless, we should pay attention to the anticipated problem of nirK primer specificity, which is limited to the amplification of nirK similar to those that were referred to in the primer design, as mentioned in a previous study (5).
Diversity of nirK and nirS from MWTSs.
Table 1 shows the phylotype richness (S), Shannon-Weaver diversity index (H), and evenness (E) of nirK and nirS in MWTSs. The Shannon-Weaver diversity indices of nirS and nirK were 1.96 and 1.22 for run 1 and 2.77 and 2.04 for run 2, respectively. Statistical analyses showed that the diversity level in run 2 was higher than that in run 1 for both nirK and nirS. Furthermore, the diversity of nirS was higher than that of nirK in both reactors. However, the diversities of nirK and nirS in MWTSs were lower than those in natural environments such as soil, in which the diversity indices of nirK and nirS are 3.55 and 5.27, respectively (19). This was probably due to the differences in salinity and carbon source between MWTSs and soil. For the latter reason, the influent of MWTSs included only acetic acid as a carbon source utilizable by bacteria, and any other organic compounds originally included in metallurgic wastewater could not be biologically utilized. Therefore, it is presumed that salinity and small carbon source variations decrease the diversity of microbial communities. In fact, the microbial community in a methanol-fed denitrification process (13) was completely different from that in MWTSs (31). Labbe et al. reported that members of the genus Methylophaga are predominant in a methanol-fed denitrification system for treating seawater (13). However, it was suggested that members of the genus Halomonas are dominant denitrifying bacteria in an acetate-fed saline wastewater treatment system in this study. Therefore, the diversity of nitrite reductase genes may also vary depending on the types of carbon sources used. Moreover, the nirK and nirS diversities in run 2 were higher than those in run 1 (Table 1). This difference in nitrite reductase gene diversity between runs 1 and 2 may arise from the differences between the biofilm community and the suspended microbial community caused by fluidity conditions. It was observed that most of the microorganisms in run 1 formed a biofilm in and on the medium, while those in run 2 were suspended regardless of the medium in the reactor. Other researchers reported that the difference in dissolved-oxygen concentration is one of the key factors in controlling the denitrifying community structure (15). In this study, although dissolved oxygen was almost 0 mg liter−1 in run 2, the microbial community may have more chances to come in contact with oxygen on the water surface of the fluidized bed than on that of the packed bed, suggesting that the difference in the frequency of oxygen contact affects the diversity of nitrite reductase genes. These results indicate that fluidity conditions control denitrifying communities. Comparing the nitrogen removal efficiencies of runs 1 and 2, the removal efficiency of run 1 was found to be more stable than that of run 2, suggesting that a biofilm community is more suitable for MWTSs than is a suspended microbial community. Furthermore, the expression of genes associated with denitrification is sensitive to the presence of oxygen (10). Therefore, the expression of the nitrite reductase gene may differ between a biofilm community and a suspended microbial community. More research studies that will introduce some methods of investigating bacterial activity, such as analysis of the expression of nirK and nirS, combination of fluorescence in situ hybridization and microautoradiography (14), and stable isotope probing analysis (20), will clarify the relationship between the ecology of an active denitrifying community and nitrogen removal efficiency and lead to the improvement of nitrogen removal performance in MWTSs.
TABLE 1.
Diversity analysis of nirK and nirS gene fragments from anaerobic packed bed and anaerobic fluidized bed of MWTS
| Gene and reactor | Sa | Hb | Ec |
|---|---|---|---|
| nirS | |||
| Packed bed (run 1) | 6 | 1.96 | 0.76 |
| Fluidized bed (run 2) | 10 | 2.77 | 0.84 |
| nirK | |||
| Packed bed (run 1) | 5 | 1.22 | 0.52 |
| Fluidized bed (run 2) | 7 | 2.04 | 0.82 |
Phylotype richness, S was the total number of OTUs in a sludge.
The Shannon-Weaver diversity index (25) was calculated as follows: H = −Σ(pi)(log2pi), where pi is the proportion of each phylogenetic groups to the number of all detected clones.
Evenness (16) was calculated as follows from the Shannon-Weaver index: E = H/log2S.
