Abstract
Currently there is only limited understanding of the reservoirs for Clostridium perfringens type A food poisoning. A recent survey (Y.-T. Lin and R. Labbe, Appl. Environ. Microbiol. 69:1642-1646, 2003) of non-outbreak American retail foods did not identify the presence of a single C. perfringens isolate carrying the enterotoxin gene (cpe) necessary for causing food poisoning. The present study revisited this issue, using revised methodology and food sampling strategies. In our survey, cpe-positive C. perfringens isolates were detected in ∼1.4% of ∼900 surveyed non-outbreak American retail foods. Interestingly, those enterotoxigenic isolates in non-outbreak foods appear indistinguishable from C. perfringens isolates known to cause food poisoning outbreaks: i.e., the enterotoxigenic retail food isolates all carry a chromosomal cpe gene, are classified as type A, and exhibit exceptional heat resistance. Collectively, these findings indicate that some American foods are contaminated, at the time of retail purchase, with C. perfringens isolates having full potential to cause food poisoning. Furthermore, demonstrating that type A isolates carrying a chromosomal cpe gene are the enterotoxigenic isolates most commonly present in foods helps to explain why these isolates (rather than type A isolates carrying a plasmid cpe gene or cpe-positive type C or D isolates) are strongly associated with food poisoning outbreaks. Finally, since type A chromosomal cpe isolates present in the surveyed raw foods exhibited strong heat resistance, it appears that exceptional heat resistance is not a survivor trait selected for by cooking but is instead an intrinsic trait possessed by many type A chromosomal cpe isolates.
Clostridium perfringens type A food poisoning currently ranks as the third most commonly identified foodborne illness in the United States (13). The Centers for Disease Control and Prevention conservatively estimates that this food poisoning sickens ∼250,000 Americans annually (14). Deaths from C. perfringens type A food poisoning are not common but do occur in the elderly and debilitated. Per year this food poisoning is estimated to kill ∼7 people in the United States and 50 to 100 people in the United Kingdom (1, 14).
The diarrheic and cramping symptoms of C. perfringens type A food poisoning result from C. perfringens enterotoxin (CPE). This toxin is both necessary and sufficient for the enteric virulence of C. perfringens type A food poisoning isolates. Specific inactivation of the enterotoxin gene (cpe) by allelic exchange was shown to render a C. perfringens food poisoning isolate avirulent in rabbit ileal loops, and intestinal virulence could be restored by complementing that mutant with a cloned copy of the wild-type cpe gene (16). Furthermore, ingestion of purified CPE by human volunteers was determined to be sufficient for reproducing the cramping and diarrheic symptoms of the natural food poisoning (18).
Only a small fraction (∼1 to 5%) of all C. perfringens isolates, mainly belonging to type A, carry the cpe gene (6, 10). The cpe gene can have either a chromosomal or plasmid-borne location but is nearly always present on the chromosome of food poisoning isolates (3, 4, 15, 20, 21). A recent study (17) suggests that the strong association between type A isolates carrying a chromosomal cpe gene and C. perfringens type A food poisoning is attributable (at least in part) to the exceptional heat resistance of those isolates, which should favor their survival in incompletely cooked or improperly held foods. Why the type A isolates carrying a chromosomal cpe gene are so much more heat resistant than type A isolates carrying a plasmid cpe gene is unknown at present.
To reduce the occurrence of C. perfringens type A food poisoning outbreaks, it is necessary to determine how and when C. perfringens isolates with food poisoning potential enter the American food supply. Surveys conducted more than 10 years ago clearly demonstrated that C. perfringens isolates are often present in foods, particularly raw meats and poultry, sold in the United States (see reference 11 for review). However, those findings have limited significance since whether C. perfringens food isolates include the 1 to 5% of type A isolates carrying the cpe gene that is essential for causing food poisoning was not evaluated.
A recent study (12) did address this important question by surveying 131 American retail foods. However, not a single 1 of 39 C. perfringens isolates obtained from those non-outbreak foods was found to be cpe positive. Those findings raise questions as to how C. perfringens type A food poisoning can be so common in the United States if cpe-positive isolates are so rarely present (if not completely absent) from retail foods. If correct, those findings could indicate that American foods do not contain cpe-positive C. perfringens at the time of retail purchase but instead become contaminated during food preparation in the home or institution. Since establishing the frequency of retail food contamination by cpe-positive C. perfringens holds such obvious epidemiologic importance, our present study reexamined this important issue, using different methodologies and food sampling strategies.
MATERIALS AND METHODS
Collection of food samples. Nearly 900 non-outbreak food samples were obtained from retail outlets in Pittsburgh, Pa. These stores included a mix of neighborhood grocery stores, large supermarkets, and small butcher shops. A breakdown of these samples by food type is shown in Table 1.
TABLE 1.
