Abstract
Autonomous, field-deployable molecular detection systems require seamless integration of complex biochemical solutions and physical or mechanical processing steps. In an attempt to simplify the fluidic requirements for integrated biodetection systems, we used tunable surface microparticles both as an rRNA affinity purification resin in a renewable microcolumn sample preparation system and as the sensor surface in a flow cytometer detector. The tunable surface detection limits in both low- and high-salt buffers were 1 ng of total RNA (∼104 cell equivalents) in 15-min test tube hybridizations and 10 ng of total RNA (∼105 cell equivalents) in hybridizations with the automated system (30-s contact time). RNA fragmentation was essential for achieving tunable surface suspension array specificity. Chaperone probes reduced but did not completely eliminate cross-hybridization, even with probes sharing <50% identity to target sequences. Nonpurified environmental extracts did not irreparably affect our ability to classify color-coded microparticles, but residual environmental constituents significantly quenched the Alexa-532 reporter fluor. Modulating surface charge did not influence the interaction of soluble environmental contaminants with conjugated beads. The automated system greatly reduced the effects of fluorescence quenching, especially in the soil background. The automated system was as efficacious as manual methods for simultaneous sample purification, hybridization, and washing prior to flow cytometry detection. The implications of unexpected target cross-hybridization and fluorescence quenching are discussed relative to the design and implementation of an integrated microbial monitoring system.
Molecular analysis of microbial communities typically requires some combination of sample collection, concentration, cell lysis, nucleic acid purification, PCR amplification, and specific detection in order to address fundamental questions of microbial community dynamics, activity, and function in the environment. Obviously, such molecular methods have made a substantial contribution to microbial ecology research, but they continue to be relatively ineffective for microbial monitoring due to the cost, technical difficulty, and/or retrospective nature of the analyses (7). One of the primary objectives of the Natural and Accelerated Bioremediation Research (NABIR) Program of the U.S. Department of Energy (U.S. DOE) is to develop innovative methods for measuring microbial community dynamics in (nearly) real time, in the field, ostensibly to monitor and assess bioremediation performance for the purpose of making more informed and verifiable system operating decisions. Any method developed to meet this objective, however, must also address microbial diversity and quantitation, two fundamental ecology issues that are continually identified as primary challenges facing genomic technologies and their application to bioremediation (J. Zhou, D. P. Chandler, and F. J. Brockman, Report on the NABIR workshop: application of genomic technology to bioremediation. http://www.lbl.gov/NABIR/generalinfo/workshop_reports/Genom_Tech.pdf).
For advanced genomic technologies (3, 4, 13, 15, 20, 25, 29) to fulfill engineering demands on the one hand (real-time, in-field detection) and microbiology requirements on the other hand (diversity and quantitation), it must be recognized that molecular microbial monitoring is an analytical process that begins with the environment, not a detector or a sensor. Within this context, nucleic acid sample preparation is still a limited technology for real-time, in-field bioremediation monitoring, due in large part to the coextraction of soluble soil constituents that interfere with molecular techniques (26). Continued reliance (and dependence) on PCR for assessing microbial diversity and/or abundance is also noteworthy, given the known amplification biases and inhibitors of PCR in an environmental context (28). To overcome many of the limitations associated with indirect detection methods, new techniques for the sensitive, specific, and direct detection of nucleic acids are required in order to accurately and quantitatively ascribe phylotype, phenotype, or function to in situ microorganisms.
It must also be acknowledged that the processes of molecular microbial monitoring and detection embody physical hardware components that are inextricably linked to each other via biochemistry. Indeed, most environmental monitoring scenarios require the processing of milliliter to liter volumes of environmental samples down to microliter volumes simply in order to present a nucleic acid sample to a detector. Without developing and understanding integrated biochemistry in communion with advanced detection devices specifically from an environmental perspective, it becomes difficult to understand microbial community dynamics and activity in the actual environment. Thus, there is a continued need to develop molecular methods (sample preparation and detection) that are compatible with in-field microbial detection methods while avoiding the pitfalls of PCR.
In order to address the limitations of current sample preparation technology and the difficulties associated with the automation and field deployment of benchtop nucleic acid manipulations (e.g., centrifugation, precipitation, and organic substance extraction), we have developed a suite of affinity purification techniques, renewable surface microfluidic columns, and bead or planar array detection methods for the automated isolation, purification, or detection of rRNA from environmental samples (8, 10). We hypothesize that automated rRNA capture will be more sensitive than equivalent test tube reactions. However, at low target concentrations and for nucleic acid targets containing significant secondary and tertiary structures (i.e., 16S rRNA and ribosomal DNA), solution or solid-phase hybridizations are constrained by the thermodynamic, kinetic, and equilibrium binding properties of DNA probes, potentially limiting their efficiency in a hybridization or capture format (i.e., purification or detection). The tunable surface hypothesis states that low-salt (LS) buffers and pH modulation can greatly enhance the efficiency of binding of structured nucleic acids to immobilized DNA oligonucleotides (5); we therefore also hypothesize that tunable surface chemistry will enhance rRNA detection limits in both model systems and amended environmental samples.
The objectives of this study were to test these hypotheses; to develop field-deployable rRNA detection methods; to biochemically integrate automated rRNA sample preparation functions with microarray detection by using color-coded microparticles and a simple rRNA-targeted oligonucleotide array (suspension array [14, 24]) as a unifying analytical principle; and to evaluate how the combined (automated) sample preparation and detection methods influence the fluorescent reporter systems at the point of suspension array detection.
MATERIALS AND METHODS
Bacterial strains.
Cultures of Desulfovibrio desulfuricans and Geobacter chapellei were obtained from the U.S. DOE Subsurface Microbial Culture Collection.
G. chapellei was grown anaerobically in 100-ml serum bottles containing an 80% N2-20% CO2 gas headspace and the following components (per liter): 5 mg of tryptone, 3 mg of yeast extract, 1 mg of glucose, 420 mg of KH2PO4, 220 mg of K2HPO4, 200 mg of NH4Cl, 380 mg of KCl, 360 mg of NaCl, 40 mg of CaCl2 · 2H2O, 100 mg of MgSO4 · 7H2O, 1.8 g of NaHCO3, 500 mg of Na2CO3, 8.0 g of fumarate, 10 ml of mineral elixir (see below), 25 ml of 2 M lactate, and 1 ml of 1 mM Na2SeO3. Mineral elixir contained the following components (per liter): 2.14 g of nitrilotriacetic acid, 100 mg of MnCl2 · 4H2O, 300 mg of FeSO4 · 7H2O, 170 mg of CoCl2 · 6H2O, 200 mg of ZnSO4 · 7H2O, 30 mg of CuCl2 · 2H2O, 5 mg of AlK(SO4)2 · 12H2O, 5 mg of H3BO3, 90 mg of Na2MoO4, 110 mg of NiSO4 · 6H2O, and 20 mg of Na2WO4 · 2H2O. After sterilization, 150 μl of vitamin mixture (see below) was added anaerobically to each serum bottle. Vitamin mixture contained the following components (per liter): 2 mg of biotin, 2 mg of folic acid, 10 mg of pyridoxine HCl, 5 mg of riboflavin, 5 mg of thiamine, 5 mg of nicotinic acid, 5 mg of pantothenic acid, 0.1 mg of cyanocobalamin, 5 mg of p-aminobenzoic acid, and 5 mg of thioctic acid. The bottles were inoculated with 1 ml of log-phase culture and grown in the dark at an ambient temperature for 2 weeks prior to RNA isolation.
