Abstract
A facultatively anaerobic, acid-resistant bacterium, designated strain FRCl, was isolated from a low-pH, nitrate- and U(VI)-contaminated subsurface sediment at site FW-024 at the Natural and Accelerated Bioremediation Research Field Research Center in Oak Ridge, Tenn. Strain FRCl was enriched at pH 4.5 in minimal medium with nitrate as the electron acceptor, hydrogen as the electron donor, and acetate as the carbon source. Clones with 16S ribosomal DNA (rDNA) sequences identical to the sequence of strain FRCl were also detected in a U(VI)-reducing enrichment culture derived from the same sediment. Cells of strain FRCl were gram-negative motile regular rods 2.0 to 3.4 μm long and 0.7 to 0.9 μm in diameter. Strain FRCl was positive for indole production, by the methyl red test, and for ornithine decarboxylase; it was negative by the Voges-Proskauer test (for acetylmethylcarbinol production), for urea hydrolysis, for arginine dihydrolase, for lysine decarboxylase, for phenylalanine deaminase, for H2S production, and for gelatin hydrolysis. Strain FRCl was capable of using O2, NO3−, S2O32−, fumarate, and malate as terminal electron acceptors and of reducing U(VI) in the cell suspension. Analysis of the 16S rDNA sequence of the isolate indicated that this strain was 96.4% similar to Salmonella bongori and 96.3% similar to Enterobacter cloacae. Physiological and phylogenetic analyses suggested that strain FRCl belongs to the genus Salmonella and represents a new species, Salmonella subterranea sp. nov.
Microbial immobilization of uranium has been intensively studied (3, 29). Uranium is a long-lived radionuclide that poses ecological and human health hazards. The mining and processing of uranium for nuclear fuel and nuclear weapon production have resulted in the generation of significant amounts of radioactive waste. It is critical that the uranium in radioactive wastes be effectively immobilized in order to prevent groundwater contamination (26, 45, 50). Microbial reduction of soluble hexavalent uranium U(VI) to tetravalent uranium U(IV), which precipitates as the mineral uraninite, has been proposed as one of the methods for uranium immobilization (33).
Two major groups of microorganisms reported to reduce U(VI) at a near-neutral pH are dissimilatory Fe(III)-reducing microorganisms (16, 24, 33, 43) and sulfate-reducing microorganisms (6, 32, 34, 37, 46). Batch, column, and field experiments conducted with U(VI)-contaminated groundwater and sediments have shown that indigenous metal-reducing microorganisms can be stimulated and effectively used for uranium immobilization (1, 2, 4, 13-15). In studies in which dissimilatory metal reduction was stimulated in sediments at a near-neutral pH, molecular analysis revealed that indigenous Fe(III)-reducing bacteria belonging to the Geobacteraceae family predominated during uranium reduction in laboratory incubations (20) and in an in situ uranium bioremediation field trial (4). While microorganisms as potential agents for U(VI) bioremediation at a near-neutral pH have been rather well studied, organisms that might participate in U(VI) bioremediation at a moderately acidic pH (pH 4) have not previously been described.
In this paper, we describe the isolation of a facultatively anaerobic, acidotolerant bacterium, strain FRCl, from a low-pH, nitrate- and uranium-contaminated sediment at site FW-024 at the Natural and Accelerated Bioremediation Research (NABIR) Field Research Center in Oak Ridge, Tenn. Uranium-contaminated groundwater plumes at the NABIR Field Research Center originate from the former S-3 Waste Disposal Ponds, covering a total area of about 1.44 ha, which received liquid wastes from 1951 until 1983. These liquid wastes had a pH of <2 and consisted primarily of nitric acid plating wastes, which were the source of the nitrate, various metals, and radionuclides (7, 42). Uranium is the contaminant of primary concern in this aquifer. Strain FRCl has been enriched from this site at pH 4.5 with nitrate as the electron acceptor, hydrogen as the electron donor, and acetate as the carbon source. Additionally, strain FRCl was capable of reducing U(VI) in a cell suspension and was a component of the U(VI)-reducing enrichment culture FRCk, derived from the same sediment (44). Characteristics of strain FRCl are discussed in relation to its ecological niche. Phylogenetic analysis revealed that this strain represents a new species in the genus Salmonella.
MATERIALS AND METHODS
Sediment sampling.
Sediment was collected from the saturated zone of a low-pH area of the aquifer at site FW-024 at the NABIR Field Research Center in Oak Ridge, Tenn. This site has previously been described in detail (44). The sediment was aerobic and contained 65 μmol of bicarbonate-extractable U(VI) per kg. The pore water contained 0.81 μM U(VI), 8.5 mM nitrate, 10.9 mM sulfate, 0.49 mM acetate, and 0.11 mM citrate and had a pH of 4.0. Sediment was collected on 3 April 2001 from horizons A1 to C1 below the water table. After collection, the sediment was stored at 4°C before it was shipped to the laboratory on 11 April 2001. In the laboratory, the sediment was stored aerobically at 16°C for 2 weeks before enrichment cultures were initiated.