Presence of C. perfringens isolates in American retail foods
| Fooda | No. of samples tested | MPN/g range | No. (%) of samples contaminated with C. perfringens | No. of samples withb:
|
||
|---|---|---|---|---|---|---|
| Type A | Type A-β2 | Type A-cpe | ||||
| Turkey | 68 | 1-19 | 19 (28) | 10 | 6 | 2 (1) |
| Chicken | 147 | 2-5 | 56 (38) | 24 (2) | 27 (2) | 1 |
| Pork | 144 | 3-8 | 39 (27) | 25 (5) | 5 | 3 (1) |
| Ground pork | 99 | 2-23 | 60 (61) | 47 (3) | 7 (1) | 1 (1) |
| Ham | 12 | 0 | 0 | 0 | 0 | 0 |
| Beef | 83 | 1-10 | 17 (21) | 16 | 1 | 0 |
| Ground beef | 108 | 3-32 | 25 (23) | 23 (2) | 0 | 0 |
| Fish | 113 | 0-5 | 34 (30) | 31 | 0 | 3 |
| Shrimp | 30 | 1-8 | 5 (17) | 5 | 0 | 0 |
| Lamb | 24 | 1-7 | 9 (38) | 8 (1) | 0 | 0 |
| Sausage | 59 | 0-2 | 14 (24) | 12 (1) | 1 | 0 |
| Total | 887 | 278 | 201 (14) | 47 (3) | 10 (3) | |
All foods except for ham and a few turkey products were raw when tested.
Numbers in parentheses indicate the number of food items clearly contaminated with spores, while numbers outside parentheses indicate the number of food items apparently contaminated with vegetative cells (see the text). Therefore, the total contamination found for each food equals the sum of the numbers inside and outside the parentheses in these columns.
Preparation of food samples.
A sample (∼10 g) was collected aseptically from each food item and placed into a 50-ml sterile plastic tube for transfer to the laboratory. The only exception occurred with large packages of ground meats, where two samples were removed from opposite portions of the package. Food samples were typically processed immediately but were never stored at 4°C for more than 24 h before testing.
Processing of each food sample started with a homogenization step using sterilized surgical scissors. Ten milliliters of sterile fluid thioglycolate (FTG) medium was then added to the 50-ml tube containing the minced food. An aliquot (1 ml) of each FTG food suspension was added to each of two tubes containing 10 ml of sterile FTG. To enrich for any C. perfringens spores present in the food sample, one of those two tubes was heat shocked at 72°C for 20 min before incubation at 37°C for 18 to 24 h. The other tube was directly incubated at 37°C for 18 to 24 h to enrich primarily for C. perfringens vegetative cells present in that food sample.
Each FTG enrichment culture showing growth was streaked onto one plate of tryptose-sulfite-cycloserine agar containing 10% egg yolk (TSC with egg yolk) and a second plate of brain heart infusion agar containing 10% sheep blood and 40-μg/ml neomycin (blood agar with neomycin). Both plates were then incubated for ∼18 h at 37°C in an anaerobic jar (BBL). The majority of food samples yielded no presumptive C. perfringens colonies on either the plates containing TSC with egg yolk or those containing blood agar with neomycin: i.e., those food samples failed to grow either black colonies or colonies showing lecithinase activity on plates containing TSC with egg yolk and they also failed to grow colonies surrounded by a zone of hemolysis on blood agar with neomycin. When a food sample did grow presumptive C. perfringens, those colonies were inoculated into 10 ml of FTG medium, which was then incubated overnight at 37°C. To confirm the identity of those presumptive FTG cultures as C. perfringens, standard methods were used (7). A loopful of each culture was stabbed into a tube of motility nitrate and lactose-gelatin media. Those tubes were then incubated at 37°C for 18 to 24 h. Results were interpreted as described previously (7).
Determination of MPN of C. perfringens per gram in foods.
A three-tube most probable number (MPN) method was used to investigate C. perfringens levels in food samples (12). Briefly, a 10-g aliquot of a food suspension (prepared as described above) was diluted by 10-fold increments (from 10−1 to 10−5) in FTG, and then 1-ml aliquots of each dilution from a single sample were inoculated into three tubes containing 10 ml of differential reinforced clostridial broth medium (DRCM; Merck Co.). After incubation at 37°C for 24 h, cultures testing positive for C. perfringens produced a unique black precipitation in this DRCM. Statistical analyses were performed as previously described (9).
Multiplex PCR assay to determine the toxin genotype of C. perfringens isolates from food samples.
To prepare template DNA for the multiplex PCR toxin genotyping assay, a loopful of a pure FTG culture from each isolate confirmed as C. perfringens by biochemical tests was streaked onto a brain heart infusion (BHI) agar plate. After incubation in an anaerobic jar (BBL) at 37°C for 18 h, four to five colonies from each BHI plate were suspended in 200 μl of sterilized phosphate-buffered saline (PBS). Those cell suspensions were microcentrifuged at 14,000 × g for 5 min, and the resultant pellet was similarly washed twice more with PBS before final resuspension in 100 μl of PCR-grade H2O (Sigma). Microcentrifuge tubes containing each preparation of washed C. perfringens cells were then sealed with Parafilm and subjected to heating in a microwave (700 W) for a total of 20 min (administered as four separate 5-min heat treatments with 1-min cooling intervals) to induce bacterial lysis. The resultant lysates were cleared by microcentrifugation at 14,000 × g for 5 min, and 5 μl of each supernatant was then used directly as template DNA in a multiplex PCR assay.