Culture conditions for D. desulfuricans are described at the U.S. DOE Subsurface Microbial Culture Collection website (http://caddis.esr.pdx.edu/smccw/) and in reference 6. Briefly, cells (10% inoculum) were cultivated in 100 ml of medium C. This medium (pH 7.2) contained the following components (per liter): 7.9 ml of sodium lactate syrup (60%), 4.5 g of Na2SO4, 0.06 g of CaCl2 · 2H2O, 0.3 g of sodium citrate, 1.0 g of NH4Cl, 0.5 g of KH2PO4, 2.0 g of MgSO4 · 7H2O, and 1.0 g of yeast extract (Difco); the medium was degassed with N2 and sterilized by autoclaving. Sterilized growth medium was supplemented with 0.8 ml of FeSO4 solution (see below) and 1 ml of reductant solution (see below). FeSO4 solution contained (per 50 ml) 0.025 g of FeSO4 · 7H2O and 5 ml of 1 M H2SO4 and was degassed with N2 and filter sterilized; reductant solution contained (per 50 ml) 0.5 g of sodium thioglycolate and 0.5 g of ascorbic acid and was degassed with N2 and filter sterilized. Complete medium was anaerobically inoculated (10% [vol/vol]) with starter cultures. Cultures were incubated on a shaker platform in the dark at 30°C for 4 days (log-phase cultures) prior to RNA extraction.
Shewanella oneidensis MR-1 (a generous gift from Jim Fredrickson, Pacific Northwest National Laboratory) and Escherichia coli were both cultivated in Luria broth at 30°C.
RNA purification.
Cells were harvested by centrifugation at 8,000 × g, and total RNA was extracted from cell pellets by using an RNeasy minikit according to the manufacturer's instructions (Ambion Inc., Austin, Tex.). Purified RNA was eluted from the RNeasy minicolumn with two successive applications of 50 and 30 μl of RNase-free water. Purified nucleic acids were quantified by measuring the UV absorbance, and the presence of 16S rRNA was confirmed by gel electrophoresis with 2% agarose and Tris-acetate-EDTA running buffer.
RNA fragmentation and labeling.
Total RNA was labeled directly with a Ulysis kit (Alexa-532 fluor) according to the manufacturer's instructions (Molecular Probes, Eugene, Oreg.) and hybridized to DNA-conjugated beads without further purification or manipulation. Fragmented RNA was prepared from total, labeled RNA according to standard methods (2). Briefly, 6 μg of labeled RNA was diluted to 150 μl in fragmentation buffer to achieve a final buffer composition (pH 8.4) of 40 mM Tris, 100 mM potassium acetate, and 30 mM magnesium acetate. RNA was incubated for 30 min at 95°C and cooled on ice, and 100 ng was diluted to a 17-μl total volume in diethylpyrocarbonate (DEPC)-treated water containing 1 μg of sheared salmon sperm DNA (Sigma). The target RNA then was added to 33 μl of bead array solution in 1.5× high-salt (HS) hybridization buffer to achieve a 1× final concentration of HS phosphate hybridization buffer (200 mM NaPO4, 100 mM disodium EDTA, 0.25% Tween 20; adjusted to pH 5, 6, 7, or 8 with concentrated phosphoric acid). LS hybridization buffer was prepared from HS hybridization buffer by 1:20 dilution with DEPC-treated water.
Environmental extracts.
A shallow subsurface sediment sample was obtained from David Watson of the U.S. DOE NABIR Field Research Center (FRC) located at Oak Ridge National Laboratory (http://www.esd.ornl.gov/nabirfrc/). The FRC sample represents the uncontaminated FRC background site and contains approximately 54% sand, 35% silt, 10% clay, and 0.89% organic C. A high-biomass soil sample (a generous gift from Vanessa Bailey, Pacific Northwest National Laboratory) is from a tallgrass prairie restoration experiment being conducted at the Fermi National Laboratory in Batavia, Ill. The soil is a fine-silt, mixed, mesic Typic Haplaquoll with an estimated organic carbon content of 0.14%.
Total nucleic acids were extracted from 0.5-g aliquots of environmental samples by bead-beater extraction with 0.5 g of 0.1-mm glass beads in 1 ml of HS phosphate buffer at pH 5 or 8 for 3 min at maximum speed (∼5,000 oscillations min−1). Tubes were chilled briefly on ice and centrifuged at 10,000 × g for 2 min at 4°C to remove beads and sediment debris. The supernatants were removed, and like supernatants were combined and used directly in hybridization reactions without further manipulation.
Oligonucleotide probes.
Capture probe sequences used for this study were derived from alignments of dissimilatory metal- and sulfate-reducing bacterial 16S rRNA sequences deposited in GenBank; only full-length sequences for which an isolate is available in a public culture collection were used. Probes were designed around nucleotides 420 to 440 in the 16S rRNA (G. chapellei numbering; GenBank accession number U41561). DNA capture probes contained 5′-biotin and two C18 spacers and at least two mismatched nucleotides between all other capture probes on the array (Table 1). Capture and chaperone detector probes were synthesized and purified by high-pressure liquid chromatography at BioSource International (Camarillo, Calif.). A 15-nucleotide biotinylated fluorescent beacon nonsense probe was custom synthesized and purified by high-pressure liquid chromatography at Synthegen (Houston, Tex.). All probe stock solutions were reconstituted in DEPC-treated water, adjusted to a 200 μM concentration, and stored at −20°C until use.
TABLE 1.
Oligonucleotide probes and bead regions used in this study
| Probea | Abbreviation | Sequence (5′-3′)b | Perfect match | GenBank accession no. | Bead region |
|---|---|---|---|---|---|
| Capture | |||||
| S-G-Gbc-0825-a-A-14 | Gbc825 | TACCCGCRACACCT | Geobacter | L100-L104-01 | |
| S-S-Gbc.chap-0432-a-A-15 | Gbc446 | TTAGCCACAATACAC | Geobacter chapellei | U41561 | L100-L125-01 |
| S-S-Gbc.sulf-0382-a-A-15 | Gbs | TTAGCTCTCAATCAT | Geobacter sulfurreducens | U13928 | L100-L119-01 |
| S-S-Gbc.met-0454-a-A-15 | Gbm | TTAACCCTCAATCAC | Geobacter metallireducens | L07834 | L100-L115-01 |
| S-S-Gbc.brem-0445-a-A-15 | Gbb | ATTAGCCAGCCCCAT | Geobacter bremensis | U96917 | L100-L117-01 |
| S-S-Gbc.pel-0445-a-A-15 | Gbp | ATTAACCGCACACAT | Geobacter pelophilus | U96918 | L100-L140-01 |
| S-S-Dsv.des-0460-a-A-15 | Dvd | TTAGCACAACGTAGT | Desulfovibrio desulfuricans | M34113 | L100-L155-01 |
| S-S-Dsv.gab-0409-a-A-15 | Dvgb | TATTCGCATCCTCGG | Desulfovibrio gabonensis | U31080 | L100-L147-01 |
| S-S-Dsv.halo-0455-a-A-15 | Dvh | TTCGACTCTAATGGT | Desulfovibrio halophilus | U48243 | L100-L107-01 |
| S-S-Dsv.grac-0414-a-A-22 | Dvg | TATTCGACCTCAAGG | Desulfovibrio gracilis | U53464 | L100-L130-01 |
| Fluorescent beacon | TTGTGGTGGTGTGGT-Alexa-532 | L100-L128-01 | |||
| Chaperone | |||||
| S-S-Gbc.chap-0401-a-A-15 | GAGCTTTACGACCCG | G. chapellei, G. metallireducens, G. bremensis | |||
| S-S-Dsv.des-0429-a-A-15 | GAGGTTTACGATCCG | D. desulfuricans |
Oligonucleotide probes are named according to the guidelines of Alm et al. (1).