Enrichment and isolation of strain FRCl.
Anaerobic (5, 36) and aerobic techniques were used for the enrichment and isolation of strain FRCl. All medium additions are given as final concentrations. Low-pH anaerobic enrichments were initiated in 27-ml pressure tubes containing 9 ml of modified freshwater (MFW) medium and 0.5 g of the sediment. MFW medium contained NH4Cl (0.25 g/liter), NaH2PO4 · H2O (4.2 g/liter), KH2PO4 (0.18 g/liter), a vitamin solution (10 ml/liter), and a mineral solution (10 ml/liter) (30). The pH was adjusted to 4.5 with 1 M HCl. Nine milliliters of MFW medium was dispensed into each 27-ml anaerobic pressure tube (Bellco Glass Inc.) and bubbled with N2. The tubes were capped with butyl rubber stoppers and sterilized by autoclaving. All cultures were incubated at 30°C. A reducing agent (cysteine or FeCl2 at 0.5 or 0.7 mM, respectively) was added after the medium was autoclaved. Strain FRCl was enriched with H2 as the electron donor, acetate (10 mM) as the carbon source, and KNO3 (5 mM) as the electron acceptor. After 1 month, the culture was serially diluted with the same anaerobic medium. The culture in the last positive dilution was purified by repeated plating onto aerobic agar plates with MFW medium containing 5 mM acetate as the electron donor. Following purification, strain FRCl was maintained aerobically by using nutrient broth (Difco) supplemented with acetate or glucose to a final concentration of 5 or 20 mM, respectively, or anaerobically by using MFW medium for low-pH cultures and bicarbonate-buffered freshwater (FW) medium for circumneutral-pH cultures (30).
Isolate characterization.
Routine microbiological tests normally used to characterize strains of Enterobacteriaceae were performed according to standard methods (11, 12). The growth of the isolate was tested at temperatures between 5 and 42°C and at pHs between 4 and 10 in anaerobic medium supplemented with 5 mM acetate as the electron donor, 5 mM nitrate as the electron acceptor, and yeast extract at a final concentration of 0.02%. The following compounds were added to FW medium supplemented with acetate (20 mM) to test for potential electron acceptors: oxygen, nitrate (10 mM), nitrite (10 mM), Fe(III) nitriloacetic acid (10 mM), Fe(III) pyrophosphate (10 mM), thiosulfate (10 mM), sulfite (10 mM), sulfate (10 mM), fumarate (40 mM), malate (20 mM), and anthraquinone-2,6,disulfonate (AQDS, 5 mM). AQDS reduction was monitored as a change in the color of the medium from an opaque pink to a bright orange. Fe(III) reduction was monitored as the accumulation of Fe(II). Growth on the other electron acceptors was monitored by measuring turbidity at 540 nm. The ability of washed cell suspensions to reduce U(VI) was determined as previously described (32, 33) by using uranyl acetate as the electron acceptor, acetate as the electron donor, and a reaction buffer containing NaHCO3 (2.5 g/liter), NH4Cl (0.25 g/liter), NaH2PO4 · H2O (0.006 g/liter), and KCl (0.1 g/liter). Cells were added to the reaction buffer to give a final protein concentration of 0.01 mg/ml.
To test for potential electron donors, the following compounds were added to FW medium: hydrogen, acetate, lactate, butyrate, citrate succinate, methanol, ethanol, glycerol, and butanol. Experiments to screen potential electron donors were performed in FW medium bubbled with N2/CO2 (80:20) and supplemented with 10 mM nitrate as the electron acceptor. Nitrate reduction was monitored by measuring turbidity at 540 nm. Electron donors were added to the medium from sterile anoxic stock solutions to give a final concentration of 10 to 20 mM. Controls included no donor and donor alone.
16S rDNA sequencing and phylogenetic analysis.
Molecular analysis of strain FRCl and Trabulsiella guamensis ATCC 49490 was performed by using standard 16S ribosomal DNA (rDNA) methodology as previously described (20). The 16S rDNA was amplified by using the bacterial forward primer 8F and the universal reverse primer 1525R. These primers and the conserved internal primers 338F, 519F, 519R, 907R, and 1392R were used to obtain the nearly complete sequence. Sequences from Salmonella species and other genera were obtained from GenBank for phylogenetic analysis. Phylogenetic analysis was performed by using the PAUP programs available in the Wisconsin sequence analysis package (Accelrys Inc., San Diego, Calif.). Placement of sequences was determined by distance, parsimony, and maximum-likelihood analyses. Phylogenetic trees were constructed by distance analysis (with the Jukes-Cantor correction). Bootstrap analysis was used to confirm the placements of strain FRCl and T. guamensis.
Analytical techniques.