The multiplex PCR assay used in our study has been described previously (8) and can detect the presence of genes encoding alpha-toxin (cpa), beta-toxin (cpb), beta2-toxin (cpb2), epsilon-toxin (etx), CPE (cpe), or the A component of iota-toxin (iap). The sequences of the six primer pairs used in this multiplex PCR were published previously (8). Each PCR had a total volume of 50 μl, which contained 5 μl of the primer pair mixture (final primer concentration, 0.34 μM each cpe oligonucleotide, 0.36 μM each cpb oligonucleotide, 0.36 μM each cpb2 oligonucleotide, 0.44 μM each etx oligonucleotide, 0.50 μM each cpa oligonucleotide, and 0.52 μM each iap oligonucleotide), 40 μl of Taq Complete 1.1× Master mix (GENE CHOICE), and 5 μl of a cell lysate (prepared as described above) as a template. Each PCR mixture was placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. A 15-μl aliquot of each PCR was then electrophoresed on a 2.0% Tris-borate-EDTA-agarose gel, and PCR products from each reaction were visualized by staining with ethidium bromide.
Duplex PCR assay for cpe genotyping of cpe-positive C. perfringens type A food isolates.
Food isolates identified by multiplex PCR as being cpe-positive C. perfringens type A were then tested by a duplex PCR cpe genotyping assay to establish whether they carry a chromosomal cpe locus or a plasmid cpe locus (21). To prepare template DNA for this purpose, a 0.2-ml aliquot of a pure FTG culture of a cpe-positive type A food isolate was inoculated into 10 ml of TGY medium (3% Trypticase, 2% glucose, 1% yeast extract, 0.1% cysteine) (5). After an overnight incubation at 37°C, genomic DNA was extracted from each overnight TGY culture as described previously (21). The extracted DNA was quantified by UV absorption measurement at 280 and 260 nm using a Bio-Rad Smartspec spectrophotometer. Aliquots of each extracted DNA preparation were then diluted to 10 ng/μl for use as DNA template in the duplex PCR genotyping assay.
This duplex PCR cpe genotyping assay uses two primer pairs that amplify an ∼2.1-kb product from the chromosomal cpe locus of type A isolates or an ∼3.3-kb product from the plasmid cpe locus of type A isolates (21). The sequence of these primers was previously published (21). The two primer pairs were added (final concentrations of 0.6 μM each MET-1.5F and cpe-UP primers for amplifying the plasmid cpe locus and 0.8 μM each IS1470F and 4.5F primers for amplifying the chromosomal cpe locus) to a total 50-μl PCR volume containing 10 ng of purified DNA extracted from each C. perfringens food isolate, 0.2 μM deoxynucleotide triphosphate (Promega), 5 μl of 10× Advantage 2 PCR buffer (BD), and 1 μl of Advantage 2 polymerase mix (BD). Each PCR mixture was then placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 1st cycle, 94°C for 5 min; 2nd to 34th cycles, 94°C for 30 s, 63°C for 60 s, and 68°C for 180 s; and 35th cycle, 94°C for 90 s, 60°C for 90 s, and 68°C for 7 min. A 20-μl aliquot of each PCR mixture was then electrophoresed on a 1.5% agarose gel, and the resultant products were visualized by staining the gel with ethidium bromide.
Determination of spore heat resistance for cpe-positive C. perfringens type A food isolates.
To evaluate the heat resistance of C. perfringens food isolates, the D value at 100°C for each isolate's spores was measured, as described previously (17). Briefly, sporulating cultures of C. perfringens were prepared by inoculating a 0.2-ml aliquot of a FTG culture into 10 ml of Duncan-Strong (DS) sporulation medium (17). After overnight incubation at 37°C, the presence of sporulating cells in each DS culture was confirmed by phase-contrast microscopy. Those DS cultures were then heat shocked at 72°C for 20 min to kill any remaining vegetative cells and to facilitate spore germination (17). A 0.1-ml aliquot of each heat-shocked DS culture was then serially diluted with sterile FTG medium to obtain dilutions ranging from 10−2 to 10−7. Two 1-ml aliquots of each dilution were duplicate plated onto BHI agar plates in order to establish the number of viable spores present per milliliter of DS cultures at the start of heating (i.e., at the zero time point of the experiment).