All capture probes were synthesized with 5′-biotin and two C18 spacers. The fluorescent beacon probe was synthesized with 5′-biotin and a 3′-C7-amino modifier.
Bead conjugation.
Biotinylated DNA probes were coupled to Lumavidin beads (Table 1) as specified by Luminex (Austin, Tex.). The efficiency of probe coupling to Lumavidin beads was determined by competitive binding experiments with biotin-phycoerythrin according to Luminex instructions and ranged from 87 to 96% of available biotin sites. Conjugated beads were stored at 4°C, and all bead concentrations were verified by counting bead slurries in an improved Neubauer Brightline counting chamber (Hausser Scientific Co., Horsham, Pa.) before use.
Manual nucleic acid hybridization to bead reagents.
Five thousand DNA-conjugated beads were used for each hybridization reaction. Coupled beads were rinsed and reconstituted in 33 μl of extraction-hybridization buffer as described above. To estimate tunable surface bead array detection limits (Fig. 1), 100 ng to 100 pg of total labeled RNA (intact) was heat denatured in 17 μl of DEPC-treated water at 95°C for 5 min in the presence of 1 μg of sheared salmon sperm DNA. To estimate hybridization specificity and the effect of soluble environmental constituents on the bead array process, 100 ng of intact or fragmented RNA was heat denatured in 17 μl of DEPC-treated water at 95°C for 5 min in the presence of 1 μg of sheared salmon sperm DNA and with or without a 250 nM concentration of each proximal chaperone probe per reaction.
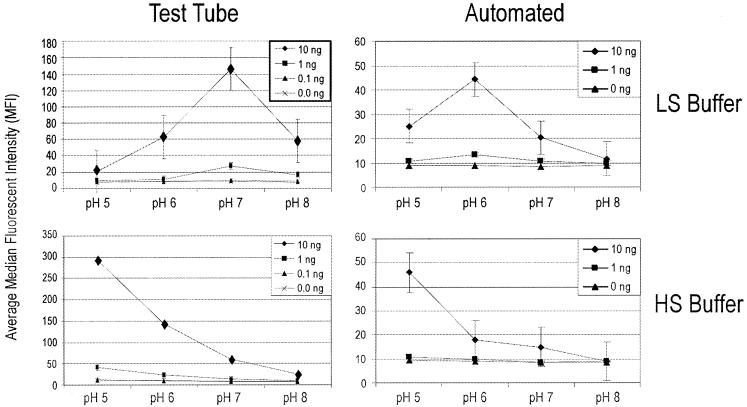
FIG. 1.
Detection limits of tunable surface microparticles and phosphate-based extraction-hybridization buffers for G. chapellei intact rRNA in both test tube and automated hybridization procedures. HS buffer, HS phosphate hybridization buffer; LS buffer, LS phosphate hybridization buffer. Test tube hybridizations were carried out for 15 min at room temperature. Total hybridization time for the automated routine was 180 s; each bolus of target rRNA was in contact with the tunable surface particles for only 30 s. Error bars indicate standard deviations.
For clarity, we use the term “proximal chaperone” in two senses. First, the probe is “proximal” to the target capture sequence and therefore is equivalent to a “stacking” probe (19). However, we borrow the term “chaperone” from the realm of protein assembly (16) to indicate that the proximal chaperone probe (presumably) serves to prevent incorrect structures (i.e., intramolecular secondary structures within the 16S rRNA target itself) from forming prior to hybridization on the array. If the chaperone probe also contains the detection label, then it serves the added function of a detector probe (11a).
The denatured target was added directly to the beads and incubated for 15 min, 2 h, or overnight at room temperature or 45°C (see Results). After hybridization, the beads were collected by low-speed centrifugation (2,200 × g), washed with 300 μl of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.02% Tween 20-0.5% Sarkosyl, collected by centrifugation, resuspended in 100 μl of 1× SSC-0.01% Tween 20-0.25% Sarkosyl (pH 7.0), and analyzed directly on the Luminex flow cytometer. At least three replicate hybridizations were performed for each treatment and used to calculate the average median fluorescence intensity (MFI) and standard deviation.
Evaluating interactions between environmental extracts and detection fluors.
The fluorescent beacon probe (5′-biotin-TTGTGGTGGTGTGGT-Alexa-532) was coupled at a 2 μM concentration to Lumavidin bead region L100-L128-01 according to the standard Luminex protocol to test for environmental background interference with the fluorescent reporter. To test for environmental background interference with the bead classification fluors, Lumavidin beads were used directly or coupled to their respective (nonfluorescent) capture probes as described above. Soil and sediment extracts (11.5 μl) in pH 5 or 8 HS phosphate buffer were added to the beads after the volume of the environmental extracts was adjusted to 17 μl with DEPC-treated water. Extracts were heat denatured at 95°C for 5 min and added to 33 μl of bead suspension (in HS phosphate buffer at pH 5 or 8, respectively) according to the standard hybridization protocol described above. Beads and heat-denatured environmental extracts were hybridized for 2 h at 45°C, washed with 2× SSC-0.02% Tween 20-0.5% Sarkosyl (pH 7.0), resuspended in 75 μl of 1× SSC-0.01% Tween 20-0.25% Sarkosyl (pH 7.0), and analyzed directly on the Luminex flow cytometer. At least three replicate hybridizations were performed for each treatment and used to calculate the average MFI and standard deviation.
Sequential-injection fluidic system.
The fluidic system was a FiaLab 3000 sequential injection system (Alitea) consisting of a 1-ml syringe pump (Cavro, Sunnyvale, Calif.), a 10-port selection valve (Valco; Cheminert, Houston, Tex.), and a holding coil. All tubing was 1-mm-inner-diameter FEP Teflon (Upchurch, Oak Harbor, Wash.). The custom renewable surface flow cell was machined from FEP fluoropolymer with an 0.89-mm-diameter nickel rod beveled to a 45° angle at one end. The beveled rod was rotated within the flow cell by using an Arsape AM1524 stepper motor and a 14:1 gear train (Donovan Micro-Tek, Inc., Simi Valley, Calif.) to achieve particle capture and release. The fluidic system and rotating rod were controlled with a laptop computer and in-house system control software written in Microsoft Visual C++ with a LabWindows/CVI (National Instruments, Austin, Tex.) user interface. In order to trap the 5.6-μm Luminex particles in the rotating-rod flow cell, we first formed a packed bed of 5 × 104 or 6.25 × 104 polystyrene beads (Bangs Laboratories, Inc., Fishers, Ind.; 19.9 ± 0.6 μm [mean and standard deviation]) and then layered 5 × 103 Luminex particles on top as described in detail elsewhere (9).
Flow cytometer detection.