Cell sizes were determined with the imaging software program SimplePC (Compix Inc., Cranberry Township, Pa.). Cells were counted by using acridine orange staining and epifluorescence microscopy as previously described (19). Organic acids were quantified by high-performance liquid chromatography. Nitrate and nitrite concentrations were measured with DX-100 ion chromatography (Dionex Corp., Sunnyvale, Calif.) with a Dionex AS4-SC IonPac column. Fe(II) was quantified by the ferrozine assay (31). U(VI) was quantified by kinetic phosphorescence analysis (Chemchec Corp., LaBrea, Calif.) after dilution in 100 mM anaerobic bicarbonate solution. In order to determine total uranium [U(VI) plus U(IV)], U(IV) in the bicarbonate solution was converted to U(VI) by bubbling with air for 15 min.
Nucleotide sequence accession numbers.
The 16S rDNA sequences of strain FRCl and T. guamensis have been deposited in GenBank under accession numbers AY373829 and AY373830, respectively.
RESULTS AND DISCUSSION
Isolate characteristics.
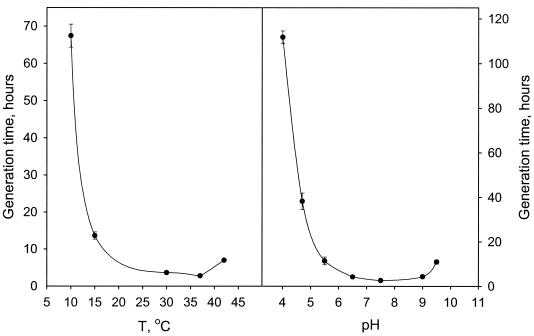
The cells of strain FRCl were motile, regular rods 2.0 to 3.4 μm long and 0.7 to 0.9 μm in diameter with rounded ends. Cells had one lateral flagellum (Fig. 1). With acetate as the electron donor and nitrate as the electron acceptor, growth was most rapid at 30 to 37°C. There was no growth at temperatures below 10°C or above 42°C (Fig. 2, left graph). Optimal growth occurred at pH 6.5 to 9.0, and no growth was observed with initial pH values lower than 4.0 and higher than 9.5 (Fig. 2, right graph). Strain FRCl was able to raise an initial pH of 4 to 4.5 to one appropriate for growth (pH 4.7). The biochemical reactions of strain FRCl in standard tests used for the Enterobacteriaceae are shown in Table 1.
FIG. 1.
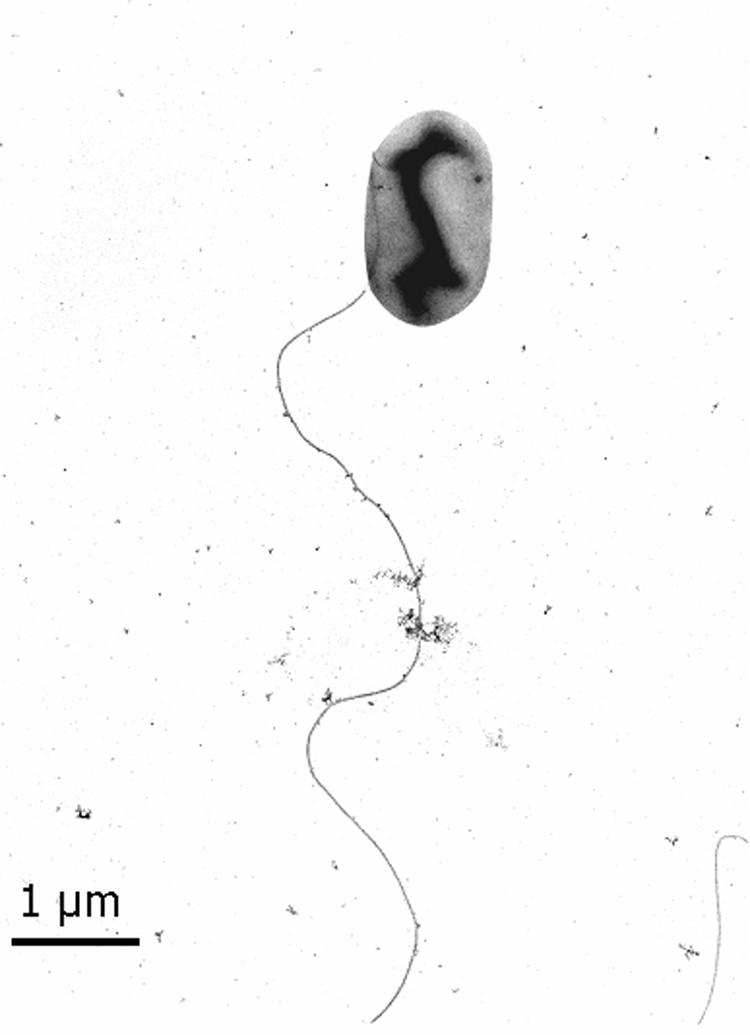
Electron micrograph of negatively stained strain FRCl grown to the mid-exponential phase with hydrogen as the electron donor and nitrate as the electron acceptor.
FIG. 2.
Effect of temperature and pH on growth of strain FRCl. (Left) Optimal growth temperature for strain FRCl. Generation times were calculated from the slopes of the growth curves (not shown) at pH 7. (Right) Influence of pH on the growth of strain FRCl. Generation times were calculated from the slopes of the growth curves (not shown) at 30°C. All growth experiments were done with acetate as the electron donor, nitrate as the electron acceptor, and 0.02% yeast extract. The results represent the means for triplicate cultures.