The remainder of each heat-shocked DS culture was then heated at 100°C for time periods ranging from 1 min to 3 h. At each time point, the boiled DS culture was mixed, and a 0.1-ml aliquot was withdrawn and diluted (dilution range, 10−2 to 10−7) with sterile FTG medium. Each dilution was then duplicate plated onto BHI agar plates, which were incubated anaerobically at 37°C for 16 to 24 h. Colonies developing from germinated spores that survived heating were then counted to determine the number of viable spores present at that time point per milliliter of each heated DS culture. Those data were graphed to determine a D value at 100°C for spores of each tested cpe-positive food isolate (17).
Western immunoblot analysis of CPE expression by cpe-positive food isolates.
A 0.2-ml aliquot of an FTG culture from each cpe-positive type A food sample was inoculated into 10 ml of DS medium. After incubation at 37°C for 8 h, those DS cultures were then examined with a phase-contrast microscope to ensure the presence of spores. At that point, the DS culture was sonicated until more than 95% of all cells had lysed (lysis was monitored by phase-contrast microscopy). The sonicated samples were centrifuged to remove debris and any unlysed cells. Each DS medium culture lysate was then analyzed for the presence of CPE by a previously described CPE Western inmmunoblot procedure (17).
RESULTS
Isolation of C. perfringens from food samples.
Our study purposely surveyed the foods most commonly implicated as vehicles for C. perfringens type A food poisoning outbreaks, namely pork, beef, poultry, seafood, and processed meat products (2). As shown in Table 1, 31.3% of all non-outbreak food samples tested in the present survey were found to be contaminated with C. perfringens isolates, which is in reasonable agreement with the food contamination frequency determined by another recent study (12). All meats and seafood items showed C. perfringens contamination rates ranging between ∼20 to 40% (Table 1), with two notable exceptions: (i) cooked ham, which consistently tested negative for the presence of C. perfringens; and (ii) ground pork, which had a much higher frequency (66%) of contamination with C. perfringens.
For most individual food samples, no more than a single C. perfringens isolate was obtained. This low food contamination frequency is consistent with MPN results indicating that the food samples tested in this survey had MPN-per-gram values ranging from 0 to 32 (Table 1). About 2% of sampled foods grew C. perfringens after heat shocking, clearly indicating they contained spores of this bacterium. Approximately 0.2% of sampled foods tested positive for C. perfringens whether or not they had been heat shocked. Those foods may either have contained a mix of both vegetative cells and spores or contained only spores that spontaneously germinated in the absence of heat shocking. About 29% of food samples grew C. perfringens only in the absence of heat shocking. It is theoretically possible that some of those food samples also contained C. perfringens spores that spontaneously germinated in the absence of heat shocking. However, the large difference in C. perfringens recovery observed between heat-shocked versus non-heat-shocked food samples (2% versus 29% positive for C. perfringens) strongly suggests that most of the non-heat-shocked food samples growing C. perfringens had been contaminated with vegetative cells, which were killed by heat shocking, rather than spores. If most of the non-heat-shocked food samples yielding C. perfringens had contained spores that spontaneously germinated into vegetative cells, those samples should also have tested positive for C. perfringens after heat shocking (i.e., the recovery rates between non-heat-shocked versus heat-shocked samples should have been much more similar than was actually observed).
Toxin genotyping of food isolates.
Multiplex PCR is now routinely used to assign C. perfringens isolates to one of five types (A to E), based upon whether an isolate carries the genes encoding alpha-, beta-, epsilon-, or iota-toxin (8). Recent versions of the multiplex PCR toxin genotyping assay can also determine whether a C. perfringens isolate possesses the cpe and cpb2 genes encoding CPE and beta2-toxin, respectively (8). Determining the toxin genotypes of C. perfringens food isolates is epidemiologically significant, since C. perfringens type A food poisoning is nearly always caused by CPE-positive type A isolates (13).
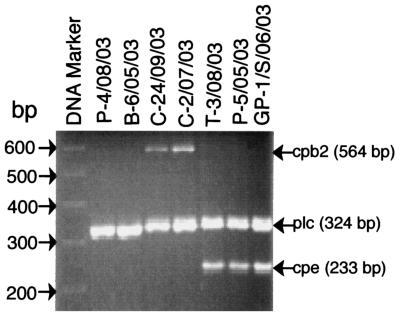
Therefore, the C. perfringens isolates collected from the 278 contaminated foods in our study were subjected to multiplex PCR to determine their toxin genotype (A to E) and whether they carry the cpe or cpb2 genes. Initial experiments (data not shown) with known type A-to-E isolates and known type A isolates carrying the cpe or cpb2 genes confirmed the reliability of this multiplex PCR assay for identifying all six toxin genes, if present. When this multiplex PCR analysis was applied to the C. perfringens food isolates collected in our survey (see Fig. 1 for representative results), these isolates were all classified as type A (i.e., all of these food isolates carry the alpha-toxin gene, but not the genes encoding beta-, epsilon-, or iota-toxin). As also indicated by the representative results shown in Fig. 1, these type A food isolates were heterogeneous with respect to subgenotype, with some carrying neither the cpe or cpb2 genes, some carrying the cpb2 gene, and some carrying the cpe gene (no type A food isolate was found to carry both the cpb2 and cpe genes). In situations where several C. perfringens isolates were obtained from a single food item, those isolates always gave identical multiplex PCR results: e.g., if two isolates were obtained from a single piece of chicken, both isolates would test as type A carrying the cpb2 gene.