A Luminex 100 flow cytometer served as the detector for all studies. This apparatus is equipped with a 635-nm diode classification laser and a 532-nm frequency-doubled diode reporter laser. Sheath fluid is passed through a flow channel measuring 200 by 200 μm at 90 μl s−1, with a 20- to 25-μm-diameter sample stream. Samples were injected at 60 μl min−1, with the cytometer set to count 100 events per bead type. A calibration run was performed with CAL1 and CAL2 calibration microspheres each day according to Luminex instructions. Bead suspensions were disaggregated by vortexing prior to sample injection into the flow cytometer. The flow cytometer was controlled with a Pentium II computer, and all Luminex data analyses were performed with LMAT version 1.7 software (Luminex).
Automated nucleic acid binding and detection.
Two automated routines were used during the course of the study as required by the transition from evaluating the tunable surface hypothesis to automating a model array and manipulating environmental extracts in the fluidic system. The tunable surface hypothesis was evaluated with the automated routine outlined in Table 2. Packed columns were formed in the appropriate HS or LS phosphate hybridization buffer. Labeled RNA was heat denatured at 95°C for 5 min off-line in the presence of 1 μg of salmon sperm DNA and perfused over the packed column in a continuous stream at 0.5 μl s−1. Reacted beads were washed with phosphate buffer, and the particles were released from the column with 2× SSC-002% Tween 20. Released particles were concentrated by low-speed centrifugation and resuspended in 1× SSC-0.01% Tween 20 for analysis on the Luminex flow cytometer as described above.
TABLE 2.
Renewable microcolumn program for evaluating the tunable surface hypothesis
| Manipulation | Solution | Vol (μl) | Flow rate (μl s−1) |
|---|---|---|---|
| Form column (rod in trap position)a | Top layer | ||
| Packing layer | Phosphate-buffered saline-Tween 20 | 100 | 10 |
| Luminex particles | HS or LS phosphate | 50 | 10 |
| Rinse beads | Phosphate buffers (see Results) | 140 | 10 |
| Perfuse with denatured rRNA target | Room temp or 45°C (see Results) | 50 | 0.5 |
| Wash beads | Phosphate buffers (see Results) | 100 | 1 |
| Release columnb | |||
| Reverse flow | 2× SSC-Tween 20 | 300 | 100 |
| Rotate rod to release | |||
| Flush column | 100 | ||
| Clean flow cell | 2× SSC-0.02% Tween 20 | 450 | 200 |
For the bead packing protocol, 5 × 104 19.9-μm beads in a 100-μl volume were injected into the rotating-rod microcolumn, followed by 100 μl of Luminex bead suspension containing 5 × 103 beads of each color.
Samples collected from the rotating rod were centrifuged and resuspended in 300 μl of 1× SSC-0.01% Tween 20 for direct analysis on the Luminex flow cytometer.
The automated nucleic acid capture and detection procedure for model arrays and environmental extracts is outlined in Table 3. Packed columns consisting of 6.25 × 104 19.9-μm packing beads and 5 × 103 Luminex beads of each color were formed as described above. Labeled RNA was heat denatured at 95°C for 5 min off-line in the presence of 1 μg of salmon sperm DNA or environmental extract and perfused over the column in 5-μl steps, with each bolus being kept in the column for 30 s. Beads (in the column) were washed with fresh hybridization buffer (at pH 5 or 8; containing 0.5% Sarkosyl) and eluted with 1× SSC-0.01% Tween 20-0.25% Sarkosyl (pH 7.0) for analysis on the Luminex flow cytometer. At least three replicate hybridizations were performed for each treatment and used to calculate the average MFI and standard deviation.
TABLE 3.
Automated renewable microcolumn procedure for rRNA capture and detection
| Manipulation | Solution | Vol (μl) | Flow rate (μl s−1) |
|---|---|---|---|
| Form column (rod in trap position)a | Top layer | ||
| Packing layer | Phosphate-buffered saline-Tween 20 | 125 | 10 |
| Luminex particles | HS phosphate | 50 | 10 |
| Rinse beads | Phosphate buffer (see Results) | 140 | 10 |
| Perfuse with denatured rRNA target | Room temp or 45°C (see Results) | 50 | 0.6b |
| Wash beads | Phosphate buffers (see Results) | 100c | 1 |
| Exchange buffer | 2× SSC-Tween 20-Sarkosyl | 340 | 1 |
| Release column | |||
| Reverse flow | 1× SSC-Tween 20-Sarkosyl | 300 | 100 |
| Rotate rod to release | |||
| Flush column | 100 | ||
| Clean flow cell | 2× SSC-0.02% Tween 20 | 250 | 20 |
For the bead packing protocol, 6.25 × 104 19.9-μm beads in a 100-μl volume were injected into the rotating-rod microcolumn, followed by 100 μl of Luminex bead suspension containing 5 × 103 beads of each color as described previously (9).
The automated hybridization routine was programmed in six steps; in each step, 5 μl of denatured target was perfused over the column, and the solution was kept in the column for 30 s before the next bolus of target solution was introduced.
For the data from some experiments (see Fig. 5), 300 μl of hybridization buffer was used to wash the column during this step.
RESULTS
Tunable surface hypothesis.
One tenet of the integrated system objective is to simplify system hardware by simplifying the embedded biochemistry of the system. In this study, we therefore used a buffer system (alkaline phosphate) that is conducive to the extraction of DNA and RNA from low-biomass environmental samples (22) and bead-based affinity purification methods (11) to test the tunable surface hypothesis. As shown in Fig. 1, the capture and detection of intact rRNA on Lumavidin beads were enhanced in both test tube and automated capture systems as the buffer pH became acidic. The detection limits in both LS and HS phosphate buffers were 1 ng of total RNA (∼104 cell equivalents) in test tube hybridizations and 10 ng of total RNA (∼105 cell equivalents) in hybridizations with the automated system. The tunable surface HS phosphate buffer condition (pH 5) provided a 10-fold increase in signal intensity and a 10-fold increase in detection limits (1 ng of total RNA versus 10 ng of total RNA) over the standard buffer condition (pH 8). Even though detection limits were identical in LS and HS phosphate buffers, buffer ionic strength (LS buffer) had an obvious and unpredictable effect on hybridization pH and signal intensity (the tunable surface response curve) in both test tube and automated formats. Further, the test tube results in the LS phosphate buffer experiments did not translate directly to automated system results as they did in the HS phosphate buffer experiments. For these reasons, all future tunable surface studies were done with the HS phosphate buffer system.
Hybridization specificity under tunable surface HS phosphate buffer conditions.
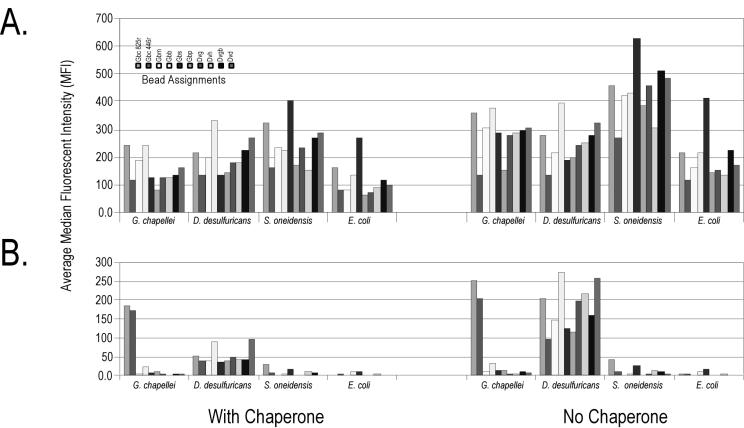
Having verified the tunable surface hypothesis with a phosphate-based extraction-hybridization buffer, we constructed a simple 10-probe bead array to assess potential (and unexpected) cross-hybridization of rRNA from metal- and sulfate-reducing bacteria and determined which additional biochemical manipulations (solutions or physical or mechanical processing) might be required to achieve hybridization specificity in the automated system. One hundred nanograms (∼106 cell equivalents of total RNA) of intact or fragmented RNA from four test organisms was hybridized to a 10-bead array at room temperature for 15 min with or without a proximal chaperone probe. As shown in Fig. 2, intact (total) RNA was not specifically detected in the model array either in the presence or in the absence of a proximal chaperone. RNA fragmentation was required to achieve any modicum of hybridization specificity, in keeping with proximal chaperone methods originally developed for planar arrays (23). The proximal chaperone did reduce nonspecific cross-hybridization of D. desulfuricans rRNA to most of the Geobacter probes, with the strongest signal generated from the D. desulfuricans capture probe. Nonetheless, the D. desulfuricans target consistently generated a significant signal (>2 times that of the background) with nontarget probes despite sharing only 50% sequence identity with the Geobacter capture probes.