TABLE 1.
Biochemical reactions of strain FRCl
| Test | Reactiona |
|---|---|
| Gram stain | − |
| Oxidase (24 h) | − |
| Indole production | + |
| Methyl red | + |
| Voges-Proskauer | − |
| Citrate (Simmons) | − |
| Hydrogen sulfide production | − |
| Urea hydrolysis | − |
| Phenylalanine deaminase (24 h) | − |
| Lysine decarboxylase | − |
| Arginine dihydrolase | − |
| Ornithine decarboxylase | + |
| Motility | + |
| Gelatin hydrolysis, 22°C | − |
| KCN, growth | + |
| Malonate utilization | − |
| d-Glucose, acid production | + |
| d-Glucose, gas production | + |
| Acid production | |
| d-Adonitol | − |
| l-Arabinose | + |
| Cellobiose | − |
| Dulcitol | + |
| Glycerol | − |
| myo-Inositol | − |
| Lactose | − |
| Maltose | + |
| d-Mannitol | + |
| d-Mannose | + |
| Melibiose | − |
| α-Methyl-d-glucoside | − |
| Raffinose | − |
| l-Rhamnose | + |
| d-Sorbitol | − |
| Sucrose | − |
| Trehalose | + |
| d-Xylose | + |
| Tartrate, Jordans | Grows but does not produce acid |
| Esculin hydrolysis | − |
| Acetate utilization | + |
| Nitrate reduction | + |
| Nitrite reduction | − |
| Deoxyribonuclease, 25°C | − |
| Lipase | − |
| ONPGb test | + |
| Pigment | +, yellow |
| Catalase production | + |
| Oxidation-fermentation | Fermentation |
−, negative at the end of the appropriate incubation period; +, positive at 24 h or at the time of the test.
ONPG, o-nitrophenyl-β-d-galactopyranoside.
With acetate serving as the electron donor, strain FRCl was capable of reducing the following electron acceptors: nitrate, thiosulfate, fumarate, and malate. Nitrate was reduced to nitrite. The following electron acceptors were tested but not utilized: sulfate, sulfite, nitrite, Fe(III) nitriloacetic acid, and Fe(III) pyrophosphate. With nitrate as the electron acceptor, strain FRCl was capable of oxidizing the following electron donors: hydrogen, acetate, lactate, citrate, butyrate, succinate, methanol, ethanol, glycerol, and butanol.
Phylogenetic analysis.
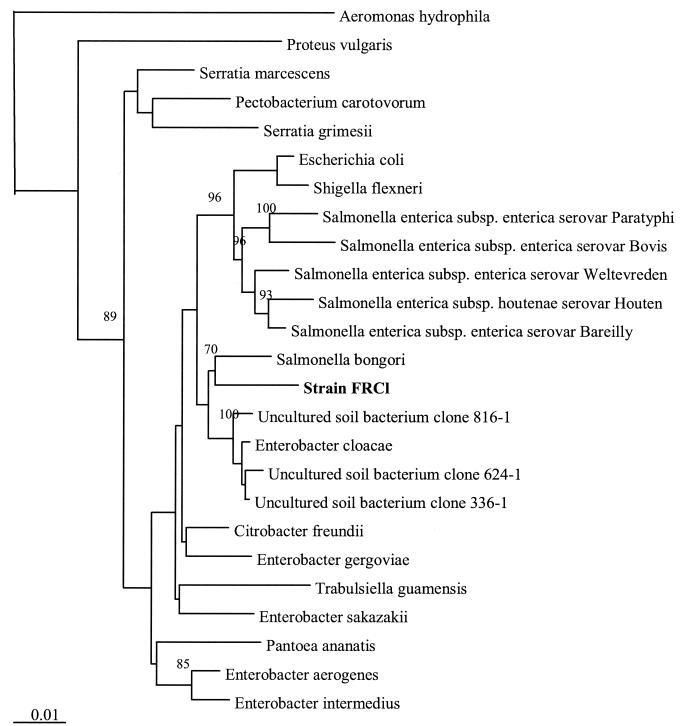
Ribosomal DNA from strain FRCl was amplified by PCR. BLAST and similarity analyses indicated that strain FRCl was similar to Salmonella bongori (nucleotide identity, 96.4%), Enterobacter cloacae (nucleotide identity, 96.3%), and uncultured soil bacteria (clones 336-1, 624-1, and 816-1) from agriculture soil (48). An environmental isolate resembling S. bongori, T. guamensis (35), for which the 16S rDNA sequence was not previously available, was sequenced. Phylogenetic analysis of these and other related sequences was performed by using 1,422 bases for comparison. The placement of strain FRCl was consistent in distance, parsimony, and maximum-likelihood analyses. The results of these analyses indicated that strain FRCl was phylogenetically most closely related to S. bongori (Fig. 3). On the basis of the phenotypic differences between strain FRCl and S. bongori (Table 2), it is suggested that strain FRCl is a representative of a novel species in the genus Salmonella. The suggested epithet is Salmonella subterranea.