FIG. 1.
Multiplex PCR analysis of representative C. perfringens food isolates. Cell lysates of confirmed C. perfringens food isolates were subjected to multiplex PCR. Aliquots of that PCR mixture were then analyzed by agarose gel electrophoresis and ethidium bromide staining. Shown are multiplex PCR results for six representative C. perfringens food isolates, including two cpe-negative, cpb2-negative type A isolates (lanes 2 and 3), two cpb2-positive type A isolates (lanes 4 and 5), and two cpe-positive type A isolates (lanes 6 and 7). Numbers to the left indicate migration of DNA markers (in base pairs). Arrows on the right indicate the expected migration and size of PCR products amplified from the beta2-toxin gene (cpb2), alpha-toxin gene (plc), and C. perfringens enterotoxin gene (cpe) in this assay.
A compilation of multiplex PCR results for all C. perfringens food isolates collected from the 278 contaminated food items is shown in Table 1. Nearly 80% of these food isolates were determined to be simple type A isolates carrying neither the cpe nor cpb2 gene. Of the nearly 20% of foods contaminated with type A isolates carrying the cpb2 gene, almost all came from poultry or pork.
The most important single result from this multiplex PCR analysis was detection of the cpe gene in ∼4.3% of all C. perfringens food isolates. Expressed another way, our results indicate that ∼1.4% of the American retail foods sampled in this survey were contaminated with cpe-positive C. perfringens type A isolates. At least ∼25% of those cpe-positive type A isolates grew from heat-shocked cultures and thus were present in foods as spores rather than as vegetative cells. One ground pork sample yielded cpe-positive isolates from both heat-shocked and non-heat-shocked cultures, suggesting either that the sample contained both vegetative cells and spores or that its spores spontaneously germinated in the non-heat-shocked sample. All other cpe-positive isolates grew from non-heat-shocked cultures, suggesting they may have been primarily present in foods as vegetative cells.
cpe genotyping analysis of food isolates.
Recent studies established that the cpe gene is almost always located on the chromosome of type A isolates causing C. perfringens type A food poisoning outbreaks (3, 4, 20, 21). Therefore, it was important to cpe genotype the 13 cpe-positive type A food isolates obtained in this study to determine whether they carry the chromosomal cpe gene so strongly associated with food poisoning.
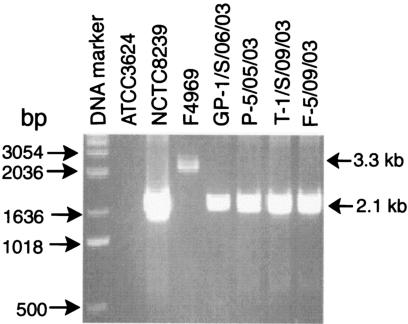
For this purpose, we employed a duplex PCR cpe genotyping assay exploiting consistent differences between the organization of the chromosomal cpe locus versus the plasmid cpe locus of type A isolates (21). To verify the reliability of this assay for cpe genotyping, initial experiments were performed with a known type A isolate carrying a chromosomal cpe gene, a known type A isolate carrying a plasmid cpe gene, and a known cpe-negative type A isolate (21). As shown in Fig. 2, the duplex PCR assay amplified, as expected, an ∼2.1-kb product from the chromosomal cpe locus of NCTC8239, an ∼3.3-kb product from the plasmid cpe locus of type A isolate F4969, and no product from cpe-negative type A isolate ATCC 3624. Previous studies (21) demonstrated that the duplex PCR patterns shown for type A isolates NCTC8239 and F4969 in Fig. 2 hold true for 82 other cpe-positive type A isolates tested whose cpe gene location (chromosomal versus plasmid) had been determined by other methods.
FIG. 2.
cpe duplex PCR genotyping analysis of representative cpe-positive C. perfringens type A food isolates. Extracted DNA from six representative type A food isolates identified as cpe positive by multiplex PCR were subjected to cpe duplex PCR genotyping to determine whether they carry their cpe gene on the chromosome or on a plasmid. Also analyzed as controls were extracted DNA from ATCC 3624 (a known cpe-negative type A isolate), NCTC8239 (a type A food poisoning isolate known to carry a chromosomal cpe gene), and F4969 (a type A isolate known to carry a plasmid cpe gene). After completion of the PCR, products were analyzed by agarose gel electrophoresis and ethidium bromide staining. On these gels, DNA from type A isolates carrying a chromosomal cpe gene should amplify an ∼2.1-kb PCR product, while DNA from type A isolates carrying a plasmid cpe gene should amplify an ∼3.3-kb PCR product (20). Numbers to the left indicate migration of DNA markers (in base pairs).