FIG. 2.
Achieving hybridization specificity on a tunable surface suspension array during manual nucleic acid hybridization. A sample of 100 ng of target RNA was heat denatured in the presence of 1 μg of salmon sperm DNA with or without a 250 nM concentration of each of the two chaperone probes in pH 5 HS phosphate buffer. Hybridization reactions proceeded for 2 h at room temperature. Beads then were washed with 2× SSC-0.02% Tween 20-0.5% Sarkosyl (pH 7) buffer at room temperature before buffer exchange into 1× SSC-0.01% Tween 20-0.25% Sarkosyl (pH 7) and analysis on the flow cytometer. (A) Intact RNA targets. (B) Fragmented RNA targets. Data are corrected for background, and error bars are omitted for presentation clarity. The order of the bead assignments (from left to right) in the key is the same as in the bar graphs.
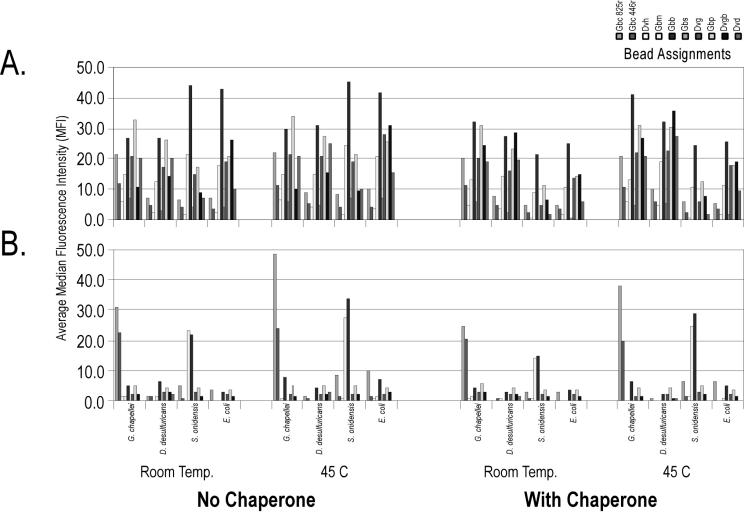
Results obtained in test tube hybridizations (Fig. 2) generally transferred to automated hybridizations (Fig. 3). There was no hybridization specificity with intact rRNA, even in the presence of a proximal chaperone or increased temperature. In contrast, fragmented RNA coupled with a proximal chaperone provided reasonable specificity in the automated system. In this instance, the D. desulfuricans target RNA hybridized very poorly, and the S. oneidensis MR-1 target RNA produced relatively strong cross-hybridization to the Gbm and Gbb probes (<50% sequence identity in the target region). Increased hybridization temperature (45°C) did not eliminate the cross-hybridization observed under room temperature hybridization conditions. These results indicate that the automated system and very short contact times can be used to analyze rRNA on a suspension array but that additional validation experiments are required in order to understand the causes of unexpected cross-hybridization or eliminate unpredictable probes. These results also indicate that fragmented rRNA and proximal chaperone probes may be the preferred bioanalytical manipulations for direct detection of rRNA on the tunable bead surface, with concomitant implications for the design, construction, and routine operation of an integrated microbial monitoring system. The implications of unpredictable cross-hybridization for the analysis of uncharacterized environmental samples are discussed below.
FIG. 3.
Achieving hybridization specificity on a tunable surface suspension array during automated nucleic acid hybridization. A sample of 100 ng of target RNA was heat denatured in the presence of 1 μg of salmon sperm DNA and a 250 nM concentration of each of the two chaperone probes in pH 5 HS phosphate buffer. Hybridization reactions proceeded as outlined in Table 2 at room temperature (Temp.) or 45°C. Beads then were washed with pH 5 HS buffer at room temperature and 2× SSC-0.02% Tween 20-0.5% Sarkosyl (pH 7) buffer before buffer exchange into 1× SSC-0.01% Tween 20-0.25% Sarkosyl (pH 7) for analysis on the flow cytometer. (A) Intact RNA targets. (B) Fragmented RNA targets. Data are corrected for background, and error bars are omitted for presentation clarity. The order of the bead assignments (from left to right) in the key is the same as in the bar graphs.
Tunable surface chemistry and environmental backgrounds.
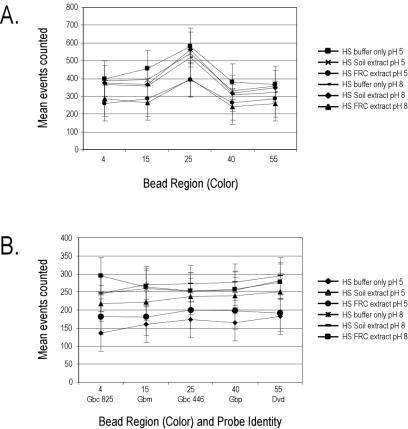
Nonpurified environmental extracts were incubated with uncoupled or coupled Lumavidin beads in a mock test tube hybridization reaction (2 h, 45°C) as described in Materials and Methods, and the number of bead counts (or events) was recorded for each bead color. As shown in Fig. 4, different buffer conditions (pH and environmental background) had significant effects on events counted for each bead color, suggesting different levels of bead aggregation in different buffers or environmental backgrounds. Within a specific buffer condition, however, all bead colors were counted with equal efficiencies (P value determined by analysis of variance, >0.05). We therefore conclude that bead classification is not adversely affected by nonpurified environmental extracts regardless of extraction buffer pH (pH 5 or 8).
FIG. 4.
Bead counting and detection efficiency after mock hybridization with nonpurified FRC and soil extracts. (A) Uncoupled Lumavidin beads. (B) Oligonucleotide-coupled Lumavidin beads. The flow cytometer was set to count 3,750 events (75% of the total beads for each color). Data are plotted as mean events counted plus or minus standard deviations.