FIG. 3.
Phylogenetic analysis of FRC isolate. The tree was constructed by distance analysis with Jukes-Cantor correction by using 1,422 bases for comparison. Numbers next to branches indicate bootstrap values.
TABLE 2.
Differentiation between strain FRCl, S. bongori, and E. cloacae
| Test | Test result
|
||
|---|---|---|---|
| Strain FRCl | S. bongoria | E. cloacaea | |
| Indole production | + | − | − |
| Methyl red | + | + | − |
| Voges-Proskauer | − | − | + |
| Urea hydrolysis | − | − | vb |
| Lysine decarboxylase | − | + | − |
| Malonate utilization | − | − | + |
| Arginine dihydrolase | − | + | + |
| H2S production | − | + | − |
| Citrate (Simmons) | − | + | + |
| Acid production | |||
| Cellobiose | − | − | + |
| Dulcitol | + | + | − |
| Lactose | − | − | + |
| Melibiose | − | + | + |
| α-Methyl-d-glucoside | − | − | + |
| Raffinose | − | − | + |
| d-Sorbitol | − | + | + |
| Sucrose | − | − | + |
| Pigment | +, yellow | − | − |
From reference 21.
v, reaction was variable.
Growth at moderately acidic pH.
Acid tolerance appears to be common among Salmonella spp. The growth of Salmonella spp. was documented at pH 3.7 to 4.4 (17, 25, 27, 51). The ability of Salmonella spp. to increase the initial pH of the medium has previously been documented (51). It was postulated that strain FRC may be able to cause a similar increase in pH.
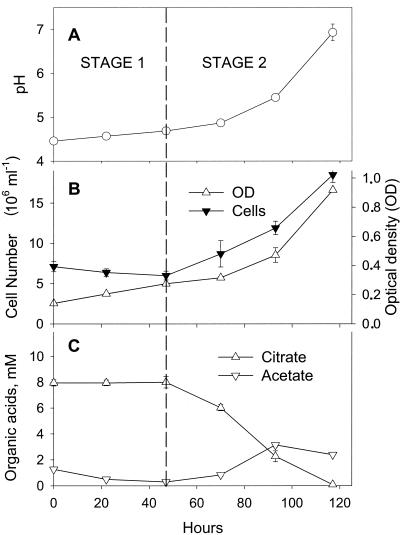
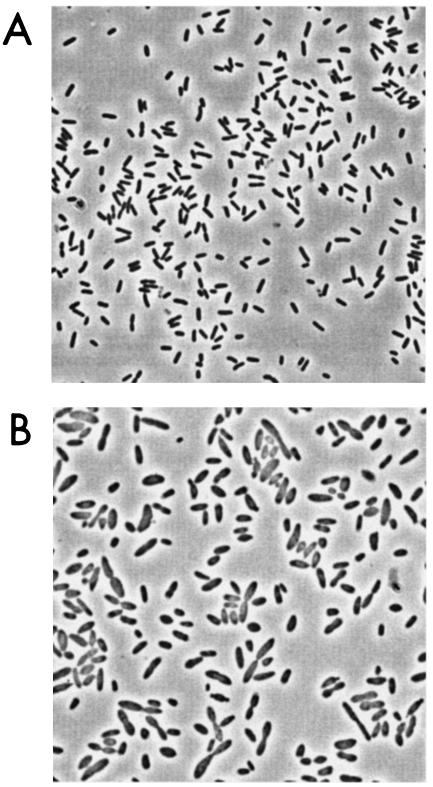
Both acetate and citrate were present in the pore water (pH 4) at the site from which FRCl was isolated. The cultivation of strain FRCl in the presence of acetate and citrate at an acidic pH can be divided into two stages (Fig. 4). In the first stage (hours 0 to 47), strain FRCl adjusted the initial pH of the medium (pH 4.4 ± 0.0) to one appropriate for growth (pH 4.7 ± 0.0) (Fig. 4A). While the pH was being adjusted (hours 0 to 47), the optical density (OD) of the culture doubled, but the number of cells decreased (from 7.11 × 106 to 6.00 × 106 cells/ml) (Fig. 4B). Although there was no growth in the first 47 h, acetate was consumed (Fig. 4C) and the morphology of the cells changed (Fig. 5). While the cells of strain FRCl grown at a near-neutral pH were 2.7 ± 0.7 μm long and 0.8 ± 0.1 μm in diameter (Fig. 5A), the cells in acidic medium were larger: they were 3.9 ± 1.2 μm long and 1.5 ± 0.3 μm in diameter (Fig. 5B). It has been shown that the peptidoglycan, a major stress-bearing structure in gram-negative bacteria, is rather flexible and can be extended fourfold the length of its most compact conformation (22, 23).
FIG. 4.
Aerobic growth of strain FRCl in the presence of citrate and acetate. The results represent the means for triplicate cultures.
FIG. 5.
Cells of strain FRCl in the low-pH aerobic culture after inoculation (A) and at 47 h (B).