The same duplex PCR assay was then applied to cpe genotype the 13 cpe-positive type A food isolates collected in our survey. The duplex PCR cpe genotyping results shown in Fig. 2 for four representative cpe-positive food isolates clearly demonstrate that these isolates all carry a chromosomal cpe locus. Similar analyses of the remaining cpe-positive type A food isolates determined that those nine isolates also carry a chromosomal cpe gene (Table 2).
TABLE 2.
Characterization of cpe-positive C. perfringens isolates obtained from food samples
| Isolatesa | Origin | Genotype by duplex PCR | D value (min) at 100°C |
|---|---|---|---|
| Control | |||
| NCTC8239 | Food poisoning | Chromosomal cpe | 93 |
| 153 | Veterinary | Plasmid cpe | 3 |
| Food | |||
| GP-1/S/06/03 | Ground pork | Chromosomal cpe | 170 |
| GP-6/06/03 | Ground pork | Chromosomal cpe | 140 |
| P-5/05/03 | Pork | Chromosomal cpe | 43 |
| P-2/S/05/03 | Pork | Chromosomal cpe | 55 |
| P-1/09/03 | Pork | Chromosomal cpe | 80 |
| P-2/09/03 | Pork | Chromosomal cpe | 65 |
| C-1/04/03 | Chicken | Chromosomal cpe | 70 |
| T-1/08/03 | Turkey | Chromosomal cpe | 43 |
| T-3/S/08/03 | Turkey | Chromosomal cpe | 113 |
| T-4/S/09/03 | Turkey | Chromosomal cpe | 95 |
| F-4/05/03 | Fish | Chromosomal cpe | 76 |
| F-3/05/03 | Fish | Chromosomal cpe | 43 |
| F-5/09/03 | Fish | Chromosomal cpe | 46 |
All isolates were CPE positive by Western blotting.
Western immunoblot analysis of CPE expression by cpe-positive C. perfringens type A food isolates.
To cause food poisoning, a type A isolate must be able to express the chromosomal cpe gene during sporulation (13). Therefore, Western immunoblot analyses were conducted to evaluate CPE expression by sporulating cultures of the 13 type A isolates carrying a chromosomal cpe gene that had been obtained from foods during our survey.
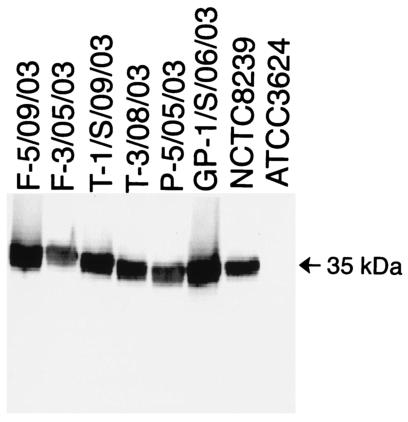
As shown in Fig. 3, the Western immunoblot assay readily detected a 35-kDa protein produced by sporulating cultures of control strain NCTC8239, which is a cpe-positive type A isolate known to express CPE. As expected, that 35-kDa protein, which comigrates with purified CPE (not shown), was absent from sporulating culture lysates of cpe-negative control strain ATCC 3624. Figure 3 also shows representative Western immunoblot analyses for six of the cpe-positive type A food isolates, which all clearly express CPE. Similar Western immunoblot analyses confirmed that the remaining seven cpe-positive food isolates also express CPE (Table 2).
FIG. 3.
Western blot analysis of CPE expression by representative cpe-positive C. perfringens type A food isolates. Six representative cpe-positive C. perfringens food isolates, cpe-positive type A isolate NCTC8239 (positive control), and cpe-negative isolate ATCC 3624 were grown in DS sporulation medium for 8 h at 37°C. After sonication, the resultant culture lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting with CPE polyclonal antibodies, as described previously (17). An arrow to the right indicates a 35-kDa migration of purified CPE on this gel.
Heat resistance properties of cpe-positive type A food isolates.
Type A food poisoning isolates carrying a chromosomal cpe gene are typically much more heat resistant than type A isolates carrying a plasmid cpe gene (17). For example, previous heat resistance determinations (17) demonstrated that sporulating cultures of type A food poisoning isolates consistently have D values at 100°C of ≥30 min, while sporulating cultures of type A, plasmid cpe isolates consistently have D values at 100°C of <2 min. Possession of this heat resistance phenotype should be favorable for causing C. perfringens type A food poisoning outbreaks, since this illness usually results from temperature abuse during cooking or holding of foods (2, 13).
Before determining whether the 13 cpe-positive type A food isolates collected in our survey possess the heat resistance phenotype, D values at 100°C were determined for sporulating cultures of type A food poisoning strain NCTC8239, which carries a chromosomal cpe gene, and veterinary type A isolate 153, which carries a plasmid cpe gene (21). As shown in Table 2, we successfully reproduced earlier results (17) indicating that sporulating cultures of NCTC8239 are very heat resistant, while sporulating cultures of type A isolate 153 are much more heat sensitive.