Microbial community analysis based on nitrite reductase genes may enable the identification of key bacteria concerned with denitrification, which was the most significant process for MWTSs. These results show that bacterial community analysis based on functional genes is important for a better understanding of microbial communities in wastewater treatment systems in addition to 16S rRNA gene analysis. Further comprehensive study of the relationships among qualitative and quantitative microbial community compositions, functions, and process stabilities will help in the design of advanced wastewater treatment systems or determination of appropriate operational conditions.
Nucleotide sequence accession numbers.
The partial nirK and nirS sequences were submitted to the DDBJ database and assigned accession numbers AB118878 to AB118904.
Acknowledgments
This study was partly supported by Matsuda Sangyo Co., Ltd., which is thanked for its assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Berendes, F., G. Gottschalk, E. Heine-Dobbernack, E. R. B. Moore, and B. J. Tindall. 1996. Halomonas desiderata sp. nov., a new alkalophilic, halotolerant and denitrifying bacterium isolated from a municipal sewage works. Syst. Appl. Microbiol. 19:158-167. [Google Scholar]
- 3.Braker, G., A. Fesefeldt, and K. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braker, G., H. L. Ayala-del-Río, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denariaz, G., W. J. Payne, and J. Le Gall. 1989. A halophilic denitrifier, Bacillus halodenitrificans sp. nov. Int. J. Syst. Bacteriol. 39:145-151. [Google Scholar]
- 7.Dennis, P. P., and L. C. Shimmin. 1997. Evolutionary divergence and salinity-mediated selection in halophilic archaea. Microbiol. Mol. Biol. Rev. 61:90-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Hallin, S., and P.-E.Lindgren. 1999. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Härting, E., and W. G. Zumft. 1999. Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system. J. Bacteriol. 181:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata, A., Y. Nakamura, and S. Tsuneda. 2001. Nitrogen removal from industrial wastewater discharged from metal recovery processes. Water Sci. Technol. 44:171-180. [PubMed] [Google Scholar]
- 12.Ichiki, H., Y. Tanaka, K. Mochizuki, K. Yoshimatsu. T. Sakurai, and T. Fujiwara. 2001. Purification, characterization, and genetic analysis of Cu-containing dissimilatory nitrite reductase from a denitrifying halophilic archaeon, Haloarcula marismortui. J. Bacteriol. 183:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labbe, N., P. Juteau, S. Parent, and R. Villemur. 2003. Bacterial diversity in a marine methanol-fed denitrification reactor at the Montreal biodome, Canada. Microb. Ecol. 46:12-21. [DOI] [PubMed] [Google Scholar]
- 14.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, X., S. M. Tiquia, G. Holguin, L. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margalef, R. 1958. Information theory in ecology. Gen. Syst. 3:36-71. [Google Scholar]
- 17.Nogales, B., K. N. Timmis, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediment after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 19.Priemé, A., G. Braker, and J. M. Tiedje. 2002. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl. Environ. Microbiol. 68:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 21.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 22.Scala, D., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 23.Scala, D., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scala, D., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana.
- 26.Smalla, K. 1995. Extraction of microbial DNA from sewage and manure slurries, p. 1.1.3. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 27.Song, B., and B. B. Ward. 2003. Nitrite reductase genes in halobenzoate degrading denitrifying bacteria. FEMS Microbiol. Ecol. 43:349-357. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vreeland, R. H., C. D. Litchfield, E. L. Martin, and E. Elliot. 1980. Halomonas elongata, a new genus and species of extremely salt-tolerant bacteria. Int. J. Syst. Bacteriol. 30:485-495. [Google Scholar]
- 31.Yoshie, S., N. Noda, T. Miyano, S. Tsuneda, A. Hirata, and Y. Inamori. 2001. Microbial community analysis in the denitrifying process of saline-wastewater by denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA and the cultivation method. J. Biosci. Bioeng. 92:346-353. [DOI] [PubMed] [Google Scholar]
- 32.Yoshie, S., N. Noda, T. Miyano, S. Tsuneda, A. Hirata, and Y. Inamori. 2002. Characterization of microbial community in nitrogen removal process of metallurgic wastewater by PCR-DGGE. Water Sci. Technol. 46:93-98. [PubMed] [Google Scholar]
- 33.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]