To test whether environmental backgrounds affect the fluorescent reporter (Alexa-532), an Alexa-532-labeled nonsense oligonucleotide was coupled through the biotin bridge to Lumavidin bead surfaces and reacted with environmental extracts under the mock test tube hybridization protocol (2 h) or the automated hybridization protocol. Data from this experiment (Table 4) generated several important results and conclusions. First, while the environmental backgrounds did not interfere with quantitative bead classification (or counting; Fig. 4), the residue from environmental backgrounds had a significant quenching effect on the Alexa-532 reporter. The extent of fluorescence quenching did depend on soil type and extraction buffer pH. Second, modulating surface charge during the wash step did little to influence the extent of fluorescence quenching of the Alexa-532 reporter, regardless of extraction or wash buffer pH. Third, the extent of fluorescence quenching was substantially lower in the automated hybridization than in the mock test tube hybridization (2 h, 45°C). The implications of fluorescence quenching at the point of detection for either an integrated system or a field-deployable analytical method are discussed below. We conclude from these data that while some nonpurified nucleic acid extracts may be amenable to direct affinity capture and detection on tunable bead surfaces (e.g., the FRC sediment), some level of prepurification is probably required for routine analysis of uncharacterized environmental samples. In addition, it will be necessary to incorporate a control for fluorescence quenching (regardless of up-front nucleic acid purification strategy) so that one can explicitly measure the effects of environmental backgrounds on the signaling system, normalize data through time and space, and (ultimately) quantify signal intensities relative to target densities in environmental samples.
TABLE 4.
Percent retention of Alexa-532 signal in model environmental backgroundsa
| Sample | pH
|
% Retention of Alexa-532 signal in the following hybridization reaction:
|
||
|---|---|---|---|---|
| Extraction | Wash | Test tube | Automated | |
| Prairie soil | 5 | 5 | 42 | 86 |
| 5 | 8 | 42 | 88 | |
| 8 | 5 | 5 | 17 | |
| 8 | 8 | 5 | 19 | |
| FRC sediment | 5 | 5 | 78 | 94 |
| 5 | 8 | 76 | 96 | |
| 8 | 5 | 91 | 90 | |
| 8 | 8 | 91 | 91 | |
A nonsense Alexa-532-labeled oligonucleotide was coupled to five different bead colors (100 events per color; three repetitions) and incubated in mock test tube or automated hybridization reactions as described in the text. Modulating surface charge did not influence the interaction of humic acids with the conjugated beads. The automated system greatly reduced the effects of fluorescence quenching, especially in the soil background.
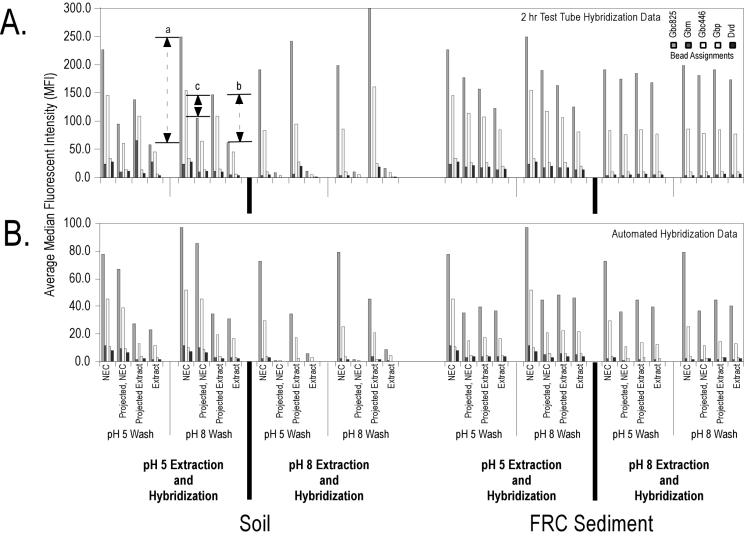
To test whether environmental backgrounds and tunable surface chemistry affect hybridization and detection efficiencies, 100 ng of G. chapellei fragmented RNA was added to environmental backgrounds and hybridized to a simple five-bead array in either a 2-h test tube hybridization protocol or an automated hybridization protocol according to the experimental procedure shown in Table 3. To separate the effects of fluorescence quenching on signal intensity (Table 3) from the effects of environmental backgrounds on hybridization efficiency, we estimated two additional signal intensity values based on the experimental data and the quenching estimates from Table 3. The first estimate (projected no-extract control) was calculated as the average MFI for the no-extract control (experimental value) × the percent reduction in signal intensity, i.e., the signal intensity one would expect if the sediment extract alone were added to the beads (i.e., the Table 3 value for the particular buffer and wash conditions). The second estimate (projected extract) was calculated as the average MFI for the amended sediment extract × (1/percent reduction in signal intensity), i.e., the expected hybridization signal assuming no quenching of or interference with the reading of the Alexa-532 reporter. As shown in Fig. 5, the difference between the average MFI values for the amended extract and the purified RNA hybridization control (interval a; shown for probe Gbc825) represents the decrease in hybridization signal due to the combined effects of fluorescence quenching and hybridization interference. The difference between the average MFI values for the amended extract and the projected extract (interval b; shown for probe Gbc825) is an estimate of the effect of the environmental background on hybridization efficiency alone. The difference between the average MFI values for the projected no-extract control and the projected extract (interval c; shown for probe Gbc825) is an estimate of the uncertainty associated with the effect of the environmental background on Alexa-532 reporting efficiency.
FIG. 5.
Integrated nucleic acid purification and flow cytometry detection in manual (A) and automated (B) formats with tunable surface extraction, hybridization, and wash conditions. A sample of 100 ng of fragmented G. chapellei target RNA and chaperone probes were added directly to each environmental extract and hybridized for 2 h in a test tube or processed through the automated routine, both at 45°C. All experimental signals are corrected for background bead fluorescence in the respective hybridization and wash conditions. Error bars are omitted for presentation clarity. NEC represents the no-extract control (experimental value); Projected, NEC represents the average MFI of the no-extract control (experimental value) times the percent reduction in signal intensity, i.e., the signal intensity one would expect if the sediment extract alone were added to the beads (i.e., the value in Table 3 for the particular buffer and wash conditions); Extract represents the amended extract (experimental value); and Projected Extract represents the average MFI of the amended extract times (1/percent reduction in signal intensity), i.e., the hybridization signal expected assuming no quenching of or interference with the reading of the Alexa-532 reporter. Interval a is the difference between the average MFIs of the amended extract and the purified RNA hybridization control and represents the decrease in hybridization signal due to the combined effects of fluorescence quenching and hybridization interference. Interval b is the difference between the average MFIs of the amended extract and the projected extract and represents an estimate of the effect of the environmental background on hybridization efficiency alone. Interval c is the difference between the average MFIs of the projected no-extract control and the projected extract and represents an estimate of the uncertainty associated with the effect of the environmental background on Alexa-532 reporting efficiency. The order of the bead assignments (from left to right) in the key is the same as in the bar graphs.
From the experimental and calculated values shown in Fig. 5, then, we can draw the following conclusions. First, the nonpurified environmental extracts not only reduced Alexa-532 detection efficiency (Table 3) but also reduced hybridization efficiency. The extent of hybridization interference was pronounced in the surface soil extracts; the pH 8 extract and background quenched fluorescence so potently that estimating the effects of soluble environmental constituents on hybridization efficiency was impossible. These data confirm that some level of sample pretreatment or purification will be required in order to develop a universal Luminex-based suspension array protocol applicable to a wider range of environmental samples. Second, the reduction in average MFI for the FRC sediment extracts could be attributed primarily to Alexa-532 quenching as opposed to a reduction in hybridization efficiency. These data show that it will be possible to directly couple nucleic acid purification and detection for at least some environmental samples and backgrounds. Third, even in the absence of information on fluorescence quenching (Table 3), the method was still able to read through the observed interferences for both backgrounds and both tunable surface buffer conditions, suggesting that the extent of sample purification prior to suspension array detection may still conform to a simple, rapid, and automated routine.