It is generally thought that undissociated acids freely diffuse across the cell membrane, which causes anion accumulation in the cytoplasm, resulting in increased internal osmotic pressure of the cell (40). According to the Henderson-Hasselbalch equation, at pH 4.4 (the starting pH for the growth of FRCl), 69% of the acetic acid (and only 4.8% of the citric acid) was in the undissociated form. This potential for an increase in the internal osmotic pressure of the cells at pH 4.4 due to the uptake of acetate correlates with both the change in the morphology of the cells, demonstrated in Fig. 5, and the discrepancy between the optical density and the cell number during hours 0 to 47 (Fig. 4B).
The second growth stage started after 47 h and represented the growth of strain FRCl concurrent with an increase in pH to a final pH of 6.9 at 117 h. The cell size of strain FRCl did not decrease during this growth phase. At this stage, strain FRCl preferentially used citrate as the electron donor (Fig. 4C). Acetate, succinate, fumarate, and a number of unidentified acids were produced as the result of citrate oxidation (Fig. 4C and data not shown). Studies with Escherichia coli and Salmonella spp. suggest that acetate is not a preferred substrate for these microorganisms and that the expression of genes for acetate utilization is usually subject to strong catabolite repression in the presence of sugars or acids of the tricarboxylic acid cycle, including citrate (9).
U(VI) reduction by cell suspension of strain FRCl.
Analysis of 16S rDNA sequences indicated that strain FRCl was a component of the U(VI)-reducing enrichment culture FRCk, which was established with sediments from the Oak Ridge study site with hydrogen and acetate as the electron donors (44). The original enrichment culture was established at pH 4.5 and then transferred to neutral-pH medium, resulting in enrichment culture FRCk. FRCk consisted of two species, a Geobacter sp. and strain FRCl (44). While U(VI) reduction by Geobacter spp. is well documented (16, 33), it was unclear what role strain FRCl might have played in the culture. Two roles were postulated for strain FRCl: (i) it could be important for mediating pH in the initial low-pH enrichment, and/or (ii) it could reduce U(VI).
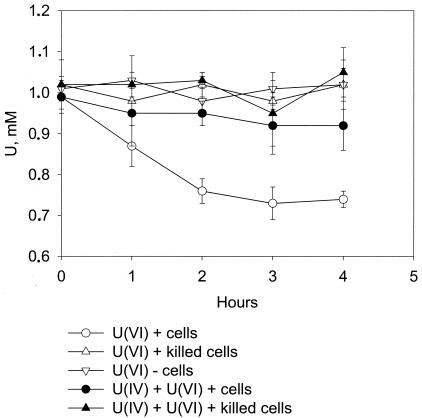
When a washed cell suspension of strain FRCl was added to buffer (pH 6.8) with acetate as the electron donor and U(VI) as the electron acceptor, U(VI) was reduced to U(IV) over time (Fig. 6). There was no U(VI) reduction in controls without cells or with killed cells. The U(VI)-reducing activity of strain FRCl [0.14 μM U(VI) per mg of protein per min] was comparable to the U(VI)-reducing activity of Geobacter sulfurreducens strain PCA [0.18 μM U(VI) per mg of protein per min] (28). This finding suggested that strain FRCl may be important for U(VI) reduction.
FIG. 6.
U(VI) reduction by the cell suspension of strain FRCl. The results represent the means for triplicate cell suspensions.
Ecological niche of S. subterranea.
Salmonella spp. are frequently isolated from aquatic (8, 39) and sedimentary environments (10, 18, 47) and are able to multiply in soil (38) and estuarine environments (41). However, soils and sediments are typically regarded as transitional environments for Salmonella spp. prior to the infection of a host (49). The finding that strain FRCl was closely related to uncultured soil bacterium clones (48) suggests that there might be a branch of Salmonella spp. that are better adapted to living in soils than most of their enteric relatives.
The ability of the members of the genus Salmonella to survive at low pH may provide them an advantage over other organisms in mildly acidic sedimentary environments, such as the acidic sediments at the NABIR Field Research Center in Oak Ridge, Tenn. Another important feature of these organisms is their ability to mediate the pH to a more neutral range in which they can grow (reference 51 and this study). The high silt (55.8%) and clay (19.3%) contents of the sediments from the FW-024 site (44) result in low water permeability, which may enhance the development of microenvironments. Despite the flux of the low-pH groundwater, the pH in such microenvironments may differ from that of the bulk sediment. Therefore, in low-permeability acidic sediments, Salmonella spp. could be responsible for the creation of microenvironments with higher pHs, in which other microorganisms may also thrive. The fact that Salmonella sp. strain FRCl was isolated from acidic uranium- and nitrate-contaminated sediment and was recovered in both uranium- and nitrate-reducing enrichment cultures, coupled with its ability to carry out uranium and nitrate reduction in pure culture, suggests that it has the potential to play a role in uranium bioremediation at mildly acidic pHs. However, whether strain FRCl is important for nitrate and U(VI) reduction as well as for mediating pH in situ remains to be investigated.
Description of S. subterranea sp. nov.