When similar heat resistance determinations were performed with sporulating cultures of the 13 cpe-positive type A food isolates obtained in our survey, all those isolates were also heat resistant (i.e., sporulating cultures of these 13 isolates had D values at 100°C exceeding 40 min).
DISCUSSION
Until now, it has been unclear when C. perfringens isolates with the potential to cause food poisoning enter the American food supply. A previous survey (12) did not detect the presence of any cpe-positive C. perfringens type A isolates in American retail foods, opening the possibility that foods might become contaminated during food preparation. However, results of our present survey now indicate that ∼1.4% of American meats, poultry, and seafoods are contaminated, at the time of retail purchase, with C. perfringens isolates indistinguishable from proven food poisoning isolates (i.e., these non-outbreak food isolates are classified as type A, carry the chromosomal cpe gene, are capable of expressing CPE, and exhibit the heat-resistant phenotype previously associated with food poisoning isolates). In this regard, it is anecdotally interesting that one laboratory worker developed symptoms of C. perfringens type A food poisoning after eating a surveyed turkey item later found to be contaminated with type A isolates carrying a chromosomal cpe gene.
As mentioned above, previous studies from our laboratory and others have shown that C. perfringens type A isolates carrying a chromosomal cpe gene are responsible for nearly all C. perfringens type A food poisoning outbreaks (3, 4, 20, 21). Subsequent studies (17) then determined that the spores and vegetative cells of six C. perfringens type A food poisoning isolates carrying a chromosomal cpe gene exhibit much greater heat resistance than the spores and vegetative cells of seven C. perfringens type A isolates carrying a plasmid-borne cpe gene. As heat resistance appears desirable for a foodborne pathogen transmitted primarily via cooked meat products, it was proposed that this exceptional heat resistance phenotype of type A food poisoning isolates carrying a chromosomal cpe gene might help explain the strong association between those isolates and C. perfringens type A food poisoning outbreaks. With respect to isolate heat resistance, it is particularly notable that our study determined that spores of type A isolates carrying a chromosomal cpe gene exhibit exceptional heat resistance even when present in raw foods (i.e., prior to cooking). This finding indicates that rather than being a property selected for isolates surviving cooking, heat resistance appears to be an intrinsic property of many C. perfringens type A isolates carrying a chromosomal cpe gene. Studies are planned to identify the physiologic and genetic bases for the heat-resistant phenotype of chromosomal cpe isolates.
However, results from our present food survey also provide a second explanation for the strong association between C. perfringens type A food poisoning outbreaks and type A isolates carrying a chromosomal cpe gene: i.e., it appears these are the cpe-positive isolates most commonly present in food vehicles. All 13 cpe-positive food isolates identified in this survey are type A isolates carrying a chromosomal cpe gene rather than type A isolates with a plasmid-borne cpe gene or cpe-positive isolates belonging to type C or D.
A combination of methodology and sampling differences probably explains why, unlike a previous survey (12), the present study found C. perfringens isolates with food poisoning potential in American retail foods. First, we surveyed considerably more food items than the previous study (i.e., nearly 900 food items were sampled during the present study versus only ∼130 food items sampled by the previous survey). Given our results indicating an ∼1.4% frequency of food contamination by potential C. perfringens food poisoning isolates, the previous survey of only ∼130 foods might have failed to detect even a single contaminated food simply on the basis of probability. In this regard, it is also notable from our survey that ∼4.3% of all C. perfringens isolates obtained from meats, poultry, and seafood were cpe-positive type A isolates. This frequency fall within the ∼1 to 5% presence of cpe-positive type A isolates present in the global C. perfringens population (6, 10, 19). Therefore, it appears that the presence in uncooked food vehicles of cpe-positive type A isolates with food poisoning potential is a random process: i.e., cpe-positive type A isolates with food poisoning potential do not appear to be overrepresented or to have any significant selective advantage in foods over other C. perfringens isolates, at least prior to cooking.
Two other significant food sampling differences also existed between the two recent surveys of cpe-positive C. perfringens isolates in American foods. First, our survey purposely focused on testing meats, poultry, seafood, while the previous survey examined a broader range of foods, including vegetables. There were two reasons for our focus on testing meats, poultry, and seafood: (i) these foods are epidemiologically important as the most common food vehicles of C. perfringens type A food poisoning outbreaks (2, 13) and (ii) the earlier survey by Lin and Labbe (12) had demonstrated these foods to be the most heavily contaminated with C. perfringens isolates. A second possibly relevant food sampling difference between the two surveys is that, with the exception of ham and a few cooked turkey items (including one contaminated with cpe-positive C. perfringens), all food items sampled in our survey were raw, which might favor detection of C. perfringens isolates. It is not clear how many meat and seafood items sampled by Lin and Labbe (12) were cooked versus raw.