DISCUSSION
Tunable surface chemistry.
The tunable surface concept described by Belosludtsev et al. (5) is based on a prototypical streptavidin-coated surface. Streptavidin is covalently attached to an array surface (typically a glass surface) in a monolayer. Biotinylated oligonucleotide probes are subsequently attached in an array. At a neutral pH, streptavidin is highly anionic and provides a bulk negative surface charge, discouraging the formation of nucleic acid duplexes at the surface. At pHs of ≤5.5, streptavidin is driven into a cationic form, providing enhanced attraction to the surface for subsequent duplex formation and shielding strand-strand charge repulsion of nucleic acid duplexes. Thus, the tunable surface hypothesis states that buffers with a low ionic strength can be effectively used for hybridization, reducing the formation of intramolecular secondary structures that might otherwise prevent the hybridization of structured molecules to a tethered probe (e.g., 16S rRNA). We tested this hypothesis with a buffer system that has also been successfully used to directly extract nucleic acids from low-biomass environmental samples in an attempt to simplify the bioanalytical process in a manner that is simultaneously conducive to integration with an autonomous environmental monitoring device.
The results in Fig. 1 show that driving a Lumavidin (streptavidin derivative) surface into a cationic state does indeed enhance the detection limit for 16S rRNA in both HS and LS phosphate buffers and in test tube and automated formats. Interestingly, the LS phosphate hybridization condition did little to enhance detection limits or average MFI values at each target concentration, a result that does not conform to the tunable surface hypothesis. Regardless, the detection limits in phosphate extraction-hybridization buffers were similar to those described in a previous study in which standard SSC hybridization buffers were used (9). The automated hybridization was approximately 10-fold less sensitive than a 15-min test tube hybridization, representing approximately 105 cell equivalents of total RNA (assuming 59 × 10−15 g of RNA for an E. coli cell grown in nutrient medium [21], of which approximately 27% is 16S rRNA). We attribute the loss in sensitivity to extremely short contact times in the microfluidic system (i.e., a 30-s residence time in the pulsed-flow renewable column versus 15 min to 2 h in the test tube). As shown in Table 3, however, the automated system was more efficient at eliminating factors that interfered with fluorescence from nonpurified environmental extracts. Thus, renewable microcolumn method detection limits may still be equivalent to or better than batch process detection limits in an environmental context. This hypothesis will need to be tested explicitly with a range of environmental samples and extracts containing known quantities of target RNA.
The “color” of soil and sediment.
One way to simplify the biochemistry and processing steps of a field-deployable monitoring device is to utilize the suspension microarray itself both as the direct affinity purification vehicle for target rRNA and as the sensing surface in a flow cytometer detector. In order for combined suspension array sample purification and detection chemistry to be practically useful for the field deployment objective and to acquire quantitative data relative to the abundance of microorganisms in the environment, however, it is necessary to understand how soluble environmental constituents affect bead classification across the range of bead colors, fluorescence reporting mechanisms, and hybridization efficiency or efficacy. Thus, a high-biomass garden soil and a low-biomass FRC sediment were extracted with HS phosphate at either pH 5 or pH 8 to test the hypotheses that a tunable surface (e.g., pH 5) hybridization method can be applied directly to nonpurified extracts without affecting fluorescence detection and that charge manipulation of the microbead surface can remove, minimize, or eliminate fluorescence quenching or autofluorescence arising from residual humic acid (or soluble environmental constituent) interactions with the suspension array.
The results in Table 3 and Fig. 4 show that the color of soluble soil and sediment constituents is (not unexpectedly) influenced by extraction buffer pH and that the current Luminex platform is challenged to read through environmental backgrounds. Indeed, continued use of the Luminex (or related) fluorescent reporters within an integrated system implies either that a universal sample purification chemistry must be developed (and formatted for autonomous applications) to remove fluorescence quenchers or that alternative colors (lasers or fluors) must be developed to retain a simplified analytical process and microfluidic architecture within an autonomous integrated system. In the meantime, the fluorescent beacon and positive control data provide a tentative normalization function and path toward quantifying target signal intensities in variable and uncharacterized environmental backgrounds.
Interestingly, the tunable surface extraction and hybridization conditions (pH 5) did not increase the average MFI values for the no-extract controls relative to the pH 8 extraction conditions (Fig. 5), and any increase in the average MFI values for the amended extracts could be attributed largely to the differential effect of extraction pH on background interference with Alexa-532 fluorescence intensity. These data imply that some level of nucleic acid purification will be required prior to bead array hybridization in order to take full advantage of the tunable surface effect and increase bead array detection limits within an automated system.
Hybridization specificity.
It is not yet known how microarray technology (in general) performs under the tunable surface hybridization conditions or in the presence of soluble environmental extracts. Earlier work with suspension array particles indicated that intact rRNA could be specifically captured in a 1,000-fold excess of salmon sperm DNA without RNA fragmentation and without the use of the secondary chaperone or stacking probes that are typically required for planar microarray analysis of rRNA (5, 11a, 23). The ability to directly and specifically detect intact rRNA would greatly simplify the bioanalytical process and attendant integrated monitoring system. We therefore constructed a simple 10-bead array to evaluate hybridization specificity under the tunable surface hybridization conditions.
As shown in Fig. 2 and 3, RNA fragmentation was essential to obtain any level of hybridization specificity, a result that is consistent with prior work on planar arrays. The chaperone probe did reduce the nonspecific hybridization of D. desulfuricans rRNA in the test tube format but did not dramatically alter hybridization profiles or cross-hybridization of Shewanella rRNA in the automated format. The differential cross-hybridization results for the test tube and automated formats are both interesting and problematic. On the one hand, the data show that short contact times (i.e., hybridization kinetics) may be used to modify or alter the extent and nature of nonspecific interactions on the array and that secondary probes may not be required in order to achieve hybridization specificity on microbead surfaces. However, the model array and target RNA set are fairly simple, and this result should be verified as the suspension array is expanded to cover more species and genera. On the other hand, inexplicable cross-hybridization of known targets to probes containing only 50% sequence identity will present a formidable challenge for the interpretation of data in the analysis of unknown or uncharacterized environmental samples. That is, traditional gene detection or expression profile microarray data analysis and interpretation are predicated on two fundamental assumptions: first, that what one knows is all that there is to know, and second, that hybridization specificity on the array is (nearly) perfect. Put another way, because only a small portion of natural microbial diversity has been identified and because microarray hybridization specificity is not perfect, it is practically and theoretically difficult to know if and when hybridization signals in a new environment result from a perfectly matched or a mismatched probe-target combination.
One strategy for addressing cross-hybridization is simply to remove unpredictable probes from the array. However, this strategy is plagued by the aforementioned uncertainties in an environmental context and leads to a never-ending (and resource-limiting) validation exercise as new organisms are isolated from the environment or new gene sequences from uncultivated organisms are recovered from clone libraries. Another common strategy for addressing unpredictable cross-hybridization is to increase the total number of probes on an array and statistically compare the signal intensities between perfectly matched and single-base-mismatched duplexes. However, how can one define a perfect match when the environmental background is uncharacterized? Indeed, recent results obtained with a high-density photolithographic phylochip (256,000 probes) and software for the analysis of perfect matches versus single-base mismatches only identified bacteria in concentrated aerosols to the third level of phylogenetic rank, as defined in the Ribosomal Database Project (18, 27).