S. subterranea (Sub.ter.ra′ne.a. L. adj. subterranea, -us, underground, subterranean). Cells are regular motile rods 2.0 to 3.4 μm long and 0.7 to 0.9 μm in diameter with rounded ends. Cells have one lateral flagellum. Optimal growth occurs at pH 6.5 to 9.0, and no growth was documented with initial pH values lower than 4.0 or higher than 9.5. Growth is most rapid at 30 to 37°C, and no growth occurs at temperatures below 10°C or above 42°C. Positive for indole production, by methyl red test, and for ornithine decarboxylase; negative by Voges-Proskauer test, for phenylalanine deaminase, for lysine decarboxylase, for arginine dihydrolase, for urea hydrolysis, for H2S production, and for gelatin hydrolysis. Uses O2, NO3−, S2O32−, fumarate, and malate as electron acceptors. Reduces U(VI) in cell suspension. Type strain FRCl was isolated from a subsurface nitrate- and U(VI)-contaminated sediment. Strain FRCl has been deposited in the American Type Culture Collection (accession number ATCC BAA-836).
Acknowledgments
This research was funded by the Natural and Accelerated Bioremediation Research program, Biological and Environmental Research, U.S. Department of Energy (grants DE-FG02-0ER62985 and DE-FG02-97ER62475).
We thank David Watson for technical assistance at the NABIR Field Research Center, Oak Ridge, Tenn. We thank L. Yin of the University of Massachusetts microscopy facility, which was supported by NSF grant BBS 8714235. We also thank the anonymous reviewers for helpful comments and suggestions.
REFERENCES
- 1.Abdelouas, A., Y. Lu, W. Lutze, and H. E. Nuttall. 1998. Reduction of U(VI) to U(IV) by indigenous bacteria in contaminated ground water. J. Contam. Hydrol. 35:217-233. [Google Scholar]
- 2.Abdelouas, A., W. Lutze, W. Gong, E. H. Nuttall, B. A. Strietelmeier, and B. J. Travis. 2000. Biological reduction of uranium in groundwater and subsurface soil. Sci. Total Environ. 250:21-35. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, R. T., and D. R. Lovley. 2002. Microbial redox interactions with uranium: an environmental perspective, p. 205-223. In M. J. Keith-Roach and F. R. Livens (ed.), Interactions of microorganisms with radionuclides. Elsevier, Amsterdam, The Netherlands.
- 4.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernard, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balch, W. E., G. E. Gox, I. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, L. L., K. Choudhury, B. M. Thomson, K. Steenhoudt, and A. R. Groffman. 1996. Bacterial reduction of soluble uranium: the first step of in situ immobilization of uranium. Radioact. Waste Manag. Environ. Restoration 20:141-151. [Google Scholar]
- 7.Brooks, S. C. 2001. Waste characteristics of the former S-3 ponds and outline of uranium chemistry relevant to NABIR Field Research Center studies. NABIR Field Research Center, Oak Ridge, Tenn.
- 8.Cherry, W. B., J. B. Hanks, B. M. Thomason, A. M. Murlin, J. W. Biddle, and J. M. Croom. 1972. Salmonellae as an index of pollution of surface waters. Appl. Microbiol. 24:334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, D. P., and J. E. Cronan. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella, vol. 1. ASM Press, Washington, D.C.
- 10.Davies, R. H., and C. Wray. 1996. Seasonal variations in the isolation of Salmonella typhimurium, Salmonella enteritidis, Bacillus cereus and Clostridium perfringens from environmental samples. J. Vet. Med. Ser. B 43:119-127. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, P. R., and W. H. Ewing. 1972. Identification of Enterobacteriaceae, 3rd ed. Burgess Publishing Co., Minneapolis, Minn.