Another reason for the present survey's success in demonstrating the presence of cpe-positive type A isolates in foods is that our methods were capable of detecting both vegetative cells and spores in foods, in contrast to the previous survey's (12) testing primarily for the presence of vegetative cells in foods by not heat shocking food samples. Our inclusion of heat shocking to help germinate spores in food samples proved important, as 30% of the cpe-positive type A isolates detected in foods during our survey were detected from heat-shocked cultures. This clear identification of spores of type A isolates carrying a chromosomal cpe gene in foods has important implications for understanding and preventing C. perfringens type A food poisoning. For example, the already mentioned exceptional heat resistance of these spores should favor their survival in temperature-abused food.
A final contributing factor to the successful identification of cpe-positive food isolates by our survey was the addition of an FTG enrichment step prior to streaking food samples onto selective agar. Control experiments (not shown) using spiked samples indicated that addition of this enrichment improved recovery of C. perfringens by ∼50-fold. The probable importance of this enrichment step receives further support from the low MPN-per-gram levels of cpe-positive type A isolates detected in our surveyed foods. The apparently low levels of C. perfringens present in most American foods may help explain why improper holding is a contributing factor to nearly 100% of C. perfringens type A food poisoning outbreaks (2, 13): i.e., American foods rarely contain the burden of cpe-positive type A isolates necessary to initiate food poisoning unless those isolates are allowed to multiply during improper holding.
This survey offers several fundamental observations regarding when American foods become contaminated with C. perfringens food poisoning isolates and why outbreaks of C. perfringens type A food poisoning so often involve type A isolates carrying a chromosomal cpe gene. However, many unanswered questions remain regarding the reservoirs and transmission of these food poisoning isolates. Further study of those questions is under way in our laboratory.
Acknowledgments
This research was generously supported by grant 2001-02517 from the Ensuring Food Safety Research Program of the U.S. Department of Agriculture and by Public Health Service grant AI19844 from NIAID.
We thank Ganes Chakrabarti, Yue Chen, Derek Fisher, Sameera Sayeed, and James G. Smedley III for helping to provide food samples for testing.
REFERENCES
- 1.Adak, G. K., S. M. Long, and S. J. O'Brien. 2002. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut 51:832-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bean, N. H., J. S. Goulding, C. Lao, and F. J. Angulo. 1996. Surveillance for foodborne-disease outbreaks—United States, 1988-1992. Morb. Mortal. Wkly. Rep. 45:1-54. [PubMed] [Google Scholar]
- 3.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 5.Czeczulin, J. R., P. C. Hanna, and B. A. McClane. 1993. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect. Immun. 61:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daube, G., P. Simon, B. Limbourg, C. Manteca, J. Mainil, and A. Kaeckenbeeck. 1996. Hybridization of 2,659 Clostridium perfringens isolates with gene probes for seven toxins (α, β, ɛ, ι, θ, μ and enterotoxin) and for sialidase. Am. J. Vet. Res. 57:496-501. [PubMed] [Google Scholar]
- 7.Food and Drug Administration. 1998. Bacteriologic analytical manual, 8th ed. Association of Official Analytical Chemists International, Gaithersburg, Md.
- 8.Garmory, H. S., N. Chanter, N. P. French, D. Busechel, J. G. Songer, and R. W. Titball. 2000. Occurrence of Clostridium perfringens β2-toxin amongst animals determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koburger, J. A. 1975. Understanding and teaching the most probable number technique. J. Milk Food Technol. 38:540-545. [Google Scholar]
- 10.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbe, R. G. 1989. Clostridium perfringens, p. 192-234. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, New York, N.Y.
- 12.Lin, Y.-T., and R. Labbe. 2003. Enterotoxigenicity and genetic relatedness of Clostridium perfringens isolates from retail foods in the United States. Appl. Environ. Microbiol. 69:1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClane, B. A. 2001. Clostridium perfringens, p. 351-372. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 14.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffen, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto, K., G. Chakrabarti, Y. Morino, and B. A. McClane. 2002. Organization of the plasmid cpe locus of Clostridium perfringens type A isolates. Infect. Immun. 70:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 17.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Skjelkvale, R., and T. Uemura. 1977. Experimental diarrhea in human volunteers following oral administration of Clostridium perfringens enterotoxin. J. Appl. Bacteriol. 46:281-286. [DOI] [PubMed] [Google Scholar]
- 19.Songer, J. G., and R. M. Meer. 1996. Genotyping of Clostridium perfringens by polymerase chain reaction is a useful adjunct to diagnosis of clostridial enteric disease in animals. Anaerobe 2:197-203. [Google Scholar]
- 20.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen, Q., K. Miyamoto, and B. A. McClane. 2003. Development of a duplex PCR genotyping assay for distinguishing Clostridium perfringens type A isolates carrying chromosomal enterotoxin (cpe) genes from those carrying plasmid-borne enterotoxin (cpe) genes. J. Clin. Microbiol. 41:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]