Developing planar array methods to unambiguously ascribe a hybridization signal to a perfect or imperfect match represents a significant technology development challenge. One precedent and path forward can be drawn from two studies in which the liquid-phase nature of three-dimensional gel element arrays was used to empirically measure thermal dissociation curves for every probe on the array (12, 17). The extent to which underlying thermal melt theory and principles can be applied to planar microarray substrates is to be determined.
In the meantime, we can offer several recommendations for the interpretation of data based on our present results and conclusions, even though this study was not designed to address microbial community profiling per se. First, we fully expect that as the array is expanded (to include more genera) and applied to environmental samples, cross-hybridization will occur. Thus, we currently restrict our interpretation of data to the genus level or above, not the species level. Second, even genus-level detection should not presently be considered absolute. If, however, we assume that any one rRNA sequence will reproducibly hybridize to an immobilized oligonucleotide probe, then we can at least be confident that changes in measured rRNA profiles through time and space represent real changes in underlying microbial community composition rather than an artifact of the analytical process (e.g., PCR bias). Thus, microarray data should not be interpreted in a vacuum; supporting biochemical, geochemical, or highly specific molecular analyses are still necessary to support or confirm ecological observations from microarray-based community profiling experiments.
Acknowledgments
This work was supported by the U.S. DOE under the Assessment Element of the NABIR Program. Argonne National Laboratory is operated for the U.S. DOE by the University of Chicago under contract W-31-109-ENG-38. Pacific Northwest National Laboratory is operated for the U.S. DOE by Battelle Memorial Institute under contract DE-AC06-76RLO-1830.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Bavykin, S. G., J. P. Akowski, V. M. Zakhariev, V. E. Barksy, A. N. Perov, and A. D. Mirzabekov. 2001. Portable system for microbial sample preparation and oligonucleotide microarray analysis. Appl. Environ. Microbiol. 67:922-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belgrader, P., W. Bennett, D. Hadley, G. Long, R. Mariella, Jr., F. Milanovich, S. Nasarabadi, W. Nelson, J. Richards, and P. Stratton. 1998. Rapid pathogen detection using a microchip PCR array instrument. Clin. Chem. 44:2191-2194. [PubMed] [Google Scholar]
- 5.Belosludtsev, Y., I. Belosludtsev, B. Iverson, S. Lemeshko, R. Wiese, M. Hogan, and T. Powdrill. 2001. Nearly instantaneous, cation-independent, high selectivity nucleic acid hybridization to DNA microarrays. Biochem. Biophys. Res. Commun. 282:1263-1267. [DOI] [PubMed] [Google Scholar]
- 6.Boone, D. R., R. L. Johnson, and Y. Liu. 1989. Diffusion of the interspecies electron carriers H2 and formate in methanogenic ecosystems and its implications in the measurement of Km for H2 or formate uptake. Appl. Environ. Microbiol. 55:1735-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brockman, F. J. 1995. Nucleic acid-based methods for monitoring the performance of in situ bioremediation. Mol. Ecol. 4:567-578. [Google Scholar]
- 8.Chandler, D. P., D. A. Holman, F. J. Brockman, J. W. Grate, and C. J. Bruckner-Lea. 2000. Renewable microcolumns for solid-phase nucleic acid separations and analysis from environmental samples. Trends Anal. Chem. 19:314-321. [Google Scholar]
- 9.Chandler, D. P., and A. E. Jarrell. 2003. Enhanced nucleic acid capture and flow cytometry detection with peptide nucleic acid probes and tunable surface microparticles. Anal. Biochem. 312:182-190. [DOI] [PubMed] [Google Scholar]
- 10.Chandler, D. P., G. J. Newton, J. A. Small, and D. S. Daly. 2003. Sequence versus structure for the direct detection of 16S rRNA on planar oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandler, D. P., J. R. Stults, S. Cebula, B. L. Schuck, D. W. Weaver, K. K. Anderson, M. Egholm, and F. J. Brockman. 2000. Affinity purification of DNA and RNA from environmental samples with peptide nucleic acid clamps. Appl. Environ. Microbiol. 66:3438-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Eggers, M. D., W. J. Balch, L. G. Mendoza, R. Gangadharan, A. K. Mallik, G. McMahon, M. E. Hogan, D. Xaio, T. R. Powdrill, B. Iverson, G. E. Fox, R. C. Willson, K. I. Maillard, J. L. Siefert, and N. Singh. 1997. Advanced approach to simultaneous monitoring of multiple bacteria in space, p. 1-8. Proceedings of the 27th International Conference on Environmental Systems, SAE International, Warrendale, Pa.
- 12.El Fantroussi, S., H. Urakawa, A. E. Bernhard, J. J. Kelly, P. A. Noble, H. Smidt, G. M. Yershov, and D. A. Stahl. 2003. Direct profiling of environmental microbial populations by thermal dissociation analysis of native ribosomal RNAs hybridized to oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2377-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilles, P. N., D. J. Wu, C. B. Foster, P. J. Dillon, and S. J. chanock. 1999. Single nucleotide polymorphic discrimination by an electronic dot blot assay on semiconductor microchips. Nat. Biotechnol. 17:365-370. [DOI] [PubMed] [Google Scholar]
- 14.Iannone, M. A., J. D. Taylor, J. Chen, M.-S. Li, P. Rivers, K. A. Slentz-Kesler, and M. P. Weiner. 2000. Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry. Cytometry 39:131-140. [PubMed] [Google Scholar]
- 15.Khandurina, J., T. E. McKnight, S. C. Jacobson, L. C. Waters, R. S. Foote, and J. M. Ramsey. 2000. Integrated system for rapid PCR-based DNA analysis in microfluidic devices. Anal. Chem. 72:2995-3000. [DOI] [PubMed] [Google Scholar]
- 16.Lewin, B. 1990. Genes IV. Oxford University Press, New York, N.Y.
- 17.Liu, W. T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 18.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maldonado-Rodriguez, R., M. Espinosa-Lara, A. Calixto-Suárez, W. G. Beattie, and K. L. Beattie. 1999. Hybridization of glass-tethered oligonucleotide probes to target strands preannealed with labeled auxiliary oligonucleotides. Mol. Biotechnol. 11:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Nasarabadi, S., F. Milanovich, J. Richards, and P. Belgrader. 1999. Simultaneous detection of TaqMan probes containing FAM and TAMRA reporter fluorophores. BioTechniques 27:1116-1118. [PubMed] [Google Scholar]
- 21.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Inc., Sunderland, Mass.
- 22.Ogram, A., W. Sun, F. J. Brockman, and J. K. Fredrickson. 1995. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl. Environ. Microbiol. 61:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small, J. A., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiro, A., and M. Lowe. 2002. Quantitation of DNA sequences in environmental PCR products by a multiplexed, bead-based method. Appl. Environ. Microbiol. 68:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiro, A., M. Lowe, and D. Brown. 2000. A bead-based method for multiplexed identification and quantitation of DNA sequences using flow cytometry. Appl. Environ. Microbiol. 66:4258-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tebbe, C. C., and W. Vahjen. 1993. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl. Environ. Microbiol. 59:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wintzingerode, F. V., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 29.Woolley, A. T., G. F. Sensabaugh, and R. A. Mathies. 1997. High-speed DNA genotyping using microfabricated capillary array electrophoresis chips. Anal. Chem. 69:2181-2186. [DOI] [PubMed] [Google Scholar]