- 12.Farmer, J. J., III, B. R. Davis, F. W. Hickman-Brenner, A. McWhorter, G. P. Huntley-Carter, M. A. Asbury, C. Riddle, H. G. Wathen-Grady, C. Elias, G. R. Fanning, A. G. Steigerwalt, C. M. O'Hara, G. K. Morris, P. B. Smith, and D. J. Brenner. 1985. Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21:46-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finneran, K. T., R. T. Anderson, K. P. Nevin, and D. R. Lovley. 2002. Potential for bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sediment Contam. 11:339-357. [Google Scholar]
- 14.Francis, A. J. 1998. Biotransformation of uranium and other actinides in radioactive wastes. J. Alloys Compounds 271-273:78-84. [Google Scholar]
- 15.Francis, A. J., C. J. Dodge, J. B. Gillow, and J. E. Cline. 1991. Microbial transformations of uranium in wastes. Radiochim. Acta 52-53:311-316. [Google Scholar]
- 16.Gorby, Y. A., and D. R. Lovley. 1992. Enzymatic uranium reduction. Environ. Sci. Technol. 26:205-207. [Google Scholar]
- 17.Goverd, K. A., F. W. Beech, R. P. Hobbs, and R. Shannon. 1979. The occurrence and survival of coliforms and salmonellas in apple juice and cider. J. Appl. Bacteriol. 46:521-530. [DOI] [PubMed] [Google Scholar]
- 18.Hendricks, C. W. 1971. Increased recovery rate of salmonellae from stream bottom sediments versus surface waters. Appl. Microbiol. 21:379-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 22.Höltje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isaac, L., and G. C. Ware. 1974. The flexibility of bacterial cell walls. J. Appl. Bacteriol. 37:335-339. [DOI] [PubMed] [Google Scholar]
- 24.Kashefi, K., and D. R. Lovley. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100°C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, C.-H., and G. M. Sapers. 2000. Attachment and growth of Salmonella Chester on apple fruits and in vivo response of attached bacteria to sanitizer treatments. J. Food Prot. 63:876-883. [DOI] [PubMed] [Google Scholar]
- 26.Lieser, K. H. 1995. Radionuclides in the geosphere: sources, mobility, reactions in natural waters and interactions with solids. Radiochim. Acta 70-71:355-375. [Google Scholar]
- 27.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, J. R., J. Chesnes, S. Glasauer, D. J. Bunker, F. R. Livens, and D. R. Lovley. 2002. Reduction of actinides and fusion products by Fe(III)-reducing bacteria. Geomicrobiol. J. 19:103-120. [Google Scholar]
- 29.Lloyd, J. R., and I. E. Macaskie. 2000. Bioremediation of radionuclide-containing wastewaters, p. 277-327. In D. R. Lovley (ed.), Environmental microbe-metal interaction. ASM Press, Washington, D.C.
- 30.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. P. Philips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 31.Lovley, D. R., and E. J. P. Phillips. 1987. Rapid assay for microbially reducible ferric iron in aquatic sediments. Appl. Environ. Microbiol. 53:1536-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovley, D. R., and E. J. P. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 34.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 35.McWhorter, A. C., R. L. Haddock, F. A. Nocon, A. G. Steigerwalt, D. J. Brenner, S. Aleksic, J. Bockemühl, and J. J. Farmer. 1991. Trabulsiella guamensis, a new genus and species of the family Enterobacteriaceae that resembles Salmonella subgroups 4 and 5. J. Clin. Microbiol. 29:1480-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, T. L., and M. J. Wolin. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietzsch, K., B. C. Hard, and W. Babel. 1999. A Desulfovibrio sp. capable of growing by reducing U(VI). J. Basic Microbiol. 39:365-372. [Google Scholar]
- 38.Pohl, P., E. Robaye, and J. Thomas. 1970. Comportement de Salmonella et Escherichia dans le milieu exterieur. Rev. Ferment. Ind. Aliment. 25:22-28. [Google Scholar]
- 39.Prokšová, M., E. Mogonová, P. Ferianc, and J. Harichová. 2002. The occurrence of Salmonella spp. in natural environment confirmed by biochemical and molecular methods. Biologia 57:797-803. [Google Scholar]
- 40.Rao, S. S. (ed.). 1989. Acid stress and aquatic microbial interactions. CRC Press, Inc., Boca Raton, Fla.
- 41.Rhodes, M. W., and H. Kator. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunders, J. A., and L. E. Toran. 1995. Modelling of radionuclide and heavy metal sorption around low- and high-pH waste disposal sites at Oak Ridge, Tennessee. Appl. Geochem. 10:673-684. [Google Scholar]
- 43.Shelobolina, E. S., C. Gaw VanPraagh, and D. R. Lovley. 2003. Use of ferric and ferrous iron containing minerals for respiration by Desulfitobacterium frappieri. Geomicrobiol. J. 20:143-156. [Google Scholar]
- 44.Shelobolina, E. S., K. O'Neill, K. T. Finneran, L. A. Hayes, and D. R. Lovley. 2003. Potential for in situ bioremediation of a low-pH, high-nitrate uranium-contaminated groundwater. Soil Sediment Contam. 12:865-884. [Google Scholar]
- 45.Silva, R. J., and H. Nitsche. 1995. Actinide environmental chemistry. Radiochim. Acta 70-71:377-396. [Google Scholar]
- 46.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 47.Thomason, B. M., D. J. Dodd, and W. B. Cherry. 1977. Increased recovery of salmonellae from environmental samples enriched with buffered peptone water. Appl. Environ. Microbiol. 34:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valinsky, L., G. D. Vedova, A. J. Scupham, S. Alvey, A. Figueroa, B. Yin, R. J. Hartin, M. Chrobak, D. E. Crowley, T. Jiang, and J. Borneman. 2002. Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl. Environ. Microbiol. 68:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wronkiewicz, D. J., and E. C. Buck. 1999. Uranium mineralogy and the geologic disposal of spent nuclear fuel, p. 475-497. In P. C. Burns and R. Finch (ed.), Uranium: mineralogy, geochemistry and the environment, vol. 38. Mineralogical Society of America, Washington, D.C.
- 51.Zhuang, R.-Y., L. R. Beuchat, and F. J. Angulo. 1995. Fate of Salmonella montevideo on and in raw tomatoes as affected by temperature and treatment with chlorine. Appl. Environ. Microbiol. 61